Abstract
Infection with pathogenic viruses is often sensed by innate receptors such as Toll-Like Receptors (TLRs) which stimulate type I and III interferons (IFNs) responses, to generate an antiviral state within many cell types. To counteract these antiviral systems, many viruses, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), encode non-structural proteins (NSPs) that mediate immune evasion. Using an overexpression system in A549 cells, we demonstrated a significant increase (p ≤ 0.0001) in Vesicular Stomatitis Virus (VSV)-EGFP reporter virus replication in cell lines overexpressing either the SARS-CoV-2 NSP1 or NSP15 when compared to control A549 cells. The increase in VSV-EGFP virus output was associated with a decrease in TLR2, TLR4 and TLR9 protein expression and a lack of antiviral protein production. Truncation of both NSP1 and NSP15 led to an increase in cellular TLR2, TLR4 and TLR9 as well as a decrease in TLR2 expression respectively. This observation can be attributed to the presence of a functional domain in NSP1 and NSP15 between amino acid (aa) 120–180 and aa 230–346, respectively. Both TLR3 and TLR9 ligands but not TLR2 ligand were highly effective at overcoming NSP1 and NSP15 functional interference based on significant decrease (p ≤ 0.0001) in VSV-EGFP virus replication. NSP1 or NSP15 intracellular interactions are likely low affinity interactions that can be easily disrupted by stimulating cells with specific TLR3 and TLR9 ligands. This report provides insights into the role of SARS-CoV-2 NSP1 and NSP15 in limiting specific TLR pathway activation, as an evasive mechanism against host innate responses.
Graphical abstract
1. Introduction
The 2019 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a beta-coronavirus, is at the center of the unprecedented Covid-19 global health pandemic (Arunachalam et al., 2020; Jiang et al., 2020). This virus is thought to have originated from wildlife, hence is regarded as a zoonotic pathogen (World Health Organization: www.who.int/csr/sars/country/en/country2003_08_15.pdf). Primary infection with SARS-CoV-2 occurs in human bronchial epithelial cells, pneumocytes and upper respiratory tract cells. Interaction between the viral glycoprotein, Spike (S), and host angiotensin converting enzyme 2 (ACE2) receptor facilitates viral entry and subsequent genome replication.
The genome of SARS-CoV-2 comprises 14 open reading frames (ORFs), two-thirds of which encode 16 non-structural proteins (NSP1–16) that not only make up the replication complex upon entry into a cell (Blanco-Melo et al., 2020; Chu et al., 2020; Setaro and Gaglia, 2021). These NSPs serve a variety of roles in virus replication, assembly, and evasion of host anti-viral sensors (Blanco-Melo et al., 2020; Setaro and Gaglia, 2021). Understanding innate immune evasion mechanisms by a virus is essential to gain an understanding of its pathogenesis (Jiang et al., 2020). Improper or delayed innate responses have been strongly associated with failure to control primary SARS-CoV-2 infection and progression to fatal Covid-19 disease (Blanco-Melo et al., 2020; Iwasaki and Yang, 2020). Type I interferon (IFN) evasion mechanisms is one of the strategies utilized by the SARS-CoV-2 virus (Chu et al., 2020). Type I and type III IFNs constitute the first lines of host defense against viral infections (Müller et al., 1994; Sadler and Williams, 2008). Following infection, pathogen-associated molecular patterns (PAMPs) of viruses are recognized by various host pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), and nucleotide binding and oligomerization domain (NOD)-like receptors (NLRs) (Meylan et al., 2006). PRRs recognition of their cognate ligand subsequently triggers adaptor proteins such as TIR-domain-containing adapter-inducing interferon-β (TRIF), Tumor necrosis factor (TNF) receptor associated factor (TRAF) 6 and myeloid differentiation primary response 88 (MyD88), and subsequent phosphorylation of transcription factors, interferon regulatory factor (IRF)1, IRF3 and IRF7 (Sadler and Williams, 2008). IRFs mediate transcription of type I (IFN-α and IFN-β) and type III IFN (IFN-λ) genes which in turn induce interferon stimulated genes (ISG) that have antiviral activity, promoting viral clearance by interfering with replication, packing, exit and spread (Der et al., 1998; Sadler and Williams, 2008). Melanoma differentiation-associated protein 5 (MDA-5), an intracellular PRR, can effectively sense the SARS-CoV-2 virus dsRNA upon infection which leads to IFN expression (Hoagland et al., 2021; Sampaio et al., 2021). However, SARS-CoV-2 infected cells lack the ability to mount an effective MDA-5 mediated antiviral response but are highly responsive to exogenous IFNs (Deng and Baker, 2018; Lokugamage et al., 2020). In contrast, sensing of viral dsRNA by TLR3 and its subsequent signalling via MyD88 is associated with greater resistance to SARS-CoV infection in mice (Sheahan et al., 2008; Totura and Baric, 2012).
Most viruses have developed mechanisms that suppress IFN expression or signaling (Hadjadj et al., 2020; Hengel et al., 2005). For instance, both the SARS-CoV and Middle East respiratory syndrome virus (MERS) possess early expressing genes, such as NSP1, NSP3, NSP8 and NSP15, which delay type I or type III IFN production or function (Chen et al., 2011; Huang et al., 2011; Kamitani et al., 2006; Kumar et al., 2007; Menachery et al., 2017). Given the pivotal role of IFN signalling in coronavirus infection, there is a need to better understand how SARS-CoV-2 can antagonize IFNs and also to develop means to neutralize this antagonistic activity. The present study aimed to demonstrate the effects of TLR ligands in overcoming antagonistic functions of SARS-CoV-2 NSP1 and NSP15. Therefore, this study investigated the potency of the SARS-CoV-2 NSP1 and NSP15 in mediating immune evasion of host antiviral responses.
2. Materials and methods
2.1. Plasmid and cloning
The 2019 SARS-CoV-2 NSP1 and NSP15 were human codon-optimized using the IDT codon-optimization tool (https://www.idtdna.com/codonopt). Start codons were added to NSP1 (accession number: YP_009725297.1) and NSP15 (accession number: YP_009725310.1), and a Kozak sequence was added before each start codon. A HindIII and EcoRV restriction sites were also added before the Kozak sequence and after the coding sequence respectively. Sequences were synthesized by ThermoFisher Scientific gene art synthesis into a pMX plasmid with ampicillin resistance. DH5α competent cells were transformed with the pMT-NSP1-NSP15. Plasmids, pMT-NSP1-NSP15 and pCDNA3.1/V5–HIS-TOPO were digested (HindIII and EcoRV restriction enzymes; New England Bioscience, ON, CA) and gel purified (Qiagen, ON, CA). NSP1and NSP15 gene fragments were subcloned (T4 ligation; New England Bioscience, ON, CA) into the pCDNA3.1/V5–HIS TOPO plasmid (lab stocks). DH5α competent cells were transformed and utilized to propagate the respective plasmids. All plasmids were linearized by BglII (New England Bioscience, ON, CA) digestion for transfection.
Truncation of NSP1 and NSP15: NSP1 and NSP15 were truncated by high fidelity PCR (GoTaq polymerase; Promega, WI, USA) using designer primers (Table 1 ) to generate NSP1-one third length (1/3), NSP1-two third length (2/3), NSP15-1/3 and NSP15-2/3 with respective 5’-HindIII and 3’-EcoRV restriction sites. Gel purified truncated DNA fragment were ligated into pDRIVE vectors (TA cloning; Qiagen, ON, CA) and subcloned (HindIII and EcoRV restriction digestion) into the pCDNA3.1/V5–HIS TOPO plasmid. DH5α competent cells were transformed and utilized to propagate the respective plasmids. All plasmids were linearized by BglII (New England Bioscience, ON, CA) digestion for transfection.
Table 1.
Primer sequences used for PCR.
| Target | Primer sequencesa | Accession No. | |
|---|---|---|---|
| NSP1 1/3 | Fwd | AAGCTTGCCACCATGGAGAGCCTTGTCC | NC_045512.2 |
| Rev | GATATCCAAGTTGAGGCAAAACGCCT | ||
| NSP1 2/3 | Fwd | AAGCTTGCCACCTGCCTCAACTTGAACAGCCC | NC_045512.2 |
| Rev | GATATCCTTGCGGTAAGCCACTGGTA | ||
| NSP15 1/3 | Fwd | AAGCTTGCCACCATGGTTTAGAAAATGTGGCT | NC_045512.2 |
| Rev | GATATCTCGTTTCAGTTGGTTTCTTGGC | ||
| NSP15 2/3 | Fwd | AAGCTTGCCACCTTGTGCACCACTCACTGTCT | NC_045512.2 |
| Rev | GATATCCAATTTCCATTTGACTCCTGGGT | ||
The boldface at 5' ends represents the HindIII and EcoRV restriction sequences inserted for restriction cloning into the pCDNA3.1/V5–HIS-TOPO plasmid.
2.2. Cell culture
-
(i)
Cell: A549 cells were cultured and maintained (37 °C and 5% CO2) in Dulbecco’s modified Eagle’s medium (DMEM; Corning, CA) supplemented with 10% fetal bovine serum (FBS; Gibco, Life Technologies, CA) and 0.1% penicillin–streptomycin (Gibco, Life Technologies, CA).
-
(ii)
Transfection/Stably expressing cell lines: One hundred thousand A549 cells were plated per well in a 24 well plates (15.6 mm wells). Overnight cells (>80% confluent) were transfected, according to manufacturer’s recommendation, using Lipofectamine stem reagent (Life Technologies, CA) with up to 1 μg of linearized pCDNA3.1/V5–HIS-TOPO empty vector or recombinant expressing NSP1, NSP1-1/3, NSP1-2/3, NSP15, NSP15-1/3 and NSP15-2/3. After 48 h (hrs), cells were passaged (0.5% trypsin; Life Technologies, CA) into T25 flasks and treated with 800 μg/mL G418 in DMEM complete medium (10% FBS and 0.1% penicillin–streptomycin). Medium were changed every 48 h until proliferating island of cells were observed. After 3 weeks of antibiotic selection, stably expressing cell lines (A549-pCDNA3.1, A549-NSP1, A549-NSP15, A549-NSP1-1/3, A549-NSP1-2/3, A549-NSP15-1/3 and A549-NSP1-2/3) were confirmed by PCR and western blot.
2.3. Anti-viral assay
To define the biological activity of NSPs, antiviral assays (AVA) are often applied whereby vesicular stomatitis virus (VSV) is employed due to its sensitivity to IFN where wildtype viruses cannot be utilized (Meager, 2002). In this assay, proteins are tested for their ability to complement the growth of a recombinant VSV-EGFP virus. As has been demonstrated with human cell lines, VSV-EGFP can be used as an effective reporter virus in overexpressing cell lines to study IFN induction and signalling (Liu et al., 2017; Zheng et al., 2020).
-
(i)
Stimulation: A549 cells and stably expressing cell lines were seeded in 96 well plates at a rate of 6.0 × 104 cells per well (37 °C and 5% CO2) in DMEM complete medium (100 μl). Next day, all adherent cells lines were visualised under a light microscope to confirm >95% cell confluency. Cells were washed with phosphate buffered saline (PBS) buffer and pre-treated for 2 h with the following TLR ligands: TLR2 ligand PAM3CSK4 (0.005, 0.05, 0.5 and 5 μg/mL; Invivogen, CA), TLR3 ligand Polyinosinic:polycytidylic acid (polyI:C) (0.005, 0.05, 0.5 and 5 μg/mL; Invivogen, CA), TLR4 ligand lipopolysaccharides from Escherichia coli O111:B4 (LPS) (0.01, 0.1, 1.0, 10 and 50 μg/mL; Millipore-Sigma, CA) and TLR9 ligand CpG ODN 2007 (0.005, 0.05, 0.5, 5 and 50 μg/mL, Life Technologies, CA).
