Abstract
Yeasts are emerging as important etiological agents of nosocomial bloodstream infections. A multiplex PCR method was developed to rapidly identify clinically important yeasts that cause fungemia. The method amplified the internal transcribed spacer 1 (ITS1) region between the 18S and 5.8S rRNA genes and a specific DNA fragment within the ITS2 region of Candida albicans. With this method, C. albicans produced two amplicons, whereas other species produced only one. Through sequence analysis, the precise lengths of the PCR products were found to be as follows: C. glabrata (482 or 483 bp), C. guilliermondii (248 bp), C. parapsilosis (229 bp), C. albicans (218 or 219 and 110 bp), C. tropicalis (218 bp), Cryptococcus neoformans (201 bp), and C. krusei (182 bp). The PCR products could be effectively separated by disk polyacrylamide gel electrophoresis. The method was used to test 249 positive blood cultures (255 isolates), from which the following species (strain number) were isolated: C. albicans (128), C. tropicalis (51), C. glabrata (28), C. parapsilosis (23), C. neoformans (9), C. krusei (5), C. guilliermondii (3), and other, minor species (8). The test sensitivity of the method was 96.9% (247 of 255 isolates). The eight minor species were either misidentified (one strain) or not identified (seven strains). From the time at which a positive bottle was found, the multiplex PCR could be completed within 8 h; the present method is simpler than any previously reported molecular method for the identification of blood yeasts.
Yeasts are emerging as important etiological agents of bloodstream infections (22, 23), a complication associated with a high mortality rate (1, 3). This problem is compounded by an increase in resistance to antifungal agents, particularly the azoles (8, 18, 22, 25, 28, 29, 31) and amphotericin B (19). Bloodstream fungal infections constitute a serious health problem because of the excessive hospital stay, added health care costs, and high morbidity and mortality attributed to the diseases (39).
Candida albicans, C. tropicalis, C. glabrata, C. parapsilosis, C. krusei, and Cryptococcus neoformans are the most common yeasts causing bloodstream infections (2, 23). These six species may account for 95 to 98% of all blood yeasts (19, 23, 27). C. guilliermondii and other, minor species may be isolated occasionally (2, 32). The rates of isolation of the major yeast species causing fungemia have been determined in several studies: C. albicans (50 to 59%), C. tropicalis (11 to 25%), C. glabrata (8 to 18%), C. parapsilosis (7 to 15%), C. krusei (2 to 4%), C. neoformans (∼2%), and other species (∼2%) (1, 19, 23, 27, 28). Therefore, rapid identification of blood yeasts could be targeted solely for these species, although the possibility of other, rarely encountered species always exists.
Fluconazole, which has a low level of toxicity, has been reported to be as effective as amphotericin B for the treatment of candidemia in patients without neutropenia (26), although C. glabrata and C. krusei are innately more resistant to fluconazole. The MIC50s of fluconazole for C. krusei and C. glabrata are 32 and 16 μg/ml, respectively; both values are much higher than those for C. albicans (0.25 μg/ml), C. tropicalis (1 μg/ml), and C. parapsilosis (1 μg/ml) (23). Therefore, earlier information regarding the species causing fungemia may help physicians to select appropriate antifungal agents and regimens to treat patients. The rate of isolation of C. glabrata from blood cultures has increased from 8% during the period from 1952 to 1992 to 18 to 20% in recent surveys (22, 23). This increase might be due to the widespread use of fluconazole for prophylaxis and treatment of candidiasis, affirming the need for more rapid and accurate identification.
At present, the identification of yeasts in positive blood cultures by use of conventional morphological and metabolic characteristics requires from one to several days after isolation. In order to decrease that time, methods devised for the rapid diagnosis of fungal infections include detection of antibody (42), cell wall mannan (5), enolase (37), and specific antibody in combination with PCR to detect C. albicans DNA (15). Efforts have been directed toward molecular testing, such as the use of rRNA genes (rDNA), for species identification. PCR followed by hybridization of the amplicons with species-specific probes has also been used to detect a variety of fungi (6, 7, 9–11, 21, 24, 30, 35, 36). Nested PCR (13, 17, 20, 33) or PCR followed by restriction enzyme analysis (16, 41) has also been used to detect several fungal pathogens. The above methods have shown promise for the diagnosis of fungal infections but have problems that prevent their routine use in a clinical laboratory. For example, the DNA hybridization technique normally involves multiple steps of incubation and washing under stringently controlled conditions, which are both time-consuming and cumbersome. The use of nested PCR or PCR in conjunction with restriction enzyme analysis, however, may add needless complexity to assay procedures.
