Abstract
Molecular surveillance of pathogens has shown the need for rapid and dependable methods for the identification of organisms of clinical and epidemiological importance. As the leading cause of community-acquired pneumonia, Streptococcus pneumoniae was used as a model organism to develop and refine a real-time fluorescence PCR assay and enhanced DNA purification method. Seventy clinical isolates of S. pneumoniae, verified by latex agglutination, were screened against 26 negative control clinical isolates employing a TaqMan assay on a thermocycler (LightCycler). The probe, constructed from the lytA gene, correctly detected all S. pneumoniae genomes without cross-reaction to negative controls. The speed and ease of this approach will make it adaptable to identification of many bacterial pathogens and provide potential for adaptation to direct detection from patient specimens.
Streptococcus pneumoniae is the leading cause of community-acquired pneumonia, meningitis, and otitis media in the United States (2). While traditional antimicrobial therapy has proven an effective treatment in the past, the emergence of penicillin- and multidrug-resistant strains has resulted in an increasing number of cases of illnesses and fatalities (4, 18). Pneumococcal isolation and identification are complicated by antimicrobial suppression of growth in culture and contamination by normal flora alpha-streptococci. Detection by classical techniques, culture, and serological methods can be time-consuming and indeterminate. Sensitive and specific assays that can be completed quickly in the clinical laboratory are essential for early diagnosis and effective therapy. Molecular assays are inherently valuable because detection can be achieved with enhanced sensitivity and specificity, and detection is not diminished with nonviable organisms.
Various molecular methods have been employed to assist investigations (8, 11, 23). These methods include restriction fragment length polymorphism (RFLP)-based protocols and fingerprinting. PCR-based assays for the detection of S. pneumoniae with primers specific to repetitive regions and genes encoding rRNA (12, 14, 21), pneumococcal surface adhesion A molecule (22), pneumolysin (20, 27, 30, 37), penicillin-binding protein (5, 6, 7, 24, 31), and autolysin (10, 25, 26) have been employed with various degrees of success. Autolysin, encoded by the lytA gene, is required for S. pneumoniae pathogenesis and is a well-characterized virulence marker (1). The lytA gene has been shown to have restricted allelic variation and therefore makes an ideal target for specific identification in clinical and epidemiological studies (9, 35). Sequencing of the lytA locus and high-resolution DNA typing of S. pneumoniae demonstrated that the gene is highly conserved within the species (32, 33), and it has been shown that lytA separates S. pneumoniae from the genotypically similiar species Streptococcus mitis and S. oralis (19, 34).
Real-time PCR with sequence-specific primers and a fluorescent TaqMan probe allows continuous monitoring of in vitro DNA amplification, eliminating the need for gel electrophoresis. The TaqMan probe is an oligonucleotide designed to a sequence within the target DNA (38). Hybridization of the probe allows continuous monitoring of the PCR through a dual fluorophore-labeled system and prevents false-positives due to the absence of nonspecific amplification product fluorescence. The reporter dye is quenched by fluorescent resonance energy transfer by a quencher dye until the reporter is released during primer elongation through 5′-to-3′ exonuclease activity of Taq polymerase. Acquisition of the resulting fluorescence measures the number of copies of target DNA product based on linear regression analysis of a standard curve generated with known genomic equivalents of the organism detected. Real-time fluorescence PCR was conducted on a LightCycler (39). Amplification and detection occur in closed glass capillaries, preventing amplicon cross-contamination. The LightCycler has a 32-sample capacity and achieves high-speed thermal cycling through fan-driven air rather than heat block conduction. In this work, results were obtained rapidly (<1 h) and were highly reproducible, and the range of detection was from fewer than 10 organisms to more than 4 million. We found that commercial, quality-controlled reagents to be proved than those that were home-brewed, more consistent and in some cases sensitivity was improved by several logs.
In this work, we describe the application of an enhanced sample preparation method and a sequence-specific fluorescent probe system that achieves rapid identification of cultured S. pneumoniae. Ultimately, it is our hope to adapt this method for direct detection from clinical specimens.
MATERIALS AND METHODS
Strain identification, cultivation, and DNA stocks.
