Abstract
Yes-associated protein (YAP) activation is crucial for tumor formation and development, and its stability is regulated by ubiquitination. ISGylation is a type of ubiquitination like post-translational modification, whereas whether YAP is ISGylated and how ISGylation influences YAP ubiquitination-related function remains uncovered. In addition, YAP can activate glucose metabolism by activating the hexosamine biosynthesis pathway (HBP) and glycolysis, and generate a large number of intermediates to promote tumor proliferation. However, whether YAP stimulates the pentose phosphate pathway (PPP), another tumor-promoting glucose metabolism pathway, and the relationship between this stimulation and ISGylation needs further investigation. Here, we found that YAP was ISGylated and this ISGylation inhibited YAP ubiquitination, proteasome degradation, interaction with-beta-transducin repeat containing E3 ubiquitin-protein ligase (βTrCP) to promote YAP stability. However, ISGylation-induced pro-YAP effects were abolished by YAP K497R (K, lysine; R, arginine) mutation, suggesting K497 could be the major YAP ISGylation site. In addition, YAP ISGylation promoted cell viability, cell-derived xenograft (CDX) and patient-derived xenograft (PDX) tumor formation. YAP ISGylation also increased downstream genes transcription, including one of the key enzymes of PPP, 6-phosphogluconolactonase (6PGL). Mechanistically, YAP promoted 6PGL transcription by simultaneously recruiting SMAD family member 2 (SMAD2) and TEA domain transcription factor 4 (TEAD4) binding to the 6PGL promoter to activate PPP. In clinical lung adenocarcinoma (LUAD) specimens, we found that YAP ISGylation degree was positively associated with 6PGL mRNA level, especially in high glucose LUAD tissues compared to low glucose LUAD tissues. Collectively, this study suggested that YAP ISGylation is critical for maintaining its stability and further activation of PPP. Targeting ISGylated YAP might be a new choice for hyperglycemia cancer treatment.
Subject terms: Cancer metabolism, Cancer microenvironment
Introduction
Yes-associated protein (YAP) is an effector downstream of the Hippo-YAP signaling pathway [1]. As an intracellular connexin and transcriptional co-activator, YAP regulates transcription and signal transduction of multiple genes by binding and activating transcription factors (i.e., TEADs, SMADs, RUNX family transcription factors (RUNXs), tumor protein 73) [2, 3]. YAP has been identified as an oncogene [4, 5]. The abnormal expression of YAP has been found in a variety of malignant tumors, and transgenic expression of YAP in mouse liver reversibly enlarged livers and eventually led to tumor formation suggesting that YAP has an important role in the occurrence and development of tumors [1, 6, 7]. As for LUAD, YAP is activated, promotes cell proliferation, and assists with tumor formation [8, 9]. High expression of YAP predicts poor prognosis in epidermal growth factor receptor (EGFR)-mutant non-small-cell lung cancer (NSCLC) patients [10]. YAP is also involved in metabolic regulation, such as promoting glucose metabolism, lipogenesis, and glutamine utilizing [11–13]. YAP promotes glucose metabolism by stimulating glycolysis and HBP to further stimulate tumorigenesis [14, 15]. These effects maintain YAP as a key oncogenic factor. However, whether YAP promotes another pro-cancer metabolism, such as another form of glucose metabolism PPP, remains uncovered.
Post-translational modification of protein is one of the main ways to regulate YAP function [16]. It has been found that YAP can undergo post-translational modifications such as phosphorylation, O-GlcNAcylation, ubiquitination, SUMOylation, methylation, acetylation, and so on [15, 17–20]. Under normal circumstances, YAP is phosphorylated by large tumor suppressor kinase (LATS) and binds with 14-3-3 protein in the cytoplasm [20]. During cytoplasmic retention, YAP can be further phosphorylated by casein kinase 1 (CK1), ubiquitinated by several E3 ligases, and further degraded in proteasome [21]. When the upstream Hippo signal regulation is abnormal, YAP is dephosphorylated, enters the nucleus, and acts as a transcriptional co-activator to promote tumorigenesis [15].