-
(ii)
Infection and Readout: A549 cells and stably expressing cell lines were infected (multiplicity of infection; MOI of 0.4) with the vesicular stomatitis virus (VSV)-EGFP and incubated (37 °C and 5% CO2) for 24 h. Infected well were viewed (GFP signal) using an inverted fluorescence microscope (Leica, DMi8 S Platform) to determine infection rate prior to readout. Cells were read using a GloMAX-Multi microplate reader (360/40 nm excitation filter, a 460/40 nm emission filter). The mean GFP-fluorescence readout in uninfected cells was used as the normalization factor for VSV-EGFP readout in infected A549 cells. Cell numbers (DAPI staining) were also used to normalize readout.
2.4. Flow cytometry
Intracellular TLR staining: Three hundred thousand A549 cells stably expressing NSP1, NSP1-1/3, NSP1-2/3, NSP15, NSP15-1/3 and NSP15-2/3 were stained (10 min in 1% bovine serum albumin buffer: BSA) with 7-AAD (ThermoFisher Scientific, Ca) and subsequently fixed for 30 min using the fixation/permeabilization kit (BD bioscience, Ca). Cells were incubated in permeabilization (perm/wash) buffer for 30 min at 4 °C, blocked (20 min at 4 °C) with 1% bovine serum albumin (BSA) followed by incubation (20 min at 4 °C) with mouse anti-TLR2-biotin (eBioscience, Life Technologies, Ca), rat anti-TLR3 (eBioscience, Life Technologies, Ca), mouse anti-TLR4-biotin (eBioscience, Life Technologies, Ca) and mouse anti-TLR9-biotin (eBioscience, Life Technologies, Ca). Cells were washed twice in staining buffer (PBS + 1% BSA) and again incubated with streptavidin-PE (Life Technologies, Ca) or Mouse anti-rat-PE for 20 min at 4 °C in staining buffer. Finally, cells were washed twice in staining buffer and acquired on a BD FACS Canto II and the data were processed by FlowJo V10 software.
2.5. RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from A549 stably expressing cell lines using TRIzol (ThermoFisher Scientific, CA) according to the manufacturer’s protocol and treated with DNA-free DNase (ThermoFisher Scientific, CA). Subsequently, 1 μg of purified RNA was reverse transcribed to cDNA using a Superscript II first-strand synthesis kit (ThermoFisher Scientific, CA) and oligo(dT) primers according to the manufacturer’s recommended protocol. The resulting cDNA was diluted 1 : 10 in diethyl pyrocarbonate-treated (DEPC) water. qRT-PCR using SYBR green was performed on diluted cDNA using a LightCycler 480 II (Roche Diagnostics GmbH, Mannheim, GER) as according to manufacturer’s recommendation. In brief, the quantitative qRT-PCR was performed with the following conditions: initial denaturation was performed at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 10 s (s), primer annealing listed in Table 2 for 15s and extension at 72 °C for 20s, with end point melt-curve analysis. The relative fold change of target genes was calculated by 2−ΔΔCT method. The Ct value for each sample was normalized against GAPDH housekeeping gene for respective sample. Data represent means from 6 biological replicates, using primers outlined in Table 1.
Table 2.
Primer sequences used for Real-Time PCR.
| Target | Primer sequence | Accession No. | Annealing temp (oC) | |
|---|---|---|---|---|
| Pan-IFNα | Fwd | CACACAGGCTTCCAGGCATTC | NC_045512.2 | 61 |
| Rev | TCTTCAGCACAAAGGACTCATCTG | |||
| IFN-β | Fwd | TGCTCTCCTGTTGTGCTTCTCCAC | NM_002176.4 | 61 |
| Rev | ATAGATGGTCAATGCGGCGTCC | |||
| IFN-λ1 | Fwd | CGCCTTGGAAGAGTCACTCA | NM_172140.2 | 61 |
| Rev | GAAGCCTCAGGTCCCAATTC | |||
| IRF-1 | Fwd | TCTTAGCATCTCGGCTGGACTTC | NM_002198.3 | 61 |
| Rev | CGATACAAAGCAGGGGAAAAGG | |||
| IRF-7 | Fwd | GATGTCGTCATAGAGGCT GTTGG | NM_004029.4 | 61 |
| Rev | TGGTCCTGGTGAAGCTGGAA | |||
| ISG15 | Fwd | GAGAGGCAGCGAACTCATCT | NM_005101.4 | 60 |
| Rev | AGGGACACCTGGAATTCGTT | |||
| ISG54 | Fwd | ATTCTATCACCAAGCCCGTGG | NM_001547.5 | 60 |
| Rev | TGGAGTCTGGAAGCCTCATCC | |||
| ISG56 | Fwd | CAGCAACCATGAGTACAAAT | NM_001548 | 60 |
| Rev | AAGTGACATCTCAATTGCTC | |||
| OAS1 | Fwd | GCCCTGGGTCAGTTGACTGG | NM_016816 | 60 |
| Rev | TGAAGCAGGTGGAGAACTCGC | |||
| PKR | Fwd | ACACTCGTCTCTGAATCATC | NM_002759 | 60 |
| Rev | GAGACCATTCATAAGCAACG | |||
| IFIT1 | Fwd | GATCTCAGAGGAGCCTGGCTAA | NM_001548.5 | 60 |
| Rev | TGATCATCACCATTTGTACTCATGG | |||
| IFITM3 | Fwd | GATGTGGATCACGGTGGAC | NM_021034.3 | 61 |
| Rev | AGATGCTCAAGGAGGAGCAC | |||
| VIPERIN | Fwd | GAGAGCCATTTCTTCAAGACC | NM_080657 | 60 |
| Rev | CTATAATCCCTACACCACCTCC | |||
| GAPDH | Fwd | TACTCCTTGGAGGCCATGTG | NM_001357943.2 | 61 |
| Rev | CACAGTCCATGCCATCACTG | |||
2.6. Western blot
Protein samples were prepared by lysing cells in RIPA lysis buffer (10 mM Tris, pH 7.5, 5 mM EDTA, 150 mM NaCl, 0.1% SDS, 1% TritonX-100, 1% sodium deoxycholate) containing protease and phosphatase inhibitors (Roche, Ca) and quantified by spectrophotometry. Lysates were boiled in Laemmli’s sample loading buffer (Millipore-Sigma, ON, CA) and loaded onto 10% SDS-PAGE precast gels (Bio-Rad, ON, CA). After semidry transfer of SDS-PAGE onto a nitrocellulose membrane, the membrane was blocked in 5% skim milk powder for 2 h. The membranes were incubated on a shaker with the anti-V5 (IgG2a isotype; life Technologies, ON, CA) primary antibody for 12 h at 4 °C, washed, and incubated on a shaker with a rabbit anti-mouse IgG2a-biotin for 12 h at 4 °C. Finally, the blots were developed with a chemiluminescence kit (Thermo Fisher Scientific, ON, CA) and imaged using a Biorad ChemiDoc XRS + reader.
2.7. Confocal microscopy
A549 (NSP1 full length, NSP1-1/3, NSP1-2/3, NSP15 full length, NSP15-1/3 and NSP15-2/3) stably expressing cell lines were prepared for imaging. Cells were trypsinized and seeded in 24-well plates that contained 12-mm-diameter round coverslips at a rate of one hundred thousand cells per well. In brief, A549 stably expressing cells were fixed with 4% paraformaldehyde for 45 min at room temperature (RT) and washed twice with PBS. Cells were subsequently permeabilized with 0.1% Triton X-100 buffer solution (15 min at RT), blocked (1 h in 0.5% BSA–PBS), and incubated overnight (4 °C) with an anti-V5 (mouse IgG2a; 0.5 μg/500 μl in 0.5% BSA–PBS) and anti-LAMP1 (mouse IgG1; 0.5 μg/500 μl in 0.5% BSA–PBS). Cells were washed twice with PBS and again incubated overnight (4 °C) with anti-IgG2a-488 nm (goat anti-mouse, 0.5 μg/500 μl in 0.5% BSA–PBS) and anti-IgG1-488 nm (goat anti-mouse, 0.5 μg/500 μl in 0.5% BSA–PBS). The next day, nuclei were labeled with DAPI. Coverslips were mounted in Vectashield mounting medium for fluorescence imaging.
-
(i)
Visualization: Cells were viewed using a Leica SP5 laser scanning confocal microscope, and optical sections were recorded using either 663 or 640 nM with a numerical aperture of 1.4 or 1.25, respectively. All data were collected sequentially to minimize cross talk between fluorescence signals. The data are presented as maximum projections of z-stacks (20–25 sections: spacing of 0.3 mm). Maximum projections of z-stacks were analyzed with LAS AF software for localization of the relative fluorescence intensity across a straight line.
-
(ii)
Manders’ colocalization coefficient: All images were processed using Fiji – ImageJ software, and Manders’ colocalization coefficient was calculated using the colocalization (COLOC) function for a specified region of interest (ROI). Manders’ M1 and M2 weighted coefficients were calculated to determine the extent of colocalization between a pair of fluorescent signals and imaged in two channels. Manders’ M1 determines the degree of channel colocalization (NSP1–V5 or NSP15–V5 with LAMP1) and M2 determines the reverse.
2.8. Protein alignment
NSP1 and NSP15 Protein sequences from SARS-CoV-2 (YP_009724392.1), SARS-CoV (NP_828854.1), MERS (YP_009047226.1) and bat SARS-like CoV (AGZ48809.1) were aligned and phylogenetic tree, 1000 boot strap, generated using MEGA X (megasoftware.net).
2.9. Statistical analysis
All data are presented as means ± standard deviations (SD) from at least three independent experiments. Quantification was performed using GraphPad Prism 7 for Windows. The differences between groups, in each experiment, were analyzed by one-way analysis of variance (ANOVA) followed by a Tukey multiple-comparison test or non-parametric Wilcoxon tests (Mann-Whitney) to identify those groups that differed if the ANOVA result was significant. Results were considered statistically significant at a P value of < 0.05 (∗).
