Abstract
Cryptosporidium parvum is usually considered the agent of human cryptosporidiosis. However, only in the last few years, molecular biology-based methods have allowed the identification of Cryptosporidium species and genotypes, and only a few data are available from France. In the present work, we collected samples of whole feces from 57 patients from France (11 immunocompetent patients, 35 human immunodeficiency virus [HIV]-infected patients, 11 immunocompromised but non-HIV-infected patients) in whom Cryptosporidium oocysts were recognized by clinical laboratories. A fragment of the Cryptosporidium 18S rRNA gene encompassing the hypervariable region was amplified by PCR and sequenced. The results revealed that the majority of the patients were infected with cattle (29 of 57) or human (18 of 57) genotypes of Cryptosporidium parvum. However, a number of immunocompromised patients were infected with C. meleagridis (3 of 57), C. felis (6 of 57), or a new genotype of C. muris (1 of 57). This is the first report of the last three species of Cryptosporidium in humans in France. These results indicate that immunocompromised individuals are susceptible to a wide range of Cryptosporidium species and genotypes.
Cryptosporidium spp. are apicomplexan protozoa that infect the gastrointestinal or respiratory tracts of humans and animals. In immunocompetent hosts, the infection is typically acute and self-limiting, whereas in immunocompromised individuals, such as persons receiving immunosuppressive drugs and AIDS patients, the infection is often chronic. Since drug therapy for the control or elimination of these organisms is not yet available, persistent infections in these patients are especially severe and can be life-threatening. The potential of Cryptosporidium as an opportunistic parasite and recent reports of major outbreaks of cryptosporidiosis in the United States, the United Kingdom, and Australia due to contamination of drinking-water supplies indicate that Cryptosporidium should be regarded as a major public health problem (11, 25).
To date, eight Cryptosporidium species have been regarded as valid on the basis of host specificity, pathogenesis, and oocyst morphology (12). These include Cryptosporidium parvum in mammals, C. muris in rodents and ruminants, C. felis in domestic cats, C. wrairi in guinea pigs, C. baileyi and C. meleagridis in birds, C. serpentis in reptiles, and C. nasorum in fish. According to this classification, the causative agent of cryptosporidiosis in humans and a range of mammalian species is the species C. parvum. However, C. parvum does not seem to be a uniform species. Indeed, numerous reports from several laboratories have identified two distinct genotypes of C. parvum isolates: the human genotype (genotype 1), which has so far been found exclusively in humans and in a single nonhuman primate, and the cattle genotype (genotype 2), which has been found in domestic livestock such as cattle, sheep, and goats but which can also infect humans (32). More recently, additional new genotypes were distinguished in C. parvum: a mouse genotype that has been found in mice from around the world and in bats, a pig genotype, a marsupial genotype that has been found in koalas and kangaroos, a dog genotype, a ferret genotype, and a monkey genotype (33). Although the human and cattle genotypes were thought to be the only two genotypes infective for human hosts, it has recently been shown that immunocompromised individuals and even immunocompetent individuals are susceptible to more than just these two genotypes of C. parvum. Indeed, C. felis, C. meleagridis, C. muris, and C. parvum dog genotype have been associated with human infections (18, 26, 27, 36, 38, 49). In the absence of effective therapeutic agents, control and treatment are dependent upon early and accurate diagnosis and an accurate understanding of the epidemiology and transmission dynamics. The identification and characterization of Cryptosporidium isolates are therefore important prerequisites for clarifying the epidemiology of Cryptosporidium and for limiting its spread.
In France, only one study on the genetic typing of Cryptosporidium has been published. In that study, Bonnin et al. typed 23 C. parvum isolates using PCR-restriction fragment length polymorphism (RFLP) analysis of a repetitive sequence and found that 10 of 10 isolates from calves and 7 of 13 isolates from human immunodeficiency virus (HIV)-infected individuals had the same profile, indicating zoonotic transmission, whereas 6 of 13 human isolates presented another pattern (3). In the present work, we studied the prevalence of genotypes of C. parvum and other Cryptosporidium species in patients with cryptosporidiosis from France. In order to advance the understanding of the presence and the circulation of these parasites, 57 isolates of Cryptosporidium spp. were characterized at the 18S rRNA gene (rDNA) locus.
MATERIALS AND METHODS
Source of parasite isolates, microscopic examination, and patient data.
A total of 57 human samples testing positive for Cryptosporidium were obtained from laboratories of medical parasitology in France (see Table 1). Isolates were obtained as unpurified fecal samples and were stored at 4°C in 2.5% potassium dichromate solutions. All samples were routinely reexamined microscopically (Axiophot 2 Zeiss microscope) from either direct fecal smears or smears of fecal concentrates (concentration in phosphate-buffered saline–ether) (46), after staining with modified Ziehl-Neelsen stain (16), or by immunofluorescence assay with the Crypto/Giardia-Cel Test IF kit (Cellabs, Biomedical Diagnostics, Marne-la-Vallée, France). Immunological, clinical, and risk factor data for the patients were collected retrospectively, when possible.
TABLE 1.