ISGylation is a ubiquitination-like post-translational modification that interferon-stimulated gene 15 (ISG15) protein covalently linked to the lysine residue of the target protein under the catalysis of E3 ligase [22]. A series of distinct enzymes are involved in the process of protein ISGylation, including ubiquitin-activating E1 enzyme (ubiquitin-activating enzyme E1-like protein, UBE1L), conjugating E2 enzyme (ubiquitin-conjugating human enzyme 8, UbCH8), and several types of ligating E3 ligases [23]. The influence between ISGylation and ubiquitination is very close. ISGylation may occur at the same K site with ubiquitination or close to the ubiquitinated K site, further affecting protein function by competing with ubiquitination [24]. In addition, following viral infection, neural precursor cell expressed developmentally downregulated protein 4 (NEDD4) is ISGylated and blocks the interaction with the ubiquitin-conjugating enzyme (E2), further inhibiting NEDD4-induced ubiquitination to promote anti-viral immune response [25]. Since the ubiquitination of YAP significantly reduces the stability of YAP [21, 26], whether YAP is ISGylated to compete with ubiquitination and the effect of YAP ISGylation on YAP function deserve further investigation.
Here, we found that YAP was ISGylated and its ISGylation increased its stability by inhibiting ubiquitination and interaction with βTrCP. Furthermore, YAP ISGylation was critical for LUAD malignant phenotypes and downstream PPP activation. Targeting YAP ISGylation might be beneficial for hyperglycemia LUAD treatment.
Results
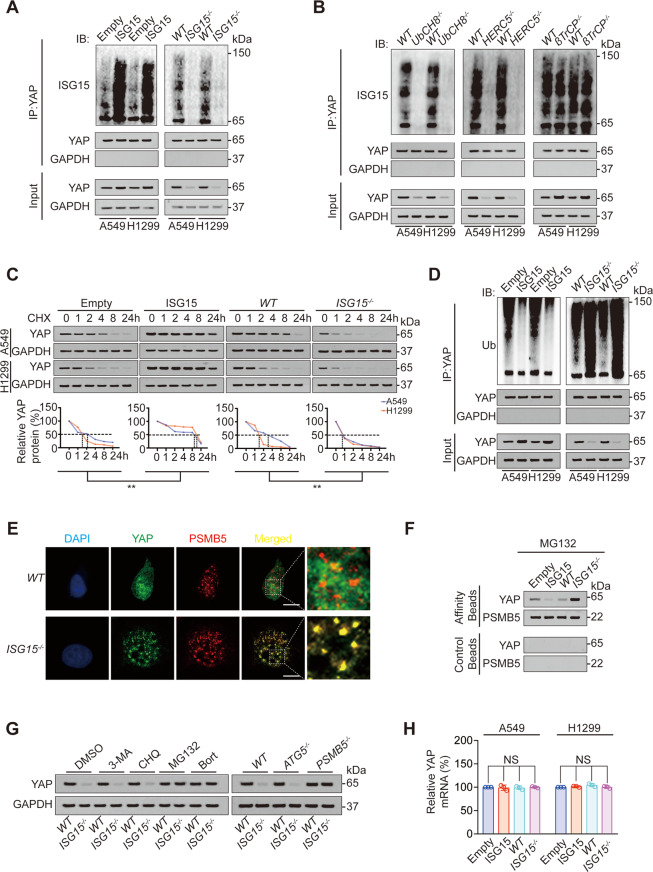
YAP ISGylation increases its stability by inhibiting its ubiquitination
Firstly, we investigated whether ISGylation regulated YAP expression. We observed that overexpression of ISG15 and interferon α (IFNα) treatment was significantly induced, whereas knockout of ISG15 significantly inhibited YAP ISGylation and expression in LUAD cell lines A549 and H1299 (Fig. 1A, Fig. S1A). Knockout of UbCH8 (E2 ligase for ISGylation) and HECT and RLD domain containing E3 ubiquitin-protein ligase 5 (HERC5, E3 ligase for ISGylation) also inhibited YAP ISGylation and expression; however, βTrCP (E3 ligase for ubiquitination) knockout had no influence on YAP ISGylation (Fig. 1B, Fig. S1B). The above findings suggested YAP ISGylation elevated YAP expression.