An S. pneumoniae type strain (ATCC 33400; American Type Culture Collection, Rockville, Md.) and 70 confirmed isolates of S. pneumoniae from patient samples were used to determine the PCR in vitro assay sensitivity and test sensitivity and to validate the enhanced DNA purification protocol. Specificity was determined with 27 negative control organisms representing oral and respiratory flora selected from laboratory bacterial stocks. Each isolate was randomly assigned a sample number (U1 to U97). Identical experiments were conducted in two separate laboratories under identical conditions. All isolates were originally identified as S. pneumoniae by latex agglutination (LA) using the Pneumoslide method (BBL Pneumoslide test for Streptococcus pneumoniae; BBL Microbiology Systems, Cockeysville, Md.). Clinical isolates were recovered from frozen cultures and restreaked to single colonies on blood agar plates. Cultures for positive-control genomic DNA isolation were grown in TSAII (Remel Inc., Lenexa, Kans.). Both plates and broth cultures were grown at 37°C under CO2 using MGC AnaeroPack System (Mitsubishi Gas Chemical Company, Inc., New York, N.Y.). Isolates showing no growth on MacConkey agar and mucoid, alpha-hemolytic colonies were found to be gram-positive cocci on Gram stain. Subsequent to the double blind study, samples with discordant PCR and LA results were identified to the species level by biochemical analyses using Phadebact, RapID Strep, and the GPI card on the Vitek system. Phadebact (Remel Inc.) is a coagglutination test using specific antipneumococcal antibodies coupled to protein A. RapID STR system (Remel Inc.) is a combination of conventional biochemical tests and single-substrate chromogenic test. The GPI card is used in the Vitek Automicrobic system (Biomerieux-Vitek Inc., Hazelwood, Mo.). Negative-control organisms were cultured under conditions appropriate for each species.
S. pneumoniae type strain (ATCC 33400) was used for the PCR in vitro assay sensitivity optimization. A cross-reactivity panel of 44 species representing diverse bacterial genera was established for specificity testing from DNA stocks previously purified in our laboratory and human genomic DNA (Roche Molecular Biochemicals, Mannheim, Germany). Quantification and determination of quality of DNA stock were conducted spectrophotometrically and by agarose gel electrophoresis. All clinical isolates and negative-control organisms were collected over several years prior to this study at either the Clinical Microbiology Laboratory, Air Force Institute for Environment and Occupational Health Risk Analysis/Epidemiology Surveillance Division, Brooks Air Force Base (AFB) or Wilford Hall Medical Center, Clinical Microbiology Laboratory, Lackland AFB. For each specimen, the source, date of collection, and location are on file.
Chromosomal DNA isolation: positive-control organism.
Genomic DNA from the positive control, S. pneumoniae ATCC 33400, was isolated from overnight cultures with a modified version of the Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.). Cell lysis was completed by adding six 1.0-ml aliquots of cell suspension (from 8.0 ml of overnight liquid culture) to six sterile 1.5-ml microcentrifuge tubes that were placed in an ice block. The tubes were placed in a tabletop microcentrifuge and spun at 12,000 × g for 60 s to pellet the cells. The supernatant was removed using a pipette, and 600 μl of sterile H2O was added to each cell pellet and gently pipetted up and down until the cells were resuspended. To each tube, 10.0 μl of lysostaphin (2 U/5 μl) was added, and the tubes were inverted 25 times to mix and incubated at 37°C for 60 min to digest the cell walls. The preceding step was added to the nominal kit protocol to augment cell lysis but has subsequently been corrected to use lysozyme (1 mg/ml) as the enzyme of choice for streptococcal cell wall digestion. The tubes were inverted occasionally during the incubation period. The samples were centrifuged at 12,000 × g for 60 s to pellet the cells, and the supernatant was removed. Cell lysis was continued by adding 600 μl of cell lysis solution to each cell pellet and gently pipetting up and down. To each cellular lysate, 6.0 μl of RNase A solution (4 mg/ml) was added, and the tubes were inverted 25 times and incubated at 37°C for 1.5 h. Following RNase A treatment, 10 μl of proteinase K (14.4 mg/ml) was added and incubated for 1 h at 37°C.
Protein precipitation was completed by cooling the samples to room temperature and adding 200 μl pf protein precipitation solution to the cell lysate. The tubes were vortexed vigorously at high speed for 20 s to mix the protein precipitation solution uniformly with the cell lysate. The samples were centrifuged at 12,000 × g for 3 min. The precipitated proteins formed a tight white pellet. Precipitation of the DNA was completed by pipetting the supernatant into six sterile 1.5-ml microcentrifuge tubes containing 600 μl of 100% isopropanol (2-propanol) and mixing the samples by inverting gently 50 times. The tubes were centrifuged at 12,000 × g for 1 min, and the supernatant was pipetted off and allowed to drain for 15 min on clean absorbent paper. To each tube, 600 μl of 70% ethanol was added, and the tubes were inverted several times to wash the DNA pellet and centrifuged at 12,000 × g for 1 min. The ethanol was carefully pipetted off the pellet, and the tubes were inverted on clean absorbent paper to air dry for 15 min. The DNA pellets were combined in 100 μl of DNA hydration solution, and the solution was placed in a 1.5-ml sterile microcentrifuge tube, hydrated overnight at 4°C, and stored at −20°C. Nucleic acid concentration was determined spectrophotometrically, and genome copy number concentration was calculated. (Refer to the Puregene DNA isolation kit for the manufacture's upgraded protocol for isolation of gram-positive DNA.)
Chromosomal DNA isolation: solid-phase DNA purification protocol.
