The COVID-19 pandemic is a global health crisis. Moreover, emerging mutated virus strains present an even greater challenge for existing vaccines and medications. One possible solution is to design drugs based on the properties of the virus epigenome.
As a Food and Drug Administration (FDA)-approved drug for myelodysplastic syndrome [1], 5-Azacytidine (5Aza) is a structural analog of cytidine (Fig. S1 online), and has been established as a potent inhibitor of DNA methylation, both in preclinical models and in cancer patients [2]. Besides, due to substitution from carbon to nitrogen in cytidine, 5Aza can inhibit cytosine methylation at the C5 position (m5C) within RNA by sequestrating m5C RNA methyltransferases (m5C-RMT) [3], [4]. It exhibits antiviral effects against several viruses [5], [6]. However, whether 5Aza inhibits SARS-CoV-2 infection by targeting m5C RNA methylation is unknown.
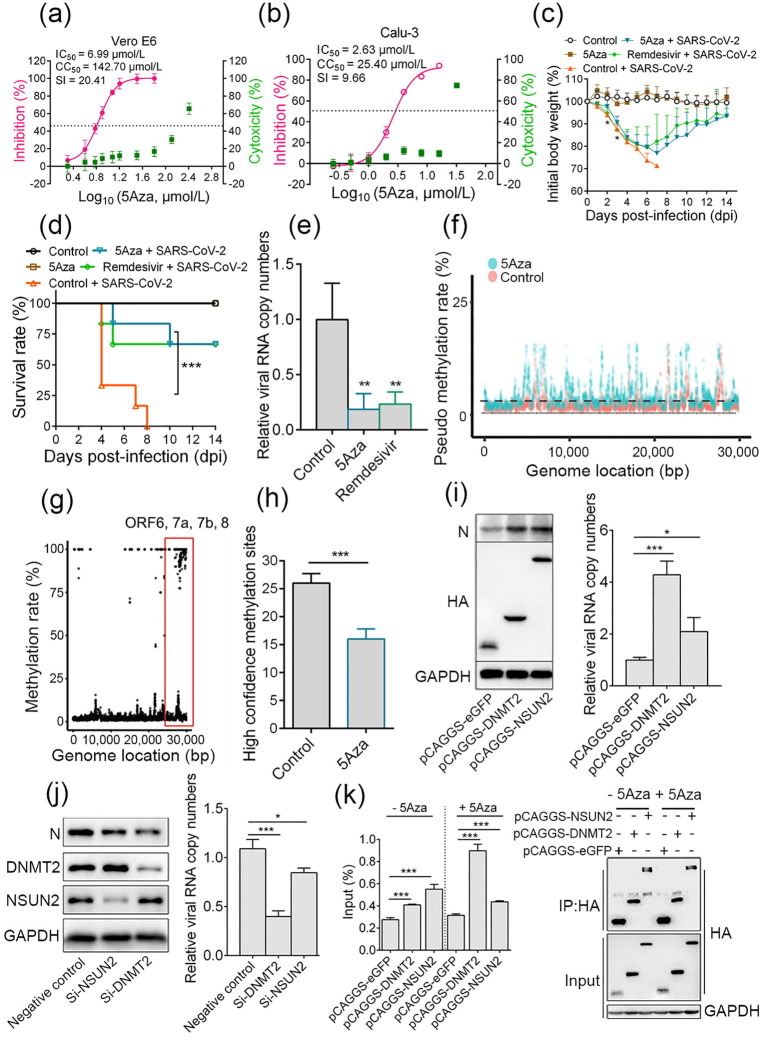
We first examined the anti-SARS-CoV-2 activity of 5Aza in Vero E6 and Calu-3 cells. The results showed that the half maximal inhibition concentration (IC50) and half cytotoxic concentration (CC50) values of 5Aza in Vero E6 cells were 6.99 and 142.70 μmol/L, respectively, with the selectivity index (SI) of 20.41 (Fig. 1 a). In Calu-3 cells, the IC50 and CC50 values of 5Aza were 2.63 and 25.40 μmol/L, respectively (SI = 9.66) (Fig. 1b). The data suggests that the IC50 values of 5Aza are higher than those of Remdesivir (IC50 = 1.70 and 2.28 μmol/L in Vero E6 and Calu-3 cells, respectively; Fig. S2 online), a known anti-SARS-CoV-2 drug. 5Aza antiviral activity was further confirmed by indirect immunofluorescence assay (IFA), with Remdesivir as a positive control (Fig. S3 online). To define the specific step of virus infection inhibited by 5Aza, a time-of-addition assay was performed (Fig. S4a online), which showed that 5Aza functioned after virus entry (Fig. S4b–d online).
