Abstract
Corona Virus Disease 2019 (COVID-19) has developed into a global pandemic in the last two years, causing significant impacts on our daily life in many countries. Rapid and accurate detection of COVID-19 is of great importance to both treatments and pandemic management. Till now, a variety of point-of-care testing (POCT) approaches devices, including nucleic acid-based test and immunological detection, have been developed and some of them has been rapidly ruled out for clinical diagnosis of COVID-19 due to the requirement of mass testing. In this review, we provide a summary and commentary on the methods and biomedical devices innovated or renovated for the quick and early diagnosis of COVID-19. In particular, some of micro and nano devices with miniaturized structures, showing outstanding analytical performances such as ultra-sensitivity, rapidness, accuracy and low cost, are discussed in this paper. We also provide our insights on the further implementation of biomedical devices using advanced micro and nano technologies to meet the demand of point-of-care diagnosis and home testing to facilitate pandemic management. In general, our paper provides a comprehensive overview of the latest advances on the POCT device for diagnosis of COVID-19, which may provide insightful knowledge for researcher to further develop novel diagnostic technologies for rapid and on-site detection of pathogens including SARS-CoV-2.
Keywords: COVID-19, Point-of-care testing, Micro/nano devices
1. Introduction
The ongoing outbreak of COVID-19 has caused a global pandemic with considerable morbidity and mortality [[1], [2], [3], [4]]. Without timely diagnosis and treatment, some COVID-19 patients may develop into worse symptoms, including pneumonia and acute respiratory distress syndrome, even deaths, especially for the seniors above 50 years old [5,6]. The fighting against COVID-19, among all infectious diseases caused by viruses, still remains challenging to the human beings, albeit tremendous efforts and technical advances in public healthcare [7]. Although various medicines or vaccines have been proved to be effective to the disease [[8], [9], [10]], advanced techniques for rapid and accurate detection of the virus still greatly contribute the control on viral spreading and early treatment [[11], [12], [13]].
SARS-CoV-2 is one of the beta coronavirus genera, which comprises a nucleocapsid (N) protein associated single-stranded positive-sense RNA (29,881 nucleotides, the genetic material) and three structural surface proteins, including the membrane (M), the spike (S) and the envelope (E) (Fig. 1 a) [[14], [15], [16], [17], [18]]. It has been demonstrated that SARS-CoV-2 is infectious in humans, animals, and herds [19]. Compared with the other emerging viruses that have caused wide epidemics in recent years, such as Middle East respiratory syndrome coronavirus (MERS), severe acute respiratory syndrome coronavirus (SARS-CoV), Ebola virus and Zika virus, SARS-CoV-2 is spreading with a significantly higher rate and wider range [[20], [21], [22], [23], [24]], making even more difficult to control. The major symptoms of infected patients include neurological and respiratory diseases, such as fever, pain in the muscles and tiredness, cough and shortness of breath [[25], [26], [27]]. Unfortunately, these symptoms are not specific for the diagnosis of the infection [28].
Fig. 1.
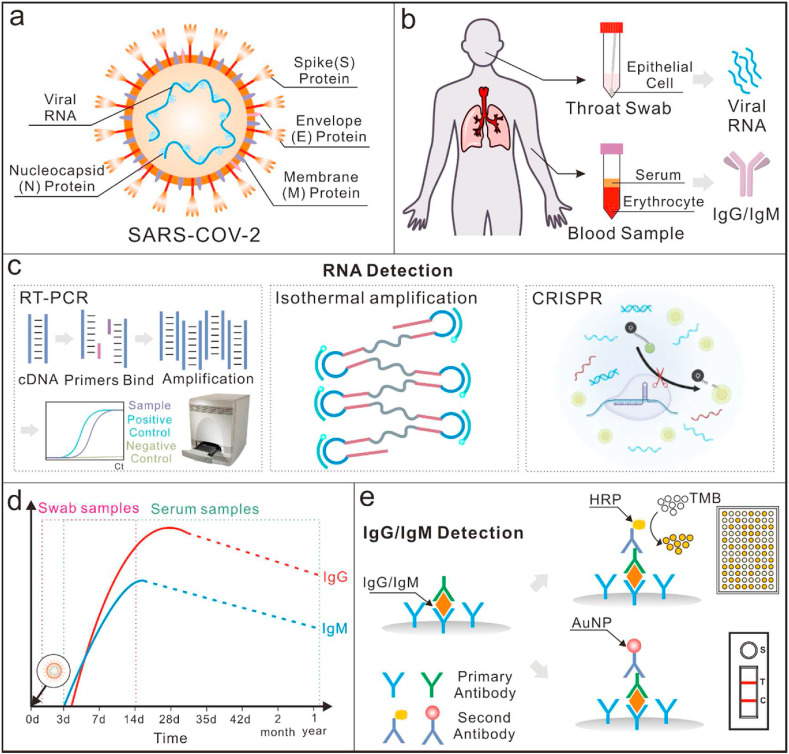
An overview of point-of-care detection methods for COVID-19. (a) Diagrammatic expression of SARS-CoV-2: a single stranded viral RNA with three surface proteins, including Spike (S), Envelop (E), and Membrane (M). (b) Real samples collected for COVID-19 detection: throat swab for virus detection and blood samples for antibody detection; (c) An overview of RNA detection methods, including reverse transcription polymerase chain reaction (RT-PCR), isothermal amplification methods and clustered regularly interspaced short palindromic repeats (CRISPR)-based methods [7]; Reproduced with permission. Copyright 2020, BioRxiv. (d) Schematic illustration of concentrations of the IgG and IgM antibodies in individuals once infected with SARS-CoV-2: IgM antibodies are first to appear in serum samples and are detectable as early as 3 days after infection. The concentration of IgM peaks between 2 and 3 weeks. IgG antibodies come after IgM but last longer. They peak after 2 weeks [49]; (e) Schematic detection mechanisms of IgG and IgM based on lateral flow immunoassays and enzyme-linked immunosorbent assays.
In the past few months, numerous scientific teams and companies have reported methods for COVID-19 detection [[29], [30], [31], [32], [33]]. In terms of their working principle, the major diagnostics methods include: immunoassay-based methods for detection of antibodies in blood serums [[34], [35], [36], [37]] and nucleic acid testing (NAT) for direct determination of the virus in nasal/throat swabs (Fig. 1b) [38,39]. The characteristics of some frequently used NAT methods and immunoassay-based methods are summarized in Table 1 . Thanks to the advantages of robust and sensitive assay, NAT has gained tremendous development with the technical innovation in molecular biology and biomedical engineering, and currently it is a gold standard for COVID-19 diagnosis [40,41]. They can identify and detect trace amounts of the specific viral genomic sequence with various amplification, e.g. reverse transcript -polymerase chain reaction (RT-PCR). In terms of the mechanism underlying the routes for identifying and amplifying the targeted nucleic acids, three methods are currently adopted, including thermo-cycling-based amplification, isothermal amplification methods and CRISPR-based methods (Fig. 1c) [[42], [43], [44]]. NAT is generally very sensitive, but it needs a central laboratory and well-trained technical to operate experiments and interpret the data. Alternatively, immunoassays can provide the information concerning any active viral infections as well as past exposures [45]. The basic mechanism of immunoassay was to detect the specific antibodies against SARS-CoV-2 produced by the immune response in blood serums [46], particularly immunoglobulin M (IgM) and immunoglobulin G (IgG) [47]. As a result, detecting the existence of IgG and IgM antibodies against SARS-CoV-2 is a feasible way to indicate infection [48]. Furthermore, due to the reason that there are a large portion of the population that have been vaccinated, it is important to distinguish between actual infection and vaccination. According to the suggestions of China Food and Drug Administration (CFDA), a combination test of IgM antibodies against the S protein and the N protein can help. Specifically, the people who has a positive IgM antibody test against the S protein needs to take an extra test against the N protein. If both tests were positive, it means that the antibodies are induced from an actual infection. Otherwise, the antibodies are induced from vaccination. Usually, SARS-CoV-2 triggered antibodies could be detected as early as on the 3rd day and decrease gradually as immune responses go on in patients’ body. The antibodies generally become undetectable in about two weeks (Fig. 1d) [49]. Typically, IgM antibodies appear in the serum samples in an earlier stage than IgG does. To date, a wide variety of immunoassays have been developed to detect SARS-CoV-2 antibodies, including enzyme-linked immunosorbent assay, chemiluminescent immunoassay [50] and even a lateral flow immunoassay (Fig. 1e) [51]. Compared with NAT, these immunoassays can provide more convenient and rapid detection of SARS-CoV-2 antibodies in human serum or blood without needing biosafety laboratories, which can also be suitable for the epidemiological of COVID-19.
Table 1.
Summary of the characteristics of some NAT methods and immunoassay-based methods.
| Detection Method | Timing | Sensitivity | Cost | Advantages | Disadvantages | |
|---|---|---|---|---|---|---|
| Nucleic-acid Testing Methods | RT-qPCR | 2–3 h | High | High | Wide detection range; High throughput | Relatively higher cost; False-negative results |
| RT-LAMP | 0.5–1 h | High | Medium | Easy opreation; Isothermal Amplification | False-positive results | |
| RPA | 10–20 min | High | High | Ultra-fast speed; Isothermal amplification (37 °C) | Lower flexibility in the kit formulation | |
| NEAR | 15–30 min | High | Low | Fast amplification speed; Isothermal amplification | False-positive results with short primers | |
| RCA | 4 h | High | Low | Simple reaction mechanism; Isothermal amplification; | Complex sample preparation; | |
| CRISPR-based | 0.5–1 h | High | High | High efficiency, High Specificity, and precision | Potential aerosol contamination | |
| Immunoassay-based Methods | LFI | 10 min | Low | Low | Easy opreation and detection; Low cost | Qualitative dtection; False-positive |
| Chemliuminescence | 0.5–1 h | Medium | Low | High-throughput; Good accuracy | Multiple opeation steps, Higher instrumentation requirements | |
| Electrochemistry | 10–30 min | Medium | Low | Fast quantitative detection | High requirements for reaction conditions | |
| FET | 10–30 min | Medium | Low | Fast quantitativedetection; Easy operation and integration | High requirements for reaction conditions |
Clinically, chest computed tomography (CT) [52,53] and transmission electron microscopy (TEM) [54] also assist in the evaluation and diagnosis of the symptomatic patients. Nevertheless, these instruments are unaffordable in most undeveloped countries. So far, noticeable reviews about SARS-CoV-2 have been provided, with special focus on the origin [55,56], transmission [57,58], clinical features [[59], [60], [61]], and even treatment methods [[62], [63], [64]]. However, a timely review comprehensively summarizing the advancements of rapid diagnostics platforms for point-of-care testing (POCT) of SARS-CoV-2 remains open, especially for those with improved performance by involving micro and nanotechnologies. In this review, we summarize the emerging micro-/nano-scale biomedical devices applied for detecting SARS-CoV-2 in the last two years, which can provide a fast turn-around and sample-to-answer assay. In particular, those micro-nano devices may provide a point-of-care testing and home diagnosis to release the burden of central hospital [65]. We hope this review offers an insight on the methodologies used in developing advanced devices for point-of-care detection of COVID-19 and inspires novel diagnostic technologies in clinical trials in the future.