Recently, a fluorescent capillary electrophoresis system was developed to identify fungi by use of the length variability of the internal transcribed spacer 2 (ITS2) genetic region (34). However, the fragment lengths of the ITS2 regions were similar in several important yeasts that cause fungemia, thereby preventing the identification of some species. The aim of the present study was to evaluate a multiplex PCR method for the identification of C. neoformans and Candida species that are frequently isolated from blood cultures. The method was based on the size variability of the ITS1 regions in different species and on the amplification of a specific DNA fragment of the ITS2 region of C. albicans.
MATERIALS AND METHODS
Yeast strains.
A total of 22 stock yeast cultures were used in this study (Table 1). Among these cultures, 18 strains were obtained from the Culture Collection and Research Center (CCRC, Hsinchu, Taiwan), and the remaining 4 strains were clinical isolates.
TABLE 1.
Pure yeast cultures used in this study and the lengths of PCR products
| Organism | Straina | PCR product (bp) |
|---|---|---|
| C. krusei | CCRC 20514 | 182 |
| CCRC 21720 | 182 | |
| CCRC 22342 | 182 | |
| Cryptococcus neoformans | CCRC 20528 | 201 |
| CCRC 22241 | 201 | |
| CCRC 20532 | 201 | |
| 9966 | 201 | |
| 7280 | 201 | |
| C. tropicalis | CCRC 20520 | 218 |
| CCRC 20521 | 218 | |
| CCRC 21436 | 218 | |
| C. albicans | CCRC 20512 | 219, 110 |
| CCRC 20513 | 218, 110 | |
| CCRC 21538 | 218, 110 | |
| C. parapsilosis | CCRC 20515 | 229 |
| CCRC 21253 | 229 | |
| CCRC 21544 | 229 | |
| C. guilliermondii | CCRC 21500 | 248 |
| CCRC 21599 | 248 | |
| C. glabrata | CCRC 20586 | 482 |
| 2332 | 483 | |
| C3-3 | 483 |
Strains without a CCRC designation were clinical isolates.
DNA extraction from pure cultures.
Stock cultures of yeasts were subcultured on Sabouraud dextrose agar (Difco, Detroit, Mich.) and incubated at 37°C. Colonies of these strains were suspended in saline to obtain the turbidity of a 0.5 McFarland standard at a 530-nm wavelength. Two microliters of cell suspension was added to 18 μl of microLYSIS solution (Microzone Limited, East Sussex, United Kingdom) in a 0.2-ml Eppendorf tube and overlaid with 20 μl of sterilized mineral oil. The tube was heated in a thermal cycler (OmniGen; Hybaid Limited, Middlesex, United Kingdom) using the following temperature profile, as recommended by the manufacturer: 65°C, 5 min; 96°C, 2 min; 65°C, 4 min; 96°C, 1 min; 65°C, 1 min; 96°C, 30 s; and 30°C, 5 min. After cycling, the lysis solution-DNA mixture was used directly for PCR amplification or stored at −20°C for further use. Escherichia coli ATCC 25922, Staphylococcus aureus 0400, Klebsiella pneumoniae 03583, Enterobacter cloacae 00109, and Streptococcus pneumoniae 0424 were cultivated on blood agar at 37°C for 18 to 24 h, and the bacterial DNA was extracted in a manner similar to that used for pure yeast cultures.
Clinical specimens.
Blood samples were collected from the National Cheng Kung University Medical Center, Tainan, Taiwan, and from Chang Gung Memorial Hospital. BACTEC blood culture bottles (Becton Dickinson Microbiology Systems, Cockeysville, Md.) were normally inoculated with 3 to 10 ml of blood from patients, inserted into the BACTEC NR660 instrument (Becton Dickinson Microbiology Systems), and incubated at 37°C. Gram stain smears of aliquots from positive bottles were prepared to check for the presence of yeasts. A total of 249 positive blood culture bottles containing yeasts were analyzed in this study. The blood yeasts isolated on subculture plates were identified by conventional procedures based on phenotypic and biochemical reactions (38).
Isolation of yeast DNA from positive blood cultures.
The method of Fujita et al. (9) was used with a small modification to extract yeast DNA from the positive culture broths. An aliquot (0.2 ml) of positive broth containing yeasts was added to 0.8 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) containing 0.05% proteinase K (Worthington Biochemical Inc., Lakewood, N.J.) and 0.05% Tween 20. The cell suspension was incubated at 55°C for 30 min and then centrifuged at 8,000 × g for 10 min in a microcentrifuge. The pellet was washed with 0.5 ml of TE buffer containing 0.5% Tween 20 and then with 0.5 ml of SE solution (1 M sorbitol, 0.1 M EDTA). After centrifugation at 8,000 × g for 10 min, the pellet was suspended in 0.5 ml of Lyticase solution (10 mg/ml; Sigma Chemical Co., St. Louis, Mo.) and incubated at 37°C for 1 h. After centrifugation, the pellet was suspended in 10 μl of TE buffer, and 1 μl of the suspension was added to 19 μl of microLYSIS solution. The suspension was heated in a thermal cycler to extract yeast DNA as previously described for pure cultures. Seven randomly selected positive blood cultures containing bacteria were processed in the same manner for DNA extraction. In addition, DNA was extracted from two blood samples from healthy individuals for PCR assay.
