Abstract
The LightCycler system (two-step reverse transcription-PCR–fluorescent hybridization [LC RT-PCR–FH]) was used to quantify Candida albicans actin mRNA as a means of assessing its viability in a reconstituted skin model of cutaneous candidiasis following the application of an antimycotic. A 192-bp ACT exon fragment was ligated into the pCR2.1 plasmid vector, and dilutions of the cloned insert (pACT; 4.092 kb) were used as the standard reference template. The LC RT-PCR–FH system could detect 1 fg of pACT, equivalent to 2.2 copies of the plasmid. The ACT exon-based PCR primers and FH probes were C. albicans specific, and electrophoretic analysis of the LC RT-PCR–FH assay product showed a 174-bp band in agarose gel. The number of copies of C. albicans ACT mRNA per milligram of tissue decreased with increasing amounts of amorolfine applied to a C. albicans-infected skin model, showing a reduction in viability. Detection and quantification of ACT mRNA in tissue by the LC RT-PCR–FH assay corresponded with cultural isolation of C. albicans from samples. The ACT mRNA-targeted LC RT-PCR–FH assay represents a sensitive, specific, rapid, and quantitative means of assessing the viability of C. albicans in infected tissue. This method may also be useful in evaluating the therapeutic efficacies of antifungal drugs in the treatment of various forms of candidiasis and other fungal diseases.
Candida species are frequently found as saprophytes that colonize the surfaces of certain mucous membranes in human. The superficial and disseminated infections caused by them are largely opportunistic, and Candida albicans accounts for most of the etiologic species isolated from cutaneous candidiasis. Microscopic demonstration of yeast elements in potassium hydroxide preparations of infected tissue and cultural isolation of yeast from skin scrapings are used to confirm clinical diagnosis of cutaneous candidiasis. Rapid and highly sensitive diagnostic assays, based on PCR of DNA, have also been used to identify candidal pathogens in tissue (3, 4, 5, 7, 8).
The amplification of a targeted DNA sequence by PCR does not, however, indicate the viability of the source organism, important information for evaluating the efficacy of a particular therapy and thus for avoiding an unnecessary prolongation of therapy. For this reason, reverse transcription (RT)-PCR of mRNA molecules has been used to assess the viability of microorganisms (9, 11, 14). However, RT-PCR is a qualitative assay, and this places a limitation on the inferences that can be drawn from the result; differences in the quantities of target mRNAs cannot easily be ascertained from the intensities of amplification bands. A determination of quantitative differences is needed in an assessment of the potencies of different concentrations of an agent or the progressive efficacy of a particular treatment. Furthermore, with the template at low levels, bands are seldom visualized in gel, resulting in false-negative interpretations (14).
In this study, we estimated the viability of C. albicans in a reconstituted-skin model of cutaneous candidiasis by detection of C. albicans ACT mRNA using the LightCycler (LC) system (a two-step RT-PCR–fluorescent hybridization [FH] assay) (10). LC DNA-based identification of fungi has been shown to be highly sensitive and rapid (10), and we applied of this system to rapidly quantify C. albicans ACT mRNA. The ACT mRNA was specifically targeted because of the constitutive nature of the gene and the major role of actin in cell division (6, 15).
MATERIALS AND METHODS
Materials.
An isolate of C. albicans (serotype A; JUH 3181) was used. The isolate was obtained from the Juntendo University Hospital culture collection. Amorolfine powder was a gift from Kyorin Pharmaceutical Co. Ltd. (Tokyo, Japan).
Isolation of fungal total RNA.
A single C. albicans colony from a 24-h Sabouraud dextrose agar culture was inoculated into 5 ml of Sabouraud broth. The culture was incubated at 30°C with orbital shaking for 18 h. The yeast cells were harvested by centrifugation at 3,000 × g and 4°C for 5 min and washed three times in phosphate-buffered saline (PBS). The cells were homogenized by grinding them with a pellet mixer and then by passaging them several times through a 23-guage needle. Total RNA was extracted by the guanidine isothiocyanate-phenol method (2) using Isogen (Wako Junyaku Kogyo, Osaka, Japan) according to the manufacturer's instructions.
Design of PCR primers and fluorescent hybridization (FH) probes.