3. Results
3.1. NSP1 and NSP15 stably expressing cells lines support an increase in RNA virus replication
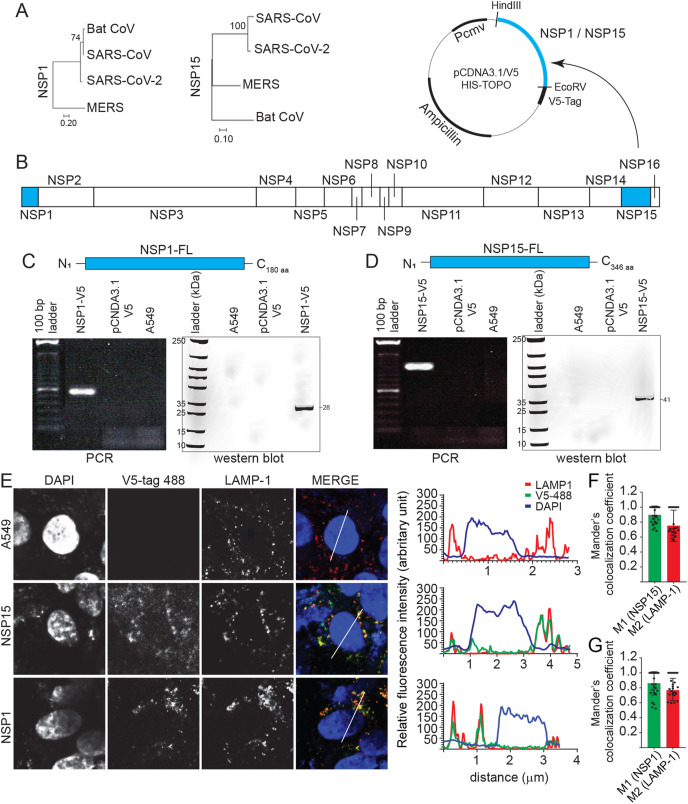
The bat coronavirus and SARS-CoV, but not MERS, shared greater genome sequence similarity (>90%) in their NSP1 and NSP15 with SARS-CoV-2 (Fig. 1 A). Sequence similarity informs of a conserved 3D structure that likely targets the ribosome at the translation initiation step (Kim et al., 2020; Schubert et al., 2020). Coding sequences of either NSP1 or NSP15 of SARS-CoV-2 were subcloned into two different pCDNA3.1/V5–HIS-TOPO plasmid (Fig. 1B) which were transfected into A549 cells, an adenocarcinomic human alveolar basal epithelial cells. Modified A549 cell lines, through antibiotic selection (G418), were confirmed to express either NSP1 (Fig. 1C) or NSP15 (Fig. 1D), at RNA and protein levels. Cells were tested, on a bi-weekly basis by PCR to confirm retention and sustained overexpression of NSP1 or NSP15. Using a light microscope or by FACS, no cytotoxic effects were observed in A549 cells as a result of overexpression of either NSP1 or NSP15. Both NSP1 and NSP15 overexpressing A549 cell lines were subsequently probed by confocal microscopy and the result demonstrate that both NSP1 (V5-488 nm) and NSP15 (V5-488 nm) localised mainly within the cytoplasm forming focal punctate as demonstrated by the relative fluorescence intensity based on the specific region of interest (ROI) identified (Fig. 1E). NSP15 more so than NSP1 was found in the nucleus (DAPI), while NSP1 showed perinuclear localization. The NSP1 and NSP15 punctate colocalized to a high degree with lysosomal vesicle as defined by lysosomal-associated membrane protein 1 (LAMP1). Manders’ coefficient for LAMP-1 and NSP15 (Fig. 1F) or NSP1 (Fig. 1G) confirms these observations suggesting the involvement of lysosomal vesicles in its subcellular trafficking.
Fig. 1.
NSP1 and NSP15 stably expressing A549 cells lines. Phylogenetic analysis of the SARS-CoV-2 (A) NSP1 and NSP15 as performed using MEGAX software. (B) NSP1 and NSP15, part of the ORF1a and 1b were subcloned into a pCDNA3.1/V5–HIS-TOPO plasmid (HindIII and EcoRV restriction sites) and transfected into A549 cells. Expression of (C) NSP1 and (D) NSP15 was confirmed by total RNA extraction (polymerase chain reaction; PCR) and total protein extraction (Western blot). (E) Representative pictures of confocal microscopy imaging with maximum projections of z-stacks for each channel demonstrating the subcellular localization of expressed NSP1 (anti-V5 antibody; 488 nm) and NSP15 (anti-V5 antibody; 488 nm) and lysosome-associated membrane protein 1 (LAMP-1) (red) in stably expressing A549 cells. Maximum projections of all combined channels (merge) were analyzed using LAS AF software to determine the relative fluorescence intensity (arbitrary units [A.U]) profiles in stably expressing cell line for anti-V5 (green) and DAPI (blue) and LAMP-1 (red) along the white line path (micrometers) indicated in the corresponding ROIs (merge) demonstrating overlapping signals. Validation of colocalization efficiencies by Manders’ colocalization coefficient as analyzed using ImageJ 72 h. Colocalizations between (F) NSP1-anti-V5 (M1) and LAMP-1 (M2) and (G) NSP15-anti-V5 (M1) and LAMP-1 (M2) represent the overlap of M1 as the denominator and vice versa (n = 50). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
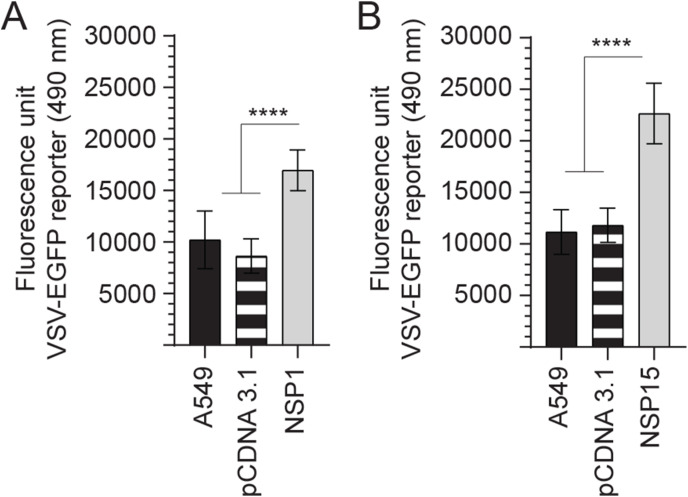
To understand the importance of either NSP1 or NSP15 overexpression for subcellular signalling or responses, a VSV-EGFP-based reporter assay was utilized to characterise differences between these cell lines (Fig. 2 ). All experiments were performed with a minimum of six replicates and repeated three times. Data were normalized to uninfected cells and analyzed by non-parametric Wilcoxon tests (Mann-Whitney). Both NSP1 and NSP15 overexpressing cell lines can effectively support higher VSV-EGFP virus replication as confirmed by GFP readout. The results demonstrated a significant (p ≤ 0.001) increase in VSV-EGFP virus output by 67.24% and 102.26% in NSP1 (Fig. 2A) and NSP15 (Fig. 2B), respectively. No effects on VSV-EGFP readouts were observed in empty vector control when compared to unmodified A549 cells.
Fig. 2.
Replication of VSV-EGFP in NSP1 and NSP15 stably expressing A549 cells lines. Replication efficiency of the interferon sensitive VSV-EGFP virus in A549 cells. Analysis of VSV-EGFP (moi = 0.4) virus replication in (A) NSP1 and (B) NSP15 stably expressing A549 cells lines compared to control transfected (pCDNA3.1/V5–HIS-TOPO) or negative (A549 cells) based on fluorescence (EGFP; excitation 490 nm). The mean fluorescence unit in uninfected cells was used as the normalization factor for VSV-EGFP readout in infected A549 cells. Non-parametric Wilcoxon tests (Mann-Whitney) was used to assess normal distribution and test significance with the results shown as mean ± SD. ∗∗∗∗ (p ≤ 0.001) indicates a statistically significant difference. All viral infection experiments were performed in 10 technical replicates, and the data are representative of results from 3 independent experiments.
3.2. Differential expression of innate sensors in both NSP1 and NSP15 stably expressing cells
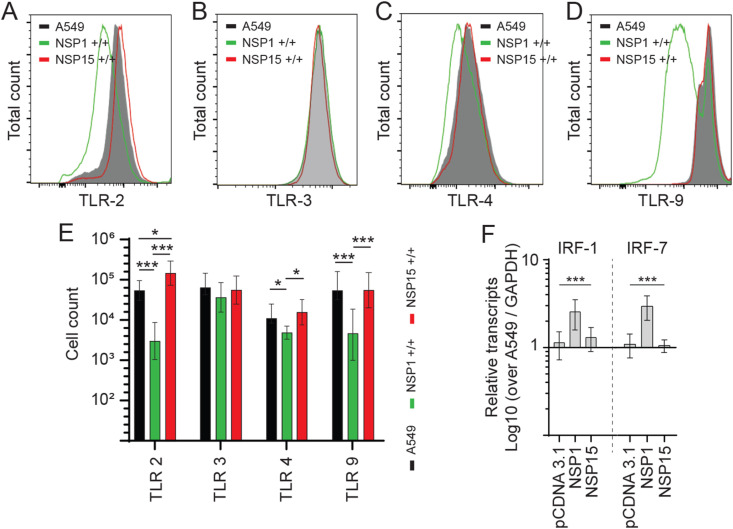
Innate sensing by TLRs and subsequent downstream signalling is essential for the host cell to mount an effective anti-viral immune response. To demonstrate how either NSP1 or NSP15 overexpression can sustain higher VSV-EGFP replication, cell lines were probed by flow cytometry for expression of TLR2, TLR3, TLR4 and TLR9 protein. Representatives of FACS histogram plots are shown demonstrating detection of TLR2 (Fig. 3 A), TLR3 (Fig. 3B), TLR4 (Fig. 3C) and TLR9 (Fig. 3D) in A549 cells. The total number of cells expressing TLR2, TLR3, TLR4 and TLR9 protein were analyzed. The results demonstrates that NSP1 overexpressing cells had altered expression levels of TLR2, TLR4 and TLR9, whereas NSP15 overexpressing cells had altered TLR2 expression based on relative cell number and fluorescence intensity (Fig. 3). Specifically, there was a significant decrease in the total number of NSP1 overexpressing A549 cells ability to express TLR2 (p ≤ 0.005) (Fig. 3E), TLR4 (p ≤ 0.05) (Fig. 3E) and TLR9 (p ≤ 0.005) (Fig. 3E) when compared to unmodified A549 cells or NSP15 overexpressing cells. There was no difference in the total number of cells expressing TLR2, TLR3, TLR4 and TLR9 protein when comparing unmodified A549 to NSP15 overexpressing cells. In contrast, there was significantly higher (p ≤ 0.005) number of TLR2 expressing cells in NSP15 overexpressing cells when compared to unmodified A549 cells or NSP1 overexpressing cells (Fig. 3E). However, there was no difference in TLR3 expression level (Fig. 3E) in all cell lines. Crosslinking of TLRs eventually leads to downstream phosphorylation of IRF and their respective nuclear translocation. At the mRNA level, NSP1 overexpressing cells had significantly (p ≤ 0.005) higher transcript levels of IRF1 and IRF7 when compared to A549 and NSP15 overexpressing cells (Fig. 3F). Similar transcript levels of IRF1 and IRF7 were observed between A549 and NSP15 overexpressing cells (Fig. 3F).
Fig. 3.
TLR expression in NSP1 and NSP15 stably expressing A549 cells. Representative overlapping FACS histogram demonstrating the expression of (A) TLR2 (B) TLR3 (C) TLR4 and (D) TLR9 gated on live (7AAD-) cells to analyze (E) the number of live cells expressing TLR2, TLR3, TLR4 and TLR9 in NSP1 and NSP15 stably expressing A549 cells lines compared to negative control (A549 cells). (F) Fold changes in gene (Interferon regulatory factor 1; IRF1 and IRF7) expression based on qRT-PCR in NSP1 and NSP15 stably expressing A549 cells lines over negative control (A549 cells). Non-parametric Wilcoxon tests (Mann-Whitney) was used to assess normal distribution and test significance with the results shown as mean ± SD. ∗ (p ≤ 0.05), and ∗∗∗ (p ≤ 0.005) indicates a statistically significant difference. All experiments were performed in 3 technical replicates, and the data are representative of results from minimum of 3 independent experiments.