Chromosomal DNA from the S. pneumoniae (ATCC 33400) positive-control organism, S. pneumoniae clinical isolates, and a panel of negative-control organisms were isolated by a modified version of the Generation Capture Disk protocol for DNA purification from 3 μl of gram-negative bacteria (Gentra Systems Inc.). The capture disk is a solid-phase DNA purification system comprised of a 3-mm-diameter paper disk shipped in a sterile 2.0-ml microcentrifuge tube (spin tube) within a removable, inner basket assembly. The nominal capture disk protocol calls for a 3-μl suspension containing at least 600,000 cells from an overnight liquid culture. In the modified protocol, colonies were selected after 18 h of growth (eliminating overnight, liquid growth) and directly transferred using a sterile loop to 50 μl of sterile H2O in a 1.5-ml microcentrifuge tube. The suspension was vortexed vigorously, and the entire volume was pipetted onto a disk in a spin tube. The disk and bacterial suspension were allowed to incubate at room temperature for approximately 10 min and centrifuged at 12,000 × g for 20 s to remove excess fluid. To each sample, 200 μl of generation DNA purification solution was carefully added down the side of the spin tube so that the disk was immersed in the entire volume. The disk was incubated for 60 s in the DNA purification solution, and the spin tube was centrifuged at 12,000 × g for 60 s. Each disk was transferred with a sterile, 20-g needle into the collar of a sterile, thermocycler capillary tube, 20 μl of the complete PCR mixture was added to the disk, and the capillary tube was capped. The capped capillary tubes containing the disk and PCR mixture were incubated approximately 10 min at room temperature. The capillary tubes were then centrifuged at 12,000 × g for 3 s, reaction volumes were verified, and placed into the thermocycler reaction carousel for PCR. The thermocycler reaction carousel accommodated 32 samples. The assay was unsuccessful when attempted with crude lysates prepared by boiling isolates and then exposing directly to the PCR.
Primer and TaqMan probe oligonucleotide design.
The lytA gene sequence was compared across 15 S. pneumoniae strains. A 901-bp highly conserved sequence was identified, and within this region a 101-bp sequence was selected to develop primers and probe. The forward primer oligonucleotide sequence was 5′-ACGCAATCTAGCAGATGAAGC-3′ at bp 306 to 326 within the lytA gene, the reverse primer sequence was 5′-TGTTTGGTTGGTTATTCGTGC-3′ at bp 386 to 406 within the lytA gene, and the TaqMan probe sequence was 5′- 6-carboxy-fluorescein (FAM)–TTTGCCGAAAACGCTTGATACAGGG–6- carboxy-tetramethyl-rhodamine (TAMRA)-3′ at bp 330 to 354. Amplicon product size was 101 bp. Optimal probe and primer sequences were computed using Primer Express software according to the manufacturer's instructions (PE Applied Biosystems, Foster City, Calif.). Primer sequences were identified with Tm values 10°C less than that of the probe. The fluorescent reporter molecule at the 5′ end of the TaqMan probe was FAM, and the quenching molecule was TAMRA. Primers and probe oligonucleotides were synthesized commercially (Synthetic Genetics, Rockville, Md.).
Real-time PCR.
A LightCycler thermocycler was used to conduct real-time PCR (Roche Molecular Biochemicals, Mannheim, Germany). Assays were carried out in LightCycler capillaries in a 20-μl reaction volume. Reaction reagents were purchased in a preformatted kit (LightCycler-DNA Master Hybridization Probes; Roche Diagnostics GmbH, Roche Molecular Biochemicals, Mannheim, Germany) containing 10× concentrations of Taq DNA polymerase, deoxynucleoside triphosphates (dNTPs) (with dUTP instead of dTTP), 10 mM MgCl2, and reaction buffer. The use of a clean room for reaction mixture preparation separate from where DNA samples were prepared and loaded into capillary tubes, sterile technique, and the closed environment of the system obviated the need for carryover prevention using the uracil-DNA glycosylase protocol described by the kit manufacturer. The following concentrations proved optimal: forward primer (F1), 0.5 μM; reverse primer (R1), 5.0 μM; Taqman probe (TM1), 0.1Μ; and MgCl2, 5.0 mM. Exogenous MgCl2 (25 mM stock) was used to bring the final concentration to 5.0 mM, and PCR-grade, sterile H2O was used to adjust the final reaction volume per the manufacturer's instructions. Each genomic equivalent of positive-control DNA was added in a 2-μl volume to 18 μl of master mix. Human DNA negative control was prepared by adding 4.4e3 genomic equivalents in a 2-μl volume to 18 μl of master mix.
