Abstract
Acute lymphocytic leukemia is the most common type of cancer in pediatric individuals. Glucose regulated protein (GRP78) is an endoplasmic reticulum chaperone that facilitates the folding and assembly of proteins and regulates the unfolded protein response pathway. GRP78 has a role in survival of cancer and metastasis and cell-surface associated GRP78 (sGRP78) is expressed on cancer cells but not in normal cells. Here, we explored the presence of sGRP78 in pediatric B-ALL at diagnosis and investigated the correlation with bona fide markers of leukemia. By using a combination of flow cytometry and high multidimensional analysis, we found a distinctive cluster containing high levels of sGRP78, CD10, CD19, and CXCR4 in bone marrow samples obtained from High-risk leukemia patients, which was absent in the compartment of Standard-risk leukemia. We confirmed that sGRP78+CXCR4+ blood-derived cells were more frequent in High-risk leukemia patients. Finally, we analyzed the dissemination capacity of sGRP78 leukemia cells in a model of xenotransplantation. sGRP78+ cells emigrated to the bone marrow and lymph nodes, maintaining the expression of CXCR4. Testing the presence of sGRP78 and CXCR4 together with conventional markers may help to achieve a better categorization of High and Standard-risk pediatric leukemia at diagnosis.
Subject terms: Biomarkers, Oncology
Introduction
The 78-kDa immunoglobulin heavy chain binding protein BIP/HSPA5, also known as glucose regulated protein (GRP78) is an endoplasmic reticulum (ER) resident molecular chaperone that belongs to the Hsp70 family of heat shock proteins and it is a component of the unfolded protein response (UPR) pathway that plays an important role in ER homeostasis1–3. In normal conditions, GRP78 interacts with PKR-like ER kinase (PERK), inositol-requiring enzyme 1 (IRE1), and activating transcription factor 6 (ATF6). On accumulation of defective proteins in ER, GRP78 dissociates from these sensors and releases them to initiate the UPR cascade4.
The activation of the UPR cascade seems to be important for integrity maintenance of hematopoietic stem cells (HSCs) by preventing the propagation of damaged cells, reducing malignancy risk5. However, UPR cascade also represents a selective advantage to preleukemic stem cells, promoting their clonal expansion6.
As a master regulator of ER stress, GRP78 has been proposed to be important for stem cell survival, since GRP78 deficient mice showed enhanced cell death leading to a loss of the HSC compartment7.
Cell surface localization of GRP78 (sGRP78) was first reported in 1997 and recent studies have shown that GRP78 can be translocated to different cell compartments8. Importantly, sGRP78 works as a multifunctional receptor at the cell surface on tumor cells, where it triggers different cellular responses associated to proliferation, invasion, apoptosis, inflammation, and immunity8–12. Furthermore, sGRP78 has been observed in a variety of tumors including glioblastoma, prostate, breast, lung, gastric, colon and leukemia13–18.
GRP78 haploinsufficiency potently suppresses leukemogenesis and AKT/mTOR signaling in PTEN null bone marrow cells and the GRP78 level expression may alter the sensitivity of human leukemic cells to Arabinoside-induced apoptosis. Furthermore, the emerging association of elevated GRP78 expression in leukemic blasts of adult patients suggests that GRP78 may be important in the field of leukemia research15, 19–21. However, information about the presence of sGRP78 in childhood leukemia is scarce. Therefore, we aimed to evaluate sGRP78 in pediatric patients with B-lineage acute lymphoblastic leukemia (B-ALL) at diagnosis and investigate its association with stem cell markers (CD34 and CD38), lineage markers (CD10 and CD19). Also, sGRP78 expression was compared with CXCR4 expression, a well-known biomarker associated with extramedullary organ infiltration in childhood leukemia22.
Materials and methods
Human samples
Patients newly diagnosed with B-ALL were recruited from the Hospital Infantil de México Federico Gómez and Hospital Pediátrico de Moctezuma in México City. Bone marrow (BM) samples were collected according to International and Institutional Guidelines from children and adolescents younger than 18 years and diagnosed with ALL before any treatment. Control samples were obtained from children undergoing orthopedic surgery without diagnosis of oncology or hematology diseases or solid tumors, according with international and institutional guidelines. National Cancer Institute (NCI)-Rome guidelines were used for risk classification. High: WBC ≥ 50 × 109/L or ≥ 10 years; standard: WBC < 50 × 109/L and age 1 < 10 years.
A total of 43 newly diagnosed patients with B-ALL and 13 controls were used in this study (Supplementary Fig. 1). 43 B-ALL samples from BM and 4 BM controls were used for multiparametric flow cytometry. 9 blood control samples and 26 B-ALL blood samples from newly diagnosed patients with B-ALL were also analyzed. (Supplementary Fig. 1)
5 Standard-risk bone marrow B-ALL samples and 6 High-risk samples (from the described 43 samples) were used for flow cytometry and FlowSOM clustering analysis. Finally, 3 High risk B-ALL samples of bone marrow were used in xenotransplant assays (from the described 43 samples).
Ethics statement
Only remnant samples were collected, and all protocols and investigations of our study were approved by the ethics committees of Hospital Infantil de México Federico Gómez (HIM-2016-023) and Hospital Pediátrico de Moctezuma in México City (2016-01-3385). Patients and their parents signed the informed consent.
The study was conducted in accordance to Declaration of Helsinki.
Cell lines and culture conditions
SUP-B15, RS4;11, and Reh ALL cell lines were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA). The cells were cultured according to ATCC guidelines and maintained in 5% CO2 incubator at 37 °C. Freshly harvested cells were counted and resuspended in PBS with 2% FBS to follow the flow cytometry staining.