PCR amplification.
The fungus-specific, universal primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS2 (5′-GCT GCG TTC TTC ATC GAT GC-3′) (40) were used to amplify a small conserved portion of the 18S rDNA region, the adjacent ITS1, and a small portion of the 28S rDNA region. In addition, C. albicans-specific primers CA3 (5′-GGT TTG CTT GAA AGA CGG TAG-3′) and CA4 (5′-AGT TTG AAG ATA TAC GTG GTA G-3′) (12) were also included in the PCR mixture to amplify a portion of the ITS2 region of C. albicans. The four primers (ITS1, ITS2, CA3, and CA4) were synthesized at TIB MOLBIOL (Berlin, Germany). Multiplex PCR was performed with 2 μl (1 to 5 ng) of template DNA in a total reaction volume of 50 μl consisting of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.8 mM deoxyribonucleoside triphosphates (0.2 mM each), 3.2 μM primers (ITS1 and ITS2, 0.4 μM each; CA3 and CA4, 1.2 μM each), Taq DNA polymerase (1.25 U), and 50 μl of a mineral oil overlay. PCR was carried out with an OmniGen thermal cycler under the following conditions: initial denaturation, 94°C, 3 min; 35 cycles of denaturation (94°C, 1 min), annealing (60°C, 1 min), and extension (72°C, 1 min); and final extension, 72°C, 5 min. A negative control run was performed with each test run by replacing the template DNA with sterilized water in the PCR mixture. A positive culture broth containing C. albicans was run in parallel with unknown samples, and this culture broth was used as a positive control.
Limit of detection of C. albicans in blood.
To determine the limit of detection of the multiplex PCR, whole blood was seeded with C. albicans CCRC 20512 to reach a concentration of 2 × 105 CFU/ml. The seeded blood was serially diluted 10-fold with whole blood, and 0.2 ml of the diluted samples was used for PCR as described above. The cell numbers (CFU per milliliter) of the diluted cell suspensions were determined by the plate count method (11) with Sabouraud dextrose agar as the culture medium. Plates were incubated at 35°C for 48 h before enumeration.
Determination of the lengths of the PCR products.
To determine the precise lengths of the PCR products amplified by primers ITS1 and ITS2, the amplicons were purified by using a PCR cleanup kit (Viogene, Sunnyvale, Calif.) and were directly cycle sequenced in both directions with an ABI Prism 377 automated system (Applied Biosystems, Taipei, Taiwan). The size of the fragment amplified from C. albicans by primers CA3 and CA4 was determined in a similar way. For each species, two to five strains were sequenced in both directions to determine the precise lengths of the amplicons (Table 1). The PCR products of several minor species (C. famata, C. lusitaniae, C. pelliculosa, Rhodotorula rubra, and Trichosporon beigelii) isolated in this study were also sequenced to determine the precise lengths of their amplicons.
Disk PAGE.
PCR products were analyzed by disk polyacrylamide gel electrophoresis (PAGE) (4) with a minigel system (Mini-Protean II; Bio-Rad, Hercules, Calif.). The running gel had an acrylamide concentration of 9% and was 0.75 mm in thickness. The time required for running electrophoresis was 3 h. After electrophoresis, the gels were stained with ethidium bromide (0.5 μg/ml) and viewed with an IS-1000 digital imaging system (Alpha Innotech Corporation, San Leandro, Calif.). In addition to the 50-bp DNA ladder, equal amounts (20 μl) of the PCR products amplified from C. glabrata CCRC 20586 (482 bp), C. guilliermondii CCRC 21500 (248 bp), C. parapsilosis CCRC 20515 (229 bp), C. albicans CCRC 20512 (219 and 110 bp), C. tropicalis CCRC 20520 (218 bp), C. neoformans CCRC 20528 (201 bp), and C. krusei CCRC 20514 (182 bp) were mixed to serve as markers for species identification. The species markers were run in parallel with the PCR products amplified from unknown samples to facilitate side-by-side comparisons between the markers and the PCR products amplified from the blood samples.
Definition of test sensitivity and specificity.
For identification of the seven yeast species (C. albicans, C. tropicalis, C. parapsilosis, C. glabrata, C. krusei, C. guilliermondii, and C. neoformans) in blood cultures, the sensitivity of the multiplex PCR was defined as the number of strains of these species correctly identified (true positives) divided by the total number of yeast strains isolated. The test specificity was defined as the number of strains which did not belong to the above seven species and were not identified as any one of the seven major species (true negatives) divided by the total number of strains not included in these seven species (14).