No-template controls (NTC) were prepared by adding a 2-μl volume of PCR-grade, sterile H2O to 18 μl of master mix. Thermocycling conditions were optimized to one cycle of denaturation at 95°C for 60 s, followed by 45 cycles of denaturation at 95°C for 0 s and amplicon extension at 60°C for 60 s, with a single fluorescence acquisition step at the end of extension. Fluorimeter settings were based on the precycling fluorescence of the probe read in the negative control sample with the real-time fluorimeter (RTF) software (Boehringer Mannheim Corporation, Indianapolis, Ind.). While running the RTF, the fluorimeter settings for FAM and TAMRA were adjusted to 10 and 60%, respectively, on the y axis fluorescence scale. This was performed using a concentration matrix of from 0.10 to 0.50 μM probe. Amplicon from the lytA gene was verified by running 5 μl of the PCR product on 2% agarose gels (data not shown).
RESULTS
Sensitivity and specificity of PCR in vitro assay.
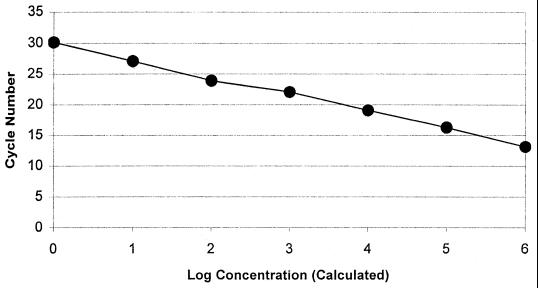
The sensitivity of the PCR in vitro assay was optimized to four genomic equivalents (10 fg) of purified S. pneumoniae DNA. Real-time PCR in vitro assay sensitivity optimization using the S. pneumoniae type strain (ATCC 33400) resulted in a linear regression curve across 4.4e6 to 4.4e0 genomic equivalents (10 ng to 10 fg of DNA) with an error of <1% and a correlation coefficient at −1.00 (Fig. 1). Agarose gel electrophoresis band intensity correlated with the calculated concentrations of amplicon. Human DNA and NTC displayed no detectable fluorescence above background, and upon agarose gel electrophoresis, the presence of amplicon was not observed. Specificity of the in vitro assay initially tested using 1.0 ng of purified DNA from each of 44 cross-reactivity panel bacterial organisms and 4.4e3 genomic equivalents of human DNA displayed no detectable fluorescence (Table 1). The presence of amplicon was not observed with agarose gel electrophoresis examination.
FIG. 1.
Serial dilutions of S. pneumoniae (ATCC 33400) DNA isolated by a standardized method and quantitated spectrophotometrically to 4.4e6 through 4.4e0 genomic equivalents were used for determination of real-time PCR assay detection limits or in vitro sensitivity testing. Cycle number plotted against the log of calculated concentration values resulted in a standard curve with an error of 0.592 and correlation coefficient at unity. Human genomic DNA at 4,500 genomic equivalents and NTC samples did not fluoresce above background signal. The detection limits of the PCR assay demonstrated similar results when the dilution series panel was run in testing of all cross-reaction panel and unknown organisms.
TABLE 1.
Cross-reactivity panel: negative-control organisms
| Genus | Species or serovar(s) (subtypes) |
|---|---|
| Homo | H. sapiens |
| Streptococcus | S. agalactiae, S. bovis, S. equi, S. equisimilis, S. pyogenes, S. sanguis (type II) |
| Campylobacter | C. coli, C. lari, C. jejuni |
| Citrobacter | C. freundii |
| Escherichia | E. coli H1, O28, O55, O111, 112, 0126, 0128, O157:H7 |
| Klebsiella | K. pneumoniae |
| Leclercia | L. adecarboxylata |
| Neisseria | N. lactamica |
| Proteus | P. vulgaris |
| Pseudomonas | P. aeruginosa |
| Salmonella | Bovis-morbificus, Choleraesuis, Cubana, enteritidis, Heidelberg, Infantis, Javiana, Lanka, Kovka, Montevideo, Newport, Paratyphi-A, Poona Typhi-1 (1078), Typhimurium (528) |
| Shigella | S. flexneri, S. boydii (type I) S. dysenteriae (type III) |
| Staphylococcus | S. aureus |
| Mycoplasma | M. pneumoniae, M. hominis |
In addition, 1.0 ng of genomic DNA from laboratory stock, as well as DNA purified by the modified capture disk method, from each of 10 S. pneumoniae strains (ATCC 6305, ATCC 49619, ATCC 33400, ATCC 51915, 1301, 1346, 1518, 1661, 1830, and 2113) were correctly identified by the PCR assay. The presence of the 101-bp amplicon was verified by agarose gel electrophoresis.
Sensitivity and specificity of PCR-based testing of clinical isolates.