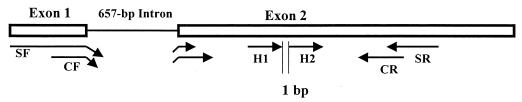
Figure 1 shows a schematic representation of the C. albicans ACT gene indicating the target areas of the primers and probes. The primers and probes were based on the published sequence of the actin genes of C. albicans (GenBank accession no. X16377). The uniqueness of the primers and probes for C. albicans ACT1 was determined using the BLAST database search program (1), courtesy of The Institute for Genomic Research (http://www.tigr.org.).
FIG. 1.
Schematic representation of the intron-containing actin gene (ACT) fragment of C. albicans showing the locations of the PCR primers and hybridization probes.
The PCR primers were 5′-ACGGTGAAGAAGTTGCTGCT-3′ (CF; bp 5 to 23) and 5′-GGGCTTCATCACCAACATAAG −3′ (CR; bp 158 to 178). The upstream primer spanned the exon-exon boundaries of ACT, thus excluding the amplification of genomic DNA. The FH probes, 5′-TAGACCAAGACATCAAGGTATCATGG-3′ (H1; bp 87 to 109) and 5′-AGCTGTTTTCCCATCTCTTGTTG-3′(H2; bp 111 to 136), were respectively labeled at the 5′ and 3′ ends with LC Red 640 fluorophore and fluorescein (Roche Diagnostics GmbH, Mannheim, Germany). The former was also modified at the 3′ end by phosphorylation to avoid extension during PCR. The probes anneal to adjacent internal sequences of the ACT, 1 bp apart, thus enabling fluorescent resonance energy transfer between the fluorophores (10).
Preparation of the pACT plasmid, a standard ACT mRNA quantification reference.
A 192-bp C. albicans ACT exon fragment containing the target ACT fragment was amplified by RT-PCR (RNA PCR kit; TaKaRa Shuzo Co. Ltd., Ohtsu, Japan) using 5′-ATGGACGGTGAAGAAGTTGC-3′ (SF; bp 1 to 20) and 5′-ACCTCTTTTGGATTGGGCTTCA-3′ (SR; bp 171 to 192) (Fig. 1). RT was performed with a 20-μl reaction mixture containing 1× RNA PCR buffer (10 mM Tris-HCl, pH 8.3); 50 mM KCl; 250 μM (each) dTTP, dUTP, dATP, and dGTP; 2.5 U of avian myeloblastosis virus reverse transcriptase, 5 mM MgCl2, 20 U of an RNase inhibitor, and a 0.125 μM concentration of the oligo(dT) adapter nucleotide primer. RT was performed in a DNA thermal cycler (model 480; Perkin-Elmer, Norwalk, Conn.) with one cycle of 30°C for 10 min, 50°C for 30 min, 99°C for 5 min, and 4°C for 5 min.
PCR was performed in the same tube, with the final 100-μl reaction mixture containing 1× RNA PCR buffer, 2.5 mM MgCl2, 2.5 U of Taq polymerase, and a 20 pM concentration of each of the primers SF and SR. PCR was performed in a DNA thermal cycler with 1 cycle of 5 min at 95°C; 30 cycles of 1 min at 95°C, 30 s at 62°C, and 30 s at 72°C; and a final extension at 72°C for 5 min. Amplification products were separated by electrophoresis through 1.5% (wt/vol) agarose gel containing 0.5 μg of ethidium bromide/ml. The band of the amplification product of the expected size was cut from the gel and purified using a MERmaid kit (Bio 101 Inc., Vista, Calif.) as instructed by the manufacturer. The amplicon was sequenced using the PCR primers and a dRhodamine terminator cycle sequencing kit (Perkin-Elmer) according to the manufacturer's instructions. Sequence analysis was performed on an ABI PRISM 310 genetic analyzer (Perkin-Elmer).
The 192-bp ACT exon fragment was ligated into the pCR 2.1 plasmid vector (3.9 kb) and transformed into One Shot cells (INVαF′) using a TA cloning kit (Invitrogen Corp., Carlsbad, Calif.) as instructed by the manufacturer. Dilutions of the plasmid containing the cloned insert (pACT; 4.092 kb) were then used as the standard reference templates for detection and quantification of ACT mRNA by the LC RT-PCR–FH assay.
Sensitivity of the LC RT-PCR–FH system.