Isolation of mononuclear cells
Mononuclear cells (MNC) were isolated from BM and peripheral blood by using Ficoll-High Paque separation media. Briefly, a centrifuge tube with a volume of 1 to 2 mL of Ficoll-High Paque separation media was used to carefully place a mixture of BM or blood with PBS1x (1:1 volume ratio) and then centrifuged at 800×g for 30 min, at RT with the brake off. Harvested cells were washed with 5 mL of PBS1x twice. Cell viability was evaluated using trypan blue dye exclusion test.
Multiparameter flow cytometry
1–2 × 106 cells were placed in 200 uL of PBS1X with 2% FBS on ice and antibodies were added, mixed, and incubated for 30 min. Then, cells were washed with PBS1X and centrifuged at 800×g for 5 min. Finally, samples were acquired in a CytoFLEX LX cytometer (Beckman Coulter). Analysis was performed using CytExpert software. The following anti human antibodies were used: AF488 anti-GRP78 (Clone: C38), AF700 anti-CD45 (Clone: HI30), APC anti-CD34 (Clone: 561), PE/Cy7 anti-CD38 (Clone: HB-7), PerCP/Cy5.5 anti-CD19 (Clone: HIN19), PE/Dazzle anti-CD10 (Clone: HI10a), Brilliant Violet 421 anti-CD184 (clone: 12G5) and the Zombie Aqua™ Fixable Viability Kit was used according to the manufacture (Biolegend).
FlowSOM clustering and tSNE representation
For the analysis, after Flow Cytometry acquisition, possible debris and doublets events were excluded, LiveCD45+ cells from each sample were gated and 40,000 events were exported using CytExpert2.0. Pre-processed and compensated FCS files were loaded and analyzed into R Studio with the packages flowCore, flowVix, flowSOM and CATALYST23, 24. tSNE is based on the arcsinh-transformed expression of Live CD45+ cells stained with the LLA lineage markers: CD38, CD19, CD10, CD34, CXCR4, and GRP78. Clustering was run with no PCA, 2000 cells were randomly selected from each sample and the represented dimensional reduction was made with tSNE.
Mice
Recipient BALB/c nude immunodeficient mice were obtained from Instituto Nacional de Nutrición Salvador Zubirán (INCMNSZ) and maintained in a specific pathogen-free environment throughout the experiments. All animal-related experimental protocols were approved by the animal research committee at Hospital Infantil de México Federico Gómez (HIM-2019-031). All methods are reported in accordance with ARRIVE guidelines. We confirm that all methods were performed in accordance with the relevant guidelines and regulations.
Xenotransplant assay
To evaluate the dissemination capacity of sGRP78 cells, 250,000 to 500,000 sGRP78 sorted cells using FACS Aria IIu, were embedded in 100 uL of RPMI and 100 uL of Matrigel and injected subcutaneously into the right flank of each mouse (n = 3). Mice were examined daily and 3 weeks upon xenotransplantation, bone marrow and inguinal lymph node cells were recovered and analyzed by flow cytometry. Due to anti-GRP78 antibody cross-reacted against mouse cells, we selected human cells by using anti human CD45 antibody (HI30), then CD45+ human cells were gated for sGRP78, CD34 (581) and CXCR4 (12G5) detection. Both anti CD34 and anti CXCR4 recognized only human cells.
Results
High proportion of bone marrow lymphoblastic leukemia cells express sGRP78
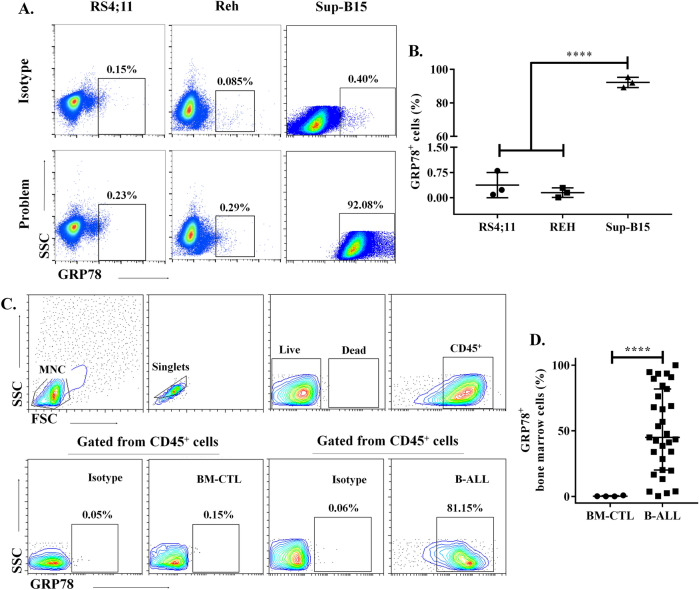
We assessed the expression of sGRP78 by flow cytometry in ALL cell lines (Fig. 1A). We found that around 90% of Sup-B15 cells, expressed sGRP78. Conversely, less than 1% of Reh and RS4;11 cell lines expressed sGRP78 (Fig. 1B). Thus, the expression of sGRP78 may be different according to the leukemic cell line tested. Then we decided to analyze sGRP78 expression in pediatric acute leukemia by flow cytometry analysis for GRP78 detection on pediatric bone marrow samples with B cell acute leukemia at diagnosis, classified in Standard and High-risk leukemia (Fig. 1C, Table 1). We observed that a high proportion of live single CD45+ leukemia cells expressed sGRP78, interestingly healthy cells barely expressed sGRP78 (Fig. 1D).