2. Detection mechanism of SARS-CoV-2
2.1. Nucleic acid testing
Polymerase chain reaction (PCR) offers ultra-high sensitivities and sequence specificities in medical and biological applications, including DNA sequencing [66], functional genes analysis [67] and infectious diseases identification [68]. Especially during the current period, a great number of efforts have been made to improve the performances of PCR systems, which have boosted the whole market. With an additional reverse transcription (RT) process that transfers RNA into complementary DNA (cDNA) strands, RT-PCR can be used for direct detection of RNA-viruses [69]. In the past few years, RT-PCR has achieved a considerable achievement that speeded up its practical applications. Basically, a standard RT-PCR test takes about 3 h from RNA extraction to the final amplification (Fig. 1c) [70]. It uses specific primer sets to hybridize and amplify the target genomic sequence [71]. With the designed primers, it is possible to test the presence of SARS-CoV-2 with a real-time PCR instrument in a Biosafety Level II Laboratory [38]. However, as the gold-standard method, RT-qPCR requires central laboratory and skilled person to operate and interpret the data. The time-consuming and high-cost facts of the thermal-cycling instruments limit their applications in most of on-site, point-of-care circumstances [72]. Moreover, the dependence on the specialized reagents further limits its universally available in those resource-limited regions [73]. Presently, the existing regents and enzymes for the PCR reactions typically require specific refrigeration for storage and transport [74]. As a result, the reagents and medical professionals may be easily constrained in those low and middle-income countries during the pandemic. To improve the testing capabilities in remote locations or at the point-of-care applications, more attention should be given to the innovations of low-cost and stable reagents for PCR with the consideration of room-temperature stable reagents [26,75].
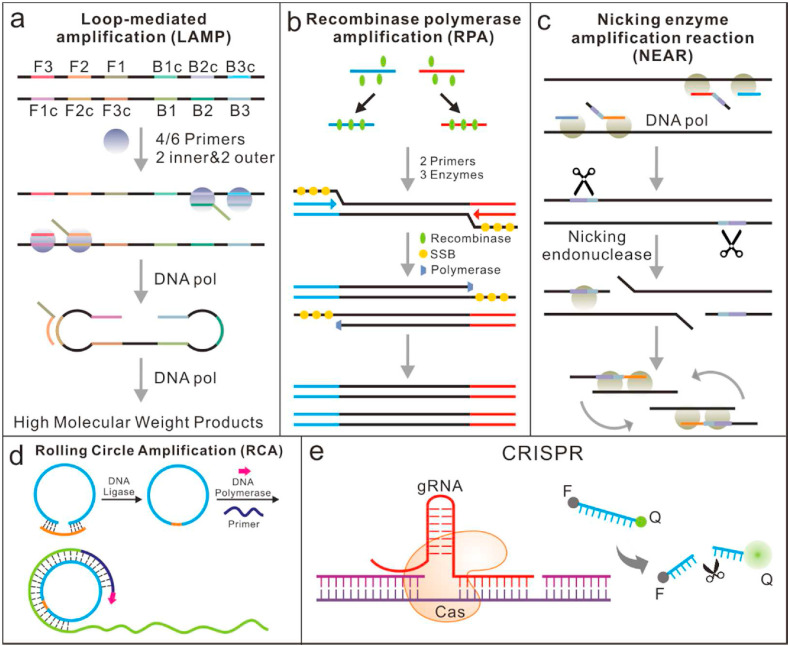
In most scenarios where the expensive thermal cycling instruments are not affordable, isothermal amplification methods, including loop-mediated isothermal amplification (LAMP) [40], recombinase polymerase amplification (RPA) [76], nicking enzyme amplification reaction (NEAR) [77] and rolling circle amplification (RCA) [78] have been developed for the point-of-care testing of SARS-CoV-2. These isothermal amplification methods, conducted with a fixed temperature [79], are independent of expensive thermal cycling units that a PCR amplification needs. Each of these methods has its unique strategy or mechanism to initiate and recycle a new round of dsDNA separation, extension and synthesis (Fig. 2 ). Among them, LAMP and RPA are initiated by a DNA target while NEAR enables direct RNA identification and amplification [40]. In addition to the above advantages, there are also actually known issues for isothermal amplification such as nonspecific or non-template amplification.
Fig. 2.
The working principle of some isothermal amplifications for the detection of COVID-19. (a) LAMP: utilize a set of four to six specially designed primers and a strand-displacement polymerase to recognize and amplify a specific nucleic acid fragment. (b) RPA: two opposing DNA primers designed to be complementary to the target sequence of interest and three enzymes are used during amplification. (c) NEAR: based on polymerase extension plus a single-strand cutting event, can be used to detect RNAs directly. (d) RCA: a short DNA or RNA primer is amplified to form a long single stranded DNA or RNA using a circular DNA template and special DNA or RNA polymerases; (e) CRISPR-based methods for COVID-19 detection. Conventional RNA extraction can be used as an input to the assay, which is visualized by a fluorescent reader.
Reverse transcription loop-mediated isothermal amplification (RT-LAMP) is one of the most common isothermal methods for RNA-based pathogen diagnosis [80,81], which consists of a reverse transcriptase process and a one-step amplification reaction where RNA plays a part as the final template (Fig. 2a). It can recognize and amplify a specific nucleic acid fragment at a constant temperature (60–65 °C) in less than 1 h with high sensitivity by utilizing a set of four to six primers and a strand-displacement polymerase [82]. The stem-loop DNAs as the final products include multiple inverted repeats of the target and exhibit a cauliflower-like appearance. In comparison with the real-time RT-PCR assay, a single protocol-based LAMP is a more straightforward method that can fade the dependence on thermocycler as well as energy consumption [83]. The RT-LAMP results from viral RNA amplification are usually read out colorimetrically or fluorescently, by adding colorimetric pH indicators or fluorescence dyes [84].
In addition, LAMP could also be integrated with sequencing infrastructures [85]. Up to now, LAMP assay kits have already been commercially available. Davidson et al. used paper based-strategy and RT-LAMP to develop an instant device, which visually detected SARS-CoV-2 in saliva in 60 min, with a detection limit of up to 200 copies/μl [86]. However, it does not show absolute superiorities over PCR in clinical scenarios due to the fact that there are limited suitable devices which can do sample processing and RNA extraction on-site [80]. Moreover, there are four to six primers used during LAMP reactions, as a result, the primers need to be strictly designed to avoid potential cross-interactions. Moreover, LAMP may cause false positive detection of samples as well as the carry-over contaminations.
Compared with LAMP, RPA is a relatively new isothermal amplification method reported firstly in 2006 [87]. However, it has experienced an exponential growth in terms of popularity and applications due to the advantages of fast reaction speed and high sensitivity [88]. During amplification, recombinases are combined with primers to form protein-DNA complexes that can bind specifically to the target genes, initiating chain exchange reactions and DNA replications. It is capable of amplifying as low as 1–10 DNA target copies within 10 min with minimal sample preparation [89]. The amplification is under control from a specific combination of enzymes and proteins below 37 °C (Fig. 2b). With a reverse transcription process, RT-RPA based assays could be easily adapted for detecting SARS-CoV-2 [90,91]. The developed assay produced 100% diagnostic sensitivity and specificity with a total run time between 15 and 20 min when compared to RT-qPCR (n = 20), indicating a viable alternative detection method [76]. Thomas et al. developed an RPA-LF based on test strips that detected 35.4 copies/μl of SARS-COV-2 cDNA nucleocapsid (N) gene [91]. However, the prices of RPA kits are relatively high, which limits its clinical applications. Moreover, the flexibility in the kit formulation, as well as the application, are highly limited compared to other isothermal methods [92].
NEAR belongs to the family of isothermal amplification detection methods, which is mainly used for detection of short oligonucleotide sequences [40]. With the help of a DNA polymerase, the primer will extend in the presence of a target template (Fig. 2c). The extended primer will be cut by the nicking endonuclease, releasing the short oligonucleotides due to insufficiently stable duplex under 55 °C. Subsequently, the primers are regenerated and undergo another round of extension and cleavage. Based on the NEAR isothermal amplification technique, a rapid detector called ID NOW has been manufactured by Abbott. Co (Chicago, IL, USA) and authorized [93,94].
RCA is an efficient isothermal enzymatic process that simulates the rolling circle replication process of natural microbial circular DNA (Fig. 2d) [33,95]. With a DNA polymerase, a single primer can trigger the strand displacement synthesis along the circular DNA template to achieve isothermal linear amplification. Owing to its isothermal nature, it is an ideal method with a simple and efficient process. Up to now, RCA-based detection systems or platforms have been successfully applied to test various types of targets [96]. Typically, in a circle-to-circle amplification process, amplicons of the first round RCA will be converted into multiple circles by monomerization (endonuclease digestion) and ligation. Subsequently, newly formed circles will act as templates for the following round RCA, thus reaching an ultra-low limit of detection [97]. To further improve the efficiency of amplification, some exponential or quadratic amplification formats are developed and combined with RCA-based approaches. As a result, it will achieve a detection limit up to sub-femtomolar level [98]. Moreover, it significantly simplifies the operation without the dependence on the time-consuming and labor-intensive operations. Tian et al. reported a typically RCA-based amplification method for quick and ultra-sensitive detection of SARS-CoV-2, achieving a limit of detection of 0.4 fM [78]. Generally, the amplification template for RCA is required to be circular DNA or a linear DNA circularized firstly. In addition, the results of RCA do not have ideal quality control goals up to now, which greatly limit their wide applications.
CRISPR/Cas-based nucleic acid detection technologies were recently developed for COVID-19 detection [99], by using a unique group of Cas-nucleases, which demonstrated noticeable advantages of sensitivity, specificity, rapidity and simplicity for detection (Fig. 2e). By using extracted nucleic acids as input, both CRISPR Cas13-and Cas12-based assays have been developed for SARS-CoV-2 detection [100]. Cas12, as an RNA guided DNase, can cleave ssDNA indiscriminately upon binding its target sequence [101]. In normal conditions, it will be combined with isothermal amplification to achieve high detection sensitivity and specificity. Broughton et al. proposed a CRISPR-Cas12-based assay, which performed RT-LAMP at 62 °C for 20–30 min followed by Cas12 detection at 37 °C for 10 min of predefined coronavirus sequences and visualized on a lateral flow strip [102]. Other isothermal amplification methods, such as recombinase aided amplification (RAA) have also been used to amplify the extracted RNAs before a CRISPR/Cas12a reaction. When SARS-CoV-2 presents in the system, a quenched green fluorescent molecule labelled ssDNA reporter will be cleaved by Cas12a, resulting in the motivation of green fluorescence, which can be observed directly with a naked eye under 485 nm light [103].
In comparison, Cas13a is a non-specific RNase that remains inactive until it binds its programmed RNA target [104]. Cas13a-based detection is highly programmable and specific, as it relies on complementary based pairing between the target RNA and the CRISPR RNA sequence [105]. A CRISPR-Cas13a-based tool named SHERLOCK (Specific High-sensitivity Enzymatic Reporter unLOCKing) was recently designed by Feng Zhang group specifically for SARS-CoV-2 diagnosis. This protocol involves an RPA and T7 transcription, followed with method Cas13-mediated collateral cleavage of a single-stranded RNA reporter. Combining with colorimetric or fluorescent readouts, the assay enabled detection of 10 copies/μL of synthetic RNA [106]. In the pursuit of less time and labor-intensive, Arizti-Sanz et al. finalize detection by combining isothermal amplification, T7 transcription and Cas13-based detection into a single step. Compared to the two-step assay, this single-step SHERLOCK assay could diagnose SARS-CoV-2 with reduced sample-to-answer time and equal sensitivity with optimized conditions [107]. Zhang et al. combined RPA with SHERLOCK to detect the S gene and Orf1ab gene of SARS-COV-2 [108].
The CRISPR-based molecular diagnostics has great potential towards POCT, quantitation and digital analysis of SARS-CoV-2 with detection sensitivity comparable to real-time RT-PCR assay. If coupled with lateral flow readouts, they will be an attractive option for easy, at-home testing scenarios. However, as newly-emerged methods, more efforts should be taken to guarantee its accuracy and prevent the issues of aerosol contamination and false positive rate in clinical trials.
