ABSTRACT
Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs), such as Gefitinib, have been recommended as the first-line treatment reagent for advanced EGFR-mutant non-small cell lung cancer (NSCLC). However, the mechanisms of drug resistance development are not fully determined. This study aimed to explore the role of circular RNA (circ_0014235) in Gefitinib-resistant NSCLC. The expression of circ_0014235, microRNA-146b-5p (miR-146b-5p) and Yes1 associated transcriptional regulator (YAP) mRNA was detected by quantitative real-time PCR (qPCR). Cell viability was detected by CCK-8 assay. Cell proliferation was assessed by colony formation assay and EdU assay. Cell cycle and cell apoptosis were determined by flow cytometry assay. The expression of marker proteins, YAP protein and programmed death ligand 1 (PD-L1) protein was detected by Western blot. The putative relationship between miR-146b-5p and circ_0014235 or YAP was ensured by dual-luciferase reporter assay and RIP assay. Animal models were established to explore the role of circ_0014235 in vivo. Circ_0014235 was highly expressed in Gefitinib-resistant NSCLC cells. Circ_0014235 downregulation reduced Gefitinib IC50, inhibited cell proliferation and induced cell apoptosis and cell cycle arrest in Gefitinib-resistant NSCLC cells, while these effects were reversed by the inhibition of miR-146b-5p, a target of circ_0014235. In addition, YAP was a target gene of miR-146b-5p, and circ_0014235 relieved miR-146b-5p-mediated inhibition on YAP by targeting miR-146b-5p. MiR-146b-5p restoration-blocked Gefitinib IC50 and cell malignant behaviors were recovered by YAP overexpression. YAP positively regulated PD-L1 expression, and YAP overexpression contributes to Gefitinib IC50 and cell malignant behaviors by upregulating PD-L1. Circ_0014235 confers Gefitinib resistance and malignant behaviors in Gefitinib-resistant NSCLC by governing the miR-146b-5p/YAP/PD-L1 pathway.
KEYWORDS: Circ_0014235, miR-146b-5p, YAP, PD-L1, Gefitinib, NSCLC
Introduction
Non-small cell lung cancer (NSCLC) is the leading subtype of lung cancer, accounting for 85% of all lung cancer cases [1]. Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) are classical small-molecule inhibitors, which have been shown to prolong the survival time of NSCLC patients with EGFR-activated mutations [2]. EGFR-TKIs, such as Gefitinib, have been widely recommended as the first-line standard treatment for EGFR-mutant advanced NSCLC [3,4]. However, patients will inevitably develop acquired resistance after treatment [5]. Tumor immunotherapy is a promising approach to tumor therapy, which is characterized by activating the immune system to induce tumor immune surveillance or reverse tumor immune escape [6]. However, the related mechanisms of immune escape in NSCLC patients with EGFR-mutation remain not fully understood.
Circular RNAs (circRNAs) are non-coding RNA molecules and characterized by closed-loop structure. A growing body of studies indicate that deregulated circRNAs are vital biological participants in the progression of various cancers. CircRNAs are involved in multiple biological processes of cancer development, such as cell proliferation, differentiation, chemoresistance, and immune escape [7–9]. For example, circ_0084003 overexpression promoted NSCLC cell proliferation, migration and immune evasion, and the resistance to anti-programmed cell death 1 (PD-1)-based therapy [9]. In addition, circSETD3 upregulation enhanced acquired resistance of NSCLC cells to Gefitinib [10]. Circ_0014235 is derived from S100 calcium-binding protein A2 (S100A2) that is a well-demonstrated cancerogenic driver in NSCLC [11]. Circ_0014235 was previously shown to be highly expressed in lung squamous cell carcinoma, a subtype of NSCLC, by circRNA expression profile analysis [12]. It was also shown to strengthen the chemoresistance of NSCLC cells to cisplatin (DDP) [13]. Nonetheless, the functions of circ_0014235 in NSCLC were not fully addressed, such as in EGFR-TKIs-targeted therapy and immune escape.
MicroRNAs (miRNAs) are important regulators in cancer development. For example, miR-146b-5p expression was decreased in NSCLC, and miR-146b-5p overexpression inhibited NSCLC cell proliferation, clonogenicity, and migration [14]. CircRNAs act as miRNA sponges to mediate gene expression [15]. The data from bioinformatics databases presented that there are binding sites between miR-146b-5p and circ_0014235 or Yes1 associated transcriptional regulator (YAP) 3ʹ untranslated region (3ʹUTR). YAP is the nuclear effector of the Hippo signaling pathway and has emerged as a therapeutic target in various cancers [16]. Emerging studies have presented that targeting programmed death ligand 1 (PD-L1) via YAP provides a promising therapeutic strategy for EGFR-TKIs-resistant lung cancer [17]. PD-L1 is a major protein that promotes immune evasion of tumor cells by suppressing T-cell activation, and its expression is positively regulated by YAP [18,19]. We speculated that circ_0014235 regulated the miR-146b-5p/YAP/PD-L1 network in EGFR-TKIs-resistant NSCLC.
Here, we constructed EGFR-TKIs-resistant NSCLC cell lines by treating NSCLC cells with Gefitinib. We investigated the expression and function of circ_0014235 in Gefitinib-resistant NSCLC cells. In addition, mechanism analysis showed that circ_0014235 regulated Gefitinib resistance and immune escape of Gefitinib-resistant NSCLC by targeting the miR-146b-5p/YAP/PD-L1 pathway. The objective of this study was to further illustrate the role of circ_0014235 in NSCLC.
Materials and methods
Cell lines and cell culture
Normal NSCLC cell lines (PC9 and HCC827) were purchased from Bena Culture Collection (Beijing, China) and cultured in RPMI-1640 medium containing 10% FBS at 37°C conditions with 5% CO2. We established PC9 and HCC827 cell lines with acquired Gefitinib resistance by gradually increasing the dosages of Gefitinib. Cells were initially treated with a low concentration of Gefitinib and continued to be treated with an appropriate increase of the concentration of Gefitinib when cells grew stably. After 6 months, PC9 and HCC827 cells were able to grow stably in complete medium with 2 μmol/L Gefitinib, and PC9 and HCC827 cells were considered to acquire Gefitinib (PC9/GR and HCC827/GR). The experimental 293 T cells (Bena Culture Collection) were cultured in DMEM containing 10% FBS at 37°C conditions with 5% CO2.