Figure 1.
sGRP78 expression on bone marrow cells of pediatric individuals with newly diagnosed B-ALL. (A) Acute lymphocytic leukemia cell lines were stained with anti-GRP78 antibody and representative experiments of each cell line are depicted. (B) Statistical representation of cell surface GRP78+ cells in leukemic cells lines is shown. Data are the mean plus SEM of n = 3. Statistical analysis was performed by One-way ANOVA, p < 0.0001. (C) Gating strategy for sGRP78+ cells analysis from bone marrow derived cells of pediatric individuals with B-ALL and controls (BM-CTL). (D) Statistical representation of cell surface GRP78+ cells in BM-CTL (n = 4) and B-ALL (n = 43). Statistical analysis was performed by two-tailed Mann–Whitney U test, *p < 0.05, **p < 0.01.
Table 1.
Patients and characteristics.
| ID | Age (years) | Sex | Leucocytes 103/uL | Blasts % | Translocations | Risk |
|---|---|---|---|---|---|---|
| 1 | 2 | F | 4.93 | 80 | – | Standard |
| 2 | 5 | F | 1.6 | 83 | – | Standard |
| 3 | 7 | F | 114 | 87 | – | High |
| 4 | 8 | F | 6.59 | 92 | – | Standard |
| 5 | 4 | M | 70.15 | 89 | – | High |
| 6 | 14 | M | 1.48 | 55 | – | High |
| 7 | 12 | M | 1.059 | 100 | – | High |
| 8 | 6 | F | 3.3 | 83 | – | Standard |
| 9 | 5 | M | 155 | 97 | – | High |
| 10 | 12 | F | 3.87 | 80 | – | High |
| 11 | 6 | M | 1.55 | 60 | – | Standard |
| 12 | 5 | M | 35 | 97 | Neg | Standard |
| 13 | 6 | M | 17.5 | 84 | – | Standard |
| 14 | 2 | F | 2.1 | 81 | – | Standard |
| 15 | 15 | M | 61.2 | 92 | – | High |
| 16 | 2 | F | 79.88 | 97 | – | High |
| 17 | 3 | F | 1.520 | 60 | – | Standard |
| 18 | 10 | M | 269 | 100 | – | High |
| 19 | 4 | F | 73 | 98 | t(4;11) | High |
| 20 | 1a | F | 3.9 | 85 | t(4;11) | Standard |
| 21 | 7 | M | 15 | 60 | Neg | Standard |
| 22 | 6 | M | 540.4 | 77 | – | High |
| 23 | 5 | F | 37.39 | 97 | – | Standard |
| 24 | 4 | M | 31 | 89.3 | – | Standard |
| 25 | 12 | M | 41 | 95 | Neg | High |
| 26 | 17 | F | 8 | 64 | – | High |
| 27 | 16 | F | 12.5 | 83 | – | High |
| 28 | 3 | F | 98 | 56 | t(1;19) | High |
| 29 | 1 | F | 29 | – | – | Standard |
| 30 | 7 | F | 25.1 | – | – | Standard |
| 31 | 15 | M | 287.7 | 94 | Neg | High |
| 32 | 5 | F | 71.4 | 98 | t(1;19) | High |
| 33 | 10 | F | – | – | Neg | High |
| 34 | 6 | F | 52.4 | 78 | – | High |
| 35 | 5 | F | 90.8 | 99.5 | Neg | High |
| 36 | 4 | M | 8.6 | 96.5 | – | Standard |
| 37 | 15 | F | 176 | 97 | t(12;21) | High |
| 38 | 4 | M | 25.7 | 90 | – | Habitual |
| 39 | 2 | M | 47.92 | 93 | – | Habitual |
| 40 | 12 | F | 185.2 | 83 | Neg | High |
| 41 | 13 | F | 36.2 | 96.5 | t(16;21) | High |
| 42 | 10 | F | 184.5 | – | – | High |
| 43 | 5 | M | 60.4 | 6 | – | High |
Risk stratification is shown.
sGRP78+ cells are associated with CD34, CD38, CD10 and CXCR4 expression in the high-risk group of B-ALL patients
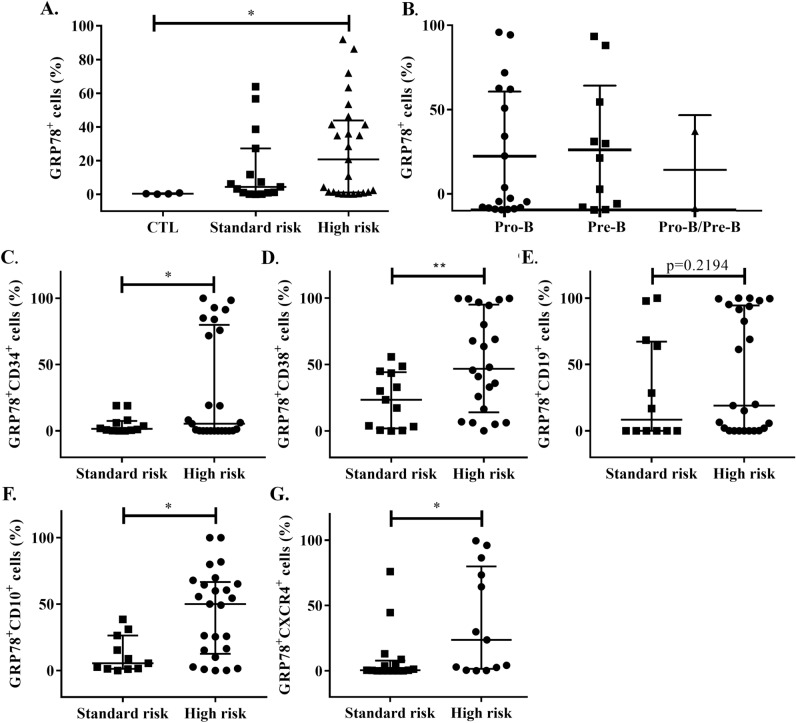
Since we observed a high proportion of sGRP78+ cells in bone marrow samples from children with newly diagnosed B acute lymphoblastic leukemia, we wanted to evaluate the association of sGRP78+ cells with other clinical criteria for leukemia classification. Analysis of the proportion of sGRP78+ cells according to the risk group of leukemia showed no differences between them (Fig. 2A), also we found no differences between the proportion of sGRP78+ cells with the stages of B cell development (ProB, PreB and ProB/PreB cells) (Fig. 2B). However, since initial analysis of leukemia cells by ImageStream showed coexpression of CD34 and sGRP78 in single cells derived from High and Standard-risk patients (Supplementary Fig. 2), we decided to analyze a set of classical surface antigens on leukemia cells. We observed a higher proportion of sGRP78+ cells coexpressing CD34, CD38 or CD10 in the High-risk group compared to the Standard-risk group, but we found no differences in the proportion of sGRP78+ expressing CD19 (Fig. 2C–F).