Isolate genotypes and clinical and epidemiological data for patients with Cryptosporidium infection analyzed in the study
| Isolate code | Sourcea | Age/sexb | Species and genotype by analysis of 18S rDNA marker | Immunological statusc | CD4 count (no./μl) | Other known concurrent infection(s) |
|---|---|---|---|---|---|---|
| H1 | CHD | 6/M | C. parvum cattle genotype (A) | Immunocompetent | Entamoeba coli infection | |
| H5 | CHD | 6/M | C. parvum cattle genotype (A) | Immunocompetent | ||
| H28 | CHD | 4/M | C. parvum cattle genotype (A) | Immunocompetent | ||
| H70 | CHD | <1/F | C. parvum human genotype | Immunocompetent | ||
| H71 | CHD | 21/F | C. parvum cattle genotype (A) | Immunocompetent | ||
| H4 | CHRUL | 3/M | C. parvum human genotype | Immunocompetent | ||
| H26 | CHRUL | 19/F | C. parvum cattle genotype (A) | Transplant | ||
| H41 | CHRUL | 15/F | C. parvum human genotype | Transplant | ||
| H72 | CHRUL | 2/M | C. parvum human genotype | Immunocompetent | ||
| H77 | CHRUL | 8/M | C. parvum cattle genotype (B) | Immunocompetent | ||
| H3 | CHUA | 28/M | C. parvum human genotype | HIV+ | 5 | Candida albicans in feces |
| H32 | CHUA | 44/M | C. parvum cattle genotype (A) | HIV+ | 23 | Giardiosis, toxoplamosis, Candida esophagitis, onychomycosis |
| H6 | HSLP | 33/F | C. felis | HIV+ | 6 | None |
| H7 | HSLP | 43/M | C. parvum human genotype | HIV+ | Microsporidiosis | |
| H8 | HSLP | 41/M | C. parvum human genotype | HIV+ | 26 | Microsporidiosis, Candida esophagitis, Kaposi's sarcoma |
| H9 | HSLP | 45/M | C. parvum human genotype | HIV+ | 158 | Microsporidiosis, Candida esophagitis, septicemic salmonellosis |
| H10 | HSLP | 32/M | C. parvum human genotype | HIV+ | 102 | Microsporidiosis |
| H11 | HSLP | 35/M | C. meleagridis | HIV+ | None | |
| H12 | HSLP | 49/M | C. parvum human genotype | HIV+ | Microsporidiosis | |
| H13 | HSLP | 31/M | C. parvum human genotype | HIV+ | None | |
| H14 | HSLP | 38/NK | C. parvum cattle genotype (A) | HIV+ | Isospora belli | |
| H34 | HSLP | 37/M | C. meleagridis | HIV+ | 100 | None |
| H35 | HSLP | 49/M | C. parvum cattle genotype (A) | Transplanted | 179–361 | |
| H36 | HSLP | 44/M | C. parvum cattle genotype (A) | HIV+ | 17 | Candida esophagitis |
| H37 | HSLP | 18/M | C. parvum human genotype | Lymphoma | ||
| H38 | HSLP | 43/M | C. parvum cattle genotype (A) | HIV+ | 176 | None |
| H39 | HSLP | 40/M | C. parvum human genotype | HIV+ | 40 | None |
| H40 | HSLP | 28/M | C. parvum cattle genotype (A) | HIV+ | ||
| H66 | HSLP | 32/M | C. parvum human genotype | HIV+ | Microsporidiosis | |
| H67 | HSLP | 33/M | C. felis | HIV+ | <50 | None |
| H68 | HSLP | 71/M | C. parvum cattle genotype (B) | Lymphoma | ||
| H78 | HSLP | 29/M | C. felis | HIV+ | 300 | Kaposi's sarcoma, E. coli and Entamoeba hartmanni infection |
| H79 | HSLP | 65/F | C. parvum cattle genotype (B) | HIV+ | 14 | Pneumocystosis |
| H80 | HSLP | 56/M | C. parvum human genotype | HIV+ | Brain toxoplamosis | |
| H81 | HSLP | 23/F | C. felis | Lymphoma | ||
| H82 | HSLP | 52/M | C. parvum cattle genotype (A) | Transplant | ||
| H83 | HSLP | 52/M | C. felis | HIV+ | <50 | Pneumocystosis, Candida albicans |
| H16 | HHMC | NK/NK | C. parvum cattle genotype (A) | Immunocompetent | Salmonellosis | |
| H15 | CHRUR | NK/NK | C. parvum cattle genotype (A) | HIV+ | NKd | |
| H18 | CHUN | 51/M | C. parvum cattle genotype (A) | HIV+ | NK | |
| H19 | CHUN | 51/M | C. parvum cattle genotype (A) | HIV+ | NK | |
| H20 | CHUN | 56/M | C. parvum cattle genotype (A) | HIV+ | NK | |
| H21 | CHUN | 57/M | C. parvum cattle genotype (A) | HIV+ | NK | |
| H22 | CHUN | 40/M | C. felis | HIV+ | NK | |
| H17 | CHUA | 3/M | C. meleagridis | Hypogammaglobulimenia | ||
| H2 | HEHL | 45/M | C. parvum cattle genotype (A) | HIV+ | 154 | None |
| H25 | HEHL | 53/M | C. muris | HIV+ | 10 | Candida esophagitis |
| H27 | HEHL | 32/M | C. parvum cattle genotype (B) | Lymphoma | ||
| H30 | HEHL | 6/M | C. parvum cattle genotype (A) | Transplant | ||
| H31 | HEHL | 51/M | C. parvum cattle genotype (A) | HIV+ | <50 | None |
| H73 | HEHL | 9/M | C. parvum human genotype | Immunocompetent | ||
| H74 | HEHL | 30/F | C. parvum human genotype | HIV+ | 38 | Pneumocystosis Lung cryptococcosis |
| H75 | HEHL | 45/M | C. parvum cattle genotype (B) | HIV+ | <50 | None |
| H23 | CHRUN | 32/F | C. parvum cattle genotype (A) | HIV+ | None | |
| H29 | CHRUN | 67/M | C. parvum cattle genotype (A) | Lymphoma | ||
| H53 | CHRUN | 28/M | C. parvum cattle genotype (A) | Immunocompetent | ||
| H69 | CHRUN | 1/M | C. parvum human genotype | HIV+ | 1,000 | Microsporidiosis |
CHD, Centre Hospitalier de Dunkerque; CHRUL, Centre Hospitalier Régional Universitaire de Lille; CHUA, Centre Hospitalier Universitaire d'Amiens; HSLP, Hôpital Saint Louis de Paris; HHMC, Hôpital Henri Mondor de Créteil; CHRUR, Centre Hospitalier Régional Universitaire de Rennes; CHUN, Centre Hospitalier Universitaire de Nantes; CHUA, Centre Hospitalier Universitaire d'Angers; HEHL, Hôpital Edouard-Herriot de Lyon; CHRUN, Centre Hospitalier Régional Universitaire de Nice.
M, male; F, female.
All patients are HIV negative except when HIV+ (HIV positive) is indicated.
NK, not known.
DNA extraction.
Genomic DNAs were prepared from either partially purified oocysts (after concentration in phosphate-buffered saline–ether) or whole feces by the method described by Saano and Lindstrom (40), with modifications. Samples (320 μl) were mixed with 40 μl of 100 mM Tris–1 mM EDTA (pH 8) and 40 μl of 10% sodium dodecyl sulfate. To disrupt the oocysts, the samples were frozen (liquid nitrogen, 3 min) and thawed (37°C, 3 min) three times. Then, proteinase K (Boehringer Mannheim, Indianapolis, Ind.) was added at a concentration of 0.2 mg/ml. Digestion was performed overnight at 55°C. In order to remove particulate matter, the samples were rapidly centrifuged (10,000 × g, 1 min) and the supernatants were collected in new tubes. NaCl (5 M) was added to give a final concentration of 0.7 M, and prewarmed cetyltrimethylammonium bromide (CTAB; Sigma, St. Louis, Mo.) and polyvinylpyrrolidone (PVP; Sigma) were added to concentrations of 1% each. Following incubation at 65°C for 20 min, a chloroform-isoamyl alcohol (24:1) extraction was performed. CTAB and PVP, each at a 1% final concentration, were then added to the aqueous phase, and the chloroform-isoamyl alcohol (24:1) extraction was repeated. The aqueous phase was extracted two more times with 1 volume of phenol and 1 volume of chloroform-isoamyl alcohol (24:1). The DNA was precipitated by the addition of 0.6 volume of isopropanol, and the DNA pellet was washed with 70% ethanol. After desiccation, the DNA pellet was resuspended in 100 μl of sterile water. This crude DNA was further purified with the DNA Clean Up Wizard kit (Promega Corporation, Madison, Wis.) according to the manufacturer's recommendations. The DNA was eluted from the minicolumn with 50 μl of sterile water.