3.3. Stimulation of TLR3 and TLR9 is effective at overcoming NSP1 and NSP15 pro-viral effects
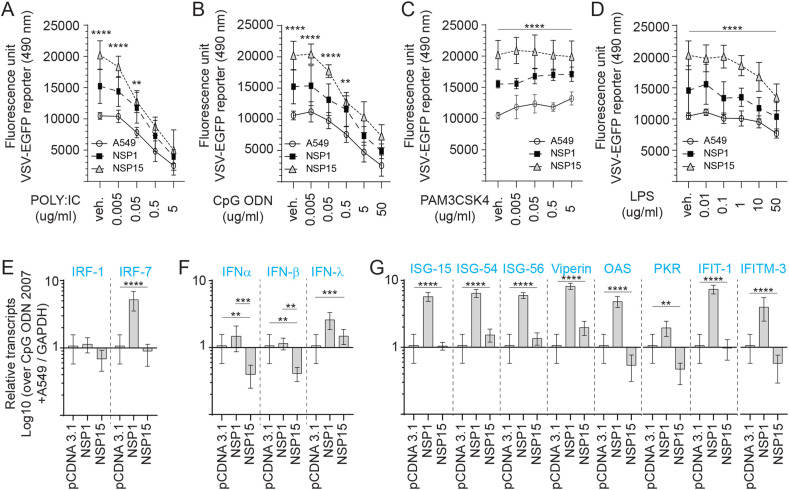
To explore whether either NSP1 or NSP15 overexpressing cells supported higher VSV-EGFP viral replication is linked to either limiting TLR sensing or subverting IFN expression, cell lines were pre-stimulated for 2 h with various concentration of specific TLR ligands; PAM3CSK4 (TLR2 ligand), polyI:C (TLR3 ligand), LPS (TLR4 ligand) and class B CpG ODN 2007 (TLR9 ligand). Post-stimulation, the cells were infected with a VSV-EGFP (MOI = 0.4) virus (Fig. 4 ). All experiments were performed with a minimum of six replicates and repeated three times. Data were analyzed by non-parametric Wilcoxon tests (Mann-Whitney). Both NSP1 and NSP15 overexpressing cells could limit polyI:C (0.005 and 0.05 μg/ml), CpG ODN 2007 (0.005, 0.05 and 0.5 μg/ml) and LPS (0.01–10 μg/ml) antiviral activity at low doses but not at the highest dose. The results demonstrate a dose dependent response that effectively limited VSV-EGFP virus replication more so in polyI:C, and CpG ODN 2007, to a lesser extent in LPS (anti-viral) but not in PAM3CSK4 pre-stimulated cell lines. The highest concentration of polyI:C (5 μg/ml; Fig. 4A) and CpG ODN 2007 (50 μg/ml; Fig. 4B) significantly limited (p ≤ 0.001) VSV-EGFP virus replication, 75.2% and 64.7% decrease respectively, in NSP1 overexpressing cells when compared to vehicle pretreated cells. Similarly, the highest concentration of polyI:C (5 μg/ml; Fig. 4A) and CpG ODN 2007 (50 μg/ml; Fig. 4B) significantly limited (p ≤ 0.001) VSV-EGFP virus replication, 69.5% and 47.7% decrease respectively, in NSP15 overexpressing cells when compared to vehicle pretreated cells. In contrast, pre-treatment with the highest concentration of PAM3CSK4 (5 μg/ml) lead to a significant increase (p ≤ 0.05) in VSV-EGFP replication (92.1% increase) when compared to vehicle treated in both unmodified and NSP1 overexpressing cells (Fig. 4C). The combination of NSP15 overexpression and stimulation with PAM3CSK4 (5 μg/ml) had no effect on VSV-EGFP virus replication indicating a maximum rate of replication had been achieved (Fig. 4A).
Fig. 4.
TLR ligand sensitivity of NSP1 and NSP15 stably expressing A549 cells. Replication efficiency of the interferon sensitive VSV-EGFP virus in A549 cells. Analysis of VSV-EGFP (moi = 0.4) virus replication in NSP1 and NSP15 stably expressing A549 cells lines compared to control transfected (pCDNA3.1/V5–HIS-TOPO) or negative (A549 cells) based on fluorescence (EGFP; excitation 490 nm). Cell lines were pre-stimulated (2 h) with (A) TLR3 ligand POLY; IC (0.005, 0.05, 0.5 and 5 μg/mL), (B) TLR9 ligand CpG ODN 2007 (0.005, 0.05, 0.5 and 5 μg/mL), (C) TLR2 ligand PAM3CSK4 (0.005, 0.05, 0.5 and 5 μg/mL), and (D) TLR4 ligand LPS (0.01, 0.1, 1.0, 10 and 50 μg/mL) prior to infection with VSV-EGFP. Fold changes in (E) IRF1 and IRF7, (F) PAN-IFN-α, IFN-β and IFN-γ, and (G) ISG-15, ISG-54, ISG-56, Viperin, OAS, PKR, IFIT1 and IFITM-3 gene expression based on qRT-PCR in NSP1 and NSP15 stably expressing A549 cells lines over negative control (A549 cells) stimulated for 2 h with 5 μg/mL CpG ODN 2007. The mean fluorescence unit in uninfected cells was used as the normalization factor for VSV-EGFP readout in infected A549 cells.Non-parametric Wilcoxon tests (Mann-Whitney) and one-way ANOVA was used to assess normal distribution and test significance with the results shown as mean ± SD. ∗∗ (p ≤ 0.01), ∗∗∗ (p ≤ 0.005) and ∗∗∗∗ (p ≤ 0.001) indicates a statistically significant difference. All viral infection experiments were performed in 10 technical replicates, and the data are representative of results from 3 independent experiments.
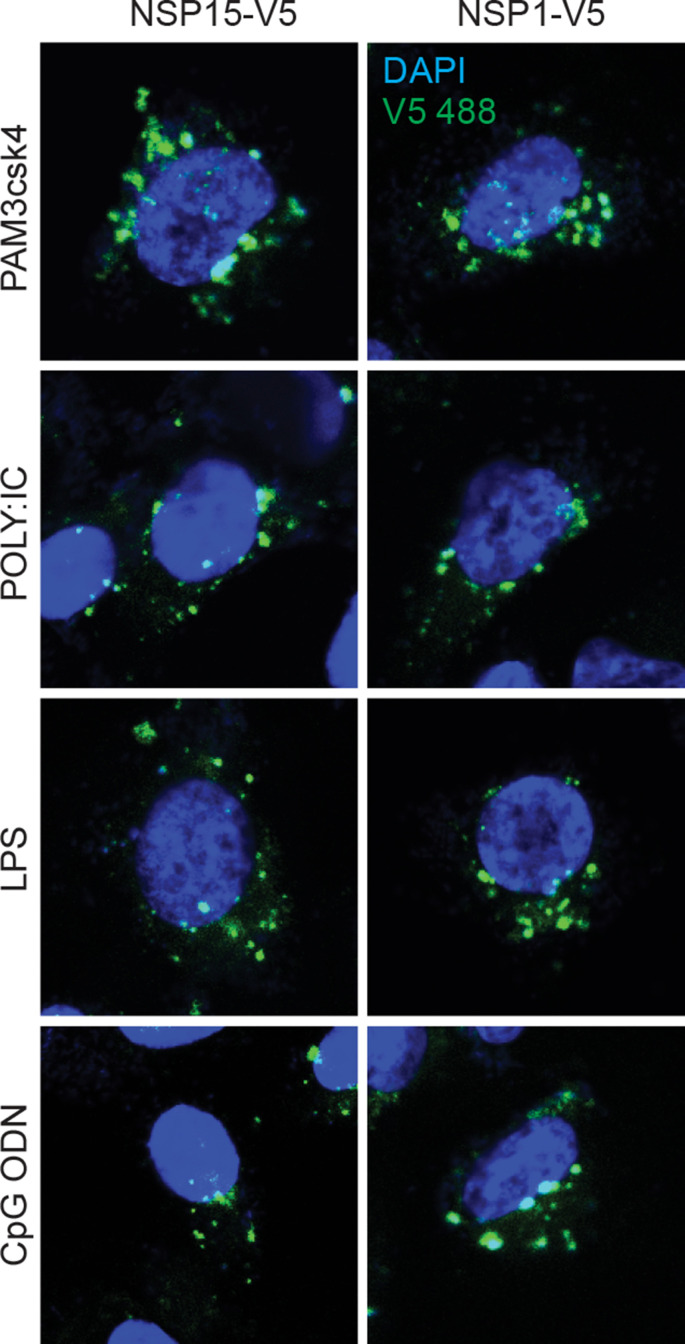
Subcellular protein localization critically influences their respective function, and cells control protein localization to regulate biological processes. Retention of proteins in specific subcellular compartments could indicate functionality and resistance to host mechanisms. To demonstrate if these TLR ligand stimulation affected NSP1 or NSP15 subcellular localization, cells were probed for confocal microscopy. Post stimulation with PAM3CSK4 (5 μg/ml), polyI:C (5 μg/ml), LPS (50 μg/ml) or CpG ODN 2007 (50 μg/ml), no changes in both NSP1 and NSP15 subcellular localization could be identified, based on punctate numbers in the cytoplasm to nucleus (Sup Fig. 1).
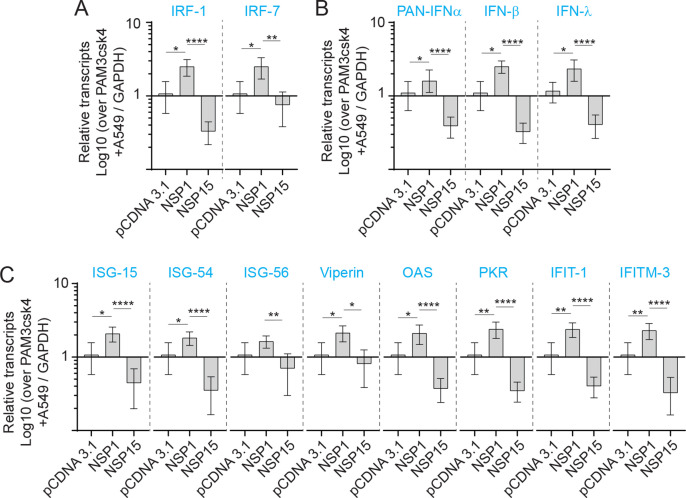
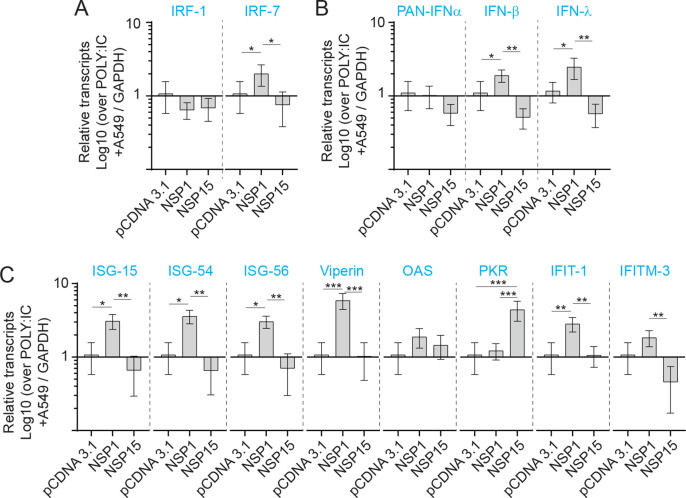
To demonstrate an association between TLR ligand stimulation and VSV-EGFP viral replication, qRT-PCR was performed post stimulation with either PAM3CSK4 (5 μg/ml) (Sup Fig. 2), LPS (50 μg/ml), polyI:C (5 μg/ml) (Sup Fig. 3) or CpG ODN 2007 (50 μg/ml) to demonstrate induction of ISGs (ISG15, ISG54, ISG56, viperin, OAS, PKR, IFIT1 and IFITM3) mRNA transcripts. An inverse relationship was observed between VSV-EGPF virus replication and detection of ISGs mRNA transcripts. Differential ISGs mRNA transcripts expression profiles were detected between NSP1 and NSP15 overexpressing cells lines in response to TLR ligand stimulation (Fig. 4E–G). Although stimulation with PAM3CSK4 did not affect VSV-EGFP viral replication, at the mRNA level, the results demonstrate higher transcript levels of IRF1 and 7 (sup Fig. 2A), IFN-α, IFN-β, IFN-λ (Sup Fig. 2B) and ISG15, ISG54, ISG56, viperin, 2′,5′-oligoadenylate synthetase (OAS), protein kinase R (PKR), Interferon Induced Protein with Tetratricopeptide Repeats 1 (IFIT1) and Interferon Induced Transmembrane Protein 3 (IFITM3) (Sup Fig. 2C) in NSP1 overexpressing cells. In contrast, NSP15 overexpressing cells had significantly lower transcript levels of IRF1 and 7 (sup Fig. 2A), IFN-α, IFN-β, IFN-λ (Sup Fig. 2B) and ISG15, ISG54, ISG56, viperin, OAS, PKR, IFIT1 and IFITM3 (Sup Fig. 2C). In CpG ODN 2007 stimulated cells, significantly (p ≤ 0.0001) higher IRF-7 but not IRF-1 transcripts levels were detected in NSP1 overexpressing cells when compared to A549 cells or NSP15 overexpressing cells (Fig. 4E). Higher IFN-α transcript levels were detected in NSP1 overexpressing cell in response to CpG ODN 2007 (Fig. 4F). In contrast, both IFN-α and IFN-β mRNA transcript were significantly (p ≤ 0.005) lower in NSP15 overexpressing cells compared to unmodified A549 or NSP1 overexpressing cell lines (Fig. 4F). IFN-λ transcript levels were significantly higher in both NSP1 and NSP15 overexpressing cells when compared to unmodified A549 cells (Fig. 4F).