2.2. Immunoassays
Serological testing of antibodies is another common method for detection of COVID-19. Compared to nucleic acid methods, it offers advantageous turn-around time, throughput and workload [109,110]. Although a wide range of doctors and experts have marked that the results from immunological methods may not be considered for the final diagnosis since they only indicate the previous infection, it does not mean that the IgM/IgG serological tests are useless since the immune status of individuals is still important to be acknowledged by the doctors in the following step of treatment (Table 2 ) [111]. The clinical interpretation of all possible scenarios that can be encountered when testing a patient with both RT-qPCR and IgM/IgG immunoassay are illustrated in Table .2. Based on the current knowledge about the rise and fall of SARS-CoV-2, the correlation of IgM level and IgG level varies during the initial time of infection, the onset of symptoms and recovery phase. The key takeaway is that the results of nucleic-acid tests and IgM/IgG serological tests do not necessarily need to agree [49]. A disagreement between the two tests, if any, can often be traced to the after-infection time points at which the tests were performed. Since the exact time of infection is often unknown, combining these two testing can further improve the accuracy of COVID-19 diagnosis [112,113].
Table 2.
The clinical significance of IgM/IgG serological test results. (”+” means “Positive”, “-” means “Negative”).
| Test Results |
Clinical Significance | ||
|---|---|---|---|
| IgG | IgM | RT-qPCR | |
| + | - | - | Patient may have had a past infection, and has recovered |
| + | - | + | Patient may be in the late or recurrent stage of infection |
| + | + | - | Patient may be in the revovery stage of an infection, or the RT-qPCR result may be false-negative |
| + | + | + | Patient is in active phase of infection |
| - | - | + | Patient may be in the window period of infection |
| - | + | - | Patient may be in the early stage of infection. RT-qPCR result may be false-negative |
| - | + | + | Patient may be in the early stage of infection. |
| - | - | - | Healthy person |
Enzyme-linked immunosorbent assay (ELISA) is one of the most widely used methods for the detection of protein-based biomarkers [114,115]. Briefly, the ELISA for total antibodies detection is developed based on double-antigens sandwich immunoassay, using mammalian cell-expressed recombinant antigens contained the receptor binding domain (RBD) of the spike protein of SARS-CoV-2 as the immobilized and HRP-conjugated antigen. According to statistics, compared with a single PCR test, the positive detection rate is significantly increased from 51.9% to 98.6% by combining the PCR with the ELISA assay for each patient [116]. Srivatsa et al. used aptamer-functionalized gold nanoparticles to identify SARS-CoV-2 spike proteins, limit of detection can up to 3540 genome copies/μl [117]. However, the cross-reactivity and low antibodies titers are among common factors that limited the detection efficacy of ELISA. It is also critical to executing the procedures of serial washings and incubation with reagents in ELISA to reduce the background noise and amplify the signal. In clinical, their results are used as an assistant to diagnose the infection.
Usually, the timing of requests for serological assays and the interpretation of antibody results are pre-requisites of crucial importance in their efficacy [118]. Combining with chemiluminescence and the immunoreactions, chemiluminescent immunoassay (CLIA) can quantitatively determine the concentrations of corresponding antigens or antibodies through the intensity of luminescence with high sensitivity and specificity [119]. Based on automatic platforms, CLIA enables high throughput detection, making an outstanding contribution to the early screening. Lyu et al. used ABEI/Co2+ dual-functionalized magnetic beads to perform rapid CLIA detection of SARS-CoV-2 nucleocapsid protein (NP) [120].
Compared with ELISA and CLIA, lateral flow immunoassay (LFI) enables direct test without extraction, which made it well fit for the large-scope, on-site screening [121]. LFI typically contain sample pad, conjugation pad, nitrocellulose membrane, test line, control line and plastic cassette [122]. The mechanism of LFI is based on the hydration and transport of reagents as the specimen across the strip via chromatographic lateral flow. If anti-SARS-CoV-2 IgG and IgM antibodies present in the sample, they will be bound by the corresponding antigen labelled gold colorimetric reagent fixed on the conjugate pad. With samples continuing to travel up the strip, the IgM antibodies are bound on the M (IgM) line, and the IgG are bound to the G (IgG) line, presenting a reddish-purple line at the test zone [112]. During the performance of all valid tests, a control line will appear whether the sample is positive or negative, demonstrating the fluid has migrated adequately through the device. In normal conditions, the detection takes at most 15 min to obtain results with one drop of various specimens such as sera, plasma of venous blood and finger stick blood. Chen et al. developed a LFIA strip based on SERS for anti-SARS-COV-2 IgM and IgG [123]. However, when the concentration of antibodies is none or at a low level, there is a risk of missed detection by the false negative results.
3. Micro/nano devices for point-of-care testing of SARS-CoV-2
Recent advances in microfluidic technology and nanotechnology have brought us closer than ever to the realization of simple yet highly sensitive and specific devices that could be used in complicated environments without central lab. Based on either nucleic acid testing or immunoassay, the micro/nano devices significantly enrich the toolset of POCT of SARS-CoV-2, which have the potential to rapidly diagnose pathogens or antibodies and efficiently monitor the infection transmission by self-tests even at home. They can act as a bridge between laboratory-based testing and home detection. Presently, more and more these devices are transformed by the companies as promising platforms for COVID-19 detection. Especially those miniaturized devices, which integrated all the steps (nucleic acid extraction, amplification and detection) by fluidic manipulation, are conducive to complex real-time diagnosis. Moreover, the introduction of nanomaterials could also significantly increase the detection sensitivity of immunoassays. In this section, we summarize the very recent progresses on the micro/nano technologies in the field of POCT, hoping to provide useful information and insight for the further researches in the area.
3.1. Micro/nano devices for nucleic acid testing
In a typical diagnostic test, there are two types of inaccuracy, including the false-negative result (FNR) and the false-positive result (FPR) [124,125]. Statistically, RT-PCR assay based on nasal or oropharyngeal swabs could produce up to 30% FNR in the clinical diagnosis of COVID-19 [[126], [127], [128]]. On the standpoint of the clinical stage of the disease and different anatomic sites in the virus-carriers, sampling strategy has a close relationship with the inconsistency of viral load which resulted in the high FNR [39]. In a traditional sampling process, regular swabs can only provide limited physical interactions with mucosal tissues. As a result, only superficial tissues could be collected, which are also readily contaminated by food and drink, resulting in a low sampling efficacy [128]. Chen et al. reported a microneedle-based oropharyngeal swab for effective and precise viral sampling. Based on the soft-tissue penetration capability of these microneedles (MNs), there would be a significant increase in sample depth (Fig. 3 a). In addition, the MNs are modified with antibodies, which will further assist in SARS-CoV-2 collection through chemical bonding [128]. Once the preparation process can be simplified, the proposed novel swab will act as a promising candidate for diverse oral or respiratory diseases sampling in the future. However, it is also worth noting that medical staffs will be in close contact with the suspected COVID-19 patients during sampling, leading to a high risk of cross-infection. In addition, the swabs themselves are potential source of infection [129]. To solve this problem, a miniature robot consisted of an active 2-degree of freedom (DOF) end-effector was proposed to assist nasopharyngeal swab collection remotely. The successful working of the miniature robot has already been verified on a pig nose. Subsequent works will focus on pursuing ethical approval for in-vivo tests. The captured nasal or oropharyngeal swabs will then be subjected to a series of standard procedures for the subsequent lysis and enrichment. Effective DNA extraction and enrichment is the premise of accurate detection [130]. However, this process not only relies on enhanced-biosafety lab, but also requires skilled personnel and mandatory instruments. Manual operation procedures may raise issues, such as artificial variants or biased data. Followed with sample preparation, a variety of nucleic-acid based methods, ranging from PCR-based methods to some isothermal amplification methods, are available to the diagnosis of infectious diseases. From sample preparation to the assay protocol, it usually requires a few hours [131]. To meet the diagnostic demands of the infectious pathogens, especially in those resource-limited settings, there is an urgent need for portable and integrated microfluidic platforms or micro/nano devices that can provide fast, accurate and even multiplex diagnosis at the point-of-care [132].
Fig. 3.
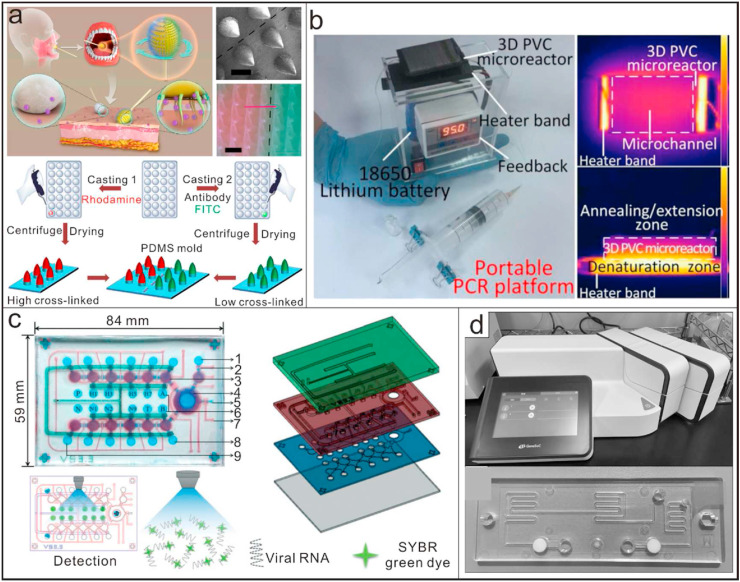
Micro/nano devices for nucleic acid testing. (a) A microneedle-based oropharyngeal swab for effective and precise viral sampling during COVID-19 detection [128]; Reproduced with permission. Copyright 2020, Elsevier. (b) A miniaturized and portable PCR platform run on batteries for multiplexed detection of clinical-level of DNA tartets. [28]; Reproduced with permission; Copyright 2020, Nature publishing group. (c) An integrated microfluidic device with functions of sample treatment, one step RT-PCR and direct fluorescence detection. The deteiled components included: 1) Wash buffer chamber, 2) Microvalve, 3) Consecutive micropumps, 4) Open-type micromixer, 5) Waste outlet, 6) RT-PCR reagent chamber, 7) Air inlet/outlet, 8) RT-PCR reagent chamber, 9) Overflow chamber [135]; Reproduced with permission. Copyright 2020, Royal Society of Chemistry. (d) A compact, reciprocal flow PCR system, GeneSoC®, with one heater for the reverse transcriptase reaction and two heaters for thermal cycling, can be used for specific gene amplification and fluorescence detection based on PCR in a very short time (within 15 min) [136]. Reproduced with permission. Copyright 2020, Elsevier.
Generally, traditional PCR instruments are practical obstacles for wide use in POCT scenarios due to the reason that they are more expensive and bigger in size [28]. They rely heavily on external electric powers to realize quick increase or decrease of the temperature. Owing to their greatly reduced size, micro/nano devices possess many unique properties compared to macro-scale devices, which opens a new perspective for field testing based on PCR. They require less reagents during amplification and have higher heat transfer efficiency. Moreover, the improved portability makes them an ideal choice for point-of-care diagnosis of COVID-19 [133,134]. Shi et al. reported a miniaturized, portable and battery-powered heater with functions of thermo-cycled control and passive continuous-flow control as the platform for PCR reactions (Fig. 3b). Integrated with a 3D microreactor, the system can be used for multiplexed detection of clinical-level DNA targets with more convenience [28]. Microfluidic systems are also well suited for highly automated processes. Recently, a microfluidic device integrated with functions of sample treatment, one step RT-PCR and direct fluorescence detection was developed and verified with influenza-A viruses (Fig. 3c). The device enabled automatic sample lysis and enrichment by using glycan-coated magnetic beads, whose capture rates could be 50% [135]. The sealed microfluidic device, with all reagents pre-loaded, could be adapted to detect SARS-CoV-2 easily. Another factor that limits the widespread use of PCR is the detection time. Under normal conditions, a standard PCR procedure takes at least 45 min. Based on microfluidic devices, a compact, high-speed reciprocal flow RT-qPCR system, GeneSoC®, has become available for specific gene amplification and detection within 15 min [136] (Fig. 3d). The system has one heater for the RT reaction and two heaters for thermal cycling, with two micro-blowers at each flow ends for the high-speed shuttle of the PCR solution. It can achieve a limit of detection (LOD) of 1.0 × 101 copies/reaction with the use of a single disposable tip per analysis. Although it has some disadvantages, such as requiring the simplification and refinement of the RNA extraction procedure, it has been demonstrated with some clinical samples that it could be used for the rapid and low-throughput selection of patients with COVID-19.