The template, pACT, was diluted to contain 20 ng, 200 pg, 2 pg, 20 fg, 2 fg, or 1 fg of DNA/μl. The LC RT-PCR–FH assay was performed with a 20-μl reaction mixture in glass capillaries which contained 2 μl of the template, 4 mM MgCl2, a 0.5 μM concentration of each of the PCR primers CF and CR, a 0.2 μM concentration of each of the FH probes H1 and H2, and 1× LC DNA master hybridization mixture (ready-to-use reaction mix containing DNA polymerase). The cycle program data of LightCycler software (version 3; Roche Diagnostics GmbH) were set at 1 cycle of denaturation at 95°C for 2 min; 60 cycles of 95°C for 0 s (once this temperature is reached, it immediately falls to the annealing temperature), annealing at 62°C for 10 s, and extension at 72°C for 7 s; and 1 cycle of cooling at 40°C for 30 s. Other program data were set as instructed by the manufacturers. PCR amplification dynamics were monitored online.
The LC RT-PCR–FH product was recovered from the capillary tubes and separated by electrophoresis on a 1.5% (wt/vol) agarose gel.
Specificities of the PCR primers and FH probes in the LC system.
The total RNA extracts of C. albicans (ATCC 76615) and the following organisms were used as templates: Candida tropicalis (ATCC 750), Candida krusei (clinical isolate), Aspergillus fumigatus (ATCC 26430), Aspergillus flavus (IFO 7540), Aspergillus niger (IFO 31628), Asperigullus terreus (IFO 31675), Cryptococcus neoformans (TIMM 3173), Trichosporon beigelli (clinical isolate), Trichosporon mucoides (clinical isolate), Microsporum canis (IFM 47138), Epidermophyton floccosum (ATCC 50266), Trichophyton mentagrophytes (IFM 45795), Trichophyton rubrum (IFM 47168), Sporothrix schenckii (clinical isolate), Nocardia asteroides (clinical isolate), Staphylococcus epidermidis (ATCC 14970), and Streptococcus sanguis (ATCC 10556). Total RNA extract of human keratinocytes was also used as a template. RT was performed, and the product was assayed by the LC RT-PCR–FH method as described above.
Infection of a reconstituted human skin model with C. albicans.
The reconstituted skin (Testskin; Toyobo Co. Ltd., Tokyo, Japan) consisted of human dermal fibroblasts in a collagen lattice overlaid by stratified human keratinocytes. Six wells of the model were set up as instructed by the manufacturer. The inoculum, C. albicans, was cultured in Sabouraud broth and adjusted to a density of 2 × 103 cells per ml of PBS. Fifty microliters of the C. albicans suspension was inoculated onto the skin surface in each of five wells. Fifty microliters of PBS was applied to the sixth skin model as a negative infection control. The skin model was maintained in serum-free Testskin assay medium (Toyobo Co. Ltd.) at 37°C for 48 h in an atmosphere of 5% CO2–95% O2.
Five, 10, 20, or 40 μg of amorolfine, in a 50-μl PBS solution, was applied to the surfaces of four C. albicans-infected skin models at 48 and 96 h postinfection, and the tissue maintenance medium was replaced with a fresh medium containing 5 μg of amorolfine/ml. Fifty microliters of PBS was also applied to the positive- and negative-infection control skin models, and the tissue maintenance medium was drug free in both cases. At 6 days postinfection, a 6-mm-diameter portion of skin was excised with a punch and the total RNA content was extracted. The remaining portion was inoculated on Sabouraud agar.
Extraction of total RNA from the skin model and quantification of C. albicans ACT mRNA.
The skin fragment was weighed, minced, and suspended in 500 μl of 10 mM Tris-HCl buffer (pH 7.8) containing 1 mM CaCl2O·2H2O, 10 mM dithiothreitol, 20 U of an RNase inhibitor, and 500 μg of proteinase K. The mixture was incubated at 37°C for 45 min and then centrifuged for 3 min at 3,000 × g and 4°C. The precipitate was washed once in 1 ml of chelating buffer containing 10 mM Tris-HCl (pH 7.8) and 2 mM EGTA and then suspended in 1 ml of Isogen (Wako Junyako Kogyo Ltd.). Total RNA was extracted as described earlier.
RT was performed as described above, and a negative control, which contained water in place of reverse transcriptase, was included for each sample. The RT product and 106, 105, and 104 copies of pACT/μl were then used as templates in the LC RT-PCR–FH. Postamplification analysis was carried out with LC data analysis software (version 3.1.102; Roche Diagnostics GmbH), and the copy number of the template was determined. The concentration of C. albicans ACT mRNA, in copy number per milligram of skin, was then calculated using the following formula: (copy number of the ACT cDNA template in the test reaction mixture − background quantity in the control reaction mixture) × dilution factor of RT reaction mixture × volume of total RNA extract/mass of skin fragment (in milligrams).