Figure 2.
Progenitor and migration markers are associated with GRP78+ cells in High-risk individuals with B-ALL. (A,B) Patients were classified by Standard risk and High risk and according to the stage of B-cell development markers, and the percentage of sGRP78+ cells was assessed. Statistical analysis was performed by two-tailed Mann–Whitney U test, *p < 0.01, **p < 0.05. (C–G) CD34, CD38, CD10, CD19 and CXCR4 were measured on GRP78+ cells by flow cytometry and the percentages are depicted for both high and standard-risk patients. Statistical analysis was performed by Mann–Whitney U test, *p < 0.01, **p < 0.05, ***p < 0.001.
sGRP78 has been associated with tumor initiating cells (TICs) in breast cancer which also express elevated levels of the chemokine receptor CXCR422, 25. Since B-ALL express CXCR4 and it has been associated to extramedullary organ infiltration correlating with unfavorable outcome26, thus, we decide to evaluate CXCR4 in sGRP78+ B-ALL samples. We observed that the percentage of sGRP78+CXCR4+ cells was associated with the High-risk group of B-ALL patients (Fig. 2G).
Interestingly, though the proportions of sGRP78+ cells coexpressing CD34, CD38, CD10 or CXCR4 were significantly higher in High-risk leukemia population, these samples showed the presence of a subgroup of double positive cells rather than a general increase of the positivity to the analyzed markers (Fig. 2C,D,F,G).
Differential distribution of sGRP78 B-ALL cells in standard vs high-risk B-ALL patients showed by high dimensional analysis
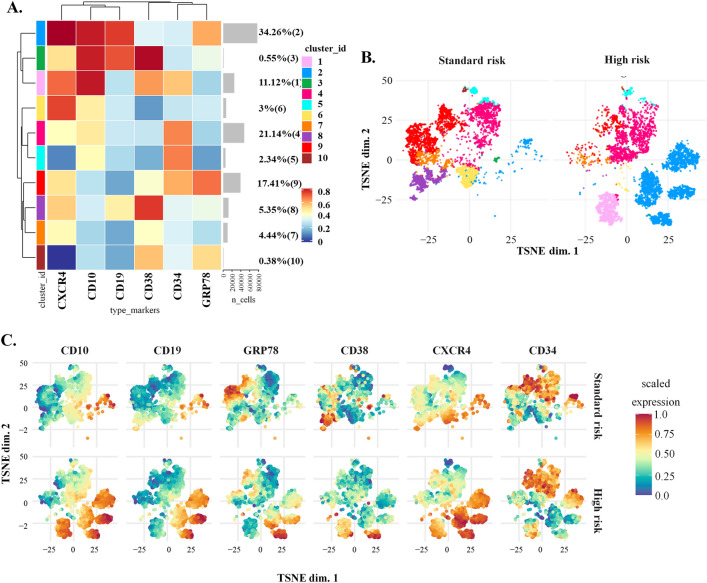
Since conventional flow cytometry evaluation showed significant differences between leukemia cells derived from High and Standard-risk patients when two surface markers were analyzed (sGRP78 in combination with CD34, CD38, CD10 or CXCR4). We wondered whether the analysis of the whole stained markers may highlight the presence of a particular sGRP78+ subset of leukemia cells. Therefore, we examined samples from five Standard-risk patients and six High-risk patients using the t-distributed stochastic neighbor embedding (tSNE). We generated a clustered organization of the samples in terms of the expression of CD34, CD38, CD10, CD19 and CXCR4. Ten clusters were generated and two of them showed the highest levels of sGRP78, cluster 2 and cluster 9, representing 34.26% and 17.41% of the whole analyzed cells respectively (Fig. 3A). Since cluster 2 was the most prominent of the 10 generated clusters, we analyzed the sample distribution and found that this cluster was associated with the High-risk group (Fig. 3B). In the other hand, cluster 9 was associated to Standard-risk samples (Fig. 3B). The distribution of the individual markers along the samples confirmed that sGRP78, CXCR4, CD10 and CD19 were enriched in the cluster 2 compared to cluster 9. In the other hand, CD34 and sGRP78 were enriched in the cluster 9, with low levels of CXCR4 (Fig. 3C). Moreover, three distinct clusters showed high levels of CD38 and low expression of CD34 (Clusters 3, 8 and 10), two of them expressed little or none sGRP78 (Cluster 3 and 8) and were only observed in the Standard risk group (Fig. 3C).
Figure 3.
Dimensional reduction of flow cytometry data from Standard and High-risk LLA patients. (A) Cluster organization and heatmap of the median marker intensities of the lineage markers and sGRP78. (B) TSNE based on the 10 clusters obtained by FlowSOM comparing LLA Standard-risk group (Standard-risk) against LLA High-risk group (High-risk). (C) TSNE showed the expression level of each marker separated by group risk. Heat maps were built by using Cytometry dATa anALYSis Tools (CATALYST). https://bioconductor.org/packages/3.10/bioc/html/CATALYST.html.