DNA amplification by PCR.
The Cryptosporidium genus-specific primer pair reported by Morgan et al. (28) was used to amplify an approximately 300-bp fragment of the Cryptosporidium 18S rRNA gene encompassing the hypervariable region. The reaction mixtures were prepared in 1× PCR buffer (50 mM KCl, 10 mM Tris HCl [pH 8.3]) and contained, per 50-μl reaction mixture, 3.5 mM MgCl2, both primers (Eurogentec, Seraing, Belgium) at a concentration of 0.5 μM, each deoxynucleoside triphosphate at a concentration of 200 μM, 2.5 U of Amplitaq Gold (Perkin-Elmer Applied Biosystems, Foster City, Calif.), and 10 μl of the purified DNA at a 1/10 dilution. A negative control, consisting of a reaction mixture with water instead of DNA template, was included in each amplification run. DNA amplification was carried out on a PTC 200 thermocycler (MJ Research). The amplification reactions were initiated by denaturation of the DNA at 94°C for 10 min; and then the mixtures were subjected to 40 cycles of denaturation at 94°C for 30 s, annealing of the primer at 58°C for 30 s, and extension at 72°C for 30s, with an additional 5-min extension at 72°C. The PCR product was analyzed by electrophoresis in a 2% agarose gel and was visualized after ethidium bromide staining.
DNA sequencing and data analysis.
Amplified PCR products were purified by filtration with a Microcon 50 concentrator (Amicon, Beverly, Mass.). They were sequenced in both directions with a model ABI 377 automated sequencer by using an ABI Prism Dye Terminator cycle sequencing kit (Perkin-Elmer Applied Biosystems) according to the manufacturer's instructions. Contiguous sequences were generated from the forward and reverse strands with Gene Jockey II software (Biosoft, Cambridge, United Kingdom). Multiple alignments of the sequences were done with the ClustalW program in the Wisconsin package (Genetics Computer Group, Madison, Wis.).
RESULTS
Microscopic examination.
Samples of feces in which the original supplier recognized Cryptosporidium oocysts were collected from a total of 57 patients. All samples were subsequently retested in our laboratory. Oocysts were detected in 54 samples by staining with modified Ziehl-Neelsen stain. In the remaining three samples, however, Cryptosporidium oocysts were detected by immunofluorescence assay. No morphological difference among parasite isolates was discernible at the light microscopic level.
18S rDNA-based molecular typing.
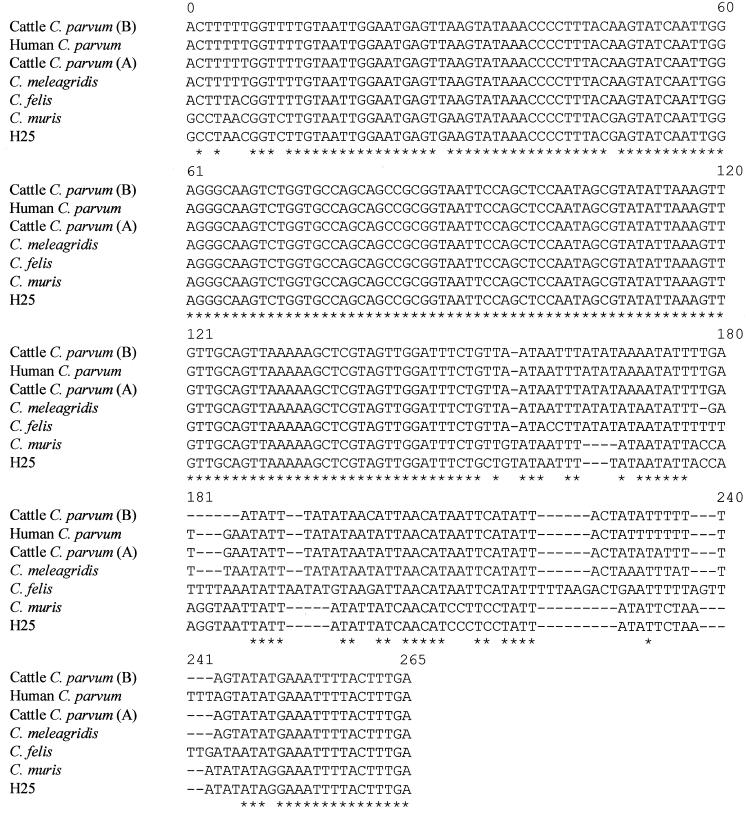
The variable region of the 18S rRNA gene of Cryptosporidium was analyzed for all samples. DNA sequence analysis at this locus identified six distinct genotype groups. Six types of sequences were identified in the GenBank database, as follows: C. parvum cattle genotypes A and B, C. parvum human genotype, C. meleagridis, C. felis, and one type that differed by three single mutations from genotype A of C. muris (34) (also called C. andersoni by Lindsay et al. [21]) (Fig. 1). Among the 57 isolates, 29 exhibited the C. parvum cattle genotype (24 type A isolates and 5 type B isolates) and 18 exhibited the C. parvum human genotype. On the whole, among the C. parvum isolates, the cattle genotype was the most common, with 62% (29 of 47) being of the cattle genotype; 38% (18 of 47) of the isolates were of the human genotype. Interestingly, six isolates exhibited the C. felis genotype, three isolates exhibited the C. meleagridis genotype, and one isolate exhibited a new genotype of C. muris.
FIG. 1.
Alignments of the Cryptosporidium 18S rRNA gene diagnostic fragments obtained with the primer pair reported by Morgan et al. (28) for C. parvum cattle genotype, C. parvum human genotype, C. meleagridis, C. felis, C. muris, and H25 human isolate. Asterisks indicate identical bases. Dashes represent alignment gaps. Numbering is arbitrary. Note that the sequence signature is unique. The GenBank accession numbers for the cattle C. parvum (A), cattle C. parvum (B), human C. parvum, C. meleagridis, C. felis, and C. muris sequences shown are AF093494, AF228682, AF093491, AF112574, AF159113, and AF093496, respectively.
Clinical and epidemiological data.
When possible, retrospective information on the patients was collected (Table 1). The 57 patients comprised 9 females and 45 males; the sexes of 3 patients were not stated. Thirty-five patients were infected with HIV. The remaining HIV-negative patients had received solid organ or bone marrow transplants (5 patients), were suffering from lymphoma (5 patients) or hypogammaglobulinemia (1 patient), or did not show any known immunocompromising condition and were therefore designated immunocompetent subjects (11 patients). Most clinical information was obtained for HIV-infected patients. They exhibited a variety of other concurrent AIDS-defining infections including pneumocystosis, microsporidiosis, toxoplasmosis, candidosis, cryptococcosis, and Kaposi's sarcoma (Table 1). When known, the CD4+ lymphocyte count was low (from 5 to 361 per μl).