In addition to PCR-based methods, micro/nano biomedical devices that utilize isothermal nucleic acid amplification methods have also been investigated. Due to the isothermal amplification does not need a thermal cycling, it's much easier to be engineered with micro and nano device, e.g. coupling with microfluidics technology [137,138]. Recent researchers have developed a range of microfluidic devices with different structures for detection of bacterial and viral pathogens, which enable multiplexed detection of different targets with corresponding primer sets deposited in each channel (Fig. 4 a, Fig. 4b) [[139], [140], [141]]. In general, compared to thermal cycling-based amplification methods, the isothermal amplification methods can usually provide lower cost, faster reaction speed, less specialized equipment and easy readout [79]. They can be combined with the application of various fully enclosed micro-structured devices, with less energy consumption to maintain a constant temperature [142]. These features greatly simplify isothermal amplification implementation in POCT platforms. Compared with those microfluidic chips based on PCR, these devices enhance their applicability for rapid detection with less requirements for instruments during amplification. The isothermal amplification can be triggered on a thermostatic heating plate or even within a thermos [143], which is very promising to provide a sample-in-result-out solution for the in-filed testing of SARS-CoV-2. By using a smartphone, the fluorescence emission generated by the dyes based on the device during amplification can be monitored in real-time [144]. The image analysis could also provide quantitative results on the time at which amplification occurred (Fig. 4c). Yang et al. reported a simple yet efficient isothermal amplification platform containing a custom-fabricated detector and a multiplexed microwell array chip to perform the RT-LAMP assay within 25 min (Fig. 4d). The platform integrated functions of sample preparation, isothermal amplification. The results could be read with naked eyes directly [145]. The system has been successfully used to detect 130 real clinical samples.
Fig. 4.
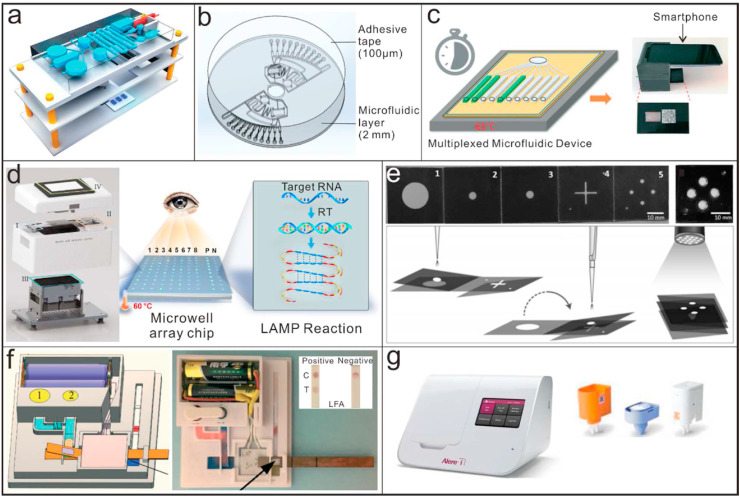
Micro/nano devices based on isothermal amplification methods for point-of-care testing applicaitons. (a) A microfluidic device using LAMP for sample-in-result-out detection of infectious pathogens [141]; Reproduced with permission. Copyright 2020, Royal Society of Chemistry. (b) A centrifugal direct microfluidic chip based on RPA for multiplexed and sensitive detection of pathogenic bacteria [139]; Reproduced with permission. Copyright 2020, Royal Society of Chemistry. (c) Workflow of a point-of-care multiplexed microfluidic device integrated with a smartphone for detecting live virus from nasal swab media [144]; Reproduced with permission. Copyright 2020, Royal Society of Chemistry. (d) A low-cost isothermal amplification platform with multiplexed microwell array biochip for rapid and visualized detection of SARS-CoV-2 [145]; Reproduced with permission. Copyright 2020, AAAS. (e) A microfluidic origami-paper-based device based on LAMP for low-cost and multiplexed detection of malaria [147]; Reproduced with permission. Copyright 2020, Wiley-VCH. (f) A paper-based device with functions of nucleic acid extraction, isothermal amplification and visual detection [148]; Reproduced with permission. Copyright 2020, Royal Society of Chemistry. (g) ID NOW instrument for point-of-care testing of SARS-CoV-2 [93].
In resource-limited settings, the lack of the required infrastructure and facilities for pathogens detection will lead to infected people either not being detected or, if identified, being diagnosed at a sufficiently late stage. With developments in paper-based microfluidic devices, some paper-based platforms based on isothermal amplification methods have been proposed for their low cost [146]. An additional feature of paper-based devices, especially for detection of infectious pathogens is that they are readily disposable, which prevents the potentials of cross-infection. Xu et al. reported a microfluidic origami-paper-based device for multiplexed detection of malaria from whole blood by using LAMP (Fig. 4e) [147]. All the required steps, including nucleic acid extraction, isothermal amplification and visual detection were integrated into the device for POCT application. Tang et al. proposed another fully functioned paper-based device (Fig. 4f). The device allowed on-chip dried reagent storage and equipment-free isothermal amplification, which further promoted their potential applications in remote settings [148]. Nguyen et al. created a sliding-paper device that combines LAMP with dopamine to detect SARS-COV-2 DNA in 25 min with a detection limit of 104 ng/μl [81].
ID NOW was launched initially in 2014 as an advanced molecular diagnostic platform for the detection of influenza A&B, streptococcus A and respiratory syncytial viruses. As to the detection of SARS-CoV-2, two primers targeting at the RdRp gene are used to trigger the amplification. Combined with fluorescence detection, it allows you to make effective clinical decisions sooner. It can enable detection of positive samples within 5 min and negative ones in 13 min (Fig. 4g) [93,149,150]. Due to the characteristics of smaller size and faster reaction speed, it has already been distributed to numerous medical centers and non-traditional places where the testee can get detection results in several minutes. However, the sensitive and specificity of the system are highly reliable on the nicking enzyme and modified primers. The disintegration difference of the enzyme will lead to different amplification efficiency, which will affect the accuracy and repeatability of the results ultimately.
3.2. Micro/nano devices for immunoassays
In addition to nucleic-acid tests, micro/nano technologies have also been extensively investigated for developing immunoassay-based POCT platforms. Presently, commercial products of IgM only and IgM-IgG combined LFI tests have been developed by a couple of In Vitro Diagnostic (IVD) companies [51]. These simple yet robust point-of-care LFI can simultaneously detect IgM and IgG antibodies of the test at different infection stages [[151], [152], [153]]. However, traditional colloidal gold-based LFI is usually limited to relatively low sensitivity and incapable of quantification measurement [154]. With the aid of nano technologies, the LFI assays enable more sensitive and rapid detection. Wang et al. reported a LFI assay based on a selenium nanoparticle-modified SARS-CoV-2 nucleoprotein to be with high sensitivity and detection speed (Fig. 5 a) [155]. The assay enabled simultaneous detection of IgG and IgM in human serum with a limit of detection of 5 ng/mL and 20 ng/mL, respectively, within 10 min. To get more quantitative results, Chen et al. reported another LFI assay for detection of IgG in human serum based on a recombinant nucleocapsid phosphoprotein and lanthanide-doped polystyrene nanoparticles (LNPs) as a fluorescent reporter [156]. Once the functionalized LNPs were captured at the control or test zone, they would produce a bright fluorescence whose excitation and emission wavelengths are 365 nm and 615 nm, respectively. The proposed assay can improve from semiquantitative to accurate quantification by using official IgG standard. Based on microfluidic devices, a novel smartphone-based POCT analyzer with microchannel capillary flow assay platform was developed for quantitation analysis of malaria biomarker (Fig. 5b) [157]. The novel analyzer integrated the ultra-high sensitivity of chemiluminescent detection, the high reaction kinetics of the microfluidic spiral chambers design and the data processing capabilities of smartphone, reaching a limit of detection (LOD) of 8 ng/mL for malaria. The terminal results derived from the positive and negative controls to decrease the risk of false diagnosis. Furthermore, the quantitative platform could easily be adapted for the detection of IgM and IgG. Essentially, the test results from this category of methods cannot confirm the existence of the target virus. Instead, it provides a piece of immunological evidence for physicians to make the correct diagnosis along with other tests, as well as establish a treatment strategy.
Fig. 5.
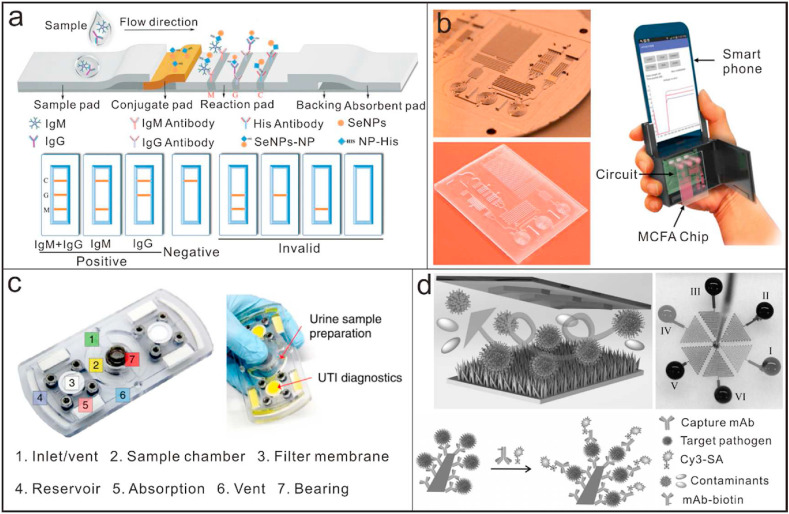
Micro/nano devices based on immunoassays for point-of-care testing applicaitons. (a) A highly sensitibe lateral flow immunoassay for detection of IgM and IgG using a selenium nanoparticle-modified SARS-CoV-2 nucleoprotein as the capture antibody [155]; Reproduced with permission. Copyright 2020, Royal Society of Chemistry. (b) A smartphone-based POCT analyzer with microchannel capillary flow assay platform for quantitative detection of malaria [157]; Reproduced with permission. Copyright 2020, Nature publishing group. (c) A diagnostic fidget spinner device as a versatile bacterial infection diagnostic platform for low-resource settings [166]; Reproduced with permission. Copyright 2020, Nature Publishing Group. (d) An immunofluorescence microdevice integrated with ZnO nanorods for highly sensitive detection of AIV [168]; Reproduced with permission. Copyright 2020, Weiley-VCH.