Double blind PCR-based testing results were concordant, and spiked negative data verified the absence of PCR inhibition (Table 2). Genomic DNA extracted from all S. pneumoniae samples fluoresced upon exposure to the real-time PCR assay, and all negative control organism DNAs failed to report fluorescence. PCR-based testing of 96 specimens from 18-h isolates by real-time fluorescence PCR demonstrated a sensitivity of 100% (70 of 70) and a specificity of 100% (26 of 26). Sensitivity of the PCR test was 8.8e6 (220 ng) to 8.8e1 genomic equivalents (220 fg) of purified S. pneumoniae. There was no indication of inhibition of the PCR in any of the negative control DNA samples. All negative control genomic DNA samples when spiked with 8.8e4 genomic equivalents of positive control DNA fluoresced. In spiked negative control experiments, 8.8e4 genomic equivalents of positive control DNA had an average Ct of ≈19 (n = 8; mean = 18.77; range, 17.63 to 19.72). Of all negative control organism DNAs spiked with 8.8e4 genomic equivalents of positive control DNA, the average Ct was ≈18 (n = 26; mean = 18.43; range, 15.74 to 20.04). Upon agarose gel electrophoresis of spiked negative control DNA, amplicon band intensity correlated with amplicon band intensity of 8.8e4 genomic equivalents of positive control DNA. Nonspiked negative control DNA, tested in parallel with the spiked samples, displayed no detectable fluorescence when exposed to the PCR assay. The presence of amplicon was not observed with agarose gel electrophoresis.
TABLE 2.
Results of double blind PCR-based testing of clinical isolates
| Sample | No. | PCR sensitivity (%) | PCR specificity (%) | Ct (spiked negative) | LA sensitivity (%) | LA specificity (%) |
|---|---|---|---|---|---|---|
| Streptococcus pneumoniae clinical isolates | 70 | 100 | 96 | |||
| Negative control organisms | 26 | 100 | 85 | |||
| Streptococcus agalactiae (ATCC 13813, Z-10) | 2 | Negative | 19.84, 16.40 | Negative | ||
| Streptococcus bovis (ATCC 9809) | 1 | Negative | 19.52 | Negative | ||
| Streptococcus equi (ATCC 9528) | 1 | Negative | 16.81 | Negative | ||
| Streptococcus intermedius (Z-13) | 1 | Negative | 18.53 | 0 | ||
| Streptococcus mitis (Z-11, Z-12) | 2 | Negative | 18.51, 16.66 | 0 | ||
| Streptococcus pyogenes (ATCC 19615) | 1 | Negative | 19.37 | Negative | ||
| Streptococcus viridans (Z-14) | 1 | Negative | 16.7 | 0 | ||
| Burkholderia cepacia (ATCC 25608) | 1 | Negative | 20.04 | Negative | ||
| Escherichia coli (ATCC 25922, 35218, A-59) | 3 | Negative | 19.45, 18.09, 18.90 | Negative | ||
| Enterococcus faecalis (ATCC 51299, 29212) | 2 | Negative | 17.15, 16.40 | Negative | ||
| Haemophilus influenzae (ATCC 49776, N-1) | 2 | Negative | 18.59, 15.74 | Negative | ||
| Haemophilus parainfluenzae (7901) | 1 | Negative | 17.53 | Negative | ||
| Neisseria gonorrhoeae (ATCC 49981) | 1 | Negative | 17.08 | Negative | ||
| Neisseria meningitidis (ATCC 13102) | 1 | Negative | 18.12 | Negative | ||
| Pseudomonas aeurginosa (ATCC 27853) | 1 | Negative | 16.54 | Negative | ||
| Pseudomonas alcalifaciens (ATCC 51902) | 1 | Negative | 18.89 | Negative | ||
| Staphylococcus liquefaciens (ATCC 27592) | 1 | Negative | 16.7 | Negative | ||
| Staphylococcus aureus (ATCC 29213) | 1 | Negative | 19.05 | Negative | ||
| Staphylococcus epidermidis (ATCC 14990) | 1 | Negative | 19.67 | Negative | ||
| Staphylococcus xylosus (ATCC 29971) | 1 | Negative | 19.07 | Negative |
For isolated colonies, the manufacturer has reported that the Pneumoslide test sensitivity is 98.4% (303 of 308) and that the test has a specificity of 93% (179 of 192) (BBL Pneumoslide test for Streptococcus pneumoniae package insert, revised October 1984). In this study, Pneumoslide tests of 96 fresh clinical isolates had a sensitivity of 96% (67 of 70) and specificity of 85% (22 of 26) (Table 2). Four false positives were identified by PCR and upon biochemical analyses proved to be two Streptococcus mitis strains, Streptococcus intermedius, and Streptococcus viridans. These data correlate with performance characteristics and limitations reported by the Pneumoslide manufacturer.