In response to CpG ODN 2007, NSP1 overexpressing cells had significantly higher (p ≤ 0.0001) transcripts of ISG15, ISG54, ISG56, viperin, OAS, PKR, IFIT1 and IFITM3 (Fig. 4G). No difference in ISG-15 and IFIT1 transcripts was detected in NSP15 overexpressing cells (Fig. 4G). In response to CpG ODN 2007 stimulation, higher (p ≤ 0.0001) ISG54, ISG56, and viperin transcripts levels were detected in NSP15 overexpressing cells (Fig. 4G). However, significantly (p ≤ 0.05) lower transcripts levels of PKR and IFITM3 were detected in CpG ODN 2007 stimulated NSP15 overexpressing cells when compared to A549 cells (Fig. 4G).
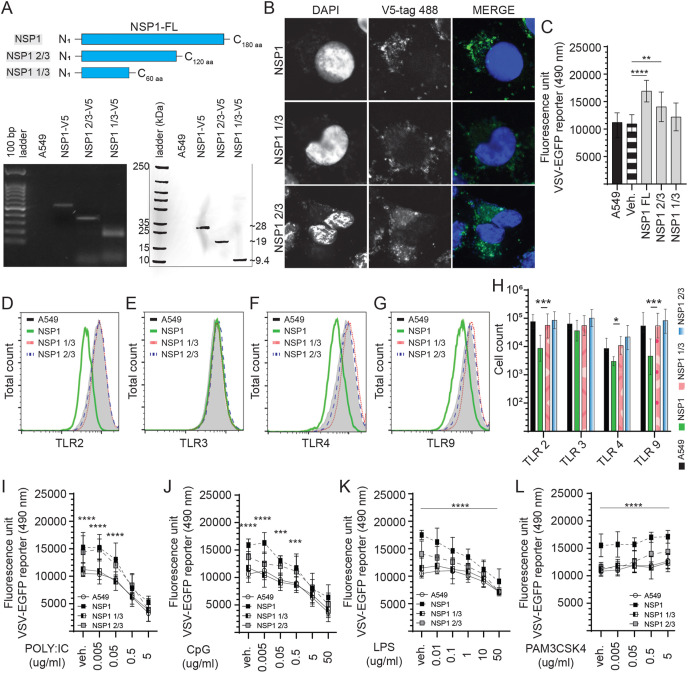
3.4. NSP1 truncation is associated with decreased VSV-EGFP replication
To define a potential functional domain, the NSP1 coding sequence was truncated by high fidelity PCR, subcloned into a pCDNA3.1/V5–HIS-TOPO plasmid (Fig. 5 ) and transfected into A549 cells. The recombinant NSP1 2/3 length (N1–C120 aa) was generated to produce a truncation lacking the C-terminal domain. In addition, the recombinant NSP1 1/3 length (N1–C60 aa) lacked both the C-terminal domain and linker sequence thereby leaving only the N-terminal domain (Fig. 5A). Such targeted domain deletion would improve the stability of the expressed truncated protein. Stably expressing cells lines were confirmed to overexpress either NSP1 full length (NSP1-FL), NSP1 2/3 or NSP1 1/3 at both RNA and protein levels (Fig. 5A). Confocal microscopy of stably expressing cell lines confirmed cytoplasmic localization of NSP1-FL, NSP1 2/3 and NSP1 1/3 with a more diffuse expression pattern observed in NSP1 2/3 and NSP1 1/3 overexpressing cells (Fig. 5B).
Fig. 5.
Identification of the NSP1 protein functional domain. Truncation of NSP1 limits VSV-EGFP virus replication. (A) Schematic diagram depicting the full length of NSP1 and truncated NSP1 1/3 and NSP1 2/3 in amino acid. The SARS-CoV-2 NSP1 was truncated by high fidelity PCR (GoTaq) and subcloned into a pCDNA3.1/V5–HIS-TOPO plasmid (HindIII and EcoRV insertion sites) which was transfected into A549 cells to generate stably expressing cell lines. Expression were confirmed by total RNA extraction (polymerase chain reaction; PCR) and total protein extraction (Western blot). (B) Representative pictures of confocal microscopy imaging with maximum projections of z-stacks for each channel demonstrating the subcellular localization of expressed full length of NSP1, NSP1 1/3 and NSP1 2/3 (anti-V5 antibody; 488 nm) in stably expressing A549 cells. Analysis of VSV-EGFP (moi = 0.4) virus replication in (C) NSP1, NSP1 1/3 and NSP1 2/3 stably expressing A549 cells lines compared to control transfected (pCDNA3.1/V5–HIS-TOPO) or negative (A549 cells) based on fluorescence (EGFP; excitation 490 nm). Representative overlapping FACS histogram demonstrating the expression of (D) TLR2 (E) TLR3, (F) TLR4 and (G) TLR9 as gated on live (7AAD-) NSP1, NSP1 1/3 and NSP1 2/3 stably expressing A549 cells to analyze (H) the number of cells expressing TLR2, TLR3, TLR4 and TLR9 compared to negative control (A549 cells). Analysis of VSV-EGFP (moi = 0.4) virus replication in NSP1, NSP1 1/3 and NSP1 2/3 stably expressing A549 cells lines compared to control transfected (pCDNA3.1/V5–HIS-TOPO) or negative (A549 cells) based on fluorescence (EGFP; excitation 490 nm) pre-stimulated (2 h) with (I) TLR3 ligand POLY; IC (0.005, 0.05, 0.5 and 5 μg/mL), (J) TLR9 ligand CpG ODN 2007 (0.005, 0.05, 0.5 and 5 μg/mL), (K) TLR4 ligand LPS (0.01, 0.1, 1.0, 10 and 50 μg/mL) and (L) TLR2 ligand PAM3CSK4 (0.005, 0.05, 0.5 and 5 μg/mL) and prior to infection with VSV-EGFP. The mean fluorescence unit in uninfected cells was used as the normalization factor for VSV-EGFP readout in infected A549 cells.Non-parametric Wilcoxon tests (Mann-Whitney) and one-way ANOVA was used to assess normal distribution and test significance with the results shown as mean ± SD. ∗ (p ≤ 0.05), ∗∗ (p ≤ 0.01), ∗∗∗ (p ≤ 0.005) and ∗∗∗∗ (p ≤ 0.001) indicates a statistically significant difference. All viral infection experiments were performed in 10 technical replicates, and the data are representative of results from 3 independent experiments.
NSP1-FL, NSP1 2/3 and NSP1 1/3 stably expressing cell lines were subsequently infected with VSV-EGFP (MOI = 0.4). All experiments were performed with a minimum of six replicates and repeated three times. Data were analyzed by non-parametric Wilcoxon tests (Mann-Whitney). The results demonstrate an association between truncation of NSP1 and VSV-EGFP virus replication in A549 overexpressing cells lines. Truncation of NSP1 to 1/3 length resulted in a 29% decrease (p ≤ 0.0001) (Fig. 5C) in VSV-EGFP virus replication when compared to NSP1-FL overexpressing cells. Truncated NSP1 1/3 expressing cells did not show an increase in VSV-EGPF virus replication unlike what was observed in NSP1-FL expressing cells compared to unmodified cells (Fig. 5C). Cells expressing truncated NSP1 2/3 had a 17.64% decrease (p ≤ 0.01) in VSV-EGFP virus replication when compared to NSP1-FL overexpressing cells (Fig. 5C). Cells expressing truncated NSP1-2/3 could support higher (p ≤ 0.01) VSV-EGFP virus replication compared to unmodified A549 cells (Fig. 5C).
To further establish a relationship between VSV-EGFP virus replication, and NSP1 expression, A549 cells were probed to define expression of various TLRs. Representatives of FACS histogram plots are shown demonstrating detection of TLR2 (Fig. 5D), TLR3 (Fig. 5E), TLR4 (Fig. 5F) and TLR9 (Fig. 5G) in A549 cells. Truncation of NSP1 affected the respective TLR expression levels in the A549 stably expressing cell lines (Fig. 5D–H). The results demonstrate, based on relative cell number and fluorescence intensity, following truncation of NSP1 by 1/3 and 2/3 length, an increase in total number of cells expressing TLR2 (p ≤ 0.001) (Fig. 5H), TLR4 (p ≤ 0.05) (Fig. 5H) and TLR9 (p ≤ 0.001) (Fig. 5H) protein with no differences observed when compared to unmodified A549 cells.
To prove that truncation of NSP1 can improve responsiveness to TLR ligand stimulation, cell lines were pre-stimulated for 2 h with various concentration of specific TLR ligands; PAM3CSK4 (TLR2 ligand), polyI:C (TLR3 ligand), LPS (TLR4 ligand) and class B CpG ODN 2007 (TLR9 ligand). Post-stimulation, cells were infected with a VSV-EGFP (MOI = 0.4) virus (Fig. 5I-L). Data were analyzed by one-way ANOVA followed by a Tukey multiple-comparison test or Mann-Whitney test. The results demonstrated a dose dependent response that effectively limited VSV-EGFP virus replication more so in polyI:C, CpG ODN 2007 and LPS (anti-viral) but not PAM3CSK4 pre-stimulated cell lines. Truncation of NSP1 to 1/3 length had no effect on negating antiviral signaling as mediated by TLR ligand stimulation at any of the tested concentrations with no differences observed in all groups when compared to A549 cells (Fig. 5I-L). The dose response curve mirrored the unmodified A549 cells. However, the specific activity of NSP1 C-terminal domain truncated mutant (NSP1 2/3 length) resulted in a decrease in VSV-EGFP virus replication when compared to that of full length NSP1 in response to CpG ODN 2007 (p ≤ 0.0001) (Fig. 5J), LPS (p ≤ 0.0001) (Fig. 5K) and PAM3CSK4 (p ≤ 0.0001) (Fig. 5J) stimulation at all concentrations. Truncated NSP1-2/3 overexpressing cells had higher VSV-EGFP virus replication compared to unmodified cells only at lower concentration of polyI:C (0.005, 0.05 μg/ml) (Fig. 5I) and CpG ODN 2007 (0.05 μg/ml) (Fig. 5J). In contrast, truncated NSP1 2/3 overexpressing cells had (p ≤ 0.0001) VSV-EGFP viral replication only at the highest concentration of PAM3CSK4 (0.5 and 5 μg/ml) when compared to unmodified or NSP1 1/3 overexpressing cells (Fig. 5L). PAM3CSK4 treatment is more likely to increase total virus yield than polyI:C, LPS or class B CpG ODN 2007.