Rapid and early identification of infectious pathogens allows for effective implementation of disease prevention and treatment measures. Based on micro/nano technologies and the principle of immunoassay, some POCT platforms are constructed directly for detection of infectious pathogens waiving nucleic acid amplifications. Integrating the detection devices into wearables can expand opportunities for long-term and noninvasive monitoring of infections [158,159]. Xue et al. reported an intelligent wearable face mask integrated with a flexible immunosensor for highly sensitive screening of exhaled coronavirus aerosols. In addition, some other kinds of on-site detection devices, such as the electrochemical biosensors, allow detection of multiple kinds of molecules, including antigens and antibodies with high sensitivity and specificity [[160], [161], [162], [163]]. Yakoh et al. reported a paper-based electrochemical biosensor for label-free detection of SARS-CoV-2 antibodies without the specific requirements of antibodies [164]. With optical assistance, Mohammad et al. designed an electro-optofluidic chip that detected target SARS-CoV-2 RNAs without amplification, the detection limits up to 104-109 copies/ml for swab samples [165]. A custom-made fidget spinner that rapidly concentrated pathogens in 1 mL samples of undiluted urine by more than 100-fold for the on-device colorimetric detection of bacterial load and pathogen identification was designed and fabricated (Fig. 5c) [166]. The device enabled on-site detection of infection with naked eyes within 50 min in urine samples from 39 patients suspected of having a urinary tract infection. Although the device is aimed to detect urinary tract infection (UTI) in their original report, we believe it will be a good, inexpensive handheld point-of-care device for the rapid concentration and detection of SARS-CoV-2 in low-resource, undeveloped countries. The use of nanomaterials or nanostructures could efficiently increase the sensitivity to meet the detection requirements. By using graphene sheets to modify the field-effect transistor (FET), Park et al. reported an ultra-sensitive sensor for detection of SARS-CoV-2 in clinical samples [167]. The limit of detection of the graphene modified FET sensor could be up to 1 fg/mL. Yu et al. reported an immunofluorescence microdevice integrated with ZnO nanorods for highly sensitive detection of avian influenza virus (AIV) [168]. The unique properties of ZnO nanorods boost the LOD of the device to an ultra-low level, which can be approximate 22 times more sensitive than conventional ELISA (Fig. 5d).
4. Summary and perspectives
The pandemic of COVID-19 has caused wide-scope outcomes to most low- and middle-income countries where the infrastructure and core-facilities for early detection are insufficient. This review provides a comprehensive summary of micro/nano biomedical devices, as well as two main categories of technologies for the rapid diagnosis of COVID-19 (including nucleic acid-based methods and immunoassays) and their working principles and targets of the virus. Among these is an immunochromatographic assay that is more applicable to primary screening in these areas. Immunoassay offers advantages such as direct detection of the serum/plasma and whole blood specimens without additional processing steps and expensive equipment. In comparison, nucleic acid-based methods are more accurate for diagnosis of COVID-19. A series of isothermal amplification techniques based on LAMP, RPA and NEAR, which do not rely on expensive thermal-cycling instruments, have shown their advantages in low-cost and simple-operations. However, after more than 20 years' development, there are still few instruments or devices based on these isothermal amplification methos that have been widely used on the market. A lot of optimization works need to be done to avoid problems such as false-positive results. Moreover, CRISPR-based techniques enable promising sensitivity and ultralow detection limit down to a few viral RNA copies, showing an emerging methodology for diagnosis. Clinicians may make a choice of selections in terms of their local circumstances. However, it's necessary to take into account of both the advantages and disadvantages of each method may be more effective and affordable in most area.
Meanwhile, it is worth noting that the false negative rate from single testing by even gold standard assay (RT-PCR) is relatively high in the clinical diagnosis of COVID-19, according to previous statistics. Further researches are still needed under the current circumstances. Till now, clinical-recognized high-throughput testing is unavailable with the infections continue to emerge, there is an urgent call for the development of multiplexed, high-throughput POCT platforms for on-site detection. The capability of early and rapid diagnosis will facilitate the chance of patients obtaining proper medical treatment, decrease the risk of medical staff infection, support therapeutic drug delivery systems. In addition, the instruments that can be used for screening a variety of viruses simultaneously should be further developed and prepared for the future contingencies.
Credit author statement
Yang Wang: Writing – original draft; Writing – review & editing; Investigation; Methodology; Huiren Xu: Writing – original draft; Zaizai Dong: Writing – original draft; Writing – review & editing; Zhiying Wang: Investigation; Zhugen Yang: Funding acquisition; Writing – review & editing; Xinge Yu: Funding acquisition; Writing – review & editing; Lingqian Chang: Funding acquisition; Investigation; Methodology; Project administration; Writing – review & editing.
Data and materials availability
All data needed to evaluate the conclusions in the paper are present in the paper.
Funding source
All sources of funding should also be acknowledged and you should declare any involvement of study sponsors in the study design; collection, analysis and interpretation of data; the writing of the manuscript; the decision to submit the manuscript for publication. If the study sponsors had no such involvement, this should be stated.
Please state any sources of funding for your research:
This work was supported by the Beijing Advanced Innovation Center for Biomedical Engineering at Beihang University, the NSFC (No. 32071407, No. 62003023, No. 61971294), the 111 Project (No. B13003), City University of Hong Kong (Grant No. 9610423, 9667199). ZY thanks UK NERC Fellowship grant (NE/R013349/2).
Ethical approval and informed consent (if applicable)
If the work involves the use of human subjects, the author should ensure that the work described has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. The manuscript should be in line with the Recommendations for the Conduct, Reporting, Editing and Publication of Scholarly Work in Medical Journals and aim for the inclusion of representative human populations (sex, age and ethnicity) as per those recommendations. The terms sex and gendershould be used correctly.
All animal experiments should comply with the ARRIVE guidelines and should be carried out in accordance with the U.K. Animals (Scientific Procedures) Act, 1986 and associated guidelines, EU Directive 2010/63/EU for animal experiments, or the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978). The sex of animals must be indicated, and where appropriate, the influence (or association) of sex on the results of the study.
The author should also clearly indicate in the Material and methods section of the manuscript that applicable guidelines, regulations and laws have been followed and required ethical approval has been obtained.
Patient consent (if applicable)
Completion of this section is mandatory for Case Reports, Clinical Pictures, and Adverse Drug Reactions. Please sign below to confirm that all necessary consents required by applicable law from any relevant patient, research participant, and/or other individual whose information is included in the article have been obtained in writing. The signed consent form(s) should be retained by the corresponding author and NOT sent to Medicine in Novel Technology and Devices.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgements
Funding sources: This work was supported by the Beijing Advanced Innovation Center for Biomedical Engineering at Beihang University, the NSFC (No. 32071407, No. 62003023, No. 61971294), the 111 Project (No. B13003), City University of Hong Kong (Grant No. 9610423, 9667199). ZY thanks UK NERC Fellowship grant (NE/R013349/2).
References
- 1.Cao W., Li T. COVID-19: towards understanding of pathogenesis. Cell Res. 2020;30:367–369. doi: 10.1038/s41422-020-0327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jee Y. WHO international Health regulations emergency committee for the COVID-19 outbreak. Epidemiol. Health. 2020;42 doi: 10.4178/epih.e2020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lotfi M., Hamblin M., Rezaei N. COVID-19: transmission, prevention, and potential therapeutic opportunities. Clin Chim Acta. 2020;508:254–266. doi: 10.1016/j.cca.2020.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pabbaraju K., Wong A., Douesnard M., Ma R., Gill K., Dieu P., Fonseca K., Zelyas N., Tipples G. A public Health laboratory response to the COVID-19 pandemic. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01110-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaul D. An overview of coronaviruses including the SARS-2 coronavirus–Molecular biology, epidemiology and clinical implications. Curr Med Res Pract. 2020;10:54–64. doi: 10.1016/j.cmrp.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udugama B., Kadhiresan P., Kozlowski H., Malekjahani A., Osborne M., Li V., Chen H., Mubareka S., Gubbay J., Chan W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 7.Shen M., Zhou Y., Ye J., Abdullah AL-maskri A.A., Kang Y., Zeng S., Cai S. Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal. 2020;10:97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen J., Kupferschmidt K. Strategies shift as coronavirus pandemic looms. Science (New York, NY) 2020;367:962–963. doi: 10.1126/science.367.6481.962. [DOI] [PubMed] [Google Scholar]
- 9.Wang L., Wang Y., Ye D., Liu Q. A review of the 2019 Novel Coronavirus (COVID-19) based on current evidence. Int J Antimicrob Agents. 2020;55:105948. doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Acquavia M.A., Foti L., Pascale R., Nicolò A., Bianco G. Detection and quantification of Covid-19 antiviral drugs in biological fluids and tissues. Talanta. 2021;224:121862. doi: 10.1016/j.talanta.2020.121862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prazuck T., Colin M., Giache S., Gubavu C., Seve A., Rzepecki V., Chevereau-Choquet M., Kiani C., Rodi V., Lyonnet E. Evaluation of performance of two SARS-CoV-2 rapid whole-blood finger-stick IgM-IgG combined antibody tests. medRxiv. 2020 doi: 10.1101/2020.05.27.20112888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang B., Bragazzi N.L., Li Q., Tang S., Xiao Y., Wu J. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov) Infect Dis Model. 2020;5:248–255. doi: 10.1016/j.idm.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong GP, Guo XJ, Sun YA, Zhang Z, Du LP, Li MY. Diagnostic techniques for COVID-19: a mini-review of early diagnostic methods. J Anal Test: 1-13 2021. [DOI] [PMC free article] [PubMed]