Fig. 1.
Azacytidine targets SARS-CoV-2 RNA to inhibit virus infection. Vero E6 and Calu-3 cells were infected with SARS-CoV-2 at a multiplicity of infection (moi) of 0.2 in the presence of different doses of azacytidine. At 24 h post-infection, cell supernatants were collected. (a, b) IC50 and CC50 were calculated by detecting viral RNA with quantitative real-time PCR (qRT-PCR) and CCK-8 assay, respectively. (c–e) The anti-SARS-CoV-2 effect of azacytidine in vivo. 8 weeks old female BALB/c mice were randomly divided into 3 groups with 10 mice per group. Mice were intranasally challenged with 4 × 103 plaque forming unit (PFU) MA-SARS-CoV-2 in 50 μL Dulbecco’s modified Eagle’s medium (DMEM) or equal DMEM. At 1 dpi, mice were intraperitoneally injected with either 2 mg/kg 5Aza, 25 mg/kg Remdesivir, or an equivalent volume of sterile saline, once daily for seven consecutive doses. (c) Body weight was measured daily for 14 d; mice with 30% body weight loss were considered moribund and euthanized. (d) The survival rates of mice (n = 6). (e) At 4 dpi, 4 mice per group were euthanized to detect viral RNA copy numbers in lungs, and data are presented as values relative to the control. (f) The pseudo m5C locations indicated incorporated azacytidine. Vero E6 cells were infected with 1 moi SARS-CoV-2 in the presence of 10 μmol/L 5Aza or saline. After 12 h, total RNA was isolated and a comparison of 5Aza-treated viral RNA with the control using bisulfite sequencing was performed; the non-overlapping points are pseudo m5C locations that indicate where 5Aza was incorporated. (g) RNA-BSseq identified m5C locations in the SARS-CoV-2 genome. There were at least 5 methylation-supporting reads per point. Data are averaged from 3 technological repeats. (h) RNAs of cells infected with SARS-CoV-2 (treated with 5Aza or untreated) were subjected to nanopore direct RNA sequencing. (i) Vero E6 cells were transiently overexpressed with DNMT2, NSUN2, or GFP; 24 h later, cells were infected with 0.1 moi SARS-CoV-2. After 24 h, the supernatant was collected for viral RNA detection by qRT-PCR, and cells were lysed and subjected to western blotting. (j) Vero E6 cells were transfected with 100 μmol/L siRNA targeting NSUN2, DNMT2, or negative control siRNA; 48 h later, cells were infected with 0.1 moi SARS-CoV-2. After 30 h, viral RNA in the supernatant was detected by RT-PCR, and cells were lysed for western blotting. (k) Vero E6 cells transiently overexpressing DNMT2, NSUN2, or GFP with a hemagglutinin (HA) tag were infected with 0.2 moi SARS-CoV-2 for 20 h, either in the presence of 16 μmol/L 5Aza or not. Lysates were prepared and split for incubation with mouse anti-HA antibody. Co-precipitated RNA was analyzed by qRT-PCR. The level of viral RNA amplicon was determined as the percentage of input (1% of lysate) (left); the expression of indicated protein and products of immunoprecipitation (IP) was validated by western blotting (right). Data are presented as mean ± standard deviation and analyzed using Student’s t-test or one-way analysis of variance (body weight change); Log-rank test was used to analyze the significance of survival differences. * P < 0.05, ** P < 0.01, *** P < 0.001.