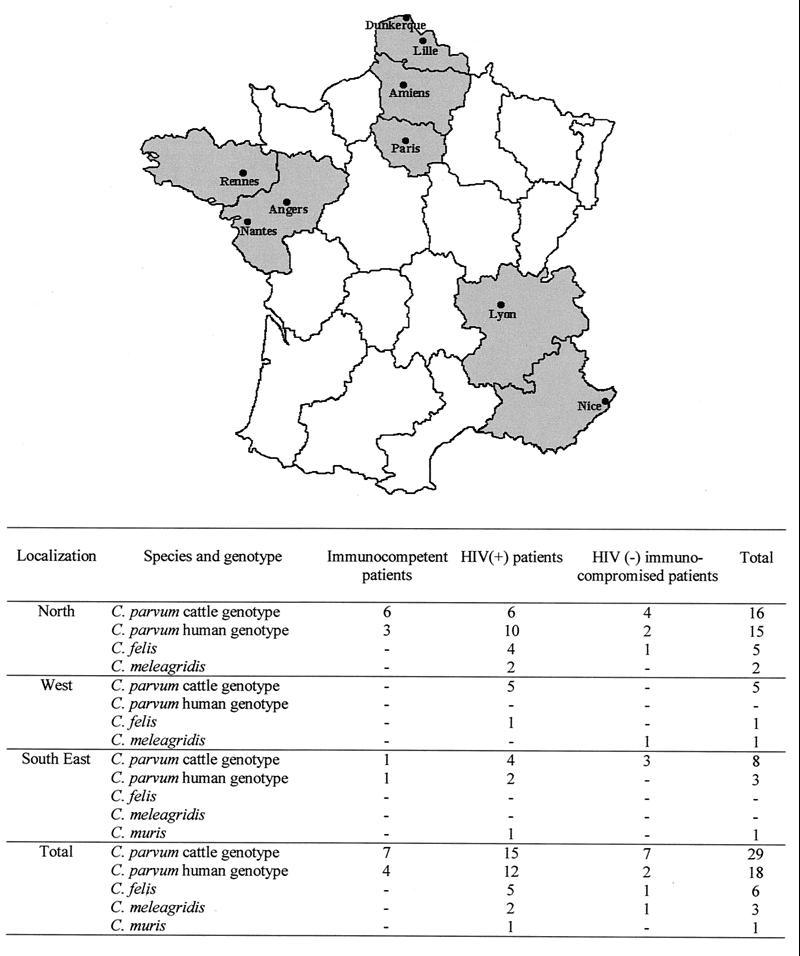
A comparison of the data in Table 1 and genotyping results is shown in Fig. 2. Among the 11 immunocompetent individuals including 8 children, 7 cattle genotypes and 4 human genotypes of C. parvum were retrieved. Of the 35 HIV-infected patients, 15 were infected with the cattle genotype of C. parvum, 12 were infected with the human genotype of C. parvum, 5 were infected with C. felis, 2 others were infected with C. meleagridis, and the last patient was infected with a new genotype of C. muris. The proportions of C. parvum cattle genotype/C. parvum human genotype strains were 4/1 for transplant recipients and 3/1 for lymphoma patients. One lymphoma patient was found to be parasitized with the C. felis genotype, and the patient with hypogammaglobulinemia was parasitized with the C. meleagridis genotype.
FIG. 2.
The map at the top shows the regions of origin of the Cryptosporidium isolates: the northern region comprises Dunkerque, Lille, Amiens, and Paris; the west region comprises Rennes, Angers, and Nantes; and the southeast region comprises Lyon and Nice. The table at the bottom gives detailed information about the Cryptosporidium species or genotypes found in each region as well as the immunological status of the patients.
DISCUSSION
Most studies on the molecular characterization of Cryptosporidium in humans have described single sporadic cases or small numbers of clustered cases. A few broader population surveys from Australia (28, 30), the United States (44), and the United Kingdom (24, 35) have appeared in the literature. A recent study reported on an analysis of 1,705 human cases in the United Kingdom (23). In France, very few data on Cryptosporidium typing are available, as only one study has been performed until now (3). In the present study, 57 Cryptosporidium isolates from French individuals were analyzed at the 18S rDNA gene locus. The results revealed that the majority of the patients were infected with the cattle (29 of 57) and human (18 of 57) genotypes of C. parvum. Twenty-four type A and 5 type B sequences were found among isolates of the cattle genotype. The differences between the C. parvum cattle type A and B 18S rRNA gene sequences reported here had previously been found by Carraway et al. (6) and Le Blancq et al. (20) in a single isolate. They found four copies of the type A rDNA unit and one copy of the type B rDNA unit. However, the type B sequence has been reported in the GenBank database (accession number AF228662) as identifying a particular strain of C. parvum from cattle (Moredun strain). This strain, which is usually referred as the MD isolate, is a parasite strain that was originally isolated from deer and that has been propagated through lambs or calves (6, 19, 43). Interestingly, in the present work, a number of patients were infected with C. meleagridis (three patients), C. felis (six patients), and a new genotype of C. muris (one patient). This is the first report of these three species in humans in France.
Early reports often described the detection of human cryptosporidiosis in adults, reflecting a high proportion of immunocompromised subjects. However, sporadic infections at the community level were also noted in the early 1980s, particularly in otherwise healthy immunocompetent children (8, 9, 15, 45). Subsequent studies have confirmed a peak incidence in children aged 1 to 5 years in most areas, but the ages are generally at the lower end of that range in developing countries. Serious Cryptosporidium infection has also been reported in other immunocompromised subjects with primary disorders such as hypogammaglobulinemia or congenital immunodeficiency or in individuals who have been treated with immunosuppressive compounds. Isolates from all these kinds of patients were represented among the 57 isolates from patients with sporadic cases of cryptosporidiosis analyzed in the present study. Eight immunocompetent children aged from 10 months to 9 years were included in the study. Half of the children were infected with the bovine genotype of C. parvum, whereas the other half were infected with the human genotype of C. parvum. Curiously, seven of the eight children were from the north of France (Dunkerque and Lille) (Fig. 2). The majority of the 35 HIV-infected patients were infected with C. parvum, and the cattle genotype was the most common (15 of 27 patients). This proportion of C. parvum infections among HIV-infected patients is in agreement with previous data from France (7 of 13 patients) (3) and Switzerland (7 of 9 patients) (26) (Table 2). Some individuals had other concurrent AIDS-defining infections, such as microsporidiosis (Table 1). Curiously, isolates from all the patients for whom microsporidioses were reported exhibited the human genotype of C. parvum. Among the HIV-infected patients for whom the CD4+ lymphocyte count was known, the majority had <180 lymphocytes/mm3. Five of the eight HIV-infected patients infected with C. felis, C. meleagridis, or the new genotype of C. muris had <100 CD4+ lymphocytes/mm3. One HIV-infected patient (patient H78, Table 1) with C. felis infection had 300 CD4+ lymphocytes/mm3. However, other immunocompromised patients such as a child with hypogammaglobulinemia (patient H17) or a young female patient with lymphoma (patient H81) were also infected with C. meleagridis and C. felis, respectively Such results confirmed those of other authors (Table 3) and indicate that immunocompromised patients are susceptible to a wide range of Cryptosporidium species. No correlation was found between the genotype and the geographic origins of the patients (Fig. 2).