Fig. 1. YAP ISGylation inhibits its ubiquitination and increases its stability.
A Co-IP was performed in control, ISG15 overexpression or knockout A549 and H1299 cells using anti-YAP antibodies. The YAP level in each co-IP sample was adjusted to the same protein content. Indicated proteins were further analyzed by IB. B Co-IP was performed in control or UbCH8, HERC5, or βTrCP knockout A549 and H1299 cells using anti-YAP antibodies. The YAP level in each co-IP sample was adjusted to the same protein content. Indicated proteins were further analyzed by IB. C CHX (10 μg/ml) chase experiments were performed in control, ISG15 overexpression or knockout A549 and H1299 cells. The relative protein levels of YAP were shown as the ratios between YAP and GAPDH, and the “0 h” points were arbitrarily set to 100%. D Co-IP was performed in control, ISG15 overexpression or knockout A549 and H1299 cells using anti-YAP antibodies. The YAP level in each co-IP sample was adjusted to the same protein content. Indicated proteins were further analyzed by IB. E Co-localization of YAP and PSMB5 was analyzed in WT or ISG15−/− A549 cells before being photographed using a confocal microscope. Scale bar, 20 μm. F Association of YAP and PSMB5 analyzed by IB in proteasomes isolated from A549 cells with or without ISG15 overexpression or knockout in the presence of MG132 (8 μM, 24 h). Samples from affinity or control beads were analyzed in parallel. G YAP expression was analyzed by IB in WT or ISG15−/− A549 cells with 3-MA, CHQ, MG132, or Bort treatment or with ATG5 or PSMB5 knockout. H YAP mRNA level was analyzed in control, ISG15 overexpression or knockout A549, and H1299 cells. The data are shown as the mean ± SD from three biological replicates (including IB). Data in C were analyzed using a two-way ANOVA test. Data in H were analyzed using a one-way ANOVA test. NS nonsignificant.
Next, we evaluated how ISGylation regulated YAP expression. Since ISGylation is a type of ubiquitination-like and ubiquitination-associated posttranslational modification [27, 28], we hypothesized that ISGylation upregulated YAP expression via competing for its ubiquitination and enhancing its stability. Using cycloheximide (CHX) chasing experiment, we observed that ISG15 overexpression increased, whereas ISG15 knockout decreased YAP half-life (Fig. 1C). YAP ubiquitination is an important reason for the decreased stability of YAP [21, 29]. As expected, ISG15 overexpression inhibited, whereas ISG15 knockout stimulated YAP ubiquitination (Fig. 1D). In addition, ISG15 knockout promoted the subcellular co-localization between YAP and proteasome 20S subunit beta 5 (PSMB5, one active site of the proteasome, Fig. 1E). In the MG132-treated isolated proteasome, we also found that ISG15 overexpression inhibited, whereas ISG15 knockout promoted the recruitment of YAP to the proteasome (Fig. 1F). The proteasome inhibitor MG132 and bortezomib (Bort) reversed ISG15 knockout-induced YAP degradation, whereas the autophagy inhibitor 3-methyladenine (3-MA) or chloroquine (CQ) had no such function (Fig. 1G). Similarly, PSMB5 knockout reversed ISG15 knockout-induced YAP degradation, whereas knockout of autophagy key factor autophagy-related 5 (ATG5) did not influence such phenomenon (Fig. 1G, Fig. S1C). We also excluded the possibility that ISG15 transcriptionally influenced YAP expression (Fig. 1H). These data suggested that the ISGylation of YAP inhibited its ubiquitination and subsequent proteasome degradation, and upregulated its stability.