We then evaluated the antiviral effect of 5Aza in BALB/c mice, using a mouse-adapted SARS-CoV-2 (MA-SARS-CoV-2) [7]. Mice were intranasally challenged with MA-SARS-CoV-2, followed by intraperitoneal administration of 5Aza at 2 mg/kg body weight at 1 d post-infection (dpi), once daily for 7 consecutive doses. Remdesivir (25 mg/kg, once daily for 7 consecutive doses) was used as a positive control. As shown in Fig. 1c, at 2 and 3 dpi, mice in the 5Aza-treated group lost less weight than the control- and Remdesivir-treated infection groups. Mice in the 5Aza- and Remdesivir-treated infection groups started to regain body weight at 8 and 7 dpi, respectively, and recovered to more than 90% of initial body weight at 14 dpi. Besides, at 4 dpi, 4 mice died in the control infection group, and 2 mice became moribund (defined as 30% loss of body mass). Comparatively, only 1 mouse died and 1 was moribund in the 5Aza- (5 dpi and 10 dpi, respectively) and Remdesivir- (4 dpi and 5 dpi, respectively) treated groups. The data indicated that 66.7% of mice survived in the 5Aza- and Remdesivir-treated groups, while none survived in the control infection group (Fig. 1d). The viral RNA copy number in the lungs of virus-infected mice was significantly decreased in 5Aza- and Remdesivir-treated groups (Fig. 1e). Moreover, histological examination revealed remarkable amelioration of lung damage at 4 dpi in the 5Aza- and Remdesivir-treated infection groups compared with that in the control infection group (Fig. S5 online). Pieces of evidence from previous studies have shown that cytokine storm may be one of the important factors in SARS-CoV-2 pathogenicity. Compared with the control group, 5Aza treatment reduced expression of several pro-inflammatory factors induced by SARS-CoV-2, such as interleukin (IL)-6, tumor necrosis factor (TNF)-α, interferon (IFN)-γ, and IL-1β, all reported to be closely related to severe SARS-CoV-2 infection (Fig. S6 online). However, certain antiviral genes, such as IFN-β, ISG15, ISG56, and RSAD2, were not remarkably altered by 5Aza treatment. Reduced pro-inflammatory response may be one of the important reasons why 5Aza treatment can protect mice from a lethal SARS-CoV-2 attack. In clinical trials, hematological toxicity (anemia, neutropenia, and thrombocytopenia) was the most commonly occurring adverse event in azacytidine recipients. However, in the present study, these adverse events were not observed (Fig. S7 online). This may be due to the shorter duration of drug treatment (tested in 4 dpi and 14 dpi) than that in clinical settings (7 d of treatment for each 28-d treatment cycle, for multiple cycles). According to the above results, 5Aza could inhibit SARS-CoV-2 infection both in vitro and in vivo, and a lower dose of 5Aza (2 mg/kg) might achieve the same therapeutic effect as a higher dose of Remdesivir (25 mg/kg).
Once 5Aza is introduced into the cells, it becomes enzymatically phosphorylated into 5Aza triphosphate (5Aza-CTP), which is randomly incorporated in the place of cytosine in nascent RNA, as RNA polymerases do not discriminate CTP from 5Aza-CTP [8]. Here, through high-resolution mass spectrometry analysis, RNA incorporated with 5Aza was found to be steadily increased around 40-fold with a short-period 5Aza treatment (Fig. S8 online). We then addressed whether 5Aza could be incorporated into viral RNA. Taking advantage of the fact that 5Aza is unable to be converted to uracil under bisulfide, we developed a new method named 5Aza-BSseq that identifies the location where 5Aza is incorporated (Fig. S9 online). By comparing the 5Aza-treated viral RNA with the non-treated control, the observed different pattern suggested efficient incorporation of 5Aza into SARS-CoV-2 RNA (Fig. 1f). It has been reported that 5Aza can lead to lethal mutagenesis to inhibit influenza virus replication. However, we found consistent mutations and comparable mutation rates in the viral RNA propagated in 5Aza-treated or control cells (Supplementary Table 1 online), excluding the role of 5Aza-induced lethal mutagenesis in anti-SARS-CoV-2.
As RNAs containing 5Aza at the precise target sites of specific m5C-RMTs will inhibit m5C RNA methylation, it was interesting to determine if 5Aza incorporation would influence m5C methylation of viral RNA. We first explored whether m5C methylation of viral RNA exists. However, it is difficult to reliably measure the m5C sites due to the mismatch errors induced by random priming in methodologies such as RNA bisulfite sequencing. We sought to develop an optimized approach to avoid the random primer insertion into the sequenced fragments, minimizing the false positive rate (Fig. S10a online). The results show that fluctuating GCTA distributes before and after a smooth distribution line, indicating few artifactual C in the modified bisulfite sequencing (Fig. S10b online). We detected more than 555 high confident m5C methylation sites in the SARS-CoV-2 genome (false positive 0.006, bisulfite conversion rate 97.37%) (Fig. 1g and Fig. S11 online), with high methylation levels detected in ORF6, 7a, 7b, and 8 (Fig. 1g, red square). High-confidence m5C methylation sites were also found in a delta variant of the SARS-CoV-2 virus (Fig. S12 online). Then, to investigate whether the 5Aza incorporation affects viral RNA methylation, we utilized the nanopore direct RNA sequencing to avoid the false-positive methylation sites caused by the unconverted azacytidine in bisulfite sequencing. The results showed that 26 and 16 high-confidence m5C sites were identified in control and azacytidine-treated viral RNAs, respectively, which indicated that 5Aza decreased the m5C methylation rate of viral RNA by 40% (Fig. 1h).
