On December 19, 2021, a 31-year-old male boarded a flight from Bhubaneswar, India to Mumbai, India, and then another flight from Mumbai to Muscat, Oman. On December 24, he returned from Muscat to Guangzhou by airplane and was transferred to a centralized isolation hotel in Foshan. He was sampled on the third day of the quarantine, and the result returned to be coronavirus disease 2019 (COVID-19) positive on December 27, 2021. He was transported to the Guangzhou Eighth People’s Hospital for isolated treatment by negative pressure ambulance on the same day. The case had two lungs focal inflamed according to the chest CT examination.
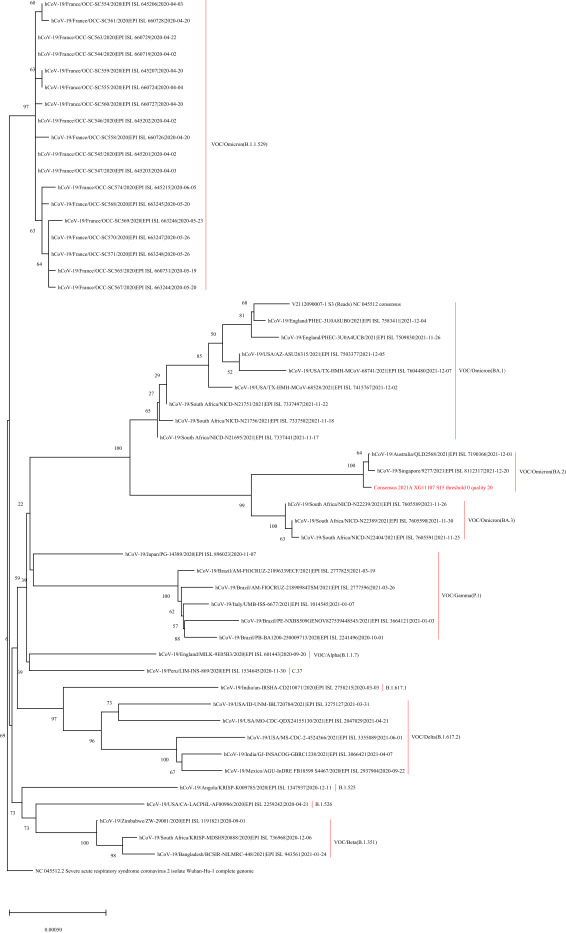
On December 28, 2021, the nasal swab of the imported case was sequenced using Illumina MiniSeq by Guangdong CDC. The viral RNA was reverse transcribed and amplified using ULSEN®2019-nCoV Whole Genome Capture Kit (Beijing MicroFuture Technology Co., Ltd, Beijing, China, V-090418), then the sequencing libraries were prepared using the Illumina Nextera® XT Library Prep Kit (Illumina, Inc., San Diego, USA, FC-131-1001) and conducted Illumina sequencing. On December 29, 2021, the sequencing analysis concluded that the virus genome belonged to lineage BA.2, VOC/Omicron (EPI_ISL_8207301). Compared with the Wuhan reference sequence (EPI_ISL_402119) (1 ), the strain from the case displayed 67 nucleotide variation sites. The sequence reported on the evolutionary branch is most similar to those reported in Singapore on December 20, 2021 and Australia on December 1, 2021 (Figure 1). In protein level, a total of 49 amino acid mutation sites and 9 amino acid deletions were observed, including 27 mutations and 3 deletions in the spike protein.
Figure 1.
Phylogenetic tree based on the full-length genome sequences of the COVID-19 virus. The Guangdong imported VOC/Omicron (BA.2) variant is highlighted in red. The Wuhan reference strain (NC_045512.2) and other variants are marked in black.
Abbreviations: COVID-19=coronavirus disease 2019; VOC=variant of concern.
On November 26, 2021, Omicron was listed by the World Health Organization (WHO) as the fifth “Variant of Concern (VOC)” (2 ). Studies showed that the VOC/Omicron strains have evolved into 3 lineages, namely as BA.1, BA.2, and BA.3, the most popular lineage is the lineage of BA.1. The BA.2 lineage has been nicknamed as “Stealth Omicron” because it differs from the “standard” variety by not having the characteristic SGTF-causing deletion (H69del and V70del) (3-6). A total of 89 sequences of BA.2 have been uploaded in GISAID, mostly comprised of sequences from South Africa, Denmark, Canada, the United Kingdom, the United States, India, and other countries. The earliest BA.2 sequence was uploaded from South Africa on November 27, 2021 (7 ). The case in this study was the first BA.2 lineage discovered in China. The biological characteristics and transmissibility may be different because of the difference of the genomes of the BA.2 lineage, so further study and long-term surveillance are urgently needed.
Contributor Information
Cao Chen, Email: chencao@ivdc.chinacdc.cn.
Baisheng Li, Email: libsn@126.com.
References
- 1.Tan WJ, Zhao X, Ma XJ, Wang WL, Niu PH, Xu WB, et al A novel coronavirus genome identified in a cluster of pneumonia cases—Wuhan, China 2019−2020. China CDC Wkly. 2020;2(4):61–2. doi: 10.46234/ccdcw2020.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern. [2021-12-29].
- 3.The Guardian. Scientists find ‘stealth’ version of Omicron that may be harder to track. https://www.theguardian.com/world/2021/dec/07/scientists-find-stealth-version-of-omicron-not-identifiable-with-pcr-test-covid-variant. [2021-12-29].
- 4.Outbreak.info. The Scripps Research Institute. “BA.1 Lineage Report”. https://outbreak.info/situation-reports?pango=BA.1. [2021-12-29].
- 5.Outbreak.info. The Scripps Research Institute. “BA.2 Lineage Report”. https://outbreak.info/situation-reports?pango=BA.2. [2021-12-29].
- 6.Outbreak.info. The Scripps Research Institute. “BA.3 Lineage Report”. https://outbreak.info/situation-reports?pango=BA.3. [2021-12-29].
- 7.GISAID. hCoV-19 tracking of variants (see menu option BA.2). https://www.epicov.org/epi3/frontend#34a5cd. [2021-12-29].