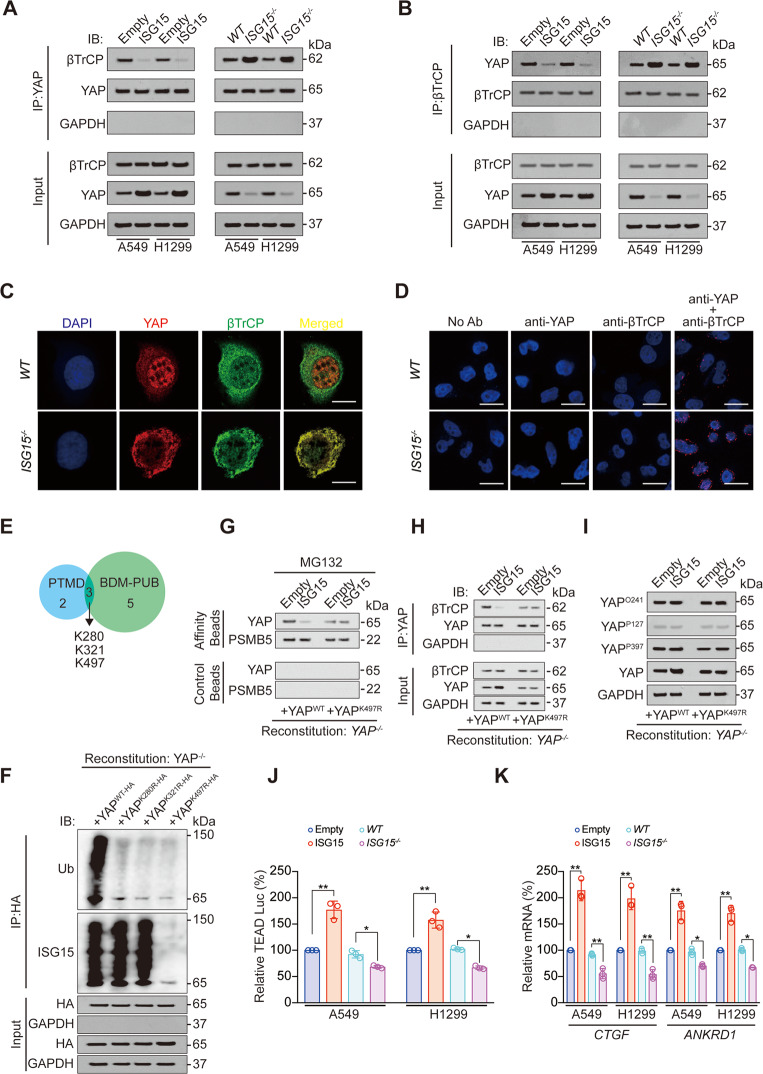
YAP ISGylation inhibits YAP-βTrCP interaction
Subsequently, we analyzed whether YAP ISGylation inhibits its interaction with βTrCP. We performed reciprocal co-IP experiments and found that overexpression of ISG15 inhibited, whereas knockout of ISG15 promoted interaction between βTrCP and YAP (Fig. 2A, B). Immunofluorescence and proximity ligation assay (PLA) ligation followed by confocal microscopy analysis revealed that ISG15 knockout stimulated the co-localization between βTrCP and YAP (Fig. 2C, D). These data suggested that YAP ISGylation inhibited YAP-βTrCP interaction.
Fig. 2. YAP ISGylation inhibits its interaction with βTrCP and promotes downstream transcription.
A, B Co-IP was performed in control, ISG15 overexpression or knockout A549 and H1299 cells using anti-YAP (A) or anti-βTrCP (B) antibodies. The YAP (A) or βTrCP (B) level in each co-IP sample was adjusted to the same protein content. Indicated proteins were further analyzed by IB. C Co-localization of YAP and βTrCP was analyzed in WT or ISG15−/− A549 cells before being photographed using a confocal microscope. Scale bar, 20 μm. D Proximal protein ligation between YAP and βTrCP, as photographed by confocal microscope in WT or ISG15−/−A549 cells Scale bar, 50 μm. E YAP ubiquitination sites analyzed by PTMD and BDM-PUB databases. F Co-IP was performed in YAP−/− reconstituted A549 cells with YAPWT-HA, YAPK280R-HA, YAPK321R-HA, or YAPK497R-HA expressed. The YAP level in each co-IP sample was adjusted to the same protein content. G Association of YAP and PSMB5 analyzed by IB in proteasomes isolated from YAP−/− reconstituted A549 cells with ISG15, YAPWT-HA or YAPK497R-HA expressed in the presence of MG132 (8 μM, 24 h). Samples from affinity or control beads were analyzed in parallel. H Co-IP was performed using anti-YAP antibodies in YAP−/− reconstituted A549 cells with ISG15, YAPWT-HA or YAPK497R-HA expressed. The YAP level in each co-IP sample was adjusted to the same protein content. Indicated proteins were further analyzed by IB. I O-GlcNAcylation of YAP at T241 (YAPO241), phosphorylation of YAP at S127 (YAPP127) and S397 (YAPP397) was analyzed in YAP−/− reconstituted A549 cells with ISG15, YAPWT-HA or YAPK497R-HA expressed by IB. J, K Relative TEAD Luc (J), CTGF and ANKRD1 mRNA level (K) were measured using a pUAS-Luc/TEAD-Gal4 system in control, ISG15 overexpression or knockout A549 and H1299 cells. The data are shown as the mean ± SD from three biological replicates (including IB). Data in J, K were analyzed using a one-way ANOVA test. **P < 0.01, *P < 0.05.