Quantitative real-time PCR (qPCR)
Total RNA was isolated using Total RNA Extractor (Sangon Biotech, Shanghai, China) and then scrambled into cDNA using the HiScript III First-Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) or Mir-X™ miRNA First-Strand Synthesis Kit (Clontech, Mountain View, CA, USA) as appropriate. Finally, qPCR procedures were performed using the Universal SYBR qPCR Master Mix (Vazyme) according to the guidelines of the protocol. Relative expression was calculated using the 2−ΔΔCt method, with β-actin or U6 as internal interferences. Primer sequences were displayed as follows:
circ_0014235, F: 5ʹ-AAGAGGGCGACAAGTTCAAG-3ʹ and R: 5ʹ-GAACTGCACATCATGGATCTGT-3ʹ; S100A2, F: 5ʹ-GGTCTGCCACAGATCCATGA-3ʹ and R: 5ʹ-CTCAGCTTGAACTTGTCGCC-3ʹ; miR-146b-5p, F: 5ʹ-CGCGTGAGAACTGAATTCCAT-3ʹ and R: 5ʹ-AGTGCAGGGTCCGAGGTATT-3ʹ; YAP, F: 5ʹ-CAACTCCAACCAGCAGCAAC-3ʹ and R: 5ʹ-GATATTCCGCATTGCCTGCC-3ʹ; β-actin, F: 5ʹ-CCATGTACGTTGCTATCCAG-3ʹ and R: 5ʹ-CTTCATGAGGTAGTCAGTCAG-3ʹ; U6, F: 5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ and R: 5ʹ-CGCTTCACGAATTTGCGTGTCAT-3ʹ.
RNase R treatment and Actinomycin D (ActD) exposure
RNase R and ActD were used to determine the stability of circ_0014235 as previously mentioned [20]. Simply put, the isolated total RNA was digested with 3 U/μg RNase R for 30 min at 37°C. The treated RNA was detected by qPCR. As for ActD treatment, cells were incubated with 2 μg/mL ActD. After culturing cells for 0, 4, 8, 12 or 24 h, cells were collected for RNA isolation and subsequent qPCR assay.
Cell transfection
Short hairpin RNA (shRNA) targeting circ_0014235 (sh-circ_0014235), shRNA targeting YAP (sh-YAP) and their negative control (sh-NC or sh-con) were synthesized by Genepharma (Shanghai, China). The mimics of miR-146b-5p (miR-146b-5p mimic), the inhibitors of miR-146b-5p (anti-miR-146b-5p) and their matched negative controls (mimic NC and anti-miR-NC) were obtained from Ribobio (Guangzhou, China). PD-L1 overexpression vector (oe-PD-L1) and blank vector (control), YAP overexpression vector (pcDNA-YAP) and blank pcDNA (pcDNA) were all provided by Sangon Biotech (Shanghai, China). Cell transfection was performed using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA).
CCK-8 assay
Cells were seeded into 96-well plates (5,000 cells/well) and cultured at 80% confluence. Cells were then treated with various dosages of Gefitinib (0, 0.01, 0.1, 1, 5, 10, 20, and 40 μM) for 24 h. Afterward, CCK-8 reagent was added to culture cells for 2 h. The absorbance at 450 nm was ascertained using a microplate reader (Bio-Rad, Hercules, CA, USA). The curve of cell viability was depicted to obtain IC50 value.
Colony formation assay
A total of 300 cells were seeded into each well of 6-well plates, and cells were cultured for 2 weeks. After fixing by fixing by methanol and then staining with 0.1% crystal violet (Sangon Biotech), cell colonies were photographed and counted using a light microscope (Nikon, Tokyo, Japan).
EdU assay
The Cell-Light EdU Apollo567 in Vitro Kit (Ribobio) was used for cell proliferation analysis. Cells after treatments were cultured in 96-well plates (5 × 103 cells/well) and labeled with EdU medium for 4 h. Then, cells were washed with PBS, fixed with 4% paraformaldehyde and permeabilized with 0.3% Triton X-100. Cell nuclei were stained with 4, 6-diamidino-2-phenylindole (DAPI) solution. EdU-positive cells were analyzed under a fluorescence microscope (Nikon).
Cell cycle analysis
Cells after treatment were seeded into 24-well plates and cultured for 48 h. Then, cells were fixed by 70% ethanol and stained with propidium iodide (PI) working buffer (containing RNase A) (Beyotime). Cell cycle distribution was analyzed using a CytoFLEX flow cytometer (Beckman Coulter, Miami, FL, USA).
Cell apoptosis analysis
Cells after treatment were seeded into 24-well plates and cultured for 48 h. Then, cells were collected and washed with PBS. Following the protocol of the cell apoptosis assay kit (Beyotime), cells were suspended into Annexin V-FITC binding buffer and stained with 5 μL Annexin V-FITC and 10 μL PI. After 20 min, the apoptotic cells were analyzed using a CytoFLEX flow cytometer.
Western blot
Total protein was extracted using RIPA lysis (Sangon Biotech) and quantified by BCA kit (Sangon Biotech). After separating by 10% SDS-PAGE, proteins were transferred to PVDF membranes and blocked by 5% skim milk. Then, proteins on the membrane were probed with the primary antibodies overnight, including anti-cyclin-dependent kinase 4 (anti-CDK4; ab108357; Abcam, Cambridge, MA, USA), ant-Bax (ab32503; Abcam), anti-pro-caspase3 (full length) (ab32351; Abcam), anti-cleaved-caspase3 (ab32042; Abcam), anti-YAP (ab52771; Abcam), anti-phosphor-YAP (p-YAP; ab76252; Abcam), anti-PD-L1 (ab205921; Abcam) and anti-β-actin (ab8227; Abcam). After incubation with Goat Anti-Rabbit IgG H&L secondary antibody (ab205718), the protein bands were clearly emerged using the ECL kit (Beyotime).
Dual-luciferase reporter assay
According to the wild-type (wt) binding sites between miR-146b-5p and circ_0014235 or YAP obtained from starbase v3.0 (http://starbase.sysu.edu.cn/), the mutant (mut) sequence fragment of circ_0014235 or YAP was designed. Luciferase reporter plasmids, including circ_0014235 wt, circ_0014235 mut, YAP 3ʹUTR wt, and YAP 3ʹUTR mut, were constructed by Sangon Biotech. For dual-luciferase reporter assay, 293 T cells (Bena Culture Collection) were transfected with miR-146b-5p or control (miR-NC) and circ_0014235 wt, circ_0014235 mut, YAP 3ʹUTR wt, or YAP 3ʹUTR mut and then incubated for 48 h. Luciferase activity in 293 T cells in different groups was examined using a Dual-luciferase Reporter Assay System (Promega, Madison, WI, USA).
RIP assay
RIP assay was performed using an EZ-Magna RIP Kit (Millipore, Billerica, MA, USA) referring to the manufacturer’s instructions. Given that Ago2 RNA binding protein is the core component of RNA-induced silencing complexes (RISC), which links miRNA and their targets. Thus, Ago2 was used for RIP assay to identify whether miR-146b-5p interacted with circ_0014235 through Ago2-mediated manner. In brief, cells were lysed in RIP lysis buffer and then incubated with magnetic beads conjugated with human Ago2 antibody (Millipore) and mouse IgG antibody (control; Millipore). Ago2-dependent immunoprecipitates were isolated and analyzed by qPCR.
Animal models
We conducted the animal study in accordance with the Animal Care and Use Committee of Gansu Provincial Cancer Hospital. Nude mice (balb/c, female, 6-week-old) were purchased from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and raised regularly. Lentivirus suspension of sh-circ_0014235 or sh-NC was provided by Genepharma and used to infect HCC827/GR cells. Nude mice were divided into 4 experimental groups (6 mice in each group) with various treatments, including sh-NC+DMSO, sh-circ_0014235+ DMSO, sh-NC+Gefitinib and sh-circ_0014235+ Gefitinib. The infected HCC827/GR cells were injected into nude mice with subcutaneous injection, and Gefitinib or control (DMSO) was injected into nude mice with intravenous injection in tail. Tumor volume (length×width2 × 1/2) was measured every 5 days. Tumor tissues were excised after growth for 30 days, photographed and preserved for further analysis.