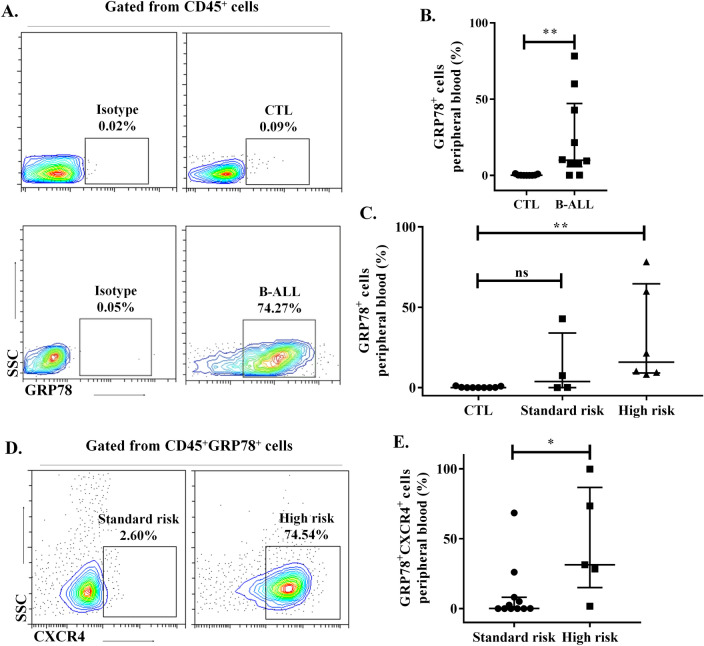
Circulating lymphoblastic leukemia cells express sGRP78 and CXCR4
Since leukemia cells have the capacity to emigrate from bone marrow to peripheral tissues, we wondered whether circulating leukemia cells also express sGRP78. We analyzed blood samples from pediatric patients with ALL at diagnostics and compared with samples from healthy donors and observed the presence of CD45+GRP78+ cells (Fig. 4A,B). Furthermore, cells from patients classified with High-risk leukemia had a higher proportion of sGRP78+ cells (Fig. 4C). Similarly, we also observed that High-risk leukemia patients had a higher proportion of sGRP78+ CXCR4+ cells (Fig. 4D,E). Thus, sGRP78+ leukemia cells can be detected in blood and together with the expression of CXCR4.
Figure 4.
sGRP78+ CXCR4+ cells are present in peripheral blood of pediatric individuals with newly diagnosed B-ALL. (A) Live cells from peripheral blood of B-ALL patients and controls (CTL) were stained with anti-CD45 and anti-GRP78. (B) Statistical representation of GRP78+ cells in B-ALL patients and controls. (C) Percentages of GRP78+ cells in high and standard risk are depicted. (D) Live cells from peripheral blood were stained with anti-CD45, anti-GRP78 and anti-CXCR4. The plots of a Standard risk patient (left) and High risk patient (right) are depicted. (E) Percentages of sGRP78+ CXCR4+ cells in high and standard risk in peripheral blood. Data are the medians plus ranks. Statistical analysis was performed by two-tailed Mann–Whitney U test, **p < 0.01, ***p < 0.001.
Analysis of sGRP78+ leukemia cells in a xenotransplantation model
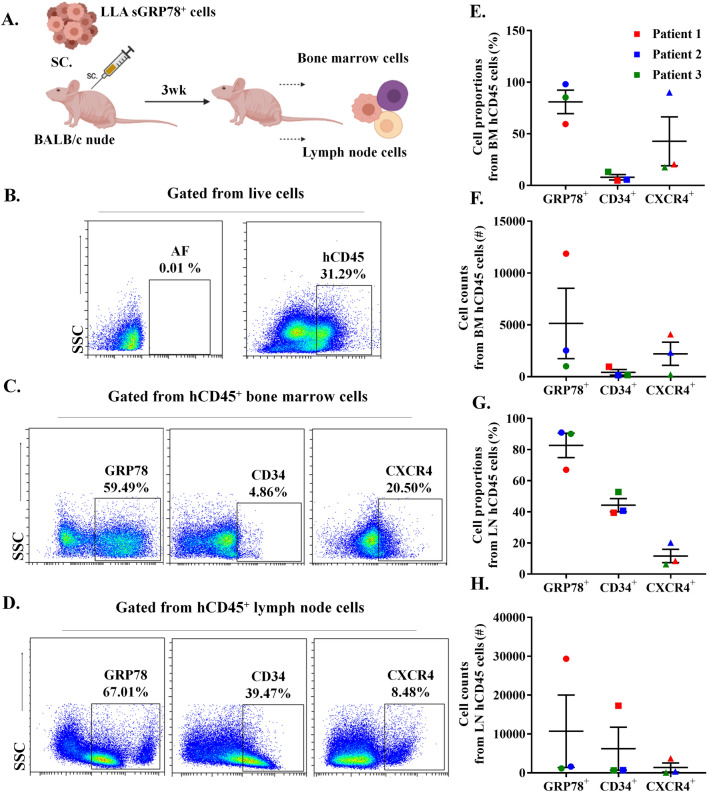
Since we observed the presence of sGRP78+CXCR4+ double positive cells in high-risk leukemia samples, both in bone marrow and peripheral blood, we wondered whether purified sGRP78+ cells could be ready to migrate and disseminate. Since whole leukemia cells were capable to infiltrate bone marrow and lymph nodes in nude mice (Supplementary Fig. 3); we subcutaneously injected purified High-risk patient-derived sGRP78+ leukemia cells (Fig. 5A) and, 3 weeks upon transplantation we detected living CD45+ human cells from bone marrow and lymph nodes expressing sGRP78, CXCR4 and CD34 (Fig. 5B–D). Around 80% of human recovered cells expressed sGRP78, both in bone in marrow and lymph node (Fig. 5E,G). Also, we recovered up to 30,000 human sGRP78+ cells in the best-case scenario (Fig. 5F,H). Compared to lymph nodes, the percentage of CXCR4+ cells were higher in bone marrow while the percentage of CD34+ cells were lower in this tissue (Fig. 5E,G). Our data suggest that sGRP78+ leukemia cells can migrate and lodge different tissues, and that some cells are able to maintain surface markers associated to the leukemic cell phenotype.