The LC RT-PCR–FH products were recovered from the capillary tubes, electrophoresed on a 1.5% (wt/vol) agarose gel, and purified. Both strands of the purified product were sequenced as described above using the PCR primers.
RESULTS
Sensitivity of the LC RT-PCR–FH system.
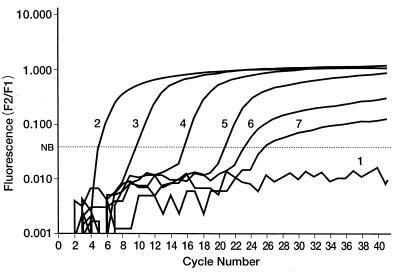
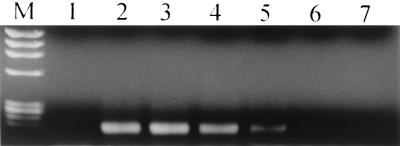
Amplification of different concentrations of pACT was monitored online by regular-log-fluorescence versus cycle number curves (Fig. 2). The irregular curve below the noise band cursor indicated nonamplification of the negative control (water). The system could detect 1 fg of pACT, and this number was equivalent to 2.2 copies of pACT. The calculation of the copy number was as described elsewhere (15): 1 mol of 4,092-bp pACT has a mass of 2.7 × 106 g, and 1 fg of pACT contains 3.7 × 10−24 mol. Multiplying this number by Avogadro's number, 6 × 1023, gives the stated copy number. Figure 3 shows the results of agarose gel electrophoresis of the amplification products from the pACT concentrations.
FIG. 2.
Sensitivity of detection and quantification of a serially diluted reference standard plasmid (pACT) by the LC system. Curves: 1, water (negative control); 2, 20 ng of DNA; 3, 200 pg of DNA/μl; 4, 2 pg of DNA/μl; 5, 20 fg of DNA/μl; 6, 2 fg of DNA/μl; 7, 1 fg of DNA/μl; NB denotes the noise band cursor.
FIG. 3.
Agarose gel electrophoresis of the serially diluted standard reference plasmid (pACT) from Fig. 2 showing a single specific band at 174 bp. Lanes: M, DNA molecular weight marker IX (Roche); 1, water (negative control); 2, 20 ng of DNA/μgl; 3, 200 pg of DNA/μl; 4, 2 pg of DNA/μl; 5, 20 fg of DNA/μl; 6, 2 fg of DNA/μl; 7, 1 fg of DNA/μl.
Specificity.
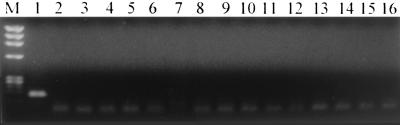
The ACT-based PCR primers and FH probes were specific for C. albicans as indicated by regular-log-fluorescence versus cycle number curves (not shown). The negative control, reference organisms, and human templates were not amplified, as is indicated by irregular curves below the noise band cursor. Electrophoresis of the LC RT-PCR–FH products showed a single band for the amplification product of the expected size of 174 bp from C. albicans; no band was observed in the reaction product of the reference organisms and human keratinocytes (Fig. 4).
FIG. 4.
Agarose gel electrophoresis images of LC-generated products of C. albicans and control organisms using C. albicans ACT-based PCR primers. A single C. albicans-specific band is shown at 174 bp. Lanes: M, DNA molecular weight marker IX (Roche); 1, C. albicans; 2, C. tropicalis; 3, C. krusei; 4, A. fumigatus; 5, A. flavus; 6, A. niger; 7, A. terreus; 8, Cryptococcus neoformans; 9, Trichosporon beigelli; 10, Trichophyton rubum; 11, Trichophyton mentagrophytes; 12, Sporothrix schenckii; 13, Staphylococcus epidermidis; 14, Streptococcus sanguis; 15, human skin; 16, water (negative control). Results for other control organisms indicated in the text are not shown.