YAP is ISGylated at K497
We subsequently explored the potential YAP ISGylation site. Post-translational modifications that are associated with human diseases and prediction of ubiquitination sites with Bayesian discriminant method database were used to predict YAP ubiquitination sites, and we found that K280, K321, and K497 were three YAP ubiquitination sites that both predicted by two databases (Fig. 2E). Since ubiquitination and ISGylation might compete for the same site [24], we performed experiments in YAP−/− reconstituted A549 cells with different YAP mutant proteins expressed, and observed that K280R, K321R, and K497R mutations all significantly declined total YAP ubiquitination, whereas only K497R significantly declined total YAP ISGylation (Fig. 2F), which suggested that K497 might be the major YAP ISGylation site (Fig. 2F). In addition, YAP K497R mutation abolished the inhibitory role of ISG15 for the entry of YAP into proteasome and the combination between YAP and βTrCP (Fig. 2G, H, Fig. S2A, B). The promotive effect of ISG15 on YAP expression was also abolished by K497R mutation (Fig. 2H, I, Fig. S2B, C). We further excluded the possibility that YAP ISGylation regulated YAP O-GlcNAcylation and phosphorylation [15, 20] (Fig. 2I, Fig. S2C). Simultaneously, we observed that ISG15 did not affect the binding of YAP to CK1 and LATS1, whereas increased YAP binding to TEAD4, a key transcription factor that promotes transcription of genes downstream of YAP [30] (Fig. S2D, E). We also found that overexpression of ISG15 stimulated, whereas ISG15 knockdown inhibited TEAD Luc activity, mRNA levels of two YAP downstream genes, connective tissue growth factor (CTGF), and ankyrin repeat domain 1 (ANKRD1) (Fig. 2J, K) [31]. Therefore, ISGylation regulated YAP independently of the classical upstream Hippo signaling pathway but promoted the downstream transcription of YAP.
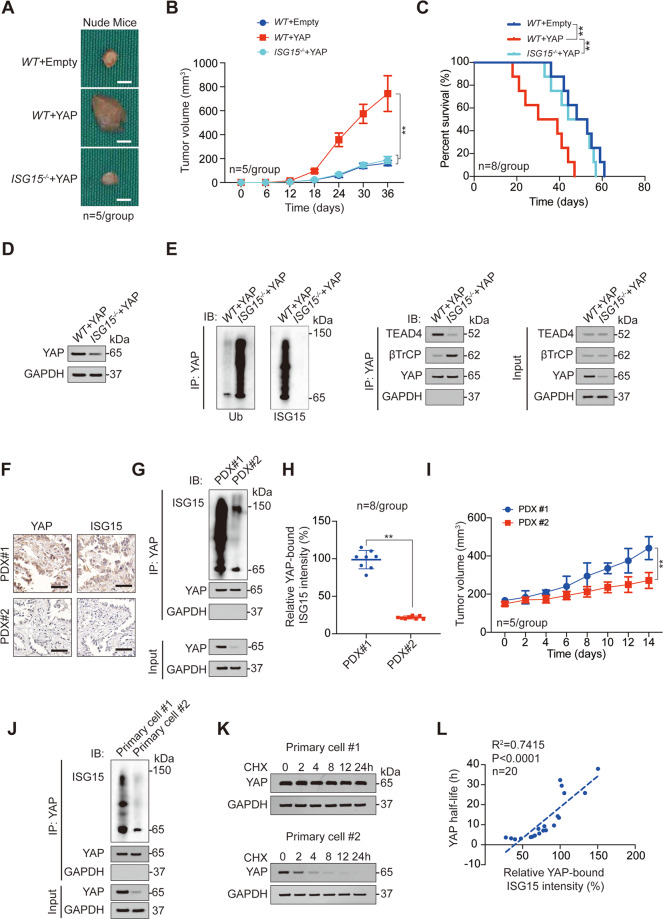
YAP ISGylation is critical for malignant phenotypes
Given that ISGylation has a great impact on the downstream transcription of YAP, we believed that ISGylation is likely to further affect YAP-related malignant phenotypes. Indeed, we observed that both increased cell viability and decreased apoptosis induced by YAP overexpression could be reversed by ISG15 knockout (Fig. S3A, B). In xenograft models, ISG15 knockdown reversed enlarged tumor size and poor mouse prognosis caused by YAP overexpression (Fig. 3A–C). Mechanistically, ISG15 knockout decreased the expression of YAP, promoted the ubiquitination of YAP, the binding of YAP to βTrCP, and inhibited the binding of YAP to TEAD4 (Fig. 3D, E). We established two series of LUAD PDX models, PDX#1 