Figure 5.
Patient-derived sGRP78+ leukemic cells disseminate in a xenotransplantation model. (A) Schematic representation of the methodology used. (B–D) sGRP78+ B-ALL human bone marrow cells were sorted from high-risk B-LLA patients and injected into a BALB/c nude immunodeficient mice. Cells from murine bone marrow (BM) and lymph node (LN) were recovered 3 weeks later and were stained with anti-human CD45 (hCD45), anti-CD34 and anti-CXCR4, representative images are depicted. (E–H) Percentages and absolute numbers of sGRP78+, CD34+ and CXCR4+ cells selected from hCD45 fraction, both in BM (E,F) and LN (G,H) are depicted. AF autofluorescence.
Discussion
Recently it has been observed that GRP78 plays a role in survival and death processes in cancer cells1, 27. GRP78 is overexpressed in different types of leukemias20, 21, 28. Interestingly, it has been pointed out the relevance of GRP78 on the cellular surface as a hallmark of a stem-like population in cancer25. sGRP78 has been associated with relapse in myeloid leukemia suggesting that its presence plays a role in the maintenance of leukemia cells7. In solid tumor studies, sGRP78 helped to select breast and prostate cancer cells lines that are chemotherapy resistant29. However, it is unknown whether sGRP78 could be related to childhood leukemia at diagnosis and more importantly, if sGRP78 may be important as an indicator of High-risk leukemia. By using a multiparametric flow cytometry approach, we investigated the presence of sGRP78 in leukemic cell lines and in a set of samples derived from bone marrow of pediatric patients with B-LLA at diagnosis. We confirmed that sGRP78 is highly enriched in the Sup-B15 cell line as was observed in proteomic analysis30, interestingly just a small population of Reh and RS4;11 expressed sGRP78, suggesting that their molecular signatures may influence the translocation of GRP78 to the membrane cell surface. Since the Sup-B15 cell line was established from a patient with second relapse, we hypothesized that sGRP78 could be associated with High-risk leukemia.
We were able to determine that the majority of the analyzed BM samples contained a fraction with a high proportion of sGRP78+ cells, both in the Standard and High-risk groups, suggesting that leukemic cells relocate GRP78 to the cell surface independently of the leukemic risk group (when number of leucocytes and age are considered; see material and methods). This may reflect an actively functioning UPR response in leukemic cells, conferring aggressive properties associated to treatment resistance and relapse as has been observed in other types of cancer31–33. Furthermore, it may also indicate that sGRP78+ cells can be selected to survive once the treatment is initiated12, 34.
Our data suggested that the presence of sGRP78 may be intrinsically associated with early stages of leukemia. Additionally, GRP78 expression is linked to hematopoietic stem cells survival and proposed as a regulator of pluripotency and oncogenesis in other cell types7, 25. We decided to analyze the presence of markers for B-cell development such as CD34, CD38 and CD10 together with sGRP78. Interestingly, the fraction of sGRP78+ cells derived from High-risk patients co-expressed CD34, CD38 or CD10 in a significant manner compared to the fraction of sGRP78+ cells obtained from Standard-risk patients, suggesting that High-risk patients have higher proportions of potential primitive sGRP78+ leukemic cells that may represent a B-cell precursor acute lymphoblastic leukemia35.
Taking advantage of flow cytometry combined with high dimensional analysis, we observed a distinctive cluster of cells that are CD10+CD19+CXCR4+sGRP78+ (cluster 2) associated with pediatric High-risk leukemia at diagnosis. Also, a distinctive cluster (cluster 9) observed in Standard-risk group shared leukemic markers like CD34 and sGRP78, with low expression of CXCR4. Thus, for the first time we are showing that the presence of both sGRP78 and CXCR4 are associated with High-risk leukemia at diagnosis but not with Standard-risk leukemia.
Additionally, two clusters with elevated CD38 but low CD34 were observed in the Standard risk group (cluster 3 and 8), interestingly none of them showed marked expression of sGRP78. This finding supports previous data indicating that the subset CD38+CD34- is associated with better prognosis in B-LLA36.
Also, we explored whether ALL-sGRP78 cells were also located in circulation. We showed that blood cells from leukemia patients also expressed sGRP78. Moreover, the sGRP78+CXCR4+ cell subset was enriched in the High-risk group similar to the findings observed in bone marrow cells. Thus, the presence of sGRP78+CXCR4+ leukemic blood cells may indicate that this subset could seed peripheral tissues as has been observed in solid tumors37, 38. On the other hand, sGRP78+ cells were quite absent in the control group, similarly to the phenotype observed in bone marrow, suggesting that the presence of sGRP78 in circulating cells reflects the presence of a leukemic niche in bone marrow in leukemic patients.
Since GRP78 facilitates cancer cell invasion and seeding39, we analyzed whether sGRP78+ leukemia cells could disseminate in a mouse model of xenotransplantation. We found that subcutaneously injected sGRP78+ cells emigrated and survived for at least 3 weeks in the transplanted mice, as we were able to detect them in bone marrow and lymph nodes. Although the percentages and absolute numbers of recovered cells from xenografts were mixed, probably because of phenotype changes of the transplanted cells or altered migratory capabilities influenced by the CXCL12/CXCR4 axis40, 41; the majority of living human CD45+ cells maintained the expression of sGRP78 and a less proportion kept the expression of CD34 and CXCR4. Although, several mouse models have been used to analyze the dissemination, homing and tumor formation capability of leukemia cells42, our data also support that BALB/c nude immunodeficient mice could be used to study the dissemination process of acute leukemia via subcutaneous injection.