DISCUSSION
This study shows that real-time fluorescence PCR, using primers and a TaqMan probe complementary to sequences within the lytA gene, is a sensitive and specific assay for the rapid identification of S. pneumoniae from isolates; however, this method's inherent value is in its potential for adaptation to direct detection from clinical specimens. Since the inception of the PCR over 15 years ago, PCR methods for the detection of infectious organisms have been recognized as increasingly valuable clinical diagnostic tools. The development of highly sensitive and specific PCR assays has alleviated problems typically associated with microorganisms that are found in low densities in tissue (or tissue fluids), difficult to culture, or serologically similar. To prevent false-negative results, the development of efficient DNA (RNA) purification methods is necessary to isolate genetic material from cellular substances found to inhibit DNA polymerase activity during the PCR (15). The efficiency of the DNA isolation protocol and level of in vitro sensitivity, test sensitivity, and specificity make the method a valid candidate for adaptation to direct detection from patient specimens and in this embodies our ultimate goal.
Our DNA purification protocol resulted in significant time, cost, and labor savings. Thirty-two samples were processed, and the PCR assay was completed in less than 2 h from time of isolate collection to completion of the report. Microbial DNA capture and PCR assay achieved a level of efficiency that allowed complete concordance in the results of double blind testing in two separate PCR laboratories. The in vitro assay sensitivity was 4 genomic equivalents, with a test sensitivity of 100% (70 of 70) and a specificity of 100% (26 of 26). In regard to quantitation, although we have developed an exquisitely sensitive assay for S. pneumoniae in vitro, we have not yet tested the capture disk protocol for its inherent limit of detection on clinical specimens. Therefore, until we test a battery of clinical specimens, we will not know whether this approach is in itself quantitative and over what sensitivity range. Clearly, any sample preparation method will impact a quantitative method. It remains to be seen what the impact of the capture disk protocol will be on our ability to quantitate. Rather, we have demonstrated an in vitro sensitivity with a minimum number of genomic equivalents detectable by the PCR assay that is very low and that test sensitivity was completely concordant. In specificity testing, 44 cross-reaction panel organisms representing diverse genera and 26 negative control organisms, including seven streptococcal species, did not react. Our method correctly detected three false-negative specimens and four false-positive organisms identified as S. mitis (two strains), S. intermedius, and S. viridans by the Pneumoslide test. As the method evolves to the next phase of development, testing with clinical specimens, additional specificity testing will be conducted as more strains become available to us, including typical and atypical oral streptococci (36), to more fully validate the PCR assay.
In its current format, our method can be used as a simple confirmatory assay due to increased sensitivity and specificity over standardized methods but is not practical for routine testing due to existing standard biological assays for S. pneumoniae that are effective, simple to apply, and inexpensive. We have implemented the method as a part of confirmatory testing to help us identify isolates that proved difficult to identify using our laboratory's routine S. pneumoniae assay, the Pneumoslide. While bile solubility and optochin tests are commonly used in many clinical laboratories for the detection of S. pneumoniae, the Pneumoslide is the assay of preference in our laboratory because it has been our experience that bile solubility, when done on a blood plate, can be difficult to interpret and, when done in a tube, requires a large amount of inoculum. We have found that difficulty with optochin disks may arise, as some viridans streptococci and aerococci may also periodically show small zones of inhibition, causing the organism to be falsely identified as S. pneumoniae. If alpha-hemolytic streptococci are Pneumoslide negative, we routinely identify the organism using the battery of tests described in the Materials and Methods section of this paper. As real-time fluorescence PCR technologies become more widely used, applications will evolve to increasingly simplified protocols more practical for routine clinical laboratory diagnosis.
Due to our success with the capture disk DNA isolation protocol and high degree of sensitivity, specificity, and rapidity achieved by the PCR assay described in this paper, we have begun collecting body fluid specimens to develop the clinical application of the method. We have compared our capture disk PCR assay to direct PCR of boiled S. pneumoniae isolates. The capture disk procedure performed as described, while the boiled lysate preparations were negative. Additionally, direct application of saliva to the capture disk produced sufficient DNA to conduct a PCR-based assay designed to screen for transgenic mice markers (16). In our laboratory, the modified capture disk protocol with a TaqMan PCR assay appears promising in isolating and detecting Campylobacter jejuni DNA directly from stool specimens (unpublished data).
In this study we have shown that real-time fluorescence PCR, using primers and a TaqMan probe complementary to sequences within the lytA gene, is a sensitive and specific assay for the rapid identification of S. pneumoniae and that with the capture disk DNA isolation protocol it provides potential for clinical applications.
ACKNOWLEDGMENTS
We thank Melisa Gaiser for technical assistance and Rebecca Medina and Andrew J. Rohrer, United States Air Force Academy, for help in preparation of the manuscript. We thank J. Peter Pelletier, Wilford Hall Medical Center, Lackland AFB, Tex., for kindly reviewing the manuscript.