RESULTS
Fragment analysis of PCR products.
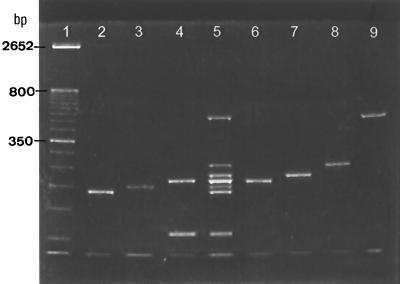
Through sequence analysis, the precise lengths of the PCR products amplified by the fungus-specific, universal primers ITS1 and ITS2 were determined: C. krusei (182 bp), C. neoformans (201 bp), C. tropicalis (218 bp), C. albicans (218 or 219 bp), C. parapsilosis (229 bp), C. guilliermondii (248 bp), and C. glabrata (482 or 483 bp) (Table 1). Different strains of the same species produced PCR products having the same length or differing in length by only 1 bp, and the PCR products of different species could be separated more easily by disk PAGE than by agarose gel electrophoresis. This was especially true for separating C. parapsilosis (229 bp) from C. tropicalis (218 bp). Another problem was that amplicons of the ITS1 regions of C. albicans (218 or 219 bp) and C. tropicalis (218 bp) had the same mobility on polyacrylamide gels (Fig. 1, lanes 4 and 6). In order to discriminate between these two species, primers CA3 and CA4, which are specific for C. albicans (12), were included in the PCR mixture, and an additional product was obtained for the organism (Fig. 1, lane 4). With the multiplex approach, C. albicans produced two amplicons (218 or 219 and 110 bp) (Fig. 1, lane 4), whereas each of the remaining six species produced only one.
FIG. 1.
Multiplex PCR using primers ITS1, ITS2, CA3, and CA4. Lane 1, 50-bp DNA ladder. Lanes 2 to 4, C. krusei CCRC 20514, C. neoformans CCRC 20528, and C. albicans CCRC 20512, respectively. Lane 5, species markers formulated from amplicons of the seven major yeast species; the bands from top to bottom were PCR products of C. glabrata, C. guilliermondii, C. parapsilosis, C. tropicalis and C. albicans, C. neoformans, C. krusei, and C. albicans, respectively. Lanes 6 to 9, C. tropicalis CCRC 20520, C. parapsilosis CCRC 20515, C. guilliermondii CCRC 21500, and C. glabrata CCRC 20586, respectively.
Species designation was possible by comparing the electrophoretic mobilities of the amplicons with the commercial 50-bp DNA ladder (Fig. 1, lane 1). Species identification was facilitated by running PCR products in parallel with the species markers containing amplicons of the seven individual species (Fig. 1, lane 5), enabling side-by-side comparisons of an unknown with the species markers.
Limit of detection of the PCR.
The limit of detection of the multiplex PCR for C. albicans CCRC 20512 artificially inoculated in whole blood was approximately 20 CFU/ml (data not shown). With serially diluted DNA in water, the limit of detection of the PCR was 4 pg of C. albicans DNA per assay. The limit of detection of yeast DNA was very close to that (10 pg DNA) reported by Jaeger et al. (13) and was approximately equal to 100 cells (37 fg of DNA per cell of C. albicans).
Identification of yeasts in positive blood cultures.
A total of 249 positive blood cultures containing yeasts were analyzed by the multiplex PCR for species identification. From these blood cultures, 255 strains of yeasts were isolated. The most frequently isolated species was C. albicans (128 strains, 50.4%), followed by C. tropicalis (51 strains, 19.7%), C. glabrata (28 strains, 11%), C. parapsilosis (23 strains, 9.1%), C. neoformans (9 strains, 3.5%), C. krusei (5 strains, 2%), C. guilliermondii (3 strains, 1.2%), and other species (8 strains, 3.1%) (Table 2). All strains of the above species, except for the eight minor species, were correctly identified, resulting in a test sensitivity of 100% for each of the above seven species (Table 2). However, the test sensitivity for the PCR assay was 96.9%, based on the total number (247 strains) of yeasts identified divided by the total number (255 strains) of yeasts isolated.
TABLE 2.