The primary writers for m5C on mRNAs have been proposed to be NSUN2 and DNMT2, which are demonstrated to contribute to m5C methylation of HIV-1 RNA and thus facilitate virus infection [9], [10]. Here, we found that overexpression of DNMT2 and NSUN2 significantly promoted the SARS-CoV-2 replication (Fig. 1i); knockdown of DNMT2 and NSUN2 resulted in about 2-fold and 20% reduction of viral RNA, respectively (Fig. 1j). The data suggest that DNMT2 plays a more important role in SARS-CoV-2 replication. To investigate the possible link between these two host factors and SARS-CoV-2 RNA, we performed immunoprecipitation (IP) assay, which showed that DNMT2 and NSUN2 could bind to SARS-CoV-2 RNA (Fig. 1k). Notably, DNMT2 binds more RNAs in the presence of 5Aza. 5Aza is a suicide mechanism-based inhibitor of m5C-RMTs [11], and therefore RNAs containing 5Aza at the precise target site will sequester the m5C-RMT by generating RNA-m5C-RMT adducts, which will result in the decreased level of active endogenous enzymes [3], [4], [12]. Consistent with this theory, we observed an obvious decrease in DNMT2, rather than in NSUN2 protein, upon 5Aza treatment in SARS-CoV-2 infected cells (Fig. S13 online). However, how m5C RNA methylation affects SARS-CoV-2 infection is still unknown. The high m5C methylation region contains ORF6, 7, and 8, which were reported to mediate immune escape by blocking IFN signaling [13]. Besides, ORF7 and 8 may contribute to the pro-inflammatory response; ORF6 can block the cellular nucleocytoplasmic transport system by hijacking Nup98-Rae1, thus rendering host cells incapable of responding to SARS-CoV-2 infection [14]. It is likely that a reduction of m5C methylation in the high methylation region might influence viral RNA stability, the splicing of viral RNA, translation [9], [10], [15], or virus-host interaction. Further investigation should focus on the mechanism of m5C methylation and how this modification regulates SARS-CoV-2 infection.
In summary, we demonstrated that 5Aza can incorporate into SARS-CoV-2 RNA and disturb m5C RNA methylation, potentially contributing to 5Aza’s anti-SARS-CoV-2 activity. Although hematological toxicity was the most commonly occurring adverse event in azacytidine recipients for myelodysplastic syndrome/acute myelocytic leukemia treatment, this can be managed by dose reduction, dose delay, or combined therapy. Therefore, 5Aza might be a promising candidate for combating COVID-19. This study also introduced the possibility of targeting viral epigenomes as a novel antiviral strategy.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
Acknowledgments
This work was supported by the Key Project of Novel Coronavirus Normalized Prevention and Control in Hubei Province (2021ACB003), the National Science and Technology Major Projects (2020ZX10001016), and the Natural Science Foundation of Fujian Province (2020J02004). We thank Prof. Meilin Jin (Huazhong Agricultural University) for kindly providing the mouse-adapted SARS-CoV-2 strain.
Author contributions
Xian Lin and Xianliang Ke investigated the antiviral effects of drugs on SARS-CoV-2, and performed the in vivo assay. Xiaoqin Jian contributed to the in vivo assay, cell culture, plasmids construction, and virus detection. Xia Lin and Yang Yang contributed to the materials preparation and data analysis. Tianying Zhang, Hualong Xiong, Binghai Zhao, and Wen Liu contributed to the drug screening and data collection. Wen Liu, Quanjiao Chen, and Chong Tang conceived and designed the study. Chong Tang performed the BSseq and data analysis. Xian Lin, Xianlinag Ke, Quanjiao Chen, and Chong Tang analyzed the data. Xian Lin and Xianliang Ke wrote the original draft, Quanjiao Chen and Chong Tang reviewed and revised the manuscript.