highly expressed YAP and ISG15, while PDX#2 lowly expressed YAP and ISG15 (Fig. 3F). The ISGylation level of YAP in PDX#1 models was also significantly higher than that in PDX#2 models (Fig. 3G-H). In addition, the transplanted tumor size of the PDX#1 model was significantly larger than that of the PDX#2 model (Fig. 3I), and the prognosis of PDX#1 model mice was worse than that of PDX#2 mice (Fig. S3C). We used the tumor cells constructing the PDX model for primary culture and found that the ISGylation of YAP in Primary cell #1 (used for constructing PDX#1) was significantly higher than that in Primary cell #2 (used for constructing PDX#2), and the half-life of YAP in Primary cell #1 was also significantly higher than that in Primary cell #2 (Fig. 3J, K). Moreover, in the total of 20 primary cell lines constructed from other fresh LUAD tissues, we found that the half-life of YAP was negatively correlated, while the cell viability was positively correlated with YAP ISGylation (Fig. 3L and S3D). These data suggested that YAP ISGylation is critical for malignant phenotypes of LUAD cells.
Fig. 3. YAP ISGylation is critical for LUAD malignant phenotype.
A–C Representative images (A), tumor volume (B), and survival analysis (C) for xenograft tumor formed by WT or ISG15−/− A549 cells with or without YAP overexpression. D YAP expression was analyzed by IB in xenograft tumors formed by WT or ISG15−/− A549 cells with YAP overexpression. E Co-IP experiments analyzing YAP ISGylation, ubiquitination, and interacted proteins in xenograft tumors formed by WT or ISG15−/− A549 cells with YAP overexpression. The YAP level in each co-IP sample was adjusted to the same protein content. F YAP and ISG15 expression in PDX#1 and PDX#2 were analyzed by IHC. Scale bar, 200 μm. G, H YAP ISGylation was analyzed by co-IP experiments and counted in PDX#1 and PDX#2 (n = 8). The YAP level in each co-IP sample was adjusted to the same protein content. I Tumor volume of PDX#1 or PDX#2 for indicated times. J YAP ISGylation was analyzed by co-IP experiments in Primary cell #1 and Primary cell #2. The YAP level in each co-IP sample was adjusted to the same protein content. K CHX (10 μg/ml) chase experiments were performed in Primary cell #1 and Primary cell #2. L Association between YAP half-life and relative YAP-bound ISG15 intensity was analyzed in 20 primary cell lines. The data are shown as the mean ± SD from three biological replicates (including IB). Data in B, I were analyzed using a two-way ANOVA test. Data in C were analyzed using a log-rank test. Data in H were analyzed using a student’s t test. Data in L were analyzed using a Spearman rank correlation analysis. **P < 0.01.