- 14.Cleemput S., Dumon W., Fonseca V., Abdool K.W., Giovanetti M., Alcantara L., Deforche K., De Oliveira T. Genome Detective Coronavirus Typing Tool for rapid identification and characterization of novel coronavirus genomes. Bioinformatics. 2020;36:3552–3555. doi: 10.1101/2020.01.31.928796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamid H., Khurshid Z., Adanir N., Zafar M.S., Zohaib S. COVID-19 pandemic and role of human saliva as a testing biofluid in point-of-care technology. Eur J Dermatol. 2020;14 doi: 10.1055/s-0040-1713020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai C., Shih T., Ko W., Tang H., Hsueh P. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yong S., Su P., Yang Y. Molecular targets for the testing of COVID-19. Biotechnol J. 2020;15:2000152. doi: 10.1002/biot.202000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou P., Yang X., Wang X., Hu B., Zhang L., Zhang W., Si H., Zhu Y., Li B., Huang C. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12:135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Gayle A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Trav Med. 2020 doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y., Schmitz J., Persing D., Stratton C. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58:10–17. doi: 10.9734/JAMMR/2020/v32i1430559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Y., Chang L., Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev Med Virol. 2020;30 doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin Y., Wunderink G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corman V., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D., Bleicker T., Brünink S., Schneider J., Schmidt M., Mulders D., Haagmans B., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J., Wang J., Zhong Z., Su X., Yang K., Chen Z., Zhang D., Li T., Wang Y., Zhang S., Ge S., Zhang J., Xia N. Room-temperature-storable PCR mixes for SARS-CoV-2 detection. Clin Biochem. 2020;84:73–78. doi: 10.1016/j.clinbiochem.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kevadiya B.D., Machhi J., Herskovitz J., Oleynikov M.D., Blomberg W.R., Bajwa N., Soni D., Das S., Hasan M., Patel M., Senan A M., Gorantla S., McMillan J., Edagwa B., Eisenberg R., Gurumurthy C.B., Reid S.P., Punyadeera C., Chang L., Gendelman H.E. Diagnostics for SARS-CoV-2 infections. Nat Mater. 2021;20:593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi B., He G., Wu W. A PCR microreactor machinery with passive micropump and battery-powered heater for thermo-cycled amplifications of clinical-level and multiplexed DNA targets. Microchim Acta. 2018;185:467. doi: 10.1007/s00604-018-3007-z. [DOI] [PubMed] [Google Scholar]
- 29.Bordi L., Piralla A., Lalle E., Giardina F., Colavita F., Tallarita M., Sberna G., Novazzi F., Meschi S., Castilletti C. Rapid and sensitive detection of SARS-CoV-2 RNA using the SimplexaTM COVID-19 direct assay. J Clin Virol. 2020;128:104416. doi: 10.1016/j.jcv.2020.104416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egger M., Bundschuh C., Wiesinger K., Gabriel C., Clodi M., Mueller T., Dieplinger B. Comparison of the Elecsys® Anti-SARS-CoV-2 immunoassay with the EDITM enzyme linked immunosorbent assays for the detection of SARS-CoV-2 antibodies in human plasma. Clin Chim Acta. 2020;508:18–21. doi: 10.1016/j.cca.2020.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theel E., Harring J., Hilgart H., Granger D. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58:e01243. doi: 10.1128/JCM.01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Kasteren P., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C., Meijer A. Comparison of commercial RT-PCR diagnostic kits for COVID-19. J Clin Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang R., Wu J., Ao H., Fu J., Qiao B., Wu Q., Ju H. A rolling circle-amplified G-quadruplex/hemin DNAzyme for chemiluminescence immunoassay of the SARS-CoV-2 protein. Anal Chem. 2021;93:9933–9938. doi: 10.1021/acs.analchem.1c02229. [DOI] [PubMed] [Google Scholar]
- 34.Hou H., Wang T., Zhang B., Luo Y., Mao L., Wang F., Wu S., Sun Z. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin Transl Immunol. 2020;9:e1136. doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q., Du Q., Guo B., Mu D., Lu X., Ma Q., Guo Y., Fang L., Zhang B., Zhang G. A method to prevent SARS-CoV-2 IgM false positives in gold immunochromatography and enzyme-linked immunosorbent assays. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00375-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rezaei M., Bazaz S.R., Zhand S., Sayyadi N., Warkiani M.E. Point of care diagnostics in the age of COVID-19. Diagnostics. 2020;11 doi: 10.3390/diagnostics11010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Won J., Lee S., Park M., Kim T., Park M., Choi B., Kim D., Chang H., Kim V., Lee C. Development of a laboratory-safe and low-cost detection protocol for SARS-CoV-2 of the coronavirus disease 2019 (COVID-19) Exp Neurobiol. 2020;29:107–119. doi: 10.5607/en20009e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y., Gao G., Wang S., Ma C., Xie R., Wang F., Tan C., Zhu L., Guo Y., Zhang F. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niemz A., Ferguson T.M., Boyle D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011;29:240–250. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ilkhani H., Hedayat N., Farhad S. Novel approaches for rapid detection of COVID-19 during the pandemic: a review. Anal Biochem. 2021:114362. doi: 10.1016/j.ab.2021.114362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding X., Yin K., Li Z., Liu C. All-in-One dual CRISPR-cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. bioRxiv. 2020 doi: 10.21203/rs.3.rs-25826/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishige T., Murata S., Taniguchi T., Miyabe A., Kitamura K., Kawasaki K., Nishimura M., Igari H., Matsushita K. Highly sensitive detection of SARS-CoV-2 RNA by multiplex rRT-PCR for molecular diagnosis of COVID-19 by clinical laboratories. Clin Chim Acta. 2020;507 doi: 10.1016/j.cca.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoepp N., Schlappi T., Curtis M., Butkovich S., Miller S., Humphries R., Ismagilov R. Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffman T., Nissen K., Krambrich J., Rönnberg B., Akaberi D., Esmaeilzadeh M., Salaneck E., Lindahl J., Å Lundkvist. Evaluation of a COVID-19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS-CoV-2. Infect Ecol Epidemiol. 2020;10:1754538. doi: 10.1126/10.1080/20008686.2020.1754538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong L., Chuan J., Gong B.O., Shuai P., Zhou Y., Zhang Y., Jiang Z., Zhang D., Liu X., Ma S. Detection of serum IgM and IgG for COVID-19 diagnosis. Sci China Life Sci. 2020;63:777–780. doi: 10.1007/s11427-020-1688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du Z., Zhu F., Guo F., Yang B., Wang T. Detection of antibodies against SARS-CoV-2 in patients with COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.25820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qu J., Wu C., Li X., Zhang G., Jiang Z., Li X., Liu L. Profile of IgG and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71 doi: 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee C., Lin R., Renia L., Ng L. Serological approaches for COVID-19: epidemiologic perspective on surveillance and control. Front Immunol. 2020;11:879. doi: 10.3389/fimmu.2020.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai X., Chen J., Hu J., Long Q., Deng H., Fan K., Liao P., Liu B., Wu G., Chen Y. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of corona virus disease 2019 (COVID-19) medRxiv. 2020 doi: 10.1101/2020.02.22.20026617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isabel M., Damien G., Benoit K., Hafid D., Hector R. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J Clin Virol. 2020;128:104413. doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacobi A., Chung M., Bernheim A., Eber C. Portable chest X-ray in coronavirus disease-19 (COVID-19): a pictorial review. Clin Imag. 2020;64:35–42. doi: 10.1016/j.clinimag.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lal A., Mishra A., Sahu K. CT chest findings in coronavirus disease-19 (COVID-19) J Formos Med Assoc. 2020;119:1000–1001. doi: 10.1016/j.jfma.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prasad S., Potdar V., Cherian S., Abraham P., Basu A., Team I. Transmission electron microscopy imaging of SARS-CoV-2. Indian J Med Res. 2020;151:241–243. doi: 10.4103/ijmr.IJMR_577_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela C.C.S., Wang Y., Wu C., Xiao Y. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shereen M., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;16(24):91–98. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen H., Guo J., Wang C., Luo F., Yu X., Zhang W., Li J., Zhao D., Xu D., Gong Q. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395:809–815. doi: 10.1016/j.jemermed.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rothan H., Byrareddy S. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020:102433. doi: 10.1016/10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang F., Deng L., Zhang L., Cai Y., Cheung C.W., Xia Z. Review of the clinical characteristics of coronavirus disease 2019 (COVID-19) J Gen Intern Med. 2020;35:1545–1549. doi: 10.1007/s11606-020-05762-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez-Morales A., Cardona-Ospina J., Gutiérrez-Ocampo E., Villamizar-Peña R., Holguin-Rivera Y., Escalera-Antezana J., Alvarado-Arnez L., Bonilla-Aldana D., Franco-Paredes C., Henao-Martinez A. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Trav Med Infect Dis. 2020;34:101623. doi: 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y., Kang H., Liu X., Tong Z. Combination of RT-qPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J Med Virol. 2020;92:538–539. doi: 10.1002/jmv.25721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J Crit Care. 2020;57 doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet (London, England) 2020;395:e30. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanders J., Monogue M., Jodlowski T., Cutrell J. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 65.Wang C., Liu M., Wang Z., Li S., Deng Y., He N. Point-of-care diagnostics for infectious diseases: from methods to devices. Nano Today. 2021;37:101092. doi: 10.1016/j.nantod.2021.101092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shendure J., Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1097/01.aog.0000463186.97632.d2. [DOI] [PubMed] [Google Scholar]
- 67.Enrico P., Tanzarella O.A., Paolacci A.R., Mario C. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol. 2009;10:11. doi: 10.1186/1471-2199-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alcoba-Florez J., González-Montelongo R., Íñigo-Campos A., Artola G.M.D., Flores C. Fast SARS-CoV-2 detection by RT-qPCR in preheated nasopharyngeal swab samples. Int J Infect Dis. 2020;97:66–68. doi: 10.1016/j.ijid.2020.05.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balboni A., Gallina L., Palladini A., Prosperi S., Battilani M. A real-time PCR assay for bat SARS-like coronavirus detection and its application to Italian greater horseshoe bat faecal sample surveys. Sci World J. 2012;2012:989514. doi: 10.1100/2012/989514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nagaraj T., Vasanth J.P., Desai A., Kamat A., Madhusudana S.N., Ravi V. Ante mortem diagnosis of human rabies using saliva samples: comparison of real time and conventional RT-PCR techniques. J Clin Virol Off Publ Pan Am Soc Clin Virol. 2006;36:17–23. doi: 10.1016/j.jcv.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 71.Bustin S., Nolan T. RT-qPCR testing of SARS-CoV-2: a primer. Int J Mol Sci. 2020;21:3004. doi: 10.3390/ijms21083004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sreejith K., Ooi C., Jin J., Dao D., Nguyen N. Digital polymerase chain reaction technology – recent advances and future perspectives. Lab Chip. 2018;18 doi: 10.1039/C8LC00990B. [DOI] [PubMed] [Google Scholar]
- 73.Lorenz T. Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies. JoVE. 2012;63:e3998. doi: 10.3791/3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hajia M. Limitations of different PCR protocols used in diagnostic laboratories: a short review. Mod Med Lab J. 2017;1:1–6. doi: 10.30699/mmlj17-01-01. [DOI] [Google Scholar]
- 75.Moiana A., Turner L.J., Ventura L., Gramegna M. A new self-containing lyophilized PCR Mix for RNA amplification. Clin Biochem. 2013;46:1156. doi: 10.1016/j.clinbiochem.2013.05.035. [DOI] [Google Scholar]