IHC assay
Tumor tissues from animal models were fixed in 4% paraformaldehyde and embedded in paraffin. Then, tissue slides (4 μm) were prepared. The sections were deparaffinized in xylene and incubated with the primary antibodies, including anti-ki-67 (ab92742; Abcam), anti-YAP (ab52771; Abcam) and anti-PD-L1 (ab205921; Abcam). Three slides per group were observed under a light microscope (Nikon).
Statistical analysis
In this study, all experiments were conducted 3 times. The data were collected and processed using GraphPad Prism 7.0 software (San Diego, CA, USA). For difference comparison in different groups, Student’s t-test and ANOVA (with Tukey’s post-hoc test) were applied as appropriate. The data were shown as mean ± standard deviation (SD). Statistical significance was defined when P value less than 0.05.
Results
Circ_0014235 was highly expressed in gefitinib-resistant NSCLC cells, and circ_0014235 was resistant to RNase R and ActD
The expression of circ_0014235 was notably elevated in PC9/GR and HCC827/GR cells compared with that in PC9 and HCC827 cells, respectively (Figure 1(a)). Circ_0014235 was produced by back-splicing from exon2 of S100A2 mRNA, with 154 bp in length (Figure 1(b)). RNase R and ActD were used to test the stability of circ_0014235. As a result, circ_0014235 was rarely degraded by RNase R compared to linear S100A2 in PC9/GR and HCC827/GR cells (Figure 1(c)). Likewise, circ_0014235 was hardly digested by ActD, while linear S100A2 was significantly digested by ActD (Figure 1(d)). In short, circ_0014235 expression was increased in gefitinib-resistant NSCLC cells, and circ_0014235 was resistant to RNase R and ActD.
Figure 1.
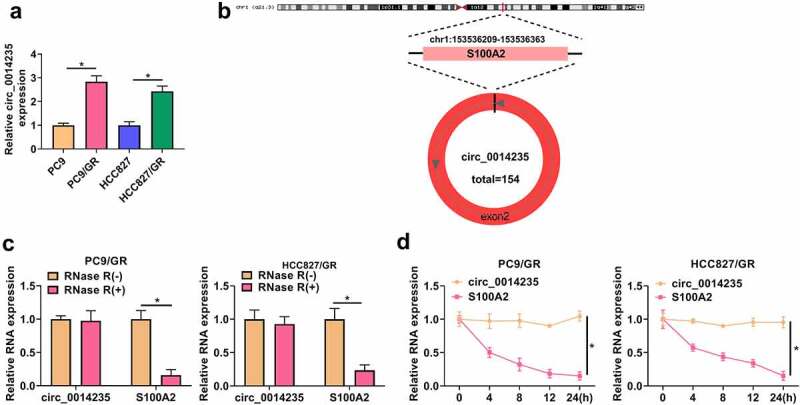
Circ_0014235 was overexpressed in Gefitinib-resistant NSCLC cells and resistant to RNase R and ActD. (a) The expression of circ_0014235 in PC9/GR, HCC827/GR and their parental cells was detected by qPCR. (b) The formation of circ_0014235. (c) The stability of circ_0014235 was tested by RNase R. (d) The stability of circ_0014235 was tested by ActD. *P < 0.05.
Circ_0014235 downregulation reduced gefitinib IC50, inhibited cell growth, and promoted cell apoptosis of PC9/GR and HCC827/GR cells
We reduced the endogenous level of circ_0014235 in PC9/GR and HCC827/GR cells by transfecting sh-circ_0014235. The data from qPCR showed that the expression of circ_0014235 was strikingly declined in PC9/GR and HCC827/GR cells transfected with sh-circ_0014235 compared to sh-NC (Figure 2(a)). After transfection, PC9/GR and HCC827/GR cells were treated with different concentrations of gefitinib, and the curve cell viability showed that IC50 in PC9/GR and HCC827/GR cells transfected with sh-circ_0014235 was lower than that in PC9/GR and HCC827/GR cells transfected with sh-NC (Figure 2(b)). Colony formation assay presented that circ_0014235 knockdown impaired the ability of colony formation of PC9/GR and HCC827/GR cells (Figure 2(c)). EdU assay showed that circ_0014235 knockdown reduced the number of EdU-positive cells, suggesting that cell proliferation was suppressed by circ_0014235 knockdown (Figure 2(d)). In the analysis of cell cycle, circ_0014235 knockdown significantly induced cell cycle arrest at the G0/G1 stage (Figure 2(e)). For cell apoptosis investigation, the data from flow cytometry showed that circ_0014235 knockdown notably promoted the apoptosis of PC9/GR and HCC827/GR cells (Figure 2(f)). Additionally, some markers of cell cycle and apoptosis were quantified by Western blot. The data showed that the expression of CDK4 was declined, while the expression of Bax and cleaved-caspase3 was increased in PC9/GR and HCC827/GR cells transfected with sh-circ_0014235 (Figure 2(g)). In short, these data indicated that circ_0014235 knockdown inhibited gefitinib resistance and the malignant behaviors of PC9/GR and HCC827/GR cells.
Figure 2.
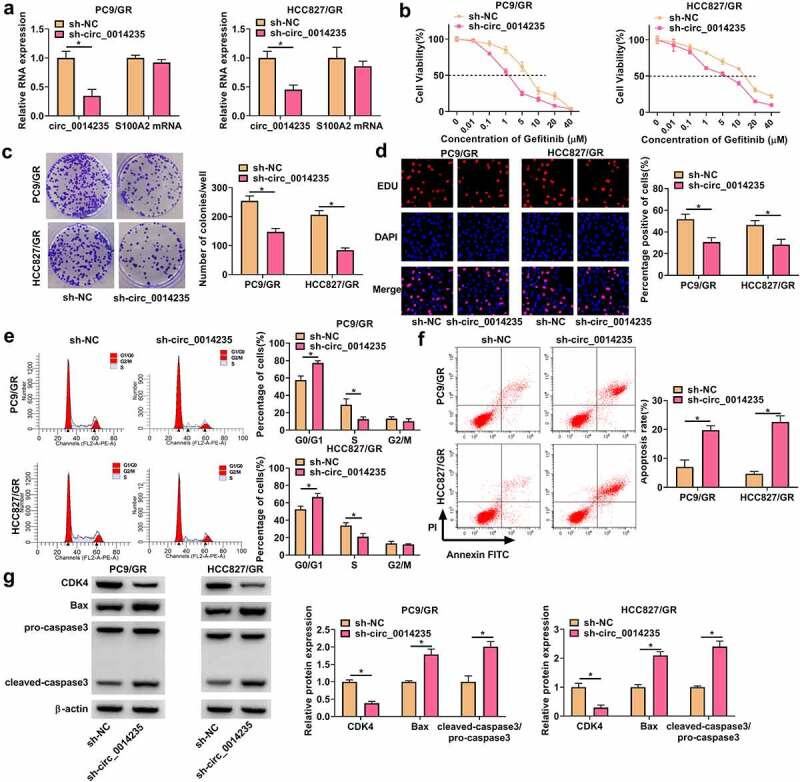
Circ_0014235 knockdown inhibited Gefitinib resistance and cell malignant behaviors in PC9/GR and HCC827/GR cells. (a) The expression of circ_0014235 and linear S100A2 mRNA in PC9/GR and HCC827/GR cells transfected with sh-circ_0014235 or sh-NC was detected by qPCR. In these transfected cells, (b) cell viability was detected by CCK-8 assay to conclude IC50. (c) Cell proliferation was assessed by colony formation assay. (d) Cell proliferation was assessed by EdU assay. (e) Cell cycle progression was determined by flow cytometry assay. (f) Cell apoptosis was determined by flow cytometry assay. (g) The protein levels of CDK4, Bax and cleaved-caspase3 were detected by Western blot. *P < 0.05.