TABLE 2.
Prevalence of C. parvum human and cattle genotypes in human isolates in various studiesa
| Marker used | Sample source | Total no. of:
|
Geographic location | Reference | ||
|---|---|---|---|---|---|---|
| Isolates (from HIV+ patients) | Human genotype | Cattle genotype | ||||
| PGM and HKb | Sporadic cases | 9 | 8 | 1 | England | 1 |
| Sporadic case | 1 | 1 | Guinea Bissau | |||
| Genomic DNAc | Sporadic cases | 14 | 12 | 2 | Australia | 29 |
| ITS 1d and 18S rDNAd | HIV+ patients | 2 | 2 | USA | 6 | |
| Repetitive DNA fragmente | HIV+ patients | 13 | 6 | 7 | France | 3 |
| Poly(T) | Sporadic cases | 6 (2) | 6g (2)h | 7 | ||
| TRAP C2d | Outbreak cases | 15 | 13c | 2 | USA | 37 |
| Outbreak case | 1 | 1 | Canada | |||
| COWPc | Sporadic cases | 3 | 1 | 2 | Wales | 43 |
| Outbreak cases | 4 | 4 | England | |||
| Genomic DNAf | Sporadic cases | 32 | 28 | 4 | Australia | 28 |
| 18s rDNAd and acetyl-CoAd | Sporadic cases | 4 | 3 | 1 | Australia | 31 |
| Sporadic cases | 1 | 1 | Wales | |||
| Outbreak cases | 2 | 2 | England | |||
| Genomic DNAf | Sporadic cases | 36 | 30 | 6 | Australia | 30 |
| Poly(T)e COWP,c and RNRe | HIV+ patients | 7 | 5 | 2 | USA | 48 |
| β-Tubulin | HIV+ patients | 3 | 3h | USA | 47 | |
| Sporadic case | 1 | 1 | USA | |||
| Sporadic case | 1 | 1 | UK | |||
| TRAP C1e | Sporadic case | 1 | 1 | Wales | 42 | |
| Outbreak cases | 2 | 2 | England | |||
| TRAP C1,e COWP,e poly(T),e RNR,e and ITSIf | Outbreak cases | 4 | 4 | England | 41 | |
| Sporadic case | 1 | 1 | Wales | |||
| Outbreak case | 1 | 1 | Wales | |||
| Sporadic cases | 6 (2) | 4j (2) | 4j | USA | ||
| Sporadic cases | 5 | 4 | 1 | Australia | ||
| Genomic DNAc | Outbreak and sporadic cases | 15 | 9 | 6 | England and Guinea- Bissau | 2 |
| TRAP C2e | Outbreak cases | 25 | 19 | 6 | USA | 44 |
| HIV+ patients | 17 | 15 | 2 | USA | ||
| Outbreak cases | 2 | 1 | 1 | Canada | ||
| Outbreak cases | 2 | 2 | India | |||
| HIV+ patients | 4 | 4 | Guatemala | |||
| β-Tubulin,e TRAP C2,e poly(T),e | Outbreak cases | 10 | 10 | The Netherlands | 5 | |
| COWPe and genomic DNAf | HIV+ patients | 4 | 4 | Italy | ||
| COWPe | Outbreak cases | 94 | 92k | 3k | England | 35 |
| Sporadic cases | 46 | 31k | 16k | England | ||
| COWP,e TRAP C1e | Sporadic cases | 194 | 74 | 120 | England | 24 |
| Sporadic cases | 3 | 3 | Scotland | |||
| Sporadic cases | 3 | 3 | Northern Ireland | |||
| COWPe | Outbreak cases | 25 | 24 | 1 | The Netherlands | 17 |
| Sporadic cases | 15 (3) | 4 (3) | 11 | The Netherlands | ||
| 18S rDNAd | HIV+ patients | 6 | 5 | 1 | USA | 38 |
| 18S rDNA,d Hsp 70,d and acetyl-CoAd | HIV+ patients | 9 | 2 | 7 | Switzerland | 26 |
| HIV+ patients | 5 | 4 | 1 | Kenya | ||
| Microsatellited | HIV+ patients | 4 | 4 | USA | 4 | |
| Outbreak cases | 8 | 8 | The Netherlands | |||
| Sporadic cases | 17 (1) | 5 (1) | 12 | The Netherlands | ||
| Outbreak cases | 5 | 5 | Japan | |||
| HIV+ patients | 8 | 8 | Italy | |||
| HIV+ patients | 6 | 6 | Northern Ireland | |||
| 18S rDNAd | Outbreak cases | 19 | 19l | USA | 39 | |
| COWPe | Outbreak cases | 522 | 238l | 288l | England | 23 |
| Sporadic cases | 1,088 | 397j | 693j | England | ||
| Sporadic cases | 11 | 1 | 10 | Scotland | ||
| Sporadic cases | 48 | 5 | 43 | Northern Ireland | ||
| Sporadic cases | 31 | 10 | 21 | Wales | ||
| 18S rDNAc | Sporadic cases | 75 | 67 | 8 | Peru | 49 |
| 18S rDNAd | Sporadic cases | 47 (27) | 18 (12) | 29 (15) | France | This study |
Abbreviations: PGM, phosphoglucomutase; HK, hexokinase; ITS 1, internal transcribed spacer 1; Poly(T), polythreonine; TRAP C2, thrombospondin-related adhesive protein 2; COWP, Cryptosporidium oocyst wall protein; acetyl-CoA, acetyl coenzyme A synthase; RNR, ribonucleotide reductase; TRAP C1, thrombospondin-related adhesive protein 1; Hsp 70, heat shock protein 70, HIV+, HIV positive; USA, United States.
Isoenzyme analysis.
Random amplified polymorphic DNA analysis.
PCR and sequencing.
PCR-RFLP analysis.
PCR.
Three isolates have hybrid genotype.
one isolate has hybrid genotype.
Two isolates have a hybrid genotype.
Two isolates have both genotypes.
One isolate has both genotypes.
Four isolates have both genotypes.
TABLE 3.