Viability assessment of C. albicans in skin samples by quantification of the ACT mRNA. Table 1 shows a summary of quantification of C. albicans ACT mRNA in the infected-skin model by the two-step LC RT-PCR–FH system. The copy numbers of C. albicans ACT mRNA per milligram of tissue decreased with an increase in the amount of drug applied. The untreated control expectedly yielded the highest copies of ACT mRNA per milligram of skin. An amorolfine control, using water containing 10 or 100 μg of drug/ml in the RT assay mixture, showed that amorolfine had no effect on the calculated amount of ACT mRNA per milligram of skin of the untreated control (data not shown). No fluorescent signal was generated by the uninfected control. With the exception of what occurred with the skin model to which was applied 20 μg of amorolfine, the presence of C. albicans ACT in the skin model corresponded with isolation of the organism in Sabouraud dextrose agar cultures of tissue samples. Electrophoretic analysis of the LC RT-PCR–FH product from the ACT mRNA-positive samples showed an amplicon of the expected size of 174 bp in agarose gel. A nucleotide sequence analysis of the product showed complete identity with the targeted fragment of the C. albicans ACT exon.
TABLE 1.
Quantification of C. albicans ACT mRNA in total RNA extract of a reconstituted skin model of cutaneous candidiasis by the LC system
| Status of skin model | Amt of drug applied (μg) | No. of copies of C. albicans ACT mRNA/mg of skin |
|---|---|---|
| Noninfected negative control | 0 | 0 |
| Infected positive control | 0 | 2.7 × 107 |
| Infected and treated sample | 5 | 1.6 × 104 |
| 10 | 1.8 × 102 | |
| 20 | 2.5 × 10 | |
| 40 | 0 |
DISCUSSION
Routine qualitative RT-PCR assay has been used in assessing the viability of microorganisms by targeting the heat shock protein 70 mRNAs of Pneumocystis carinii (11), Giardia cysts, and Cryptosporidium oocysts (9) and the actin mRNA of C. albicans (14). In this study, we quantified C. albicans actin mRNA as a means of assessing the viability of C. albicans in a reconstituted-human-skin model of cutaneous candidiasis following the application of the antimycotic. Amorolfine was used because of its effectiveness in the topical therapy of superficial mycoses caused by yeasts and dermatophytes and its high affinity to stratum corneum and nails (12, 13).
The two-step LC RT-PCR–FH system was used to detect and quantify mRNA. The system combines rapid PCR-FH in glass capillaries with online fluorescence detection of the hybridized PCR amplicon (10). We further developed a method for calculating the number of copies of C. albicans ACT mRNA per milligram of the infected skin. The sense PCR primer was designed to span an intron splice site, thus ensuring the amplification of only the reverse transcripts of the ACT mRNA.
The LC system proved itself extremely sensitive in detecting 1 fg of the standard plasmid online, with a reproducibility of 100% among five replicates. In contrast, the LC PCR products of 2 or 1 fg of pACT did not show any visible band on an electrophoretic gel (Fig. 3). This result showed that detection of amplicons by means of FH in the LC system was more sensitive than visual observation of amplicon bands in gels. Thus, the LC system has sufficient sensitivity to detect ACT mRNA expressed from a single genome of C. albicans comparable to that reported for the 5′ nuclease TaqMan PCR assay (16). Although the extreme sensitivity of the system may not be a priority in cutaneous candidiasis, our model has conveniently demonstrated that this system is applicable to other mycoses provided that the appropriate primers and hybridization probes are used.
The PCR primers and FH probes, based on the ACT exon, were specific for C. albicans. Although the ACT sequence is highly conserved among eukaryotes and much more so among Candida species (17), the combination of PCR and hybridization at a high annealing temperature enhanced the specificities of the reagents in the LC system.
The quantity of ACT mRNA is related to the residual population of metabolically active yeasts, as evidenced by a decrease in signal per milligram of skin as the amount of the antimycotic agent applied to the infected skin model increased. However, the population of C. albicans in tissue cannot not be directly estimated from the number of copies of ACT mRNA since the quantity of mRNA in a cell, unlike that of the DNA, is varied and depends on the cellular metabolic state. The absence of C. albicans colonies in Sabouraud dextrose agar cultures of the ACT mRNA-positive skin model, which was treated with 20 μg of amorolfine, may be due to the inhibitory effect of residual drug in tissue.