MiR-146b-5p was targeted by circ_0014235
Numerous miRNAs were predicted to be potential targets of circ_0014235 by starbase3.0. The expression of these miRNAs was examined in PC9/GR, HCC827/GR and their parental cells. The data showed that miR-146b-5p was one of miRNAs that were notably downregulated in PC9/GR and HCC827/GR compared to their parental cells (Figure 3(a-b). To verify the relationship between circ_0014235 and miR-146b-5p, dual-luciferase reporter plasmids of circ_0014235 wt and circ_0014235 mut were constructed (Figure 3(c)). In 293 T cells, miR-146b-5p mimic transfected with circ_0014235 wt significantly reduced luciferase activity, while miR-146b-5p mimic transfected with circ_0014235 mut rarely reduced luciferase activity (Figure 3(d)). In addition, circ_0014235 and miR-146b-5p were abundantly enriched by Ago2 compared to IgG by the analysis of RIP assay (Figure 3(e)), confirming that miR-146b-5p interacted with circ_0014235 through Ago2-mediated manner. The evidence demonstrated that miR-146b-5p was a target of circ_0014235.
Figure 3.
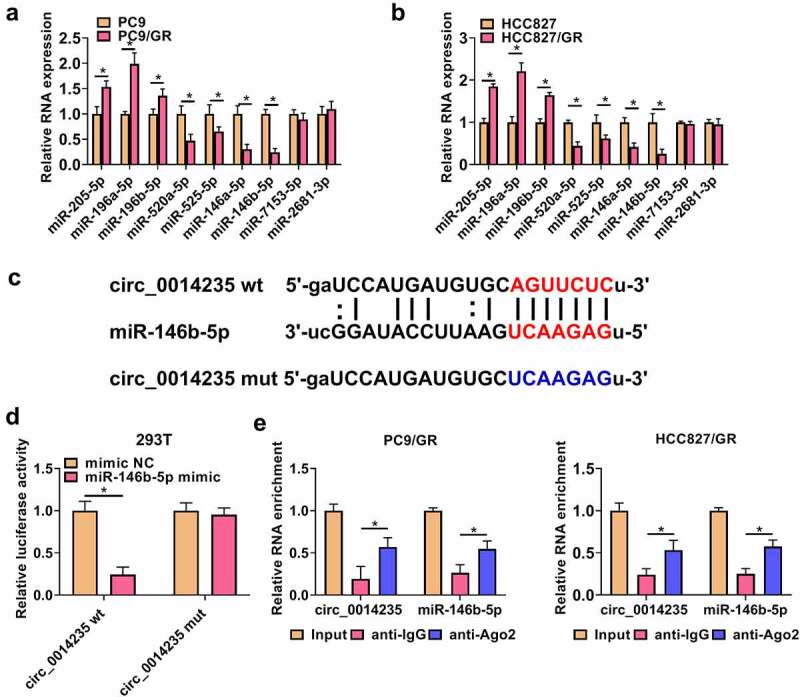
MiR-146b-5p was a target of circ_0014235. (a and b) The expression of predicted miRNAs in PC9/GR and HCC827/GR cells was detected by qPCR. (c) The sequence fragments of circ_0014235 wt or circ_0014235 mut were used for dual-luciferase reporter assay. (d) Dual-luciferase reporter assay was conducted to verify the relationship between circ_0014235 and miR-146b-5p. (e) RIP assay was conducted to verify the relationship between circ_0014235 and miR-146b-5p. *P < 0.05.
Circ_0014235 downregulation inhibited gefitinib resistance and the malignant behaviors of PC9/GR and HCC827/GR cells by releasing miR-146b-5p
The expression of miR-146b-5p was markedly decreased in PC9/GR and HCC827/GR cells transfected with anti-miR-146b-5p (Figure 4(a)). Besides, the expression of miR-146b-5p was notably enhanced in PC9/GR and HCC827/GR cells transfected with sh-circ_0014235 but repressed in cells cotransfected with sh-circ_0014235+ anti-miR-146b-5p (Figure 4(b)). Functionally, IC50 inhibited in PC9/GR and HCC827/GR cells transfected with sh-circ_0014235 was largely recovered in PC9/GR and HCC827/GR cells transfected with sh-circ_0014235+ anti-miR-146b-5p (Figure 4(c)). The number of colonies and EdU-positive cells was reduced by circ_0014235 knockdown alone was largely restored in PC9/GR and HCC827/GR cells transfected with sh-circ_0014235+ anti-miR-146b-5p (Figure 4(d-e). In addition, miR-146b-5p deficiency partially relieved circ_0014235 knockdown-induced cell cycle arrest of PC9/GR and HCC827/GR cells (Figure 4(f)), and circ_0014235 knockdown-induced PC9/GR and HCC827/GR cell apoptosis was also partially suppressed by miR-146b-5p deficiency (Figure 4(g)). The expression of CDK4 reduced by sh-circ_0014235 was recovered by sh-circ_0014235+ anti-miR-146b-5p, and the expression of Bax and cleaved-caspase3 promoted by sh-circ_0014235 was suppressed by sh-circ_0014235+ anti-miR-146b-5p in PC9/GR and HCC827/GR cells (Figure 4(h)). The data indicated that circ_0014235 knockdown inhibited gefitinib resistance and the malignant behaviors of PC9/GR and HCC827/GR cells by enriching the expression of miR-146b-5p.
Figure 4.