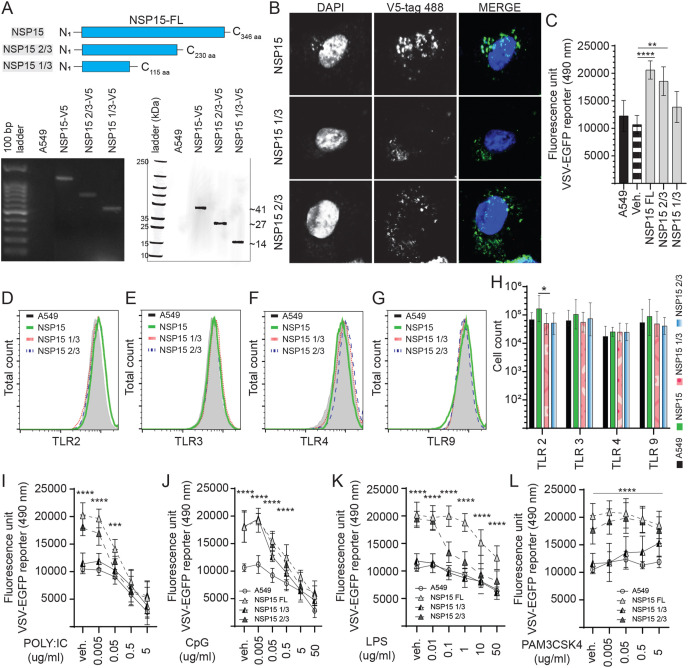
3.5. NSP15 truncation is associated with decreased VSV replication
To define a potential interacting and functional domain within NSP15, high fidelity PCR was employed to generate various truncated oligos. The truncated NSP15 oligos were subcloned into a pCDNA3.1/V5–HIS-TOPO plasmids (Fig. 6 ), transfected into A549 cells to generate stably expressing cells lines which were confirmed to express either NSP15 full length (NSP15-FL), NSP15 2/3 or NSP15 1/3 (Fig. 6A) at both an RNA level (PCR) and protein level. Truncation of NSP15 was based on interfering with the Nendo-U catalytic domain (C-terminal domain) while retaining the oligomerization domain (N-terminal). Truncation of NSP15 to 2/3 length (N1–C230 aa) resulted in the deletion of the catalytic groove defined by His235, His250, Lys290, Thr341, Tyr343, and Ser294. In addition, truncation of NSP15 to 1/3 length (N1–C115 aa) supported oligomerization (N-terminal domain) of the truncated protein without a catalytic and middle domain (Fig. 6A). Such targeted domain deletion was used to decrease the enzymatic activity of NSP15. Confocal microscopy of stably expressing cell lines demonstrated the cytoplasmic localization of NSP15-FL, NSP15 2/3 and NSP15 1/3 whereby a more diffuse expression pattern for the NSP15 2/3 and NSP15 1/3 was observed (Fig. 6B).
Fig. 6.
Identification of the NSP15 protein functional domain. Truncation of NSP15 limits VSV-EGFP virus replication. (A) Schematic diagram depicting the full length of NSP15 and truncated NSP15 1/3 and NSP15 2/3 in amino acid. The SARS-CoV-2 NSP15 was truncated by high fidelity PCR (GoTaq) and subcloned into a pCDNA3.1/V5–HIS-TOPO plasmid (HindIII and EcoRV insertion sites) which was transfected into A549 cells to generate stably expressing cell lines. Expression were confirmed by total RNA extraction (polymerase chain reaction; PCR) and total protein extraction (Western blot). (B) Representative pictures of confocal microscopy imaging with maximum projections of z-stacks for each channel demonstrating the subcellular localization of expressed full length of NSP15, NSP15 1/3 and NSP15 2/3 (anti-V5 antibody; 488 nm) in stably expressing A549 cells. Analysis of VSV-EGFP (moi = 0.4) virus replication in (C) NSP15, NSP15 1/3 and NSP15 2/3 stably expressing A549 cells lines compared to control transfected (pCDNA3.1/V5–HIS-TOPO) or negative (A549 cells) based on fluorescence (EGFP; excitation 490 nm). Representative overlapping FACS histogram demonstrating the expression of (D) TLR2 (E) TLR3, (F) TLR4 and (G) TLR9 as gated on live (7AAD-) NSP15, NSP15 1/3 and NSP15 2/3 stably expressing A549 cells lines cells to analyze (H) the number of cells expressing TLR2, TLR3, TLR4 and TLR9 compared to negative control (A549 cells). Analysis of VSV-EGFP (moi = 0.4) virus replication in NSP15, NSP15 1/3 and NSP15 2/3 stably expressing A549 cells lines compared to control transfected (pCDNA3.1/V5–HIS-TOPO) or negative (A549 cells) based on fluorescence (EGFP; excitation 490 nm) pre-stimulated (2 h) with (I) TLR3 ligand polyI:C (0.005, 0.05, 0.5 and 5 μg/mL), (J) TLR9 ligand CpG ODN 2007 (0.005, 0.05, 0.5 and 5 μg/mL), (K) TLR4 ligand LPS (0.01, 0.1, 1.0, 10 and 50 μg/mL) and (L) TLR2 ligand PAM3CSK4 (0.005, 0.05, 0.5 and 5 μg/mL) and prior to infection with VSV-EGFP. The mean fluorescence unit in uninfected cells was used as the normalization factor for VSV-EGFP readout in infected A549 cells. Non-parametric Wilcoxon tests (Mann-Whitney) or one-way ANOVA was used to assess normal distribution and test significance with the results shown as mean ± SD. ∗ (p ≤ 0.05), ∗∗ (p ≤ 0.01), ∗∗∗ (p ≤ 0.005) and ∗∗∗∗ (p ≤ 0.001) indicates a statistically significant difference. All viral infection experiments were performed in 10 technical replicates, and the data are representative of results from 3 independent experiments.
NSP15-FL, NSP15-2/3 and NSP51-1/3 overexpressing A549 cell lines were subsequently infected with VSV-EGFP (MOI = 0.4). The results demonstrated a clear association between truncation of NSP15 and VSV-EGFP virus replication. Truncated NSP15 1/3 expressing cells did not show an increase in VSV-EGPF virus replication unlike the observation in NSP1-FL expressing cells compared to unmodified cells (Fig. 6C). No difference was observed in VSV-EGFP virus between NSP15-FL and NSP15 2/3 overexpressing cells lines (Fig. 6C). Truncated NSP15-1/3 expressing cells had a 41.74% decrease (p ≤ 0.0001) (Fig. 6C) in VSV-EGFP virus replication when compared to NSP15-FL overexpressing cells. Whereas truncated NSP15-2/3 expressing cells had a 9.34% decrease (p = NS) in VSV-EGFP virus replication when compared to NSP15-FL overexpressing cells (Fig. 6C). Truncation of NSP15-FL to -2/3 length still resulted in significantly higher (p ≤ 0.01) VSV-EGFP virus replication compared to unmodified AS549 cells (Fig. 6C).
To further demonstrate an associative mechanism, cells were probed to determine expression level of various TLRs. Representatives of FACS histogram plots are shown demonstrating detection of TLR2 (Fig. 6D), TLR3 (Fig. 6E), TLR4 (Fig. 6F) and TLR9 (Fig. 6G) in A549 cells. Truncation of NSP15 affected the respective TLR expression (Fig. 6D–H). The results demonstrated, based on relative cell number and fluorescence intensity, that truncation of NSP15-FL to 1/3 and 2/3 length resulted in a decrease (p ≤ 0.05) in the total number of cells expressing TLR2 protein (Fig. 6H). No difference was observed in TLR2 expression between NSP15-1/3 and −2/3 length and unmodified A549 cells. No changes were observed in TLR3 (Fig. 6H), TLR4 (Fig. 6H) and TLR9 (Fig. 6H) expression when compared to unmodified A549 cells.
To prove that truncation of NSP15 can limit VSV-EGFP viral replication and improve sensitivity to TLR ligand stimulation, cell lines were pre-stimulated for 2 h with various concentration of specific TLR ligands; PAM3CSK4 (TLR2 ligand), polyI:C (TLR3 ligand), LPS (TLR4 ligand) and class B CpG ODN 2007 (TLR9 ligand). Post-stimulation, cells were infected with a VSV-EGFP (MOI = 0.4) virus (Fig. 6H–K). Data were analyzed by one-way ANOVA followed by a Tukey multiple-comparison test or Mann-Whitney test. The results demonstrate an association between length of NSP15 and a dose dependent response to TLR ligands that effectively limited VSV-EGFP virus replication in polyI:C, CpG ODN 2007 and LPS (anti-viral) but not PAM3CSK4 pre-stimulated cell lines. Cell overexpressing truncated NSP15-1/3 did not cause an increase in VSV-EGFP replication as did not increase cellular resistance to TLR ligand stimulation, except for CpG ODN 2007 (0.005 and 0.05 μg/ml) (Fig. 6J) and PAM3CSK4 (0.5 and 5 μg/ml) (Fig. 6L), at the tested concentrations with no differences observed in all groups when compared to A549 cells. Cells overexpressing truncated NSP15-2/3 had higher VSV-EGFP virus replication compared to NSP15-1/3 and unmodified cells. Specifically, there was an improved cellular resistance to polyI:C (0.005 and 0.05 μg/ml) (p ≤ 0.0001) (Fig. 6I), CpG ODN 2007 (0.005, 0.05 and 0.5 μg/ml) (p ≤ 0.0001) (Fig. 6J) and LPS (0.01 and 0.1 μg/ml) (p ≤ 0.0001) (Fig. 6K) stimulation based on the increase in VSV-EGFP virus compared to NSP15-1/3 and unmodified cells. A similar dose dependent curve in response to polyI:C, CpG ODN 2007, and LPS stimulation of truncated NSP15-2/3 to that of NSP15-FL overexpressing cell lines was observed compared to NSP15-1/3 and unmodified cells. In contrast, stimulation with PAM3CSK4 exceeding 0.005 μg/ml (0.05, 0.5 and 5 μg/ml) in NSP15 2/3 overexpressing cells could support higher (p ≤ 0.0001) VSV-EGFP viral replication compared to unmodified cells (Fig. 6L). No significant difference was observed in VSV-EGFP virus replication in cells overexpressing truncated NSP1-2/3 that were treated with PAM3CSK4 (0.005 μg/ml) and vehicle control cells (Fig. 6L).