Results of multiplex PCR for the identification of the seven major yeast species that cause fungemia
| Organism | No. of strains
|
%
|
|||
|---|---|---|---|---|---|
| Tested | Identified | Misidentified | Sensitivity | Specificity | |
| C. albicans | 128 | 128 | 0 | 100 | |
| C. tropicalis | 51 | 51 | 0 | 100 | |
| C. glabrata | 28 | 28 | 0 | 100 | |
| C. parapsilosis | 23 | 23 | 0 | 100 | |
| C. neoformans | 9 | 9 | 0 | 100 | |
| C. krusei | 5 | 5 | 0 | 100 | |
| C. guilliermondii | 3 | 3 | 0 | 100 | |
| Other species | 8 | 0 | 1a | 87.5 | |
R. rubra was misidentified as C. parapsilosis.
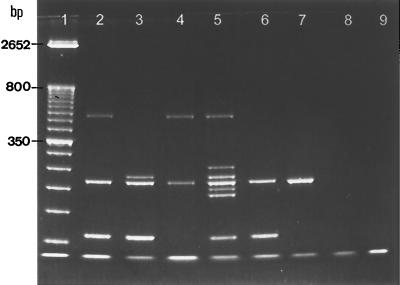
Among the 249 positive blood cultures, 16 were mixed cultures. Ten of these mixed cultures were from polymicrobial infections, with one strain of yeast and one strain of bacterium being isolated from each of the 10 blood samples. Coexisting bacteria in blood specimens did not produce any detectable PCR products and did not interfere with yeast identification (Fig. 2, lanes 6 and 7). The remaining six mixed cultures were polyfungal, with each containing two yeast strains, which could be simultaneously identified by the multiplex PCR. Three examples are shown in Fig. 2; lane 2 depicts PCR products from a mixed culture of C. albicans and C. glabrata, lane 3 contains a mixed culture of C. albicans and C. parapsilosis, and lane 4 contains a mixed culture of C. tropicalis and C. glabrata. All isolates from these mixed cultures were confirmed by conventional isolation and identification methods.
FIG. 2.
Identification of yeasts present in mixed cultures by the multiplex PCR. Lane 1, 50-bp DNA ladder. Lane 2, C. albicans and C. glabrata. Lane 3, C. albicans and C. parapsilosis. Lane 4, C. tropicalis and C. glabrata. Lane 5, species markers. Lane 6, C. albicans and E. coli. Lane 7, C. tropicalis and E. cloacae. Lane 8, sample of human blood. Lane 9, negative control.
Specificity of the multiplex PCR.
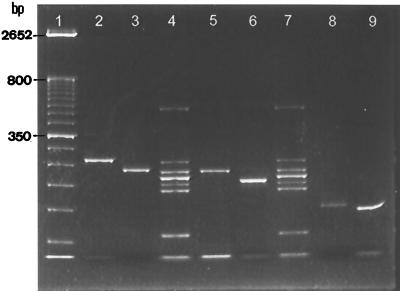
The eight miscellaneous strains included three strains of Candida spp. and one strain of each of the following species: C. famata, C. lusitaniae, C. pelliculosa, R. rubra, and T. beigelii. As shown in Fig. 3, the electrophoretic mobilities of amplicons of C. pelliculosa (262 bp, lane 2), C. famata (236 bp, lane 3), T. beigelii (196 bp, lane 6), C. lusitaniae (147 bp, lane 8), and one undetermined Candida species (lane 9) were different from those of the seven species markers; hence, the five species were not identified. However, the PCR products amplified from R. rubra (232 bp; Fig. 3, lane 5) and C. parapsilosis (229 bp, lane 4) were only 3 bp apart; therefore, R. rubra was misidentified as C. parapsilosis, resulting in a test specificity of 87.5% (seven of eight strains). The relatively low specificity was due to the limited proportion (3.1%; 8 of 255 strains) of minor yeast species recovered from positive blood cultures.
FIG. 3.
Identification of minor yeast species present in positive blood cultures by the multiplex PCR. Lane 1, 50-bp DNA ladder. Lane 2, C. pelliculosa. Lane 3, C. famata. Lane 4, species markers. Lane 5, R. rubra. Lane 6, T. beigelii. Lane 7, species markers. Lane 8, C. lusitaniae. Lane 9, Candida sp. R. rubra (lane 5) was misidentified as C. parapsilosis.
The sample of whole blood (Fig. 2, lane 8) was negative, as were seven randomly selected positive blood cultures containing the following bacteria: E. coli, S. aureus, K. pneumoniae, Pseudomonas aeruginosa, Serratia marcescens, E. cloacae, and S. pneumoniae (data not shown). Furthermore, no PCR products were obtained by using template DNA extracted from pure cultures of E. coli 03190 and ATCC 25922, S. aureus 0400, K. pneumoniae 03583, E. cloacae 00109, and S. pneumoniae 0424 (data not shown).