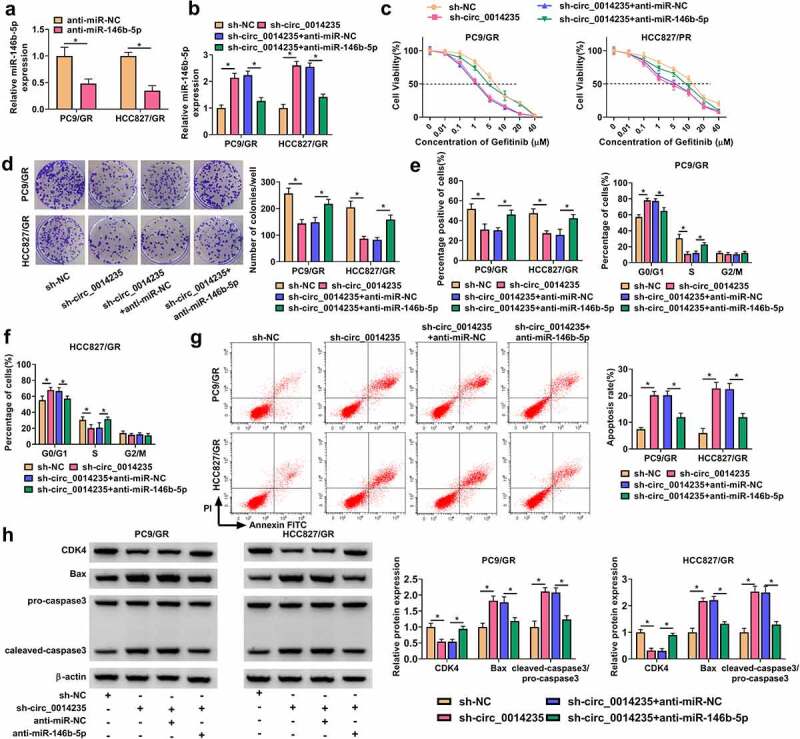
The effects of circ_0014235 downregulation were reversed by miR-146b-5p inhibition. (a) The efficiency of miR-146b-5p inhibitor was checked by qPCR. The following assays were performed in PC9/GR and HCC827/GR cells transfected with sh-circ_0014235 or sh-circ_0014235+ anti-miR-146b-5p. (b) The expression of miR-146b-5p was detected by qPCR. (c) IC50 of Gefitinib was concluded by CCK-8 assay. (d) Cell proliferation was assessed by colony formation assay. (e) Cell proliferation was also assessed by EdU assay. (f) Cell cycle progression was determined by flow cytometry assay. (g) Cell apoptosis was determined by flow cytometry assay. (h) The protein levels of CDK4, Bax and cleaved-caspase3 were measured by Western blot. *P < 0.05.
YAP was a target of miR-146b-5p, and circ_0014235 knockdown inhibited the expression of YAP by enriching miR-146b-5p
YAP was predicted to be a target of miR-146b-5p, and miR-146b-5p interacted with YAP 3ʹUTR. For dual-luciferase reporter assay, dual-luciferase reporter plasmids of YAP 3ʹUTR wt and YAP 3ʹUTR mut were constructed (Figure 5(a)). In 293 T cells, miR-146b-5p mimic transfected with YAP 3ʹUTR wt notably weakened luciferase activity (Figure 5(b)). The levels of YAP and p-YAP were shown to be markedly increased in PC9/GR and HCC827/GR cells compared with that in PC9 and HCC827 cells, respectively (Figure 5(c)). Moreover, the levels of YAP and p-YAP were strikingly reduced in PC9/GR and HCC827/GR cells transfected with sh-circ_0014235 alone, while its expression was recovered in cells transfected with sh-circ_0014235+ anti-miR-146b-5p (Figure 5(d)). The data suggested that circ_0014235 regulated the miR-146b-5p/YAP signaling.
Figure 5.
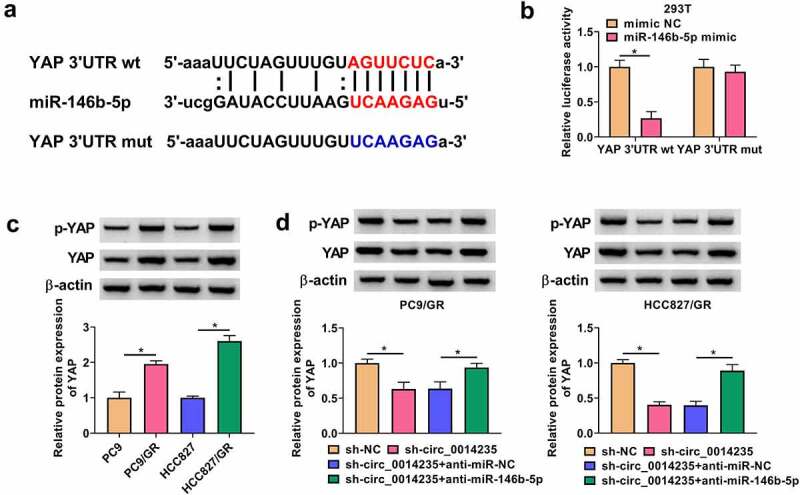
MiR-146b-5p bound to YAP 3ʹUTR, and circ_0014235 regulated YAP expression by targeting miR-146b-5p. (a) The sequence fragments of YAP 3ʹUTR wt and YAP 3ʹUTR mut were shown. (b) The relationship between miR-146b-5p and YAP was verified by dual-luciferase reporter assay. (c) The expression of YAP and p-YAP in PC9/GR and HCC827/GR, and their parental cells was detected by Western blot. (d) The expression of YAP and p-YAP in PC9/GR and HCC827/GR cells transfected with sh-circ_0014235 or sh-circ_0014235+ anti-miR-146b-5p was measured by Western blot. *P < 0.05.
MiR-146b-5p restoration inhibited gefitinib resistance and the malignant behaviors of PC9/GR and HCC827/GR cells by depleting YAP
The expression of miR-146b-5p was notably reinforced in PC9/GR and HCC827/GR cells transfected with miR-146b-5p (Figure 6(a)). The levels of YAP and p-YAP were notably increased in PC9/GR and HCC827/GR cells transfected with pcDNA-YAP (Figure 6(b)). Next, we detected that the levels of YAP and p-YAP were significantly decreased in PC9/GR and HCC827/GR cells transfected with miR-146b-5p but notably recovered in cells transfected with miR-146b-5p+pcDNA-YAP (Figure 6(c)). Subsequently, rescue experiments were performed in these transfected cells. We found that miR-146b-5p restoration notably reduced the IC50 of Gefitinib in PC9/GR and HCC827/GR cells, while YAP reintroduction restored the IC50 of Gefitinib (Figure 6(d-e). Colony formation assay showed that the ability of colony formation in PC9/GR and HCC827/GR cells was suppressed by miR-146b-5p restoration but largely promoted by YAP reintroduction (Figure 6(f)). EdU assay presented that cell proliferation capacity was significantly blocked in PC9/GR and HCC827/GR cells transfected with miR-146b-5p but largely recovered in PC9/GR and HCC827/GR cells transfected with miR-146b-5p+pcDNA-YAP (Figure 6(g)). Flow cytometry for cell cycle assay presented that miR-146b-5p restoration arrested cell cycle at the G0/G1 phase, while YAP overexpression relieved miR-146b-5p-induced cell cycle arrest (Figure 6(h)). Flow cytometry for cell apoptosis assay presented that miR-146b-5p restoration promoted the apoptosis of PC9/GR and HCC827/GR cells, while YAP overexpression suppressed the apoptosis of these cells (Figure 6(i)). Additionally, the expression of CDK4 was decreased in PC9/GR and HCC827/GR cells transfected with miR-146b-5p but enhanced in cells transfected with miR-146b-5p+pcDNA-YAP, while the expression of Bax and cleaved-caspase3 was promoted in PC9/GR and HCC827/GR cells transfected with miR-146b-5p but repressed in cells transfected with miR-146b-5p+pcDNA-YAP (Figure 6(j)). The data indicated that miR-146b-5p restoration inhibited gefitinib resistance and the malignant behaviors of PC9/GR and HCC827/GR cells by reducing YAP.