- 76.Behrmann O., Bachmann I., Spiegel M., Schramm M., El Wahed A., Dobler G., Dame G., Hufert F. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (exo-IQ) Clin Chem. 2020;8 doi: 10.1093/clinchem/hvaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smithgall M., Scherberkova I., Whittier S., Green D. Comparison of cepheid xpert xpress and abbott ID now to roche cobas for the rapid detection of SARS-CoV-2. J Clin Virol. 2020;128:104428. doi: 10.1101/2020.04.22.055327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tian B., Gao F., Fock J., Dufva M., Hansen M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens Bioelectron. 2020;165:112356. doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- 79.Zhao Y., Chen F., Li Q., Wang L., Fan C. Isothermal amplification of nucleic acids. Chem Rev. 2015;115:12491–12545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- 80.Kashir J., Yaqinuddin A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med Hypotheses. 2020;141:109786. doi: 10.1016/j.mehy.2020.109786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nguyen H.A., Lee N.Y. Polydopamine aggregation: a novel strategy for power-free readout of loop-mediated isothermal amplification integrated into a paper device for multiplex pathogens detection. Biosens Bioelectron. 2021;189:113353. doi: 10.1016/j.bios.2021.113353. [DOI] [PubMed] [Google Scholar]
- 82.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 84.Jeon S., Seo D., Oh H., Kingsley D., Choi C. Development of one-step reverse transcription loop-mediated isothermal amplification for norovirus detection in oysters. Food Control. 2016 doi: 10.1016/j.foodcont.2016.10.005. S0956713516305485. [DOI] [Google Scholar]
- 85.Dao T.V., Herbst K., Boerner K., Meurer M., Kremer L., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M., Boulant S., Klein S., Chlanda P., Khalid D., Barreto Miranda I., Schnitzler P., Kräusslich H., Knop M., Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Davidson J.L., Wang J., Maruthamuthu M.K., Dextre A., Pascual-Garrigos A., Mohan S., Putikam S.V.S., Osman F.O.I., McChesney D., Seville J., Verma M.S. A paper-based colorimetric molecular test for SARS-CoV-2 in saliva. Biosens Bioelectron X. 2021;9:100076. doi: 10.1016/j.biosx.2021.100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Daher R., Stewart G., Boissinot M., Bergeron M. Recombinase polymerase amplification for diagnostic applications. Clin Chem. 2016;62:947–958. doi: 10.1373/clinchem.2015.245829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lutz S., Weber P., Focke M., Faltin B., Hoffmann J., Müller C., Mark D., Roth G., Munday P., Armes N. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA) Lab Chip. 2010;10:887–893. doi: 10.1039/b921140c. [DOI] [PubMed] [Google Scholar]
- 89.Lobato I., O'Sullivan C. Recombinase polymerase amplification: basics, applications and recent advances. Trac Trends Anal Chem. 2018;98:19–35. doi: 10.1016/j.trac.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang T., Wang Y., Shen C., Cheng C. Point-of-care RNA-based diagnostic device for COVID-19. Diagnostics. 2020;10 doi: 10.3390/diagnostics10030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shelite T.R., Uscanga-Palomeque A.C., Castellanos-Gonzalez A., Melby P.C., Travi B. Isothermal recombinase polymerase amplification-lateral flow detection of SARS-CoV-2, the etiological agent of COVID-19. J Virol Methods. 2021;296:114227. doi: 10.21203/rs.3.rs-78408/v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stringer O.W., Andrews J.M., Greetham H.L., Forrest M.S. TwistAmp® Liquid: a versatile amplification method to replace PCR. Nat Methods. 2018;15:395. doi: 10.1038/nmeth.f.407. [DOI] [Google Scholar]
- 93.Nie S., Roth R., Stiles J., Mikhlina A., Lu X., Tang Y., Babady N. Evaluation of Alere i Influenza A&B for rapid detection of influenza viruses A and B. J Clin Microbiol. 2014;52:3339–3344. doi: 10.1128/JCM.01132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.James A.S., Alawneh J.I. COVID-19 infection diagnosis: potential impact of isothermal amplification technology to reduce community transmission of SARS-CoV-2. Diagnostics. 2020;10:399. doi: 10.3390/diagnostics10060399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ali M., Li F., Zhang Z., Zhang K., Kang D., Ankrum J., Le X., Zhao W. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem Soc Rev. 2014;43:3324–3341. doi: 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]
- 96.Gu L., Yan W., Liu L., Wang S., Zhang X., Lyu M. Research progress on rolling circle amplification (RCA)-based biomedical sensing. Pharmaceuticals. 2018;11:35. doi: 10.3390/ph11020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang B., Wang Q., Wu J., Chen Y., Wang J. Detection of nucleic acids with a novel stem-loop primer rolling circle amplification technique. Biotechniques. 2018;64:69–80. doi: 10.4155/btn-2017-0104. [DOI] [PubMed] [Google Scholar]
- 98.Zhang Q., Chen F., Xu F., Zhao Y., Fan C. Target-triggered three-way junction structure and polymerase/nicking enzyme synergetic isothermal quadratic DNA machine for highly specific, one-step, and rapid microRNA detection at attomolar level. Anal Chem. 2014;86:8098–8105. doi: 10.1021/ac501038r. [DOI] [PubMed] [Google Scholar]
- 99.Li Y., Li S., Wang J., Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 100.Mukama O., Wu J., Li Z., Liang Q., Yi Z., Lu X., Liu Y., Liu Y., Hussain M., Makafe G.G. An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids. Biosens Bioelectron. 2020;159:112143. doi: 10.1016/j.bios.2020.112143. [DOI] [PubMed] [Google Scholar]
- 101.Wang X., Shang X., Huang X. Next-generation pathogen diagnosis with CRISPR/Cas-based detection methods. Emerg Microb Infect. 2020:1–23. doi: 10.1080/22221751.2020.1793689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Broughton J., Deng X., Yu G., Fasching C., Servellita V., Singh J., Miao X., Streithorst J., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C., Guevara H., Wadford D., Chen J., Chiu C. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang X., Zhong M., Liu Y., Ma P., Dang L., Meng Q., Wan W., Ma X., Liu J., Yang G. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci Bull. 2020;65(17):1436–1439. doi: 10.1016/j.scib.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abudayyeh O., Gootenberg J., Essletzbichler P., Han S., Joung J., Belanto J., Verdine V., Cox D., Kellner M., Regev A. RNA targeting with CRISPR–Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu L., Li X., Ma J., Li Z., You L., Wang J., Wang M., Zhang X., Wang Y. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell. 2017;170:714–726. doi: 10.1016/j.cell.2017.06.050. [DOI] [PubMed] [Google Scholar]
- 106.Joung J., Ladha A., Saito M., Segel M., Bruneau R., Huang M., Kim N., Yu X., Li J., Walker B., Greninger A., Jerome R., Gootenberg S., Abudayyeh O., Zhang F. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv Prepr Serv Heal Sci. 2020 doi: 10.1101/2020.05.04.20091231. [DOI] [Google Scholar]
- 107.Arizti-Sanz J., Freije C., Stanton A., Boehm C., Petros B., Siddiqui S., Shaw B., Adams G., Kosoko-Thoroddsen T., Kemball M. Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat Commun. 2020;11:5921. doi: 10.1101/2020.05.28.119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aquino-Jarquin G. Recent progress on rapid SARS-CoV-2/COVID-19 detection by CRISPR-Cas13-based platforms. Drug Discov Today. 2021;26:2025–2035. doi: 10.1016/j.drudis.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jacofsky D., Jacofsky E.M., Jacofsky M. Understanding antibody testing for covid-19. J Arthroplasty. 2020;35(7):S74–S81. doi: 10.1016/j.arth.2020.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jin Y., Wang M., Zuo Z., Fan C., Ye F., Cai Z., Wang Y., Cui H., Pan K., Xu A. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49–52. doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jia X., Zhang P., Tian Y., Wang Junli, Zeng H., Wang Jun, Jiao L., Chen Z., Zhang L., He H., He K., Liu Y. Clinical significance of IgM and IgG test for diagnosis of highly suspected COVID-19 infection. Front Med. 2021 doi: 10.1101/2020.02.28.20029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu T., Hsiung J., Zhao S., Kost J., Sreedhar D., Hanson C.V., Olson K., Keare D., Chang S.T., Bliden K.P., Gurbel P.A., Tantry U.S., Roche J., Press C., Boggs J., Rodriguez-Soto J.P., Montoya J.G., Tang M., Dai H. Quantification of antibody avidities and accurate detection of SARS-CoV-2 antibodies in serum and saliva on plasmonic substrates. Nat Biomed Eng. 2020;4:1188–1196. doi: 10.1038/s41551-020-00642-4. [DOI] [PubMed] [Google Scholar]
- 114.Cheng C., Martinez A., Gong J., Mace C.R., Phillips S., Carrilho E., Mirica K., Whitesides G., Paper-based E.L.I.S.A. Angew Chem Int Ed. 2010;49:4771–4774. doi: 10.1002/anie.201001005. [DOI] [PubMed] [Google Scholar]
- 115.Wu L., Li G., Xu X., Zhu L., Huang R., Chen X. Application of nano-ELISA in food analysis: recent advances and challenges. TrAC Trends Anal Chem (Reference Ed) 2019;113:140–156. doi: 10.1016/j.trac.2019.02.002. [DOI] [Google Scholar]
- 116.Guo Y., Cao Q., Hong Z., Tan Y., Chen S., Jin H., Tan K., Wang D., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil Med Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aithal S., Mishriki S., Gupta R., P Sahu R., Botos G., Tanvir S., Hanson R.W., Puri I.K. SARS-CoV-2 detection with aptamer-functionalized gold nanoparticles. Talanta. 2021;236:122841. doi: 10.1016/j.talanta.2021.122841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Padoan A., Cosma C., Sciacovelli L., Faggian D., Plebani M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin Chem Lab Med. 2020;58:1081–1088. doi: 10.1515/cclm-2020-0443. [DOI] [PubMed] [Google Scholar]
- 119.Liu B., Su X., Yu G., Yang S., Wang F., Huang T., Zhou L., Hui Z., Liao Y., Qiu Y., Huang J., Gao H., Liu J., Zhong Y. An automated chemiluminescent immunoassay (CLIA) detects SARS-CoV-2 Neutralizing antibody levels in COVID-19 patients and vaccinees. Int J Infect Dis. 2021 doi: 10.1016/j.ijid.2021.12.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lyu A., Jin T., Wang S., Huang X., Zeng W., Yang R., Cui H. Automatic label-free immunoassay with high sensitivity for rapid detection of SARS-CoV-2 nucleocapsid protein based on chemiluminescent magnetic beads. Sensor Actuator B Chem. 2021;349:130739. doi: 10.1016/j.snb.2021.130739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Demey B., Daher N., François C., Lanoix J.-P., Duverlie G., Castelain S., Brochot E. Dynamic profile for the detection of anti-SARS-CoV-2 antibodies using four immunochromatographic assays. J Infect. 2020:81. doi: 10.1016/j.jinf.2020.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Koczula K., Gallotta A. Lateral flow assays. Essays Biochem. 2011;60:111–120. doi: 10.1042/EBC20150012. https://doi.org/US20110086359 A1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen S., Meng L., Wang L., Huang X., Ali S., Chen X., Yu M., Yi M., Li L., Chen X., Yuan L., Shi W., Huang G. SERS-based lateral flow immunoassay for sensitive and simultaneous detection of anti-SARS-CoV-2 IgM and IgG antibodies by using gap-enhanced Raman nanotags. Sensor Actuator B Chem. 2021;348:130706. doi: 10.1016/j.snb.2021.130706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hadjinicolaou A., Farcas G., Demetriou V., Mazzulli T., Poutanen S., Willey B., Low D., Butany J., Asa S., Kain K., Kostrikis G. Development of a molecular-beacon-based multi-allelic real-time RT-PCR assay for the detection of human coronavirus causing severe acute respiratory syndrome (SARS-CoV): a general methodology for detecting rapidly mutating viruses. Arch Virol. 2011;156:671–680. doi: 10.1007/s00705-010-0906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yu X., Pan J., Ye R., Xiang H., Kou Y., Huang Z. Preparation of armored RNA as a control for multiplex real-time reverse transcription-PCR detection of influenza virus and severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2008;46:837LP–841. doi: 10.1128/JCM.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang Y., Wang Y., Chen Y., Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol. 2020;92:568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ysa B., Chen C., Yu W.A. Early and consecutive RT-PCR tests with both oropharyngeal swabs and sputum could improve testing yield for patients with COVID-19: an Observation Cohort Study in China. Int J Infect Dis. 2021 doi: 10.1016/j.ijid.2021.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen W., Cai B., Geng Z., Chen F., Wang Z., Wang L., Chen X. Reducing false negatives in COVID-19 testing by using microneedle-based oropharyngeal swabs. Matter. 2020;3:1589–1600. doi: 10.1016/j.matt.2020.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang S., Wang K., Liu H., Hou Z. 2020. Design of a low-cost miniature robot to assist the COVID-19 nasopharyngeal swab sampling. arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Krych Ł., Castro-Mejía J.L., Forero-Junco L., Moesby D., Mikkelsen M., Rasmussen M., Sykulski M., Nielsen D. DNA enrichment and tagmentation method for species-level identification and strain-level differentiation using ON-rep-seq. Commun Biol. 2019;2:369. doi: 10.1038/s42003-019-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Karami A., Gill P., Motamedi M., Saghafinia M. A review of the current isothermal amplification techniques: applications, advantages and disadvantages. J Global Infect Dis. 2011;3:292–293. doi: 10.4103/0974-777X.83538. [DOI] [Google Scholar]