4. Discussion
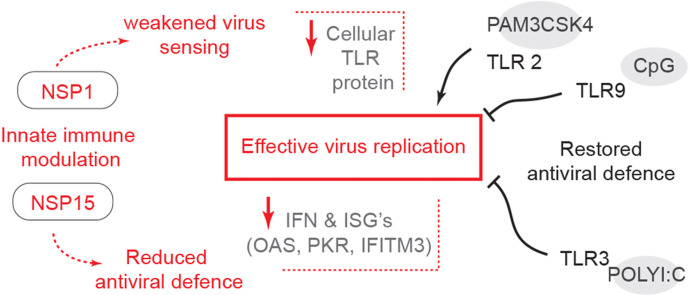
RNA viruses that replicate via dsRNA intermediates can be detected as “nonself” by host PRR. These PRR include several cytoplasmic receptors like TLRs and RIG-I. Importantly, sensors such as TLRs in the lungs are expressed by immune system cells and airway epithelial cells, where they mediate activation of these cells, including antigen-presenting cells (APCs) (Iwasaki et al., 2017). Activation of TLRs among other PRR, stimulates the production of type I IFN, which in turn up-regulates additional dsRNA sensors thereby inducing the expression of ISGs and a subsequent antiviral state. Many elements of the regulatory pathways governing both the signaling pathway and IFN gene expression are themselves regulated by IFN, providing an intricate network of overlapping feed-forward and feedback regulatory loops. To counteract these systems, many viruses have evolved strategies to evade sensing and subsequent early activation of dsRNA sensors. The coronavirus polycistronic genome encodes NSPs which have been reported to antagonize IFN responses by either supressing transcription or translation but are highly sensitive to direct treatment with recombinant IFNs indicating a threshold mechanism to overcome SARS-CoV-2 IFN antagonistic effects. Both NSP1 and NSP15 were chosen for their broad effects on mRNA sensing, translocation as well as their ability to negate viral sensors or interferon activity to boost viral replication (Deng et al., 2017; Huang et al., 2011; Sampaio et al., 2021; Yuen et al., 2020). In addition, SARS-CoV-2 can modulate the host cell activity by engaging TLR4 though its spike glycoprotein which leads to an aberrant immune response. While SARS-CoV-2 selectively engages TLR4, its innate antagonistic activity is broad (Zhao et al., 2021). Identifying potential mechanisms to interfere with viral antagonistic activity and subsequently increase host innate responses can result in enhanced immunity (Chan et al., 2020; Hoagland et al., 2021; Sampaio et al., 2021). The work presented here identifies that this IFN antagonising mechanism could in part be related to NSPs modulation of TLR (TLR2, TLR3, TLR4 and TLR9) expression, the respective cell sensitivity to TLR ligands stimulation and subsequent induction of IFN and ISGs. Crosslinking TLR3, followed by TLR9 and TLR4 with their respective ligands may be potential targets to bypass the antagonistic activity of either NSP1 or NSP15. Treatment with polyI:C was highly effective at eliciting an antiviral response. PolyI:C and CpG ODN, but not LPS, may disrupt the subcellular interactions of NSP1 or NSP15, which enable an effective antiviral response. The fold increase in IFN and ISGs transcripts in NSP1 overexpressing cells did not result in a decrease in VSV-EGFP virus replication. Furthermore, in NSP15 overexpressing cells, there was a decrease in IFN and ISGs such as OAS, PKR and IFITM-3 mRNA transcripts which resulted in an increase in VSV-EGFP virus replication. The functional roles played by NSP1 and NSP15 can effectively promote virus replication and interfere with IFN protein production and their subsequent signalling.
Shutdown of host protein synthesis is a common feature of viral infection. The first protein encoded in the SARS-CoV-2 polycistronic ORF1a, NSP1, has been shown to directly target the human ribosome subunit to inhibit protein synthesis (Thoms et al., 2020). NSP1 is a virulence determinant that plays a critical role during the SARS-CoV-2 virus life cycle. Structural analysis has demonstrated that NSP1 directly forms a complex with the ribosomal 40S and 80S subunits. This complex obstructs mRNA entry and subsequent translation resulting in shutdown of host messenger RNA (mRNA) translation (Thoms et al., 2020). Consistent with this conclusion, the present study demonstrates that NSP1 overexpressing cells had lower TLR2, TLR4 and TLR9 protein levels when compared to control A549 and NSP15 overexpressing cells. In addition, NSP1 can be detected intracytoplasmic and intranuclear. We speculate that NSP1 circulates intracellularly via lysosomal vesicles thereby linking the endoplasmic reticulum and the trans-Golgi network, two sites important for mRNA translation and protein trafficking, respectively. In contrast, it has been suggested that the SARS-CoV-2 NSP1 can block mRNA export function, from the nucleus to the cytoplasm, by interfering with mRNA export adaptor complex formation (Zhang et al., 2021). Based on confocal microscopy results showing intranuclear presence of NSP1 punctate, even in TLR stimulated cells, it cannot be ruled out that NSP1 actively shuttles between the nucleus and cytoplasm. As has been suggested (Thoms et al., 2020), during this transient localization, NSP1 can interact and or interfere with the host mRNA export or translation altogether independent of the ribosomal blocking effects. NSP1 can form a complex with viral RNA to regulate kinetic of viral replication while limiting host specific mRNA translation, which likely includes transcription of the innate immune system sensors (Tanaka et al., 2012). TLRs are normally localised in the lysosomal vesicle, LAMP-1 positive vesicles, and act as a critical primary line of sensing against invading pathogens (Sanjuan et al., 2009). In fact, our result demonstrates that NSP1 overexpressing cells could support higher (67.24% increase, less than 1 log increase) virus replication indicating an improper antiviral response. NSP1 lacks the ability to bind directly to viral RNA (Tidu et al., 2021). Under unstimulated conditions, the disrupted IRF, IFN and ISG gene-mediated antiviral pathways in NSP1 overexpressing A549 cells resulted in higher VSV-EGFP virus replication. The increase in intracellular IRFs, IFN and ISG mRNA transcripts does not translate into an antiviral state in A549 cells, based on the observation of higher VSV viral replication. Rather, the increase in mRNA transcripts may suggest intracellular accumulation due to shutoff of host translation machinery. These observations provide an insight into the SARS-CoV-2 NSP1 distinct immunoregulatory domain that potentially interacts with the ribosomal 40S and 80S subunits.
In the present study, NSP1 was truncated to 1/3 and 2/3 length by PCR and overexpressed in A549 cells to demonstrate the location of its functional domain. NSP1 truncation to 2/3 length preserved the linker sequence to the N-terminal region while truncation to 1/3 length eliminated the N-terminal linker sequence. The results obtained with truncated NSP1 2/3 in polyI:C and CpG ODN stimulated cells, at the lowest concentrations, question the minimum docking required to partially impair the ribosomal system and point to a potential function of the N-terminal domain and linker sequence independent of other SARS-CoV-2 viral proteins. Using this approach, we predicted that the distinct minimum translation inhibition domain of the SARS-CoV-2 NSP1 is likely located between amino acid (aa) 60–120. Based on our findings and recent structural studies this specific interaction, NSP1 C-terminal domain and ribosome mRNA channel, is not of high affinity (Banerjee et al., 2020). This is because, unlike SARS-CoV, the SARS-CoV-2 virus is highly susceptible to an IFN induced antiviral state (Lokugamage et al., 2020). In fact, stimulation with either the TLR3 or TLR9 ligands, at the highest concentrations, demonstrates that this translation interference mechanism can be disrupted in a similar manner as IFN stimulated cells. In CpG ODN 2007 and polyI:C stimulated cells, the results indicate that NSP1 overexpressing cells had up to 10-fold higher mRNA levels compared to control A549 cells and NSP15 overexpressing cells. TLR4 and TLR2 ligands were least effective as antiviral treatments. Truncation of NSP1 to 1/3 length but not 2/3 length led to abrogation of an antiviral status and a decrease in VSV-EGFP virus replication. The results presented here cannot differentiate between either ribosomal activity or potentially the formation of a complex with viral RNA to impair TLR sensing. Truncation of NSP1 to 2/3 length, resulted in an intermediate inhibitory effect whereby VSV-EGFP replication was significantly higher when compared to control cells or NSP1 1/3 overexpressing cells. Whereas truncation to 1/3 length led to a complete loss of TLR-mediated IFN production blocking and a decrease in VSV-EGFP production when compared to cells expressing the full length NSP1 or 2/3 length. Recently, several NSP1 deletion mutants have been documented (Benedetti et al., 2020; Islam et al., 2020). These deletions fall outside the predicted minimum functional domain suggesting that the NSP1 aa 60–120 region contains conserved motifs required for productive functional interactions and blocking translation processes. Such functional interactions are not limited to docking on the ribosomal subunit or viral RNA. The N-terminal domain, as per findings with NSP1 1/3, is thought to be dispensable for this innate immune interaction but binds directly to viral 5’-UTR region leading to an increase in viral mRNA stability and efficiency in translation. In this study, truncation of NSP1 to 1/3 length did not lead to an increase in VSV-EGFP replication nor affected host TLR protein expression in A549 cells (Benedetti et al., 2020; Islam et al., 2020). Taken together, these results suggest that wildtype NSP1 antagonistic activity can be overcome in cells treated with TLR3 and TLR9 ligands, and to a lesser extent, with a TLR4 ligand. Additionally, molecular insights into the NSP1-mediated translation inhibition could prove valuable for the design and selection of potential small-molecule inhibitors.
Aside from NSP1, most coronaviruses also encode a wide variety of NSPs that selectively impair other aspects of innate immunity such as IFN signalling (Deng and Baker, 2018). The SARS-CoV-2 NSP15, an orthologue of a similar protein encoded by SARS-CoV, is a conserved poly(U) specific endoribonuclease that form a hexameric structure and has been demonstrated to be an IFN antagonist (Yuen et al., 2020). Furthermore, NSP15 is likely a potent mediator of viral immune evasion by impairing dsRNA sensors that facilitate IFN expression and promote cellular apoptosis (Deng et al., 2017). While MDA5/MAVS can sense dsRNA, the SARS-CoV-2 NSP15 likely negates signalling which impairs an effective IFN response, as is well documented by a lack of IFN expression during viral infection (Hoagland et al., 2021; Sampaio et al., 2021). We established A549 cells stably expressing NSP15 to ascertain the extent of its IFN antagonism. We found that VSV-EGFP virus replication was significantly higher (102.26% or 2-fold increase) in NSP15 overexpressing cells when compared to both A549 and NSP1 overexpressing cells. These cells expressed similar levels of TLR3, TLR4 and TLR9 when compared to control A549 cells. However, higher TLR2 protein expression was observed in NSP15 overexpressing cells.. It is common for viruses to subvert and redirect innate immune responses enabling viral replication, dissemination, and shedding. This mechanism has been further explored in human immunodeficiency virus (HIV) 1 infection whereby induction of TLR2 and TLR4 enables a proinflammatory milieu and higher viral replication (Hernández et al., 2012). Therefore, the SARS-CoV-2 NSP15 could favor a TLR2 dependent virus replication mechanism, as demonstrated by PAM3CSK4 treatment which could increase VSV-EGFP virus replication. Gene expression analysis in TLR stimulated NSP15 overexpressing cells demonstrated a decrease in IFN-α and IFN-β transcript levels when compared to control A549 cells. Furthermore, ISGs such as OAS, PKR and IFITIM-3 were downregulated in CpG ODN 2007 stimulated cells. It has been demonstrated that the mouse hepatitis virus (MHV) and human coronavirus (HCoV)-229E NSP15 endoribonuclease activity is key to evade dsRNA sensing by OAS and PKR (Cao and Zhang, 2012; Kindler et al., 2017). Our results are in accordance with these reports demonstrating a mechanism exploited by the virus to limit induction of ISGs. NSP15 further mediates evasion by OAS and PKR detection in favor of proinflammatory conditions.