Taken together, we propose that sGRP78 is a surface marker likely associated with stem-like and migratory phenotype of leukemia cells from pediatric patients with B-ALL at diagnosis. Thus, the evaluation of sGRP78+CXCR4+ markers along with CD10 and CD19 in leukemic pediatric samples at diagnosis could contribute to a better categorization of High and Standard-risk pediatric leukemia. Whether the presence of sGRP78+CXCR4+ leukemic cells could lead to an increased risk of relapse, which is presumed to occur in patients with myeloid leukemia that express high levels of GRP78 transcripts7, 39 should be further evaluated. Finally, the presence of sGRP78 in cancer cells and its absence or low expression in normal cells, makes it a potential targetable marker. There is evidence showing that GRP78 confers multidrug resistance properties to cancer cells, thus elimination of sGRP78+ may improve treatments for childhood leukemia.
Supplementary Information
Acknowledgements
CONACYT Grants: Problemas Nacionales-3385 and Fosiss-261227. Fondos Federales Grants: HIM SSA 1372 and HIM SSA 1263 to RVR. CONACYT Grants to TAF: 309157 and GRT: 587971. Programa de Maestría y Doctorado en Ciencias Médicas y Odontológicas y de la Salud, Investigación Clínica Experimental en Salud. UNAM.
Author contributions
R.V.R. and T.A.F. conceived and designed the experiments. T.A.F., G.R.T., G.M.P., J.E. and G.Y. Performed the experiments. I.P.O., A.V.O., L.M.B., O.D., R.J., L.S.F. and B.A. Organized human samples. A.L., L.L.P., A.A.L. and R.V.R. supervised and validated data: R.V.R., T.A.F. and G.R.T. wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-05857-w.
References
- 1.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32(7):805–818. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee AS. GRP78 induction in cancer: Therapeutic and prognostic implications: Figure 1. Cancer Res. 2007;67(8):3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 3.Lee E, Nichols P, Spicer D, Groshen S, Yu MC, Lee AS. GRP78 as a novel predictor of responsiveness to chemotherapy in breast cancer. Cancer Res. 2006;66(16):7849–7853. doi: 10.1158/0008-5472.CAN-06-1660. [DOI] [PubMed] [Google Scholar]
- 4.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat. Rev. Immunol. 2008;8(9):663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 5.van Galen P, et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510(7504):268–272. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, et al. Adaptive endoplasmic reticulum stress signalling via IRE1α–XBP1 preserves self-renewal of haematopoietic and pre-leukaemic stem cells. Nat. Cell Biol. 2019;21(3):328–337. doi: 10.1038/s41556-019-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wey S, Luo B, Lee AS. Acute inducible ablation of GRP78 reveals its role in hematopoietic stem cell survival, lymphogenesis and regulation of stress signaling. PLoS ONE. 2012 doi: 10.1371/journal.pone.0039047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munro S, Pelham HRB. An hsp70-like protein in the ER: Identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46(2):291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- 9.Misra UK, Gonzalez-Gronow M, Gawdi G, Hart JP, Johnson CE, Pizzo SV. The role of Grp 78 in α2-macroglobulin-induced signal transduction: Evidence from RNA interference that the low density lipoprotein receptor-related protein is associated with, but not necessary for, GRP 78-mediated signal transduction. J. Biol. Chem. 2002;277(44):42082–42087. doi: 10.1074/jbc.M206174200. [DOI] [PubMed] [Google Scholar]
- 10.Ni M, Zhang Y, Lee AS. Beyond the endoplasmic reticulum: Atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem. J. 2011;434(2):181–188. doi: 10.1042/BJ20101569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Ni M, Lee B, Barron E, Hinton DR, Lee AS. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 2008;15(9):1460–1471. doi: 10.1038/cdd.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai YL, Zhang Y, Tseng CC, Stanciauskas R, Pinaud F, Lee AS. Characterization and mechanism of stress-induced translocation of 78-kilodalton glucose-regulated protein (GRP78) to the cell surface. J. Biol. Chem. 2015;290(13):8049–8064. doi: 10.1074/jbc.M114.618736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee HK, et al. GRP78 is overexpressed in glioblastomas and regulates glioma cell growth and apoptosis. Neuro Oncol. 2008;10(3):236–243. doi: 10.1215/15228517-2008-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun C, Rosendahl AH, Ansari D, Andersson R. Proteome-based biomarkers in pancreatic cancer. World J. Gastroenterol. 2011;17(44):4845–4852. doi: 10.3748/wjg.v17.i44.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staquicini DI, et al. Therapeutic targeting of membrane-associated GRP78 in leukemia and lymphoma: Preclinical efficacy in vitro and formal toxicity study of BMTP-78 in rodents and primates. Pharmacogenomics J. 2019;18(3):436–443. doi: 10.1038/tpj.2017.46.Therapeutic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Ridder G, Ray R, Misra UK, Pizzo SV. Modulation of the Unfolded Protein Response by GRP78 in Prostate Cancer. 1. Elsevier Inc.; 2011. [DOI] [PubMed] [Google Scholar]