DISCUSSION
This report describes the use of multiplex PCR to identify the most frequently encountered yeasts in blood cultures. The method used universal fungal primers ITS1 and ITS2 to amplify a conserved portion of the 18S rDNA region, the adjacent ITS1 region, and a small portion of the 5.8S rDNA region, yielding products with variable sizes among the major species causing fungemia. Another primer pair (CA3 and CA4) was used to amplify a specific DNA fragment of the ITS2 region of C. albicans. With disk PAGE, the PCR products could be effectively separated and recognized, even though they differed by a few base pairs. From the time at which a blood culture become positive, the multiplex method reduced the identification time for yeasts from approximately 1 to 3 days by routine identification methods to about 8 h. Another advantage of the method was that multiple yeast species coexisting in a blood culture could be detected at the same time (Fig. 2, lanes 2 to 4).
It is generally perceived that DNA extraction from yeasts either by lysis of enzymes (9, 21, 36) or by bead sonication (34) followed by phenol-chloroform extraction is the most tedious and cumbersome step of a PCR-based identification method and limits its use in a routine clinical laboratory. We found that a commercial extraction kit (microLYSIS) was an effective and simple method for extracting DNA from pure yeast cultures within 30 min. The only step used for DNA extraction with this kit was heating of yeast cell suspensions in the lysis solution in a thermal cycler, eliminating the use of phenol-chloroform and alcohol for DNA purification and precipitation, respectively. However, for extraction of yeast DNA from blood cultures, we found that a prior step of lysing blood cells with proteinase K followed by Lyticase digestion of the yeast cell walls was necessary to obtain good results.
The efficacy of the multiplex method relies on several factors. First, the concentration of yeast cells in positive blood cultures normally exceeds 105 CFU/ml (2). Second, fungal rDNA has a high copy number (40 to 80 copies per haploid genome) (40). Third, fungemia is usually caused by a limited number of fungal species. The most commonly encountered yeast species (C. albicans, C. tropicalis, C. glabrata, C. parapsilosis, C. krusei, and C. neoformans) may represent ≥95% of the total yeasts recovered from blood cultures (19, 23, 26).
Finally, Turenne et al. determined that universal fungal primers ITS3 and ITS4 and an automated system of fluorescent capillary electrophoresis could be used to determine the sizes of amplicons of the ITS2 genetic regions of some fungi (34). However, even with this sophisticated technique, it was still difficult to differentiate the PCR products of C. albicans (279 bp) and C. krusei (282 bp) by chromatographic retention times. Adapting this approach to the multiplex PCR method developed in this study, however, produced a method with a high sensitivity (96.9%). The relatively low specificity (87.5%) was due to a limited proportion (3.1%; 8 of 255 strains) of the minor yeast species recovered from our blood cultures. Starting from a positive blood culture, this method can be completed within 8 h and is simpler than any previously reported molecular method for the identification of blood yeasts.
ACKNOWLEDGMENTS
This work was supported by grant NSC 89-2314-B-006-101 from the National Science Council, Taipei, Taiwan, Republic of China.
We thank the medical technicians at the Department of Pathology, National Cheng Kung University Medical Center, for help in identifying all the clinical blood isolates.
REFERENCES
- 1.Beck-Sague C, Jarvis W R. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. National Nosocomial Infections Surveillance System. J Infect Dis. 1993;167:1247–1251. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 2.Chang H C, Chang J J, Huang A H, Chang T C. Evaluation of a capacitance method for direct antifungal susceptibility testing of yeasts in positive blood cultures. J Clin Microbiol. 2000;38:971–976. doi: 10.1128/jcm.38.3.971-976.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y C, Chang S C, Sun C C, Yang L S, Hseih W C, Luh K T. Secular trends in the epidemiology of nosocomial fungal infections at a teaching hospital in Taiwan, 1981 to 1993. Infect Control Hosp Epidemiol. 1997;18:369–375. doi: 10.1086/647628. [DOI] [PubMed] [Google Scholar]
- 4.Cooper T G. The tools of biochemistry. New York, N.Y: John Wiley & Sons, Inc.; 1977. pp. 228–231. [Google Scholar]