Cases of non-C. parvum infections in humans reported in various studies
| No. of infections caused by:
|
Patient | Geographic location | Reference | |||
|---|---|---|---|---|---|---|
| C. meleagridis | C. felis | C. parvum dog genotype | C. muris | |||
| 3 | 1 | HIV-infected patients | United States | 38 | ||
| 1 | 3 | HIV-infected patients | Switzerland | 26 | ||
| 1 | HIV-infected patient | Kenya | ||||
| 3 | HIV-infected patients | United States | ||||
| 2 | Immunocompetent children | Indonesia | 18 | |||
| 4 | Not known | England | 23 | |||
| 6 | Immunocompetent patients | England | 36 | |||
| 7 | 1 | 2 | Non-HIV-infected children | Peru | 49 | |
| 3 | 6 | 1 | Immunocompromised patients | France | This study | |
Molecular data for oocysts of human origin reported by different laboratories from tests with numerous markers revealed that two genotypes are dominant (Table 2). The human genotype (genotype 1) was detected in humans and in a single nonhuman primate. The cattle genotype (genotype 2) was detected in both animals and humans. Geographic variations in the repartition of C. parvum human and bovine genotypes seem to exist (Table 2). In Australia, anthroponotic organisms account for the majority of the cases of C. parvum infection, with infections with C. parvum human genotype comprising 85% of infections (51). In the United States, the human genotype seems to be associated with the majority of isolates obtained from individuals in nonoutbreak situations. We have recently confirmed a higher occurrence of this anthroponotic genotype in the New World by analyzing isolates from Haiti (unpublished data). In contrast, the C. parvum bovine genotype seems to be dominant in Europe (Table 2). In regard to cryptosporidiosis outbreaks (Table 2), it can be speculated that the C. parvum human genotype is more infective for humans and is therefore better adapted to this host species. Indeed, the human genotype of C. parvum has largely been responsible for most cryptosporidiosis outbreaks in North America. Similarly, strains of the C. parvum human genotype caused outbreaks in the United Kingdom and a possible outbreak in The Netherlands, countries with higher rates of background transmission of the bovine genotype. In fact, it is not clear why the C. parvum human genotype has been found to be associated with most outbreaks, even in countries where infection with the C. parvum bovine genotype is dominant (Table 2). This could suggest either that the human genotype is intrinsically more virulent than the bovine genotype or that the human genotype is more easily transmitted among humans than the bovine genotype.
Until now, no cryptosporidiosis outbreak has been reported in France. Likewise, very few outbreaks have been reported on the European continent, whereas they have been frequently reported in the United States, Canada, and the United Kingdom (25). The reasons for this are unclear. It is likely that cryptosporidiosis is underdiagnosed because clinicians fail to consider this diagnosis in patients with diarrheal illnesses (particularly immunocompetent adults and children) and do not request stool analysis for Cryptosporidium, a test not normally included in routine stool analyses. Ideally, laboratories should have ongoing communication with public health services and water utilities in order to recognize outbreaks and be able to screen patient and environmental samples by performing molecular biology-based identification and typing analyses. Another possible explanation for the unbalanced frequency of outbreaks between the European continent and the other countries cited above could be an immunological protection against Cryptosporidium in individuals on the European continent. Indeed, a recent study has shown a high prevalence of serological response to Cryptosporidium in Italian individuals, which could explain the infrequent occurrence of clinically detectable cryptosporidiosis in an Italian city (13).
For a long time, C. parvum had been considered the only Cryptosporidium species that infects humans. Whereas until very recently only C. parvum was found in immunocompetent individuals, it has been shown that immunocompromised individuals can be infected with other species or genotypes of Cryptosporidium. Indeed, we and several other groups of investigators have identified C. felis (26, 38) and C. meleagridis (26) in AIDS or other immunocompromised patients. The C. parvum dog genotype has also been detected in an HIV-infected patient (38). Recently, Pedraza-Diaz et al. reported the first cases of Cryptosporidium infection in six immunocompetent humans due to C. meleagridis (36). The case of human infection with a new genotype of C. muris in the present study was in an AIDS patient. Nevertheless, oocysts morphologically similar to C. muris and for which PCR with a C. parvum-specific primer was negative were also found in the stools of two healthy girls in Indonesia (18). Another recent publication reported other infections caused by C. meleagridis and C. felis in immunocompetent children (49). Our 10 cases of non-C. parvum infection in humans and those reported by other investigators (Table 3) are probably not the only ones. They have been detected because they had been sought. In one of the first genotyping studies on Cryptosporidium from individuals with AIDS, Bonnin et al. did not succeed in obtaining a positive PCR result for one patient, despite repeated attempts and performance of the tests in the absence of an inhibitor (3). Likewise, Widmer et al. failed to amplify the DNA fragment from two isolates (48). For one isolate the fragment could not be amplified with any of the PCR primers used, and for the second one the fragment was amplified with only the 18S rDNA-specific primers. This problem was also reported by McLauchlin et al. (24). In their study, DNA from seven samples in which oocysts were seen by microscopy was not amplified with any of the three primer pairs. A possible explanation for this may be that the oocysts detected in these patients were not of the C. parvum human or cattle genotype. Indeed, most of the typing studies carried out so far have used PCR-based methods and have analyzed single genetic loci that were all shown to be dimorphic. However, the specificities of these genotyping tools for other species of Cryptosporidium or genotypes are not always known. It is clear that the staining method and direct immunofluorescence could detect all genotypes, but this may not be true for primers that may not recognize the hybridization site on DNA and that therefore directly affect the PCR results. It is therefore particularly important to use generic primers at first for PCR detection. Then, typing could be done. In the present study, we chose to sequence the product amplified from 18S rDNA because sequence analysis of this locus produces the most complete and reliable data set, as all bases are examined. The 18S rDNA sequence is available for seven of the eight Cryptosporidium species (it is not available for C. nasorum) and for the eight genotypes of C. parvum, and the 18S rDNA sequences have been defined on the basis of the 18S rDNA sequence for C. parvum. However, RFLP analysis can be a less costly and less time-consuming alternative (50, 52). By this technique, it could be possible to distinguish either species or groups of species or genotypes.
In conclusion, the results of the present study indicate that immunocompromised humans are susceptible to a wide range of Cryptosporidium species. Even immunocompetent individuals can also be infected with species other than C. parvum. Because of these new data, the question of the public health impacts of different Cryptosporidium species and genotypes is emerging. For this reason, there is an urgent need to determine the extent of genetic diversity within Cryptosporidium strains affecting humans or animals in order to understand the molecular epidemiology of cryptosporidiosis. Additional studies with larger number of patients for whom extensive clinical information is available are required in order to understand both the public health impact of Cryptosporidium species and genotypes and the dynamics of parasite transmission. Prevention of human cryptosporidiosis would be accomplished by a thorough understanding and appreciation of its complex natural history (10) and epidemiology. Studies should ideally be done with samples from the environment (14, 22) in order to evaluate the circulation of the parasite in various ecosystems. In light of the known resistance of this parasite to both conventional water treatment methods and effective therapeutic agents (25), an intensive effort to control the exposure of humans, particularly immunocompromised populations, to this organism appears to be the best prevention strategy at this time.
ACKNOWLEDGMENTS
We thank the following people for kindly providing the fecal samples and clinical data on the patients: J. Poirriez (Centre Hospitalier de Dunkerque), E. Dutoit and J. M. Dewitte (Centre Hospitalier Régional Universitaire de Lille), M. Miegeville (Centre Hospitalier Universitaire de Nantes), Y. Le Fichoux (Centre Hospitalier Régional Universitaire de Nice), L. De Gentille (Centre Hospitalier Universitaire d'Angers), B. Degeilh (Centre Hospitalier Régional Universitaire de Rennes), and M. Deniau (Hôpital Henri Mondor de Créteil).