- 17.Yao X, et al. Cell surface GRP78 accelerated breast cancer cell proliferation and migration by activating STAT3. PLoS ONE. 2015;10(5):1–17. doi: 10.1371/journal.pone.0125634. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Uramoto H, et al. Expression of endoplasmic reticulum molecular chaperone Grp78 in human lung cancer and its clinical significance. Lung Cancer. 2005;49(1):55–62. doi: 10.1016/j.lungcan.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Wey S, et al. Inducible knockout of GRP78/BiP in the hematopoietic system suppresses Pten-null leukemogenesis and AKT oncogenic signaling. Blood. 2012;119(3):817–825. doi: 10.1182/blood-2011-06-357384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wróbel T, et al. Significance of GRP78 expression in acute myeloid leukemias. Cent. Eur. J. Med. 2014;9(2):204–209. doi: 10.2478/s11536-013-0200-7. [DOI] [Google Scholar]
- 21.Huergo-Zapico L, et al. Expression of ERp5 and GRP78 on the membrane of chronic lymphocytic leukemia cells: Association with soluble MICA shedding. Cancer Immunol. Immunother. 2012;61(8):1201–1210. doi: 10.1007/s00262-011-1195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crazzolara R, et al. High expression of the chemokine receptor CXCR4 predicts extramedullary organ infiltration in childhood acute lymphoblastic leukaemia. Br. J. Haematol. 2001;115(3):545–553. doi: 10.1046/j.1365-2141.2001.03164.x. [DOI] [PubMed] [Google Scholar]
- 23.Robinson MD, et al. CyTOF workflow: Differential discovery in high-throughput high-dimensional cytometry datasets. F1000Research. 2017;6:1–53. doi: 10.12688/f1000research.11622.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crowell HL, Zanotelli VRT, Chevrier S, Robinson MD. Catalyst: Cytometry dATa anALYSis tools. Bioconductor. 2020 doi: 10.18129/B9.bioc.CATALYST. [DOI] [Google Scholar]
- 25.Conner C, et al. Cell surface GRP78 promotes stemness in normal and neoplastic cells. Sci. Rep. 2020;10(1):1–11. doi: 10.1038/s41598-020-60269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Berk LCJ, et al. Disturbed CXCR4/CXCL12 axis in paediatric precursor B-cell acute lymphoblastic leukaemia. Br. J. Haematol. 2014;166(2):240–249. doi: 10.1111/bjh.12883. [DOI] [PubMed] [Google Scholar]
- 27.Park KW, et al. The endoplasmic reticulum chaperone GRP78/BiP modulates prion propagation in vitro and in vivo. Sci. Rep. 2017 doi: 10.1038/srep44723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steiner N, et al. Expression and release of glucose-regulated protein-78 (GRP78) in multiple myeloma. Oncotarget. 2017;8(34):56243–56254. doi: 10.18632/oncotarget.17353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng YZCC, Tsai YL, Fu X, Schiff R, Lee AS. Cancer cells resistant to therapy promote cell surface relocalization of GRP78 which complexes with PI3K and enhances PI(3,4,5)P3 production. PLoS ONE. 2013 doi: 10.1371/journal.pone.0080071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malta TM, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173(2):338–354. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan SS, et al. GRP78 up-regulation is associated with androgen receptor status, Hsp70-Hsp90 client proteins and castrate-resistant prostate cancer. J. Pathol. 2011;223(1):81–87. doi: 10.1002/path.2795. [DOI] [PubMed] [Google Scholar]
- 32.Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors. Role of ATP binding site in suppression of caspase-7 activation. J. Biol. Chem. 2003;278(23):20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Steiniger SCJ, Kim YS, Kaufmann GF, Felding-Habermann B, Janda KD. Mechanistic studies of a peptidic GRP78 ligand for cancer cell-specific drug delivery. Mol. Pharm. 2007;4(3):435–447. doi: 10.1021/mp060122j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roué G, et al. The Hsp90 inhibitor IPI-504 overcomes bortezomib resistance in mantle cell lymphoma in vitro and in vivo by down-regulation of the prosurvival ER chaperone BiP/Grp78. Blood. 2011;117(4):1270–1279. doi: 10.1182/blood-2010-04-278853. [DOI] [PubMed] [Google Scholar]
- 35.Good Z, et al. Single-cell developmental classification of B cell precursor acute lymphoblastic leukemia at diagnosis reveals predictors of relapse. Nat. Med. 2018;24(4):474–483. doi: 10.1038/nm.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Z, Wu D, Lin S, Li P. CD34 and CD38 are prognostic biomarkers for acute B lymphoblastic leukemia. Biomark. Res. 2016;4:80–83. doi: 10.1186/s40364-016-0080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller A, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 38.Prud’homme GJ. Cancer stem cells and novel targets for antitumor strategies. Curr. Pharm. Des. 2012 doi: 10.2174/138161212800626120. [DOI] [PubMed] [Google Scholar]
- 39.Chiu CC, et al. Grp78 as a therapeutic target for refractory head-neck cancer with CD24- CD44+ stemness phenotype. Cancer Gene Ther. 2013;20(11):606–615. doi: 10.1038/cgt.2013.64. [DOI] [PubMed] [Google Scholar]
- 40.Ninomiya M, et al. Homing, proliferation and survival sites of human leukemia cells in vivo in immunodeficient mice. Leukemia. 2007;21(1):136–142. doi: 10.1038/sj.leu.2404432. [DOI] [PubMed] [Google Scholar]
- 41.Tavor S, et al. CXCR4 regulates migration and development of human acute myelogenous leukemia stem cells in transplanted NOD/SCID mice. Cancer Res. 2004;64(8):2817–2824. doi: 10.1158/0008-5472.CAN-03-3693. [DOI] [PubMed] [Google Scholar]
- 42.Jacoby E, Chien CD, Fry TJ. Murine models of acute leukemia: Important tools in current pediatric leukemia research. Front. Oncol. 2014;4:1–14. doi: 10.3389/fonc.2014.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.