- 132.Dong X., Liu L., Tu Y., Zhang J., Miao G., Zhang L., Ge S., Xia N., Yu D., Qiu X. Rapid PCR powered by microfluidics: a quick review under the background of COVID-19 pandemic. Trends Anal Chem. 2021;143:116377. doi: 10.1016/j.trac.2021.116377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ding S., Chen G., Wei Y., Dong J., Du F., Cui X., Huang X., Tang Z. Sequence-specific and multiplex detection of COVID-19 virus (SARS-CoV-2) using proofreading enzyme-mediated probe cleavage coupled with isothermal amplification. Biosens Bioelectron. 2021;178:113041. doi: 10.1016/j.bios.2021.113041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Song Q., Sun X., Dai Z., Gao Y., Gong X., Zhou B., Wu J., Wen W. Point-of-care testing detection methods for COVID-19. Lab Chip. 2021;21:1634–1660. doi: 10.1039/D0LC01156H. [DOI] [PubMed] [Google Scholar]
- 135.Shen K., Sabbavarapu N., Fu C., Jan J., Wang J., Hung S., Lee G. An integrated microfluidic system for rapid detection and multiple subtyping of influenza A viruses by using glycan-coated magnetic beads and RT-PCR. Lab Chip. 2019;19:1277–1286. doi: 10.1039/C8LC01369A. [DOI] [PubMed] [Google Scholar]
- 136.Sakai J., Tarumoto N., Orihara Y., Kawamura R., Kodana M., Matsuzaki N., Matsumura R., Okane K., Kawamura T., Takeuchi S. Evaluation of a high-speed but low-throughput RT-qPCR system for SARS-CoV-2 detection. J Hosp Infect. 2020;105 doi: 10.1016/j.jhin.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oliveira K.G., Estrela P.F.N., Mendes G.D., dos Santos C.A., Silveira-Lacerda E.D., Duarte G.R.M. Rapid molecular diagnostics of COVID-19 by RT-LAMP in a centrifugal polystyrene-toner based microdevice with end-point visual detection. Analyst. 2021;146 doi: 10.1039/D0AN02066D. [DOI] [PubMed] [Google Scholar]
- 138.Sivakumar R., Dinh V.P., Lee N.Y. Ultraviolet-induced in situ gold nanoparticles for point-of-care testing of infectious diseases in loop-mediated isothermal amplification. Lab Chip. 2021;21:700–709. doi: 10.1039/D1LC00019E. [DOI] [PubMed] [Google Scholar]
- 139.Choi G., Jung J., Park B., Oh S., Seo J., Choi J., Seo T. A centrifugal direct recombinase polymerase amplification (direct-RPA) microdevice for multiplex and real-time identification of food poisoning bacteria. Lab Chip. 2016;16:2309–2316. doi: 10.1039/c6lc00329j. [DOI] [PubMed] [Google Scholar]
- 140.Loo J., Kwok H., Leung C., Wu S., Law I., Cheung Y., Cheung Y., Chin M., Kwan P., Hui M. Sample-to-answer on molecular diagnosis of bacterial infection using integrated lab-on-a-disc. Biosens Bioelectron. 2017;93:212–219. doi: 10.1016/j.bios.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 141.Wang S., Liu N., Zheng L., Cai G., Lin J. A lab-on-chip device for the sample-in-result-out detection of viable Salmonella using loop-mediated isothermal amplification and real-time turbidity monitoring. Lab Chip. 2020;20:2296–2305. doi: 10.1039/D0LC00290A. [DOI] [PubMed] [Google Scholar]
- 142.Yin J., Zou Z., Hu Z., Zhang S., Zhang F., Wang B., Lv S., Mu Y. A “sample-in-multiplex-digital-answer-out” chip for fast detection of pathogens. Lab Chip. 2020;20:979–986. doi: 10.1039/C9LC01143A. [DOI] [PubMed] [Google Scholar]
- 143.Xu G., Yang Z., Reboud J., Kasprzyk-Hordern B., Cooper J.M. Monitoring genetic population biomarkers for wastewater-based epidemiology. Anal Chem. 2017;89 doi: 10.1021/acs.analchem.7b02257. [DOI] [PubMed] [Google Scholar]
- 144.Sun F., Ganguli A., Nguyen J., Brisbin R., Shanmugam K., Hirschberg D.L., Wheeler M.B., Bashir R., Nash D., Cunningham B. Smartphone-based multiplex 30-minute nucleic acid test of live virus from nasal swab extract. Lab Chip. 2020;20:1621–1627. doi: 10.1039/D0LC00304B. [DOI] [PubMed] [Google Scholar]
- 145.Wang Y., Li K., Xu G., Chen C., Song G., Dong Z., Lin L., Wang Y., Xu Z., Yu M., Yu X., Ying B., Fan Y., Chang L., Geng J. Low-cost and scalable platform with multiplexed microwell Array biochip for rapid diagnosis of COVID-19. Research. 2020:1813643. doi: 10.34133/2021/2813643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang Z., Xu G., Reboud J., Ali S.A., Kaur G., McGiven J., Boby N., Gupta P.K., Chaudhuri P., Cooper J.M. Rapid veterinary diagnosis of bovine reproductive infectious diseases from semen using paper-origami DNA microfluidics. ACS Sens. 2018;3:403–409. doi: 10.1021/acssensors.7b00825. [DOI] [PubMed] [Google Scholar]
- 147.Xu G., Nolder D., Reboud J., Oguike M.C., van Schalkwyk D.A., Sutherland C.J., Cooper J.M. Paper-origami-based multiplexed malaria diagnostics from whole blood. Angew Chem Int Ed. 2016;55:15250–15253. doi: 10.1002/anie.201606060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tang R., Yang H., Gong Y., You M., Liu Z., Choi J.R., Wen T., Qu Z., Mei Q., Xu F. A fully disposable and integrated paper-based device for nucleic acid extraction, amplification and detection. Lab Chip. 2017;17:1270–1279. doi: 10.1039/c6lc01586g. [DOI] [PubMed] [Google Scholar]
- 149.Hassan F., Hays L., Bonner A., Bradford B., Franklin R., Hendry P., Kaminetsky J., Vaughn M., Cieslak K., Moffatt E. Multicenter clinical evaluation of the Alere i respiratory syncytial virus isothermal nucleic acid amplification assay. J Clin Microbiol. 2018;56 doi: 10.1128/JCM.01777-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Krause E., Puyskens A., Bourquain D., Brinkmann A., Biere B., Schaade L., Michel J., Nitsche A. Sensitive on-site detection of SARS-CoV-2 by ID NOW COVID-19. Mol Cell Probes. 2021;58:101742. doi: 10.1101/2021.04.18.21255688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Djaileb A., Jodaylami M.H., Coutu J., Ricard P., Lamarre M., Rochet L., Cellier-Goetghebeur S., Macaulay D., Charron B., Lavallee E., Thibault V., Stevenson K., Forest S., Live L.S., Abonnenc N., Guedon A., Quessy P., Lemay J.F., Farnos O., Kamen A., Stuible M., Gervais C., Durocher Y., Cholette F., Mesa C., Kim J., Cayer M.P., De Grandmont M.J., Brouard D., Trottier S., Boudreau D., Pelletier J.N., Masson J.F. Cross-validation of ELISA and a portable surface plasmon resonance instrument for IgG antibody serology with SARS-CoV-2 positive individuals. Analyst. 2021;146:4905–4917. doi: 10.1039/D1AN00893E. [DOI] [PubMed] [Google Scholar]
- 152.Wang C.H., Wang C., Qiu J.Y., Gao J.W., Liu H., Zhang Y., Han L. Ultrasensitive, high-throughput, and rapid simultaneous detection of SARS-CoV-2 antigens and IgG/IgM antibodies within 10 min through an immunoassay biochip. Microchim Acta. 2021;188 doi: 10.1007/s00604-021-04896-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xu W.J., Liu J.Y., Song D., Li C.S., Zhu A.N., Long F. Rapid, label-free, and sensitive point-of-care testing of anti-SARS-CoV-2 IgM/IgG using all-fiber Fresnel reflection microfluidic biosensor. Microchim Acta. 2021;188 doi: 10.1007/s00604-021-04911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Choe J., Kim J., Kwon H.H., Hong H., Jung C., Jeon C., Park E., Kim S. Diagnostic performance of immunochromatography assay for rapid detection of IgM and IgG in coronavirus disease 2019. J Med Virol. 2020 doi: 10.1002/jmv.26060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang Z., Zheng Z., Hu H., Zhou Q., Liu W., Li X., Liu Z., Wang Y., Ma Y. A point-of-care selenium nanoparticle-based test for the combined detection of anti-SARS-CoV-2 IgM and IgG in human serum and blood. Lab Chip. 2020;20:4255–4261. doi: 10.1039/D0LC00828A. [DOI] [PubMed] [Google Scholar]
- 156.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 157.Ghosh S., Aggarwal K., Nguyen T., Han J., Ahn C. A new microchannel capillary flow assay (MCFA) platform with lyophilized chemiluminescence reagents for a smartphone-based POCT detecting malaria. Microsystems Nanoeng. 2020;6:5. doi: 10.1038/s41378-019-0108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Xue Q., Kan X., Pan Z., Li Z., Pan W., Zhou F., Duan X. An intelligent face mask integrated with high density conductive nanowire array for directly exhaled coronavirus aerosols screening. Biosens Bioelectron. 2021;3:113286. doi: 10.1016/j.bios.2021.113286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nguyen P., Soenksen L., Donghia N., Angenent-Mari N., Puig H., Huang A., Lee R., Slomovic S., Galbersanini T., Lansberry G., Sallum H., Zhao E., Niemi J., Collins J. Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nat Biotechnol. 2021;39:1366–1374. doi: 10.1038/s41587-021-00950-3. [DOI] [PubMed] [Google Scholar]
- 160.Song Z., Ma Y., Chen M., Ambrosi A., Ding C., Luo X. Electrochemical biosensor with enhanced antifouling capability for COVID-19 nucleic acid detection in complex biological media. Anal Chem. 2021;93:5963–5971. doi: 10.1021/acs.analchem.1c00724. [DOI] [PubMed] [Google Scholar]
- 161.Guo K., Wustoni S., Koklu A., Diaz-Galicia E., Moser M., Hama A., Alqahtani A., Ahmad A., Alhamlan F., Shuaib M., Pain A., McCulloch I., Arold S., Grunberg R., Inal S. Rapid single-molecule detection of COVID-19 and MERS antigens via nanobody-functionalized organic electrochemical transistors. Nat Biomed Eng. 2021;5:666–677. doi: 10.1038/s41551-021-00734-9. [DOI] [PubMed] [Google Scholar]
- 162.Liu H., Yang A., Song J., Wang N., Lam P., Li Y., Law H., Yan F. Ultrafast, sensitive, and portable detection of COVID-19 IgG using flexible organic electrochemical transistors. Sci Adv. 2021;7 doi: 10.1126/sciadv.abg8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Alafee M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2021;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yakoh A., Pimpitak U., Rengpipat S., Hriankarn N., Chailapakul P., CHaiyo S. Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens Bioelectron. 2021;176:112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sampad M., Zhang H., Yuzvinsky T., Stott M., Hawkins A., Schmidt H. Optical trapping assisted label-free and amplification-free detection of SARS-CoV-2 RNAs with an optofluidic nanopore sensor. Biosens Bioelectron. 2021;194:113588. doi: 10.1016/j.bios.2021.113588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Michael I., Kim D., Gulenko O., Kumar S., Kumar S., Clara J., Ki D.Y., Park J., Jeong H.Y., Kim T.S. A fidget spinner for the point-of-care diagnosis of urinary tract infection. Nat Biomed Eng. 2020:1–10. doi: 10.1038/s41551-020-0557-2. [DOI] [PubMed] [Google Scholar]
- 167.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C., Jun S., Park D., Kim H., Kim S., Lee J., Kim B., Park C., Kim S. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c06726. [DOI] [PubMed] [Google Scholar]
- 168.Yu X., Xia Y., Tang Y., Zhang W., Yeh Y., Lu H., Zheng S. A nanostructured microfluidic immunoassay platform for highly sensitive infectious pathogen detection. Small. 2017;13:1700425. doi: 10.1002/smll.201700425. [DOI] [PMC free article] [PubMed] [Google Scholar]