In conclusion, quantification of ACT mRNA by the LC system represents a sensitive, specific, rapid, and quantitative means of assessing the viability of fungi in an infected tissue. The fact that the average assay time from sample preparation through postamplification analysis was less than 4 h, compared to 24 to 48 h for traditional Candida culture results, underscores the ACT-LC combination potential for rapid evaluation of the viability of fungi in tissue and assessment of the therapeutic efficacies of antifungal drugs in the treatment of fungal diseases.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkan A. Tissue-dependent plastid mRNA splicing in maize: transcripts from four plastid genes are predominantly unspliced in leaf meristems and roots. Plant Cell. 1989;1:437–445. doi: 10.1105/tpc.1.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donnelly S M, Sullivan D J, Shanley D B, Coleman D C. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1 intron and exon sequences. Microbiology. 1999;145:1871–1882. doi: 10.1099/13500872-145-8-1871. [DOI] [PubMed] [Google Scholar]
- 4.Einsele H, Herbert H, Roller G, Loeffler J, Rothenhofer I, Muller C A, Bowden R A, van Burik J-A, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flauhaut M, Sanglard D, Monod M, Bille J, Rossier M. Rapid detection of Candida albicans in clinical samples by DNA amplification of common regions from C. albicans-secreted aspartic proteinase genes. J Clin Microbiol. 1998;36:395–401. doi: 10.1128/jcm.36.2.395-401.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hennessey D, Drummond D R, Sparrow J C. Molecular genetics of actin function. Biochem J. 1993;282:657–671. doi: 10.1042/bj2910657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kan V L. Polymerase chain reaction for the diagnosis of candidemia. J Infect Dis. 1993;168:779–783. doi: 10.1093/infdis/168.3.779. [DOI] [PubMed] [Google Scholar]
- 8.Kappe R, Okeke C N, Fauser C, Maiwald M, Sonntag H-G. Molecular probes for the detection of pathogenic fungi in the presence of human tissue. J Med Microbiol. 1998;47:811–820. doi: 10.1099/00222615-47-9-811. [DOI] [PubMed] [Google Scholar]
- 9.Kaucner C, Stinear T. Sensitive and rapid detection of viable Giardia cysts and Cryptosporidium parvum oocysts in large-volume water samples with wound fiberglass cartridge filters and reverse transcription-PCR. Appl Environ Microbiol. 1998;64:1743–1749. doi: 10.1128/aem.64.5.1743-1749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeffler J, Henke N, Herbert H, Schmidt D, Hagemeyer L, Schumacher U, Einsele H. Quantification of fungal DNA by using fluorescence resonance energy transfer and the light cycler system. J Clin Microbiol. 2000;38:586–590. doi: 10.1128/jcm.38.2.586-590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maher N, Vermund S, Lasbury M, Lee C-H, Bartlett M, Unnasch T R. Development and evaluation of a molecular viability assay for Pneumocystis carinii. J Clin Microbiol. 2000;38:1947–1952. doi: 10.1128/jcm.38.5.1947-1952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melchinger W, Polak A, Müller J. The effects of amorolfine and oxiconazole on the ultrastructure of Trichophyton mentagrophytes. A comparison. Mycoses. 1988;33:393–404. doi: 10.1111/myc.1990.33.7-8.393. [DOI] [PubMed] [Google Scholar]
- 13.Okeke C N, Tsuboi R, Kawai M, Ogawa H. Fluorometric assessment of in vitro antidermatophytic activities of antimycotics based on their keratin-penetrating power. J Clin Microbiol. 2000;38:489–491. doi: 10.1128/jcm.38.2.489-491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okeke C N, Tsuboi R, Kawai M, Yamazaki M, Reangchainam S, Ogawa H. Reverse transcription-3′ rapid amplification of cDNA ends-nested PCR of ACT1 and SAP2 as a means of detecting viable Candida albicans in an in vitro cutaneous candidiasis model. J Investig Dermatol. 2000;114:95–100. doi: 10.1046/j.1523-1747.2000.00845.x. [DOI] [PubMed] [Google Scholar]
- 15.Reisler E. Actin molecular structures and functions. Curr Opin Cell Biol. 1993;5:41–47. doi: 10.1016/s0955-0674(05)80006-7. [DOI] [PubMed] [Google Scholar]
- 16.Sen K. Rapid identification of Yersinia enterocolitica in blood by the 5′ nuclease PCR assay. J Clin Microbiol. 2000;38:1953–1958. doi: 10.1128/jcm.38.5.1953-1958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welch M D, Holtzman D A, Drubin D G. The yeast actin cytoskeleton. Curr Opin Cell Biol. 1994;6:110–119. doi: 10.1016/0955-0674(94)90124-4. [DOI] [PubMed] [Google Scholar]