LAMP1 can be used to track vesicle movement and presence of NSP15 in LAMP-1 positive vesicle indicates a functional relationship between limiting cellular stress and viral replication. In chickens, the infectious bronchitis virus (IBV) NSP15 acts as both stress and IFN responses antagonist, thereby efficiently evading innate anti-viral and host stress responses (Gao et al., 2021). The SARS-CoV-2 NSP15 could well have multiple intracellular targets (Gordon et al., 2020). Further, we demonstrated here that the minimum NSP15 functional domain is located between aa230-346. These results are in accordance with NSP15 crystal structural analysis demonstrating a multimeric domain, and a functional interaction domain (Pillon et al., 2021). Overexpression of truncated NSP15 in A549 cells supported less viral replication and restored antiviral based on an increased sensitivity to TLR3, TL4 and TL9 ligand stimulation. In support of this, it was found that the infection of NSP15-deficient coronaviruses led to the activation of the PKR and OAS-RNase L pathways (Deng et al., 2017). It is likely that coronaviruses are highly susceptible to PKR and OAS-RNase L activity. Taken together, these results suggest again that multiple antiviral pathways, including IRF-1/IRF-7, IFN, PKR, and the OAS-RNase L system which are activated during infection with VSV-EGFP, were impaired in NSP15 overexpressing cells.
This report describes the role of SARS-CoV-2 NSP1 or NSP15 in limiting specific TLR pathway activation in favor of an anti-innate immune response. In summary, this study provides new insights into the role of NSP1 or NSP15 as antagonists of the host innate sensors of viruses. While NSP1 specifically limits antiviral responses, we propose that NSP15, by limiting IFN expression and directing host cellular responses in a TLR2-dependent manner, can promote viral replication.
Author contributions
Conceptualization, N.B. and S.S.; methodology, N.B.; validation, N.B.; formal analysis, N.B.; investigation, N.A., A.M., B.S.; writing—original draft preparation, N.B.; writing—review and editing, A.M., B.S., S.S.; visualization, N.B.; funding acquisition S.S.
Conflicts of interest
The authors declare no conflict of interest.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This research was supported with funds from the Canadian Poultry Research Council, Natural Sciences and Engineering Research Council and the Ontario Ministry of Agriculture, Food and Rural Affairs. This research was supported in part by the University of Guelph's Food from Thought initiative, thanks to funding from the Canada First Research Excellence Fund.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crviro.2022.100021.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
References
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tsang O.T.Y., Wagh D., Coller J., Pellegrini K.L., Kazmin D., Alaaeddine G., Leung W.S., Chan J.M.C., Chik T.S.H., Choi C.Y.C., CHuerta H., McCullough M.P., Lv H., Anderson E., Edupuganti S., Upadhyay A.A., Bosinger S.E., Terry H., Khatri P., Rouphael N., Peiris M., Pulendran B. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020 doi: 10.1126/SCIENCE.ABC6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., Thai J., Miller Z.D., Lin A.E., Schmidt M.M., Stewart D.G., Goldfarb D., de Lorenzo G., Rihn S.J., Voorhees R.M., Botten J.W., Majumdar D., Guttman M. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020 doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti F., Snyder G.A., Giovanetti M., Angeletti S., Gallo R.C., Ciccozzi M., Zella D. Emerging of a SARS-CoV-2 viral strain with a deletion in nsp1. J. Transl. Med. 2020 doi: 10.1186/s12967-020-02507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020 doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Zhang X. Comparative in vivo analysis of the nsp15 endoribonuclease of murine, porcine and severe acute respiratory syndrome coronaviruses. Virus Res. 2012 doi: 10.1016/j.virusres.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020 doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., Ahola T., Liang Y., Liu X., Guo D. Biochemical and structural insights into the mechanisms of sars coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011 doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.F.W., Wang Y., Yuen T.T.T., Chai Y., Hou Y., Shuai H., Yang D., Hu B., Huang X., Zhang X., Cai J.P., Zhou J., Yuan S., Kok K.H., To K.K.W., Chan I.H.Y., Zhang A.J., Sit K.Y., Au W.K., Yuen K.Y. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Baker S.C. An “Old” protein with a new story: coronavirus endoribonuclease is important for evading host antiviral defenses. Virology. 2018 doi: 10.1016/j.virol.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Hackbart M., Mettelman R.C., O’Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Proceedings of the National Academy of Sciences of the United States of America. 2017. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der S.D., Zhou A., Williams B.R.G., Silverman R.H. Proceedings of the National Academy of Sciences of the United States of America. 1998. Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B., Gong X., Fang S., Weng W., Wang H., Chu H., Sun Y., Meng C., Tan L., Song C., Qiu X., Liu W., Forlenza M., Ding C., Liao Y. Inhibition of anti-viral stress granule formation by coronavirus endoribonuclease nsp15 ensures efficient virus replication. PLoS Pathog. 2021 doi: 10.1371/JOURNAL.PPAT.1008690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V. v, Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra ., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y.F., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pène F., Marin N., Roche N., Szwebel T.A., Merkling S.H., Treluyer J.M., Veyer D., Mouthon L., Blanc C., Tharaux P.L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kernéis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020 doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengel H., Koszinowski U.H., Conzelmann K.K. Viruses know it all: new insights into IFN networks. Trends Immunol. 2005 doi: 10.1016/j.it.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Hernández J.C., Stevenson M., Latz E., Urcuqui-Inchima S. HIV type 1 infection up-regulates TLR2 and TLR4 expression and function in vivo and in vitro. AIDS Res. Hum. Retrovir. 2012 doi: 10.1089/aid.2011.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D.A., Møller R., Uhl S.A., Oishi K., Frere J., Golynker I., Horiuchi S., Panis M., Blanco-Melo D., Sachs D., Arkun K., Lim J.K., tenOever B.R. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity. 2021;54 doi: 10.1016/j.immuni.2021.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011 doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.R., Hoque M.N., Rahman M.S., Alam A.S.M.R.U., Akther M., Puspo J.A., Akter S., Sultana M., Crandall K.A., Hossain M.A. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020 doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Foxman E.F., Molony R.D. Early local immune defences in the respiratory tract. Nat. Rev. Immunol. 2017 doi: 10.1038/nri.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Du L., Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies. Emerg. Microb. Infect. 2020 doi: 10.1080/22221751.2020.1723441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Proceedings of the National Academy of Sciences of the United States of America. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29 doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Gil-Cruz C., Spanier J., Li Y., Wilhelm J., Rabouw H.H., Züst R., Hwang M., V’kovski P., Stalder H., Marti S., Habjan M., Cervantes-Barragan L., Elliot R., Karl N., Gaughan C., van Kuppeveld F.J.M., Silverman R.H., Keller M., Ludewig B., Bergmann C.C., Ziebuhr J., Weiss S.R., Kalinke U., Thiel V. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017 doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Gunalan V., Liu B., Chow V.T.K., Druce J., Birch C., Catton M., Fielding B.C., Tan Y.J., Lal S.K. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology. 2007 doi: 10.1016/j.virol.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zhang M., Chu H., Zhang H., Wu H., Song G., Wang P., Zhao K., Hou J., Wang X., Zhang L., Gao C. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat. Immunol. 2017;18 doi: 10.1038/ni.3641. [DOI] [PubMed] [Google Scholar]
- Lokugamage K.G., Hage A., de Vries M., Valero-Jimenez A.M., Schindewolf C., Dittmann M., Rajsbaum R., Menachery V.D. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020 doi: 10.1128/jvi.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meager A. Biological assays for interferons. J. Immunol. Methods. 2002 doi: 10.1016/S0022-1759(01)00570-1. [DOI] [PubMed] [Google Scholar]
- Menachery V.D., Gralinski L.E., Mitchell H.D., Dinnon K.H., Leist S.R., Yount B.L., Graham R.L., McAnarney E.T., Stratton K.G., Cockrell A.S., Debbink K., Sims A.C., Waters K.M., Baric R.S. mSphere; 2017. Middle East Respiratory Syndrome Coronavirus Nonstructural Protein 16 Is Necessary for Interferon Resistance and Viral Pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E., Tschopp J., Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006 doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- Müller U., Steinhoff U., Reis L.F.L., Hemmi S., Pavlovic J., Zinkernagel R.M., Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994 doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Pillon M.C., Frazier M.N., Dillard L.B., Williams J.G., Kocaman S., Krahn J.M., Perera L., Hayne C.K., Gordon J., Stewart Z.D., Sobhany M., Deterding L.J., Hsu A.L., Dandey V.P., Borgnia M.J., Stanley R.E. Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat. Commun. 2021 doi: 10.1038/s41467-020-20608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R.G. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008 doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio N.G., Chauveau L., Hertzog J., Bridgeman A., Fowler G., Moonen J.P., Dupont M., Russell R.A., Noerenberg M., Rehwinkel J. The RNA sensor MDA5 detects SARS-CoV-2 infection. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-92940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan M.A., Milasta S., Green D.R. Toll-like receptor signaling in the lysosomal pathways. Immunol. Rev. 2009 doi: 10.1111/j.1600-065X.2008.00732.x. [DOI] [PubMed] [Google Scholar]
- Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.A., Leibundgut M., Thiel V., Mühlemann O., Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27 doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- Setaro A.C., Gaglia M.M. All hands on deck: SARS-CoV-2 proteins that block early anti-viral interferon responses. Curr. Res. Virol. Sci. 2021;2 doi: 10.1016/j.crviro.2021.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T., Morrison T.E., Funkhouser W., Uematsu S., Akira S., Baric R.S., Heise M.T. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kamitani W., DeDiego M.L., Enjuanes L., Matsuura Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 2012 doi: 10.1128/jvi.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., MacKens-Kiani T., Cheng J., Straub J.H., Stürzel C.M., Fröhlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020 doi: 10.1126/SCIENCE.ABC8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidu A., Janvier A., Schaeffer L., Sosnowski P., Kuhn L., Hammann P., Westhof E., Eriani G., Martin F. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA. 2021;27 doi: 10.1261/rna.078121.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012 doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.K., Lam J.Y., Wong W.M., Mak L.F., Wang X., Chu H., Cai J.P., Jin D.Y., To K.K.W., Chan J.F.W., Yuen K.Y., Kok K.H. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microb. Infect. 2020 doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Miorin L., Makio T., Dehghan I., Gao S., Xie Y., Zhong H., Esparza M., Kehrer T., Kumar A., Hobman T.C., Ptak C., Gao B., Minna J.D., Chen Z., García-Sastre A., Ren Y., Wozniak R.W., Fontoura B.M.A. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci. Adv. 2021 doi: 10.1126/sciadv.abe7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Kuang M., Li J., Zhu L., Jia Z., Guo X., Hu Y., Kong J., Yin H., Wang X., You F. Publisher Correction: SARS-CoV-2 spike protein interacts with and activates TLR4. Cell Res. 2021;31(7):818–820. doi: 10.1038/s41422-021-00501-0. 10.1038/s41422-021-00495-9. Cell Research, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Zhuang M.W., Han L., Zhang J., Nan M.L., Zhan P., Kang D., Liu X., Gao C., Wang P.H. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduction and Targeted Therapy 5. 2020. [DOI] [PMC free article] [PubMed]