Figure 6.
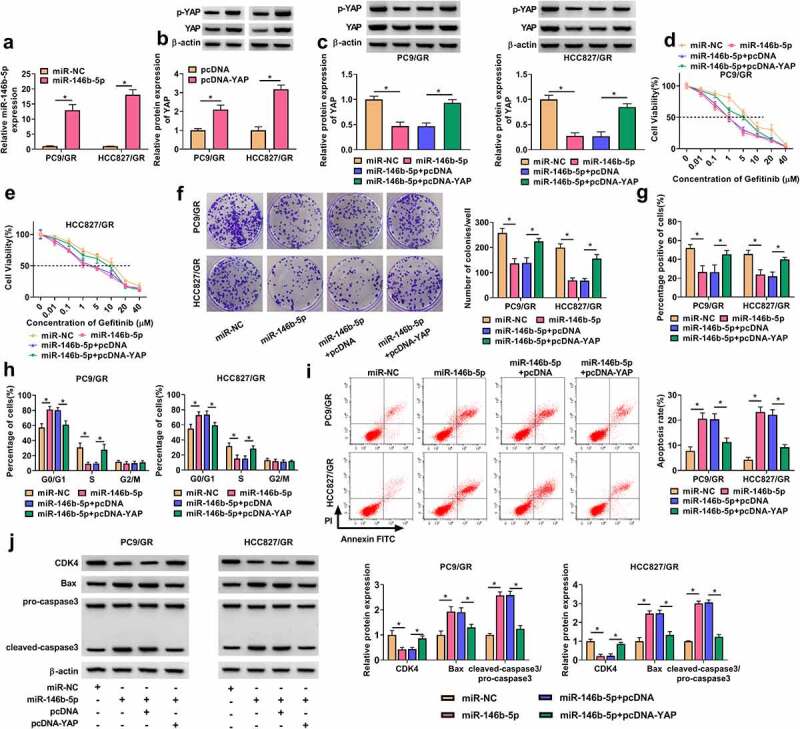
MiR-146b-5p restoration inhibited Gefitinib resistance and cell malignant behaviors in PC9/GR and HCC827/GR cells by depleting YAP. (a) The efficiency of miR-146b-5p mimic was checked by qPCR. (b) The efficiency of pcDNA-YAP was checked by Western blot. In PC9/GR and HCC827/GR cells transfected with miR-146b-5p or miR-146b-5p+pcDNA-YAP, (c) the protein levels of YAP and p-YAP were detected by Western blot. (d and e) IC50 of Gefitinib was concluded by CCK-8 assay. (f) Cell proliferation was assessed by colony formation assay. (g) Cell proliferation was also assessed by EdU assay. (h) Cell cycle progression was determined by flow cytometry assay. (i) Cell apoptosis was determined by flow cytometry assay. (j) The protein levels of CDK4, Bax and cleaved-caspase3 were measured by Western blot. *P < 0.05.
YAP depletion inhibited gefitinib resistance and the malignant behaviors of PC9/GR and HCC827/GR cells by downregulating PD-L1
It is well-known that PDL1 binds to the PD-1 receptor expressed on immune cells, and YAP-induced PD-L1 expression triggers immune evasion of tumor cells [21]. We discovered that the expression of PD-L1 protein was strikingly increased in PC9/GR and HCC827/GR cells compared to their parental cells (Figure 7(a)). Besides, the expression of PD-L1 was strikingly reduced in PC9/GR and HCC827/GR cells transfected with sh-YAP compared to sh-con (Figure 7(b)). The expression of PD-L1 protein was notably decreased in PC9/GR and HCC827/GR cells transfected with sh-YAP alone but largely recovered in PC9/GR and HCC827/GR cells transfected with sh-YAP+oe-PD-L1 (Figure 7(c)). In terms of function, the IC50 of PC9/GR and HCC827/GR cells was remarkably inhibited by YAP downregulation but recovered by the overexpression of PD-L1 (Figure 7(d-e). Colony formation assay presented that YAP knockdown repressed the colony formation ability of PC9/GR and HCC827/GR cells, while additional PD-L1 overexpression restored the colony formation ability of PC9/GR and HCC827/GR cells (Figure 7(f)). EdU assay presented that YAP knockdown suppressed PC9/GR and HCC827/GR cell proliferation, while PD-L1 overexpression promoted cell proliferation (Figure 7(g)). In addition, YAP downregulation induced cell cycle arrest at the G0/G1 phase, while further PD-L1 overexpression largely relieved cell cycle arrest (Figure 7(h)). YAP knockdown-induced PC9/GR and HCC827/GR cell apoptosis was also suppressed by PD-L1 overexpression (Figure 7(i)). Additionally, the expression of CDK4 was decreased in PC9/GR and HCC827/GR cells transfected with sh-YAP but recovered in cells transfected with sh-YAP+oe-PD-L1, and the expression of Bax and the ratio of cleaved-caspase 3/pro-caspase 3 were promoted in PC9/GR and HCC827/GR cells transfected with sh-YAP but repressed in cells transfected with sh-YAP+oe-PD-L1 (Figure 7(j)). These findings suggested that YAP knockdown inhibited gefitinib resistance and the malignant behaviors of PC9/GR and HCC827/GR cells by downregulating PD-L1.
Figure 7.
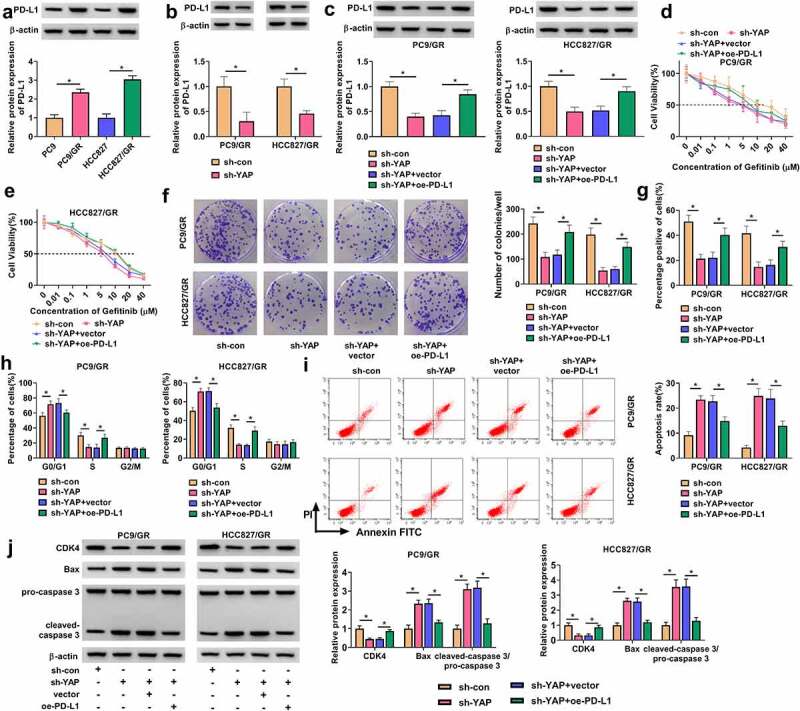
YAP knockdown reduced Gefitinib resistance and suppresed cell malignant behaviors in PC9/GR and HCC827/GR cells by upregulating PD-L1. (a) The expression of PD-L1 protein in PC9/GR, HCC827/GR, and their parental cells was detected by Western blot. (b) The efficiency of sh-YAP was checked by Western blot. In PC9/GR and HCC827/GR cells transfected with sh-YAP or sh-YAP+oe-PD-L1, (c) the expression of PD-L1 protein was detected by Western blot. (d and e) IC50 of Gefitinib was concluded by CCK-8 assay. (f) Cell proliferation was assessed by colony formation assay. (g) Cell proliferation was also assessed by EdU assay. (h) Cell cycle progression was determined by flow cytometry assay. (i) Cell apoptosis was determined by flow cytometry assay. (j) The protein levels of CDK4, Bax and cleaved-caspase3 were measured by Western blot. *P < 0.05.