REFERENCES
- 1.Berry A M, Lock R A, Hansman D, Paton J C. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect Immun. 1989;57:2324–2330. doi: 10.1128/iai.57.8.2324-2330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown P D, Lerner S A. Community-acquired pneumonia. Lancet. 1998;352:1295–1302. doi: 10.1016/S0140-6736(98)02239-9. [DOI] [PubMed] [Google Scholar]
- 3.Cherian T, Lalitha M K, Manoharan A, Thomas K, Yolken R H, Steinhoff M C. PCR-enzyme immunoassay for detection of Streptococcus pneumoniae DNA in cerebrospinal fluid samples from patients with culture-negative meningitis. J Clin Microbiol. 1998;36:3605–3608. doi: 10.1128/jcm.36.12.3605-3608.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doern G V, Brueggemann A B, Huynh H, Wingert E. Antimicrobial resistance with Streptococcus pneumoniae in the United States, 1997–98. Emerg Infect Dis. 1999;5:757–765. doi: 10.3201/eid0506.990603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.du Plessis M, Smith A M, Klugman K P. Application of pbp1A PCR in identification of penicillin-resistant Streptococcus pneumoniae. J Clin Microbiol. 1999;37:628–632. doi: 10.1128/jcm.37.3.628-632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.du Plessis M, Smith A M, Klugman K P. Rapid detection of penicillin-resistant Streptococcus pneumoniae in cerebrospinal fluid by a seminested-PCR strategy. J Clin Microbiol. 1998;36:453–457. doi: 10.1128/jcm.36.2.453-457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia A, Roson B, Perez J L, Verdaguer R, Dorca J, Carratala J, Casanova A, Manresa F, Gudiol R. Usefulness of PCR and antigen latex agglutination test samples obtained by transthoracic needle aspiration for diagnosis of pneumococcal pneumonia. J Clin Microbiol. 1999;37:709–714. doi: 10.1128/jcm.37.3.709-714.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillespie S H. The role of the molecular laboratory in the investigation of Streptococcus pneumoniae infections. Semin Respiratory Infect. 1999;14:269–275. [PubMed] [Google Scholar]
- 9.Gillespie S H, McHugh T D, Ayres H, Dickens A, Efstratiou A, Whiting G C. Allelic variation in Streptococcus pneumoniae autolysin (N-acetyl muramoyl-l-alanine amidase) Infect Immun. 1997;65:3936–3938. doi: 10.1128/iai.65.9.3936-3938.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillespie S H, Ullman C, Smith M D, Emery V. Detection of Streptococcus pneumoniae in sputum samples by PCR. J Clin Microbiol. 1994;32:1308–1311. doi: 10.1128/jcm.32.5.1308-1311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall L M C. Application of molecular typing the epidemiology of Streptococcus pneumoniae. J Clin Pathol. 1998;51:270–274. doi: 10.1136/jcp.51.4.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall L M, Duke B, Urwin G. An approach to the identification of the pathogens of bacterial meningitis by the polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1995;14:1090–1094. doi: 10.1007/BF01590946. [DOI] [PubMed] [Google Scholar]
- 13.Hassan-King M, Baldeh I, Secka O, Falade A, Greenwood B. Detection of Streptococcus pneumoniae DNA in blood cultures by PCR. J Clin Microbiol. 1994;32:1721–1724. doi: 10.1128/jcm.32.7.1721-1724.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendolin P H, Markkanen A, Ylikoski J, Wahlfors J J. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J Clin Microbiol. 1997;35:2854–2858. doi: 10.1128/jcm.35.11.2854-2858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higuchi R. Simple and rapid preparation of samples for PCR. In: Erlich H A, editor. PCR technology: principles and application for DNA amplification. New York, N.Y: Stockton Press; 1989. pp. 31–38. [Google Scholar]
- 16.Irwin M H, Moffatt R J, Pinkert C A. Identification of transgenic mice by PCR analysis of saliva. Nat Biotechnol. 1999;14:1146–1148. doi: 10.1038/nbt0996-1146. [DOI] [PubMed] [Google Scholar]
- 17.Isaacman D J, Zhang Y, Reynolds E A, Ehrlich G D. Accuracy of a polymerase chain reaction-based assay for detection of pneumococcal bacteremia in children. Pediatrics. 1998;101:813–816. doi: 10.1542/peds.101.5.813. [DOI] [PubMed] [Google Scholar]
- 18.Jacoby G A. Antimicrobial-resistant pathogens in the 1990s. Annu Rev Med. 1996;47:169–179. doi: 10.1146/annurev.med.47.1.169. [DOI] [PubMed] [Google Scholar]
- 19.Kawamura Y, Whiley R A, Shu S E, Ezaki T, Hardie J M. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology. 1999;145:2605–2613. doi: 10.1099/00221287-145-9-2605. [DOI] [PubMed] [Google Scholar]