Biographies

Xian Lin received his Ph.D. degree from Huazhong Agricultural University in 2016, and he is now an assistant researcher at Wuhan Institute of Virology. His main interest is the epigenetic regulation mechanism of virus replication and pathogenesis.

Xianliang Ke received his Ph.D. degree from Wuhan Institute of Virology in 2016, and is now an assistant researcher in the institute. His research interests include the interaction between host and Nipah virus and enterovirus, and novel antivirus strategies against important human viruses.

Wen Liu received his Ph.D. degree from the University of California, San Diego, and he is now a full professor at the School of Pharmaceutical Sciences, Xiamen University. His research interest mainly focuses on the molecular mechanisms underlying epigenetic regulation of gene transcription and splicing, and the implications in human diseases.

Quanjiao Chen received her Ph.D. degree from Wuhan Institute of Virology in 2007, and is now a professor in the institute. Her work focuses on host-virus interaction, including influenza A virus and Nipah virus, and molecular epidemiology of avian influenza virus.

Chong Tang got his Ph.D. degree from the University of Nevada, Reno. He is the director of technology in BGI and the adjunct professor in three universities. His research interest is developing new molecular biology tools to study spermiogenesis, cell transcription translation regulation, and genome conformation.
Footnotes
Supplementary materials to this article can be found online at https://doi.org/10.1016/j.scib.2022.02.002.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Kaminskas E., Farrell A.T., Wang Y.-C., Sridhara R., Pazdur R. FDA drug approval summary: Azacitidine (5-azacytidine, vidaza) for injectable suspension. Oncologist. 2005;10:176–182. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- 2.Stresemann C., Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 3.Schaefer M., Hagemann S., Hanna K., Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–8132. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- 4.Lu L.J., Randerath K. Effects of 5-azacytidine on transfer RNA methyltransferases. Cancer Res. 1979;39:940–949. [PubMed] [Google Scholar]
- 5.Diamantopoulos P.T., Michael M., Benopoulou O., et al. Antiretroviral activity of 5-azacytidine during treatment of a HTLV-1 positive myelodysplastic syndrome with autoimmune manifestations. Virol J. 2012;9:1. doi: 10.1186/1743-422X-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawson J.M.O., Daly M.B., Xie J., et al. 5-azacytidine enhances the mutagenesis of HIV-1 by reduction to 5-aza-2'-deoxycytidine. Antimicrob Agents Chemother. 2016;60:2318–2325. doi: 10.1128/AAC.03084-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang K., Zhang Y., Hui X., et al. Q493K and Q498H substitutions in spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine. 2021;67:103381. doi: 10.1016/j.ebiom.2021.103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oki Y., Aoki E., Issa J.P. Decitabine-bedside to bench. Crit Rev Oncol Hematol. 2007;61:140–152. doi: 10.1016/j.critrevonc.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Dev R.R., Ganji R., Singh S.P., et al. Cytosine methylation by DNMT2 facilitates stability and survival of HIV-1 RNA in the host cell during infection. Biochem J. 2017;474:2009–2026. doi: 10.1042/BCJ20170258. [DOI] [PubMed] [Google Scholar]
- 10.Courtney D.G., Tsai K., Bogerd H.P., et al. Epitranscriptomic addition of m5C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe. 2019;26:217–227. doi: 10.1016/j.chom.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell 1983; 33:9–10. [DOI] [PubMed]
- 12.Lu L.W., Chiang G.H., Medina D., et al. Drug effects on nucleic acid modification. I. A specific effect of 5-azacytidine on mammalian transfer RNA methylation in vivo. Biochem Biophys Res Commun. 1976;68:1094–1101. doi: 10.1016/0006-291x(76)90308-9. [DOI] [PubMed] [Google Scholar]
- 13.Redondo N., Zaldivar-Lopez S., Garrido J.J., et al. SARS-CoV-2 accessory proteins in viral pathogenesis: Knowns and unknowns. Front Immunol. 2021;12:708264. doi: 10.3389/fimmu.2021.708264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Addetia A., Lieberman N.A.P., Phung Q., et al. SARS-CoV-2 ORF6 disrupts bidirectional nucleocytoplasmic transport through interactions with Rae1 and Nup98. mBio. 2021;12:e00065-21. doi: 10.1128/mBio.00065-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S., Mason C.E. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet. 2014;15:127–150. doi: 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.