Circ_0014235 knockdown enhanced Gefitinib sensitivity in vivo
The role of circ_0014234 was further determined in animal models. The results showed that circ_0014235 knockdown significantly inhibited tumor growth, leading to smaller tumor size, tumor volume and tumor weight (Figure 8(a,c)). In Genfitinib-administered groups, we found that circ_0014235 knockdown promoted Gefitinib sensitivity, leading further decrease of tumor size, tumor volume and tumor weight (Figure 8(a-c). The data from qPCR showed that the expression of circ_0014235 and YAP mRNA was decreased, while the expression of miR-146b-5p was increased in the sh-circ_0014235+ DMSO group or sh-circ_0014235+ Gefitinib group compared with that in sh-NC+DMSO group or sh-NC+Gefitinib group, respectively (Figure 8(d)). The expression levels of YAP and PD-L1 protein were significantly declined in the sh-circ_0014235+ DMSO group or sh-circ_0014235+ Gefitinib group (Figure 8(e–f)). Moreover, IHC staining presented that the abundance of ki-67, YAP and PD-L1 was strikingly decreased in the sh-circ_0014235+ DMSO group or sh-circ_0014235+ Gefitinib group compared with that in sh-NC+DMSO group or sh-NC+Gefitinib group (Figure 8(g)). The data suggested that circ_0014235 knockdown promoted Gefitinib sensitivity and inhibited tumor growth in vivo.
Figure 8.
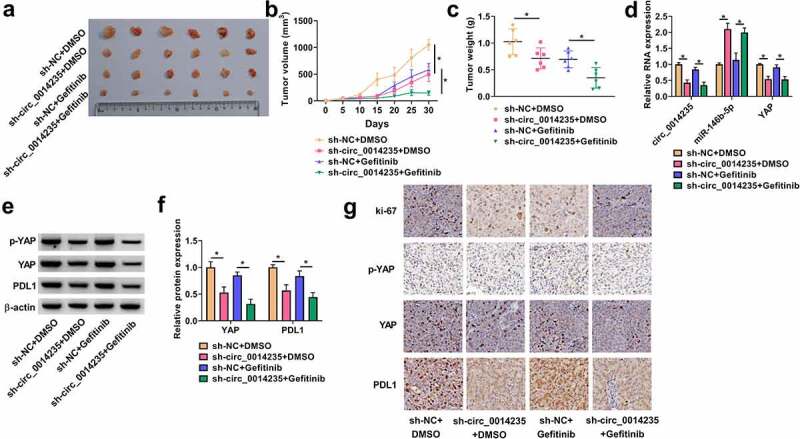
Circ_0014235 knockdown strengthened Gefitinib sensitivity and inhibited tumor growth in vivo. (a) The representative images of tumor tissues. (b and c) Tumor volume and tumor weight were measured to monitor tumor growth. (d) The expression of circ_0014235, miR-146b-5p and YAP mRNA in tumor tissues was detected by qPCR. (e and f) The protein levels of YAP and PD-L1 in tumor tissues were quantified by Western blot. (g) The expression levels of Ki-67, YAP and PD-L1 in tumor tissues were also assessed by IHC analysis. *P < 0.05.
Discussion
Our study was the first to investigate the role of circ_0014235 in Gefitinib-resistant NSCLC. We mainly discovered that the expression of circ_0014235 was increased in PC9/GR and HCC827/GR cells compared to their parental cells. The knockdown of circ_00142135 inhibited Gefitinib IC50 and cell malignant behaviors in PC9/GR and HCC827/GR cells. Mechanically, we proposed that circ_0014235 promoted the development of Gefitinib-resistant NSCLC by targeting the miR-146b-5p/YAP/PD-L1 network. This study provided new insights into the understanding of the development of NSCLC with Gefitinib chemoresistance from circ_0014235.
Increasing circRNAs have been identified to play crucial roles in Gefitinib-resistant NSCLC, such as circ_0004015 and circ_0000567 [22,23]. Nevertheless, studies on the role of circRNAs in NSCLC with chemoresistance are still limited. RNA sequencing provided a circRNA expression profile and displayed numerous differently expressed circRNAs in lung squamous cell carcinoma (LUSC) [12]. The data published that circ_0014235 was strikingly upregulated in plasma exosomes of LUSC patients [12], suggesting that circ_0014235 might participate in LUSC development. A recent study also reported that circ_0014235 was overexpressed in plasma exosomes, tumor tissues and cell lines of NSCLC [13]. Interestingly, circ_0014235 was published to promote the resistance of NSCLC to DDP [13]. Similar to these findings, we showed that circ_0014235 expression was notably elevated in Gefitinib-resistant NSCLC cells, and circ_0014235 knockdown in Gefitinib-resistant NSCLC cells inhibited drug IC50, cell proliferation and promoted cell apoptosis and cell cycle arrest. In addition, circ_0014235 knockdown strengthened Gefitinib sensitivity of tumors in vivo, thereby inhibiting tumor growth. Combined our study and previous studies, we speculated that circ_0014235 played a vital role in multi-drug resistance in NSCLC, which needed to be further demonstrated.
To determine the regulatory mechanism of circ_0014235, we screened the target miRNAs of circ_0014235 and selected miR-146b-5p whose functions had been partly investigated in NSCLC in previous studies [14,24]. The results revealed that low expression of miR-146b-5p was linked to shorter median and mean survival time of NSCLC patients, and miR-146b-5p restoration inhibited NSCLC cell growth and invasion [14]. Besides, miR-146b-5p was shown to be poorly expressed in Gefitinib-resistant NSCLC cells, and miR-146b-5p overexpression strengthened the sensitivity of NSCLC cells to Gefitinib [24]. Considering the important role of miR-146b-5p in NSCLC, we chose miR-146b-5p as a target of circ_0014235 and found that miR-146b-5p downregulation abolished the effects of circ_0014235 knockdown, leading to increased IC50 of Gefitinib, cell growth and survival in Gefitinib-resistant NSCLC cells. On the contrary, miR-146b-5p restoration inhibited the malignant behaviors of Gefitinib-resistant NSCLC cells.