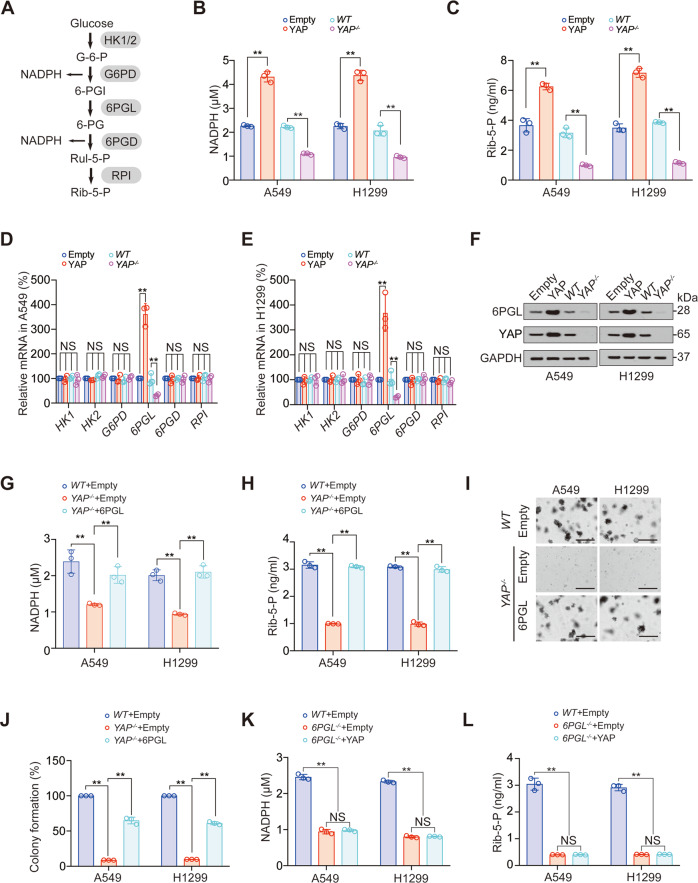
YAP stimulates PPP by elevating 6PGL
Existing studies have shown that YAP can activate glycolysis and hexosamine biosynthesis pathways in tumors [14, 15], while whether PPP, another cancer-promoting glucose metabolism pathway, is regulated by YAP has not been clarified. We observed that NADPH and ribose-5-phosphate (Rib-5-P), two key metabolites of PPP, were positively regulated by YAP overexpression (Fig. 4A–C). Furthermore, YAP upregulated the expression of 6PGL, but had no effect on other key metabolic enzymes in PPP pathway (Fig. 4D–F). Overexpression of 6PGL could recover the inhibition of NADPH and Rib-5-P by YAP knockout (Fig. 4G, H, Fig. S4A). The above data suggested that the promotion role of YAP to PPP was exerted through 6PGL. We also revealed that 6PGL overexpression partially reversed YAP knockout-resulted decreased colony formation, cell viability, 3D cell growth, and increased cell apoptosis (Fig. 4I, J, Fig. S4B–D). In addition, 6PGL knockout-induced NADPH and Rib-5-P decline could not be prevented by YAP overexpression (Fig. 4K, L, Fig. S4E). These data suggested YAP acted as the upstream of 6PGL to stimulate PPP.
Fig. 4. YAP stimulates PPP by activating 6PGL.
A Schematic presentation of PPP from glucose to Rib-5-P. B, C NADPH (B) and Rib-5-P (C) concentration in control, YAP overexpression or knockout A549 and H1299 cells. D, E Indicated mRNA levels in control, YAP overexpression or knockout A549 (D) and H1299 (E) cells. F 6PGL and YAP protein levels in control, YAP overexpression or knockout A549 and H1299 cells. G–J NADPH (G), Rib-5-P concentration (H), and colony formation (I, J) were measured in WT or YAP−/− A549 and H1299 cells with or without 6PGL overexpression. Scale bar, 200 μm. K, L NADPH (K) and Rib-5-P concentration (L) were measured in WT or 6PGL−/− A549 and H1299 cells