- 5.de Repentigny L, Marr L D, Keller J W, Carter A W, Kuykendall R J, Kaufman L, Reiss E. Comparison of enzyme immunoassay and gas-liquid chromatography for the rapid diagnosis of invasive candidiasis in cancer patients. J Clin Microbiol. 1985;21:972–979. doi: 10.1128/jcm.21.6.972-979.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Einsele H, Hebart H, Roller G, Löffler J, Rothenhöfer I, Müller C A, Bowden R A, van Burik J-A, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elie C M, Lott T J, Reiss E, Morrison C J. Rapid identification of Candida species with species-specific DNA probes. J Clin Microbiol. 1998;36:3260–3265. doi: 10.1128/jcm.36.11.3260-3265.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espinel-Ingroff A, Kish C W, Jr, Kerkering T M, Fromtling R A, Bartizal K, Galgiani J N, Villareal K, Pfaller M A, Gerarden T, Rinaldi M G, Fothergill A. Collaborative comparison of broth macrodilution and microdilution antifungal susceptibility tests. J Clin Microbiol. 1992;30:3138–3145. doi: 10.1128/jcm.30.12.3138-3145.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita S-I, Lasker B A, Lott T J, Reiss E, Morrison C J. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J Clin Microbiol. 1995;33:962–967. doi: 10.1128/jcm.33.4.962-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hee Shin J, Nolte F S, Morrison C J. Rapid identification of Candida species in blood cultures by a clinically useful PCR method. J Clin Microbiol. 1997;35:1454–1459. doi: 10.1128/jcm.35.6.1454-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry T, Iwen P C, Hinrichs S H. Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J Clin Microbiol. 2000;38:1510–1515. doi: 10.1128/jcm.38.4.1510-1515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang C H. Specific identification of yeasts on the bases of PCR-amplified ribosomal DNA immobilized by covalent bond on the piezoelectric quartz crystal. Ph.D. thesis. Taipei, Taiwan: National Taiwan University; 1996. [Google Scholar]
- 13.Jaeger E E M, Carroll N M, Choudhury S, Dunlop A A S, Towler H M A, Matheson M M, Adamson P, Okhravi N, Lightman S. Rapid detection and identification of Candida, Aspergillus, and Fusarium species in ocular samples using nested PCR. J Clin Microbiol. 2000;38:2902–2908. doi: 10.1128/jcm.38.8.2902-2908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClure F D. Design and analysis of quantitative collaborative studies: minimum collaborative program. J Assoc Off Anal Chem. 1990;73:953–960. [PubMed] [Google Scholar]
- 15.Miyakawa Y, Mabuchi T, Fukazawa Y. New method for detection of Candida albicans in human blood by polymerase chain reaction. J Clin Microbiol. 1993;31:3344–3347. doi: 10.1128/jcm.31.12.3344-3347.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morace G, Pagano L, Sanguinetti M, Posteraro B, Mele L, Equitani F, D'Amore G, Leone G, Fadda G. PCR-restriction enzyme analysis for detection of Candida DNA in blood from febrile patients with hematological malignancies. J Clin Microbiol. 1999;37:1871–1875. doi: 10.1128/jcm.37.6.1871-1875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagai H, Yamakami Y, Hashimoto A, Tokimatsu I, Nasu M. PCR detection of DAN specific for Trichosporon species in serum of patients with disseminated trichosporonosis. J Clin Microbiol. 1999;37:694–699. doi: 10.1128/jcm.37.3.694-699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman S L, Flanigan T P, Fisher A, Rinaldi M G, Stein M, Vigilante K. Clinically significant mucosal candidiasis resistant to fluconazole treatment in patiens with AIDS. Clin Infect Dis. 1994;19:684–686. doi: 10.1093/clinids/19.4.684. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen M H, Peacock J E, Jr, Morris A J, Tanner D C, Nguyen M L, Snydman D R, Wagener M M, Rinaldi M G, Yu V L. The changing face of candidemia: emergence of non-Candida albicans species and antifungal resistance. Am J Med. 1996;100:617–623. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 20.Niesters H G M, Goessens W H F, Meis J F M G, Quint W G V. Rapid, polymerase chain reaction-based identification assays for Candida species. J Clin Microbiol. 1993;31:904–910. doi: 10.1128/jcm.31.4.904-910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S, Wong M, Marras S A E, Cross E W, Kiehn T E, Chaturvedi V, Tyagi S, Perlin D S. Rapid identification of Candida dubliniensis using a species-specific molecular beacon. J Clin Microbiol. 2000;38:2829–2836. doi: 10.1128/jcm.38.8.2829-2836.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaller M A, Jones R N, Messer S A, Edmond M B, Wenzel R P. National surveillance of nosocomial blood stream infection due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE Program. Diagn Microbiol Infect Dis. 1998;30:121–129. doi: 10.1016/s0732-8893(97)00240-x. [DOI] [PubMed] [Google Scholar]
- 23.Pfaller M A, Messer S A, Hollis R J, Jones R N, Doern G V, Brandt M E, Hajjeh R A. Trends in species distribution and susceptibility to fluconazole among blood stream isolates of Candida species in the United States. Mycology. 1999;33:217–222. doi: 10.1016/s0732-8893(98)00160-6. [DOI] [PubMed] [Google Scholar]