- 20.Kearns A M, Freeman R, Murphy O M, Seiders P R, Steward M, Wheeler J. Rapid PCR-based detection of Streptococcus pneumoniae DNA in cerebrospinal fluid. J Clin Microbiol. 1999;37:3434. doi: 10.1128/jcm.37.10.3434-3434.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu J-J, Perng C-L, Lee S-Y, Wan C-C. Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J Clin Microbiol. 2000;38:2076–2080. doi: 10.1128/jcm.38.6.2076-2080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison K E, Lake D, Crook J, Carlone G M, Ades E, Facklam R, Sampson J S. Confirmation of psaA in all 90 serotypes of Streptococcus pneumoniae by PCR and potential of this assay for identification and diagnosis. J Clin Microbiol. 2000;38:434–437. doi: 10.1128/jcm.38.1.434-437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olive D M, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37:1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Neill A M, Gillespie S H, Whiting G C. Detection of penicillin susceptibility in Streptococcus pneumoniae by pbp2b PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 1999;37:157–160. doi: 10.1128/jcm.37.1.157-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pozzi G, Oggioni M R, Tomasz A. DNA probe for identification of Streptococcus pneumoniae. J Clin Microbiol. 1989;27:370–372. doi: 10.1128/jcm.27.2.370-372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudolph K M, Parkinson A J, Black C M, Mayer L W. Evaluation of polymerase chain reaction for diagnosis of pneumococcal pneumonia. J Clin Microbiol. 1993;31:2661–2666. doi: 10.1128/jcm.31.10.2661-2666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salo P, Laitinen K, Leinonen M. Detection of pneumococcus from whole blood, buffy coat and serum samples by PCR during bacteremia in mice. APMIS. 1999;107:601–605. doi: 10.1111/j.1699-0463.1999.tb01598.x. [DOI] [PubMed] [Google Scholar]
- 28.Salo P, Ortqvist A, Leinonen M. Diagnosis of bacteremic pneumococcal pneumonia by amplification of pneumolysin gene fragment in serum. J Infect Dis. 1995;171:479–482. doi: 10.1093/infdis/171.2.479. [DOI] [PubMed] [Google Scholar]
- 29.Smith A M, Klugman K P. Alterations in PBP 1A essential for high-level penicillin resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:1329–1333. doi: 10.1128/aac.42.6.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toikka P, Nikkari S, Ruuskanen O, Leinonen M, Mertsola J. Pneumolysin PCR-based diagnosis of invasive pneumococcal infection in children. J Clin Microbiol. 1999;37:633–637. doi: 10.1128/jcm.37.3.633-637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ubukata K, Asahi Y, Yamane A, Konno M. Combinational detection of autolysin and penicillin-binding protein 2B genes of Streptococcus pneumoniae by PCR. J Clin Microbiol. 1996;34:592–596. doi: 10.1128/jcm.34.3.592-596.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Belkum A, Sluijter M, De Groot R, Verbrugh H, Hermans P W M. Novel BOX repeat PCR assay for high-resolution typing of Streptococcus pneumoniae strains. J Clin Microbiol. 1996;34:1176–1179. doi: 10.1128/jcm.34.5.1176-1179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watson D A, Kapur V, Musher D M, Jacobson J W, Musser J M. Identification, cloning and sequencing of DNA essential for encapsulation of Streptococcus pneumoniae. Curr Microbiol. 1995;31:251–259. doi: 10.1007/BF00298383. [DOI] [PubMed] [Google Scholar]
- 34.Whatmore A M, King S J, Doherty N C, Sturgeon D, Chanter N, Dowson C G. Molecular characterization of equine isolates of Streptococcus pneumoniae: natural disruption of genes encoding the virulence factors pneumolysin and autolysin. Infect Immun. 1999;67:2776–2782. doi: 10.1128/iai.67.6.2776-2782.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whatmore A M, Dowson C G. The autolysin-encoding gene (lytA) of Streptococcus pneumoniae displays restricted allelic variation despite localized recombination events with genes of pneumococcal bacteriophage encoding cell wall lytic enzymes. Infect Immun. 1999;67:4551–4556. doi: 10.1128/iai.67.9.4551-4556.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whatmore A M, Efstratiou A, Pickerill A P, Broughton K, Woodard G, Sturgeon D, George R, Dowson C G. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect Immun. 2000;68:1374–1382. doi: 10.1128/iai.68.3.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheeler J, Freeman R, Steward M, Henderson K, Lee M J, Piggott N H, Eltringham G J, Galloway A. Detection of pneumolysin in sputum. J Med Microbiol. 1999;48:863–866. doi: 10.1099/00222615-48-9-863. [DOI] [PubMed] [Google Scholar]
- 38.Wittwer C T, Herrmann M G, Moss A A, Rasmussen R P. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
- 39.Wittwer C T, Ririe K M, Andrew R V, David D A, Gundry R A, Balis U J. The Lightcycler™: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques. 1997;22:176–181. doi: 10.2144/97221pf02. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Isaacman D J, Wadowsky R M, Rydquist-White J, Post J C, Ehrlich G D. Detection of Streptococcus pneumoniae in whole blood by PCR. J Clin Microbiol. 1995;33:596–601. doi: 10.1128/jcm.33.3.596-601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]