A. Follet-Dumoulin was supported by a grant from the Catholic University of Lille. This work was developed in part in the framework of the “Agence Nationale de Recherche sur le SIDA”-supported VIH-PAL program.
REFERENCES
- 1.Awad-el-Kariem F M, Robinson H A, Dyson D A, Evans D, Wright S, Fox M T, McDonald V. Differentiation between human and animal strains of Cryptosporidium parvum using isoenzyme typing. Parasitology. 1995;110:129–132. doi: 10.1017/s0031182000063885. [DOI] [PubMed] [Google Scholar]
- 2.Awad-El-Kariem F M, Robinson H A, Petry F, McDonald V, Evans D, Casemore D. Differentiation between human and animal isolates of Cryptosporidium parvum using molecular and biological markers. Parasitol Res. 1998;84:297–301. doi: 10.1007/s004360050399. [DOI] [PubMed] [Google Scholar]
- 3.Bonnin A, Fourmaux M N, Dubremetz J F, Nelson R G, Gobet P, Harly G, Buisson M, Puygauthier-Toubas D, Gabriel-Pospisil G, Naciri M, Camerlynck P. Genotyping human and bovine isolates of Cryptosporidium parvum by polymerase chain reaction-restriction fragment length polymorphism analysis of a repetitive DNA sequence. FEMS Microbiol Lett. 1996;137:207–211. doi: 10.1111/j.1574-6968.1996.tb08107.x. [DOI] [PubMed] [Google Scholar]
- 4.Caccio S, Homan W, Camilli R, Traldi G, Kortbeek T, Pozio E. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology. 2000;120:237–244. doi: 10.1017/s0031182099005508. [DOI] [PubMed] [Google Scholar]
- 5.Caccio S, Homan W, van Dijk K, Pozio E. Genetic polymorphism at the beta-tubulin locus among human and animal isolates of Cryptosporidium parvum. FEMS Microbiol Lett. 1999;170:173–179. doi: 10.1111/j.1574-6968.1999.tb13371.x. . (Erratum, 173:273.) [DOI] [PubMed] [Google Scholar]
- 6.Carraway M, Tzipori S, Widmer G. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl Environ Microbiol. 1996;62:712–716. doi: 10.1128/aem.62.2.712-716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carraway M, Tzipori S, Widmer G. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect Immun. 1997;65:3958–3960. doi: 10.1128/iai.65.9.3958-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casemore D P, Jackson B. Sporadic cryptosporidiosis in children. Lancet. 1983;ii:679. doi: 10.1016/s0140-6736(83)92552-7. [DOI] [PubMed] [Google Scholar]
- 9.Casemore D P, Sands R L, Curry A. Cryptosporidium species a “new”human pathogen. J Clin Pathol. 1985;38:1321–1336. doi: 10.1136/jcp.38.12.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumoulin A, Guyot K, Lelievre E, Dei-Cas E, Cailliez J C. Cryptosporidium and wildlife: a risk for humans? Parasite. 2000;7:167–172. doi: 10.1051/parasite/2000073167. [DOI] [PubMed] [Google Scholar]
- 11.Fayer R, Morgan U, Upton S J. Epidemiology of Cryptosporidium: transmission, detection and identification. Int J Parasitol. 2000;30:1305–1322. doi: 10.1016/s0020-7519(00)00135-1. [DOI] [PubMed] [Google Scholar]
- 12.Fayer R, Speer C A, Dubey J P. The general biology of Cryptosporidium. In: Fayer R E, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press, Inc.; 1997. pp. 1–41. [Google Scholar]
- 13.Frost F J, Muller T, Craun G F, Fraser D, Thompson D, Notenboom R, Calderon R L. Serological analysis of a cryptosporidiosis epidemic. Int J Epidemiol. 2000;29:376–379. doi: 10.1093/ije/29.2.376. [DOI] [PubMed] [Google Scholar]
- 14.Guyot K, Gireaudot-Liepmann M F, Cabon A, Riveau-Ricard I, Lange M, Delattre J M, Dei-Cas E. Influence of US EPA 1622 method successive steps on the viability of Cryptosporidium oocysts. Wat Sci Technol. 2000;41:189–196. [Google Scholar]
- 15.Hart C A, Baxby D, Blundell N. Gastroenteritis due to Cryptosporidium: a prospective survey in a children's hospital. J Infect. 1984;9:264–270. doi: 10.1016/s0163-4453(84)90574-7. [DOI] [PubMed] [Google Scholar]
- 16.Henriksen S A, Pohlenz J F L. Staining cryptosporidia by a modified Ziehl-Neelson technique. Acta Vet Scand. 1981;22:594–596. doi: 10.1186/BF03548684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homan W, van Gorkom T, Kan Y Y, Hepener J. Characterization of Cryptosporidium parvum in human and animal feces by single-tube nested polymerase chain reaction and restriction analysis. Parasitol Res. 1999;85:707–712. doi: 10.1007/s004360050619. [DOI] [PubMed] [Google Scholar]
- 18.Katsumata T, Hosea D, Ranuh I G, Uga S, Yanagi T, Kohno S. Short report: possible Cryptosporidium muris infection in humans. Am J Trop Med Hyg. 2000;62:70–72. doi: 10.4269/ajtmh.2000.62.70. [DOI] [PubMed] [Google Scholar]
- 19.Lally N C, Baird G D, McQuay S J, Wright F, Oliver J J. A 2359-base pair DNA fragment from Cryptosporidium parvum encoding a repetitive oocyst protein. Mol Biochem Parasitol. 1992;56:69–78. doi: 10.1016/0166-6851(92)90155-d. [DOI] [PubMed] [Google Scholar]
- 20.Le Blancq S M, Khramtsov N V, Zamani F, Upton S J, Wu T W. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol Biochem Parasitol. 1997;90:463–478. doi: 10.1016/s0166-6851(97)00181-3. [DOI] [PubMed] [Google Scholar]
- 21.Lindsay D S, Upton S J, Owens D S, Morgan U M, Mead J R, Blagburn B L. Cryptosporidium andersoni n. sp. (Apicomplexa: Cryptosporiidae) from cattle, Bos taurus. J Eukaryot Microbiol. 2000;47:91–95. doi: 10.1111/j.1550-7408.2000.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 22.Matheson Z, Hargy T M, McCuin R M, Clancy J L, Fricker C R. An evaluation of the Gelman Envirochek capsule for the simultaneous concentration of Cryptosporidium and Giardia from water. J Appl Microbiol. 1998;85:755–761. doi: 10.1111/j.1365-2672.1998.00588.x. [DOI] [PubMed] [Google Scholar]
- 23.McLauchlin J, Amar C, Pedraza-Diaz S, Nichols G L. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J Clin Microbiol. 2000;38:3984–3990. doi: 10.1128/jcm.38.11.3984-3990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLauchlin J, Pedraza-Diaz S, Amar-Hoetzeneder C, Nichols G L. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J Clin Microbiol. 1999;37:3153–3158. doi: 10.1128/jcm.37.10.3153-3158.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meinhardt P L, Casemore D P, Miller K B. Epidemiologic aspects of human cryptosporidiosis and the role of waterborne transmission. Epidemiol Rev. 1996;18:118–136. doi: 10.1093/oxfordjournals.epirev.a017920. [DOI] [PubMed] [Google Scholar]