- 24.Posteraro B, Sanguinetti M, Masucci L, Romano L, Morace G, Fadda G. Reverse cross blot hybridization assay for rapid detection of PCR-amplified DNA from Candida species, Cryptococcus neoformans, and Saccharomyces cerevisiae in clinical samples. J Clin Microbiol. 2000;38:1609–1614. doi: 10.1128/jcm.38.4.1609-1614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price M F, LaRocco M T, Gentry L O. Fluconazole susceptibilities of Candida species and distribution of species recovered from blood cultures over a 5-year period. Antimicrob Agents Chemother. 1994;38:1422–1424. doi: 10.1128/aac.38.6.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rex J H, Bennett J E, Sugar A M, Pappas P G C, van der Horst M, Edwards J E, Washburn R G, Scheld W M, Karchmer A W, Dine A P, Levenstein M J, Webb C D. A randomized trial comparing fluconazole with amphotericin B for the treatment of candidemia in patients without neutropenia. N Engl J Med. 1994;331:1325–1330. doi: 10.1056/NEJM199411173312001. [DOI] [PubMed] [Google Scholar]
- 27.Rex J H, Pfaller M A, Barry A L, Nelson P W, Webb C D. Antifungal susceptibility testing of isolates from a randomized multicenter trial of fluconazole versus amphotericin B as treatment of non-neutropenic patients with candidemia. Antimicrob Agents Chemother. 1995;39:40–44. doi: 10.1128/aac.39.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruhnke M, Eigler A, Tennagen I, Geiseler B, Engelmann E, Trautmann M. Emergence of fluconazole-resistant strains of Candida albicans in patients with recurrent oropharyngeal candidosis and human immunodeficiency virus infection. J Clin Microbiol. 1994;32:2092–2098. doi: 10.1128/jcm.32.9.2092-2098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandhu G S, Kline B C, Stockman L, Roberts G D. Molecular probes for diagnosis of fungal infections. J Clin Microbiol. 1995;33:2913–2919. doi: 10.1128/jcm.33.11.2913-2919.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanguineti A, Carmichael J K, Campbell K. Fluconazole-resistant Candida albicans after long-term suppressive therapy. Arch Intern Med. 1993;153:1122–1124. [PubMed] [Google Scholar]
- 32.Simor A E, Goswell G, Louie L, Lee M, Louie M. Antifungal susceptibility testing of yeast isolates from blood cultures by microbroth dilution and the E Test. Eur J Clin Microbiol Infect Dis. 1997;16:693–697. doi: 10.1007/BF01708563. [DOI] [PubMed] [Google Scholar]
- 33.Skladny H, Buchheidt D, Baust C, Krieg-Schneider F, Seifarth W, Leib-Mösch C, Hehlmann R. Specific detection of Aspergillus species in blood and bronchoalveolar lavage samples of immunocompromised patients by two-step PCR. J Clin Microbiol. 1999;37:3865–3871. doi: 10.1128/jcm.37.12.3865-3871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turenne C Y, Sanche S E, Hoban D J, Karlowsky J A, Kabani A M. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol. 1999;37:1846–1851. doi: 10.1128/jcm.37.6.1846-1851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Burik J-A, Myerson D, Schreckhise R W, Bowden R A. Panfungal PCR assay for detection of fungal infection in human blood specimens. J Clin Microbiol. 1998;36:1169–1175. doi: 10.1128/jcm.36.5.1169-1175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wahyuningsih R, Freisleben H-J, Sonntag H-G, Schnitzler P. Simple and rapid detection of Candida albicans DNA in serum by PCR for diagnosis of invasive candidiasis. J Clin Microbiol. 2000;38:3016–3021. doi: 10.1128/jcm.38.8.3016-3021.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walsh T J, Hathorn W, Sobel J D, Merz W G, Sanchez V, Maret S M, Muckley H R, Pfaller M A, Schanfele R, Silvia C, Navarron E, Lecciones J, Chandrasekar P, Lee J, Pizzu P A. Detection of circulating Candida enolase by immunoassay in patients with cancer and invasive candidiasis. N Engl J Med. 1991;324:1026–1031. doi: 10.1056/NEJM199104113241504. [DOI] [PubMed] [Google Scholar]
- 38.Warren N G, Hazen K C. Candida, Cryptococcus, and other yeasts of medical importance. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 1184–1199. [Google Scholar]
- 39.Weinstein M P, Towns M L, Quartey S M, Mirrett S, Reimer L G, Parmigiani G, Reller L B. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin Infect Dis. 1997;24:584–602. doi: 10.1093/clind/24.4.584. [DOI] [PubMed] [Google Scholar]
- 40.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gefland D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 315–322. [Google Scholar]
- 41.Williams D W, Wilson M J, Lewis M A O, Potts A J C. Identification of Candida species by PCR and restriction fragment length polymorphism analysis of intergenic spacer regions of ribosomal DNA. J Clin Microbiol. 1995;33:2476–2479. doi: 10.1128/jcm.33.9.2476-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young R, Bennett J. Invasive aspergillosis. Absence of detectable antibody response. Am Rev Respir Dis. 1971;104:710–716. doi: 10.1164/arrd.1971.104.5.710. [DOI] [PubMed] [Google Scholar]