Further analysis in this study discovered that YAP was one of the target genes of miR-146b-5p. YAP was a well-documented oncogene in various cancers and contributed to cancer cell growth, migration and invasion [25–27]. In NSCLC, YAP was reported to promote multi-drug resistance by targeting CD74-related pathways [26]. Besides, ectopic expression of YAP facilitated NSCLC cell migration and invasion [28]. Consistent with these studies, we showed that YAP upregulation abolished the effects of miR-146b-5p restoration, suggesting that miR-146b-5p restoration inhibited Gefitinib resistance and NSCLC development by depleting YAP. Additionally, YAP overexpression promoted the expression of PD-L1 in EGFR-TKIs-resistant lung cancer cells, and PD-L1 knockdown suppressed PC9/GR cell proliferation and migration [17]. Aberrant expression of PD-L1 on the surface of cancer cells is a key event in the immune escape of various cancers [29]. For instance, YAP-induced PD-L1 expression drove immune escape in BRAF inhibitor-resistant melanoma [30]. Here, we found that Gefitinib resistance and cell growth promoted by YAP overexpression were largely weakened by PD-L1 knockdown. We inferred that circ_0014235 governed the miR-146b-5p/YAP axis, and in turn regulated PD-L1 expression, thus playing effects on immune escape.
Taken together, circ_0014235 was overexpressed in Gefitinib-resistant NSCLC cells. Circ_0014235 knockdown suppressed IC50 of Gefitinib, cell proliferation and colony formation and induced cell apoptosis and cell cycle arrest in Gefitinib-resistant NSCLC cells. Circ_0014235 governed the miR-146b-5p/YAP axis and then regulated the expression of PD-L1, thereby regulating Gefitinib resistance, cancer development and immune escape in Gefitinib-resistant NSCLC. This study first revealed the role of circ_0014235 in NSCLC with Gefitinib resistance, which provided novel insights into the understanding of the development of chemoresistance in NSCLC.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- [1].Siegel RL, Miller KD, Jemal A.. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- [2].Recondo G, Facchinetti F, Olaussen KA, et al. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nature Reviews Clinical Oncology. 2018;15(11):694–708. [DOI] [PubMed] [Google Scholar]
- [3].Ettinger DS, Wood DE, Aisner DL, et al. Non-small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15:(4):504–535. [DOI] [PubMed] [Google Scholar]
- [4].Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv192–iv237. [DOI] [PubMed] [Google Scholar]
- [5].Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of Non–Small-Cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–792. [DOI] [PubMed] [Google Scholar]
- [6].Masucci GV, Cesano A, Eggermont A, et al. The need for a network to establish and validate predictive biomarkers in cancer immunotherapy. J Transl Med. 2017;15(1):223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li B, Zhu L, Lu C, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12(1):295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Di Timoteo G, Rossi F, Bozzoni I. Circular RNAs in cell differentiation and development. Development 2020;147(16):dev182725. [DOI] [PubMed] [Google Scholar]
- [9].Zhang PF, Pei X, Li KS, et al. Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Molecular Cancer. 2019;18(1):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Huang Y, Dai Y, Wen C, et al. circSETD3 contributes to acquired resistance to gefitinib in non-small-cell lung cancer by targeting the miR-520h/ABCG2 pathway. Molecular Therapy - Nucleic Acids. 2020;21:885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bulk E, Sargin B, Krug U, et al. S100A2 induces metastasis in non-small cell lung cancer. Clinical Cancer Research. 2009;15(1):22–29. [DOI] [PubMed] [Google Scholar]
- [12].Wang Y, Zhang H, Wang J, et al. Circular RNA expression profile of lung squamous cell carcinoma: identification of potential biomarkers and therapeutic targets. Biosci Rep. 2020;40(4):BSR20194512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Xu X, Tao R, Sun L, et al. Exosome-transferred hsa_circ_0014235 promotes DDP chemoresistance and deteriorates the development of non-small cell lung cancer by mediating the miR-520a-5p/CDK4 pathway. Cancer Cell Int. 2020;20(1):552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li Y, Zhang H, Dong Y, et al. MiR-146b-5p functions as a suppressor miRNA and prognosis predictor in non-small cell lung cancer. J Cancer. 2017;8(9):1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67–79. [DOI] [PubMed] [Google Scholar]
- [16].Garcia P, Rosa L, Vargas S, et al. Hippo-YAP1 is a prognosis marker and potentially targetable pathway in advanced gallbladder cancer. Cancers (Basel). 2020;12(4):778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lee BS, Park DI, Lee DH, et al. Hippo effector YAP directly regulates the expression of PD-L1 transcripts in EGFR-TKI-resistant lung adenocarcinoma. Biochem Biophys Res Commun. 2017;491(2):493–499. [DOI] [PubMed] [Google Scholar]
- [18].Hong S-H. Hippo pathway as another oncogenic mediator to promote immune evasion by PD-L1 signaling.Journal of Thoracic Disease. 2019;11(S3):S318–S321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shibata M, Ham K, Hoque MO. A time for YAP1: tumorigenesis, immunosuppression and targeted therapy. Int J Cancer. 2018;143(9):2133–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Huang G, Zhu H, Shi Y, et al. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10(6):e0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Miao J, Hsu PC, Yang YL, et al. YAP regulates PD-L1 expression in human NSCLC cells. Oncotarget 2017;8(70):114576–114587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhou Y, Zheng X, Xu B, et al. Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem Biophys Res Commun. 2019;508(2):527–535. [DOI] [PubMed] [Google Scholar]
- [23].Wen C, Xu G, He S, et al. Screening circular rnas related to acquired gefitinib resistance in non-small cell lung cancer cell lines. J Cancer. 2020;11(13):3816–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu YN, Tsai MF, Wu SG, et al. miR-146b-5p enhances the sensitivity of NSCLC to EGFR tyrosine kinase inhibitors by regulating the IRAK1/NF-kappaB pathway. Molecular Therapy - Nucleic Acids. 2020;22:471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Guo L, Chen Y, Luo J, et al. YAP 1 overexpression is associated with poor prognosis of breast cancer patients and induces breast cancer cell growth by inhibiting PTEN. FEBS Open Bio. 2019;9(3):437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Song Y, Sun Y, Lei Y, et al. YAP1 promotes multidrug resistance of small cell lung cancer by CD74-related signaling pathways. Cancer Med. 2020;9(1):259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Muhammad JS, Guimei M, Jayakumar MN, et al. Estrogen-induced hypomethylation and overexpression of YAP1 facilitate breast cancer cell growth and survival. Neoplasia 2021;23(1):68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yu M, Chen Y, Li X, et al. YAP1 contributes to NSCLC invasion and migration by promoting Slug transcription via the transcription co-factor TEAD. Cell Death Dis. 2018;9(5):464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kim MH, Kim CG, Kim SK, et al. YAP-induced PD-L1 expression drives immune evasion in BRAFi-resistant melanoma. Cancer Immunol Res. 2018;6(3):255–266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