- 26.Morgan U, Weber R, Xiao L, Sulaiman I, Thompson R C, Ndiritu W, Lal A, Moore A, Deplazes P. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J Clin Microbiol. 2000;38:1180–1183. doi: 10.1128/jcm.38.3.1180-1183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan U, Xiao L, Sulaiman I, Weber R, Lal A A, Thompson R C, Deplazes P. Which genotypes/species of Cryptosporidium are humans susceptible to? J Eukaryot Microbiol. 1999;46:42S–43S. [PubMed] [Google Scholar]
- 28.Morgan U M, Constantine C C, Forbes D A, Thompson R C. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J Parasitol. 1997;83:825–830. [PubMed] [Google Scholar]
- 29.Morgan U M, Constantine C C, O'Donoghue P, Meloni B P, O'Brien P A, Thompson R C. Molecular characterization of Cryptosporidium isolates from humans and other animals using random amplified polymorphic DNA analysis. Am J Trop Med Hyg. 1995;52:559–564. doi: 10.4269/ajtmh.1995.52.559. [DOI] [PubMed] [Google Scholar]
- 30.Morgan U M, Pallant L, Dwyer B W, Forbes D A, Rich G, Thompson R C. Comparison of PCR and microscopy for detection of Cryptosporidium parvum in human fecal specimens: clinical trial. J Clin Microbiol. 1998;36:995–998. doi: 10.1128/jcm.36.4.995-998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan U M, Sargent K D, Deplazes P, Forbes D A, Spano F, Hertzberg H, Elliot A, Thompson R C. Molecular characterization of Cryptosporidium from various hosts. Parasitology. 1998;117:31–37. doi: 10.1017/s0031182098002765. [DOI] [PubMed] [Google Scholar]
- 32.Morgan U M, Xiao L, Fayer R, Lal A A, Thompson R C. Epidemiology and strain variation of Cryptosporidium parvum. Contrib Microbiol. 2000;6:116–139. doi: 10.1159/000060369. [DOI] [PubMed] [Google Scholar]
- 33.Morgan U M, Xiao L, Fayer R, Lal A A, Thompson R C. Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int J Parasitol. 1999;29:1733–1751. doi: 10.1016/s0020-7519(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 34.Morgan U M, Xiao L, Monis P, Sulaiman I, Pavlasek I, Blagburn B, Olson M, Upton S J, Khramtsov N V, Lal A, Elliot A, Thompson R C. Molecular and phylogenetic analysis of Cryptosporidium muris from various hosts. Parasitology. 2000;120:457–464. doi: 10.1017/s0031182099005703. [DOI] [PubMed] [Google Scholar]
- 35.Patel S, Pedraza-Diaz S, McLauchlin J, Casemore D P. Molecular characterisation of Cryptosporidium parvum from two large suspected waterborne outbreaks. Commun Dis Public Health. 1998;1:231–233. [PubMed] [Google Scholar]
- 36.Pedraza-Diaz S, Amar C, McLauchlin J. The identification and characterisation of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol Lett. 2000;189:189–194. doi: 10.1111/j.1574-6968.2000.tb09228.x. [DOI] [PubMed] [Google Scholar]
- 37.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C, Ong C S, MacKenzie W R, Lal A A, Beard C B. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pieniazek N J, Bornay-Llinares F J, Slemenda S B, da Silva A J, Moura I N, Arrowood M J, Ditrich O, Addiss D G. New Cryptosporidium genotypes in HIV-infected persons. Emerg Infect Dis. 1999;5:444–449. doi: 10.3201/eid0503.990318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quiroz E S, Bern C, MacArthur J R, Xiao L, Fletcher M, Arrowood M J, Shay D K, Levy M E, Glass R I, Lal A. An outbreak of cryptosporidiosis linked to a foodhandler. J Infect Dis. 2000;181:695–700. doi: 10.1086/315279. [DOI] [PubMed] [Google Scholar]
- 40.Saano A, Lindstrom K. Small scale extraction with spun column clean up. In: Akkermans A, de Bruijn F, van Elsas J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer; 1994. [Google Scholar]
- 41.Spano F, Putignani L, Crisanti A, Sallicandro P, Morgan U M, Le Blancq S M, Tchack L, Tzipori S, Widmer G. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J Clin Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spano F, Putignani L, Guida S, Crisanti A. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp Parasitol. 1998;90:195–198. doi: 10.1006/expr.1998.4324. [DOI] [PubMed] [Google Scholar]
- 43.Spano F, Putignani L, McLauchlin J, Casemore D P, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 44.Sulaiman I M, Xiao L, Yang C, Escalante L, Moore A, Beard C B, Arrowood M J, Lal A A. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg Infect Dis. 1998;4:681–685. doi: 10.3201/eid0404.980424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tzipori S, Smith M, Birch C, Barnes G, Bishop R. Cryptosporidiosis in hospital patients with gastroenteritis. Am J Trop Med Hyg. 1983;32:931–934. doi: 10.4269/ajtmh.1983.32.931. [DOI] [PubMed] [Google Scholar]
- 46.Waldman E, Tzipori S, Forsyth J R. Separation of Cryptosporidium species oocysts from feces by using a Percoll discontinuous density gradient. J Clin Microbiol. 1986;23:199–200. doi: 10.1128/jcm.23.1.199-200.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Widmer G, Tchack L, Chappell C L, Tzipori S. Sequence polymorphism in the beta-tubulin gene reveals heterogeneous and variable population structures in Cryptosporidium parvum. Appl Environ Microbiol. 1998;64:4477–4481. doi: 10.1128/aem.64.11.4477-4481.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Widmer G, Tzipori S, Fichtenbaum C J, Griffiths J K. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J Infect Dis. 1998;178:834–840. doi: 10.1086/515373. [DOI] [PubMed] [Google Scholar]
- 49.Xiao L, Bern C, Limor J, Sulaiman I, Roberts J, Checkley W, Cabrera L, Gilman R H, Lal A A. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J Infect Dis. 2001;183:492–497. doi: 10.1086/318090. [DOI] [PubMed] [Google Scholar]
- 50.Xiao L, Escalante L, Yang C, Sulaiman I, Escalante A A, Montali R J, Fayer R, Lal A A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao L, Morgan U M, Fayer R, Thompson R C, Lal A A. Cryptosporidium systematics and implications for public health. Parasitol Today. 2000;16:287–292. doi: 10.1016/s0169-4758(00)01699-9. [DOI] [PubMed] [Google Scholar]
- 52.Xiao L, Morgan U M, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson R C, Fayer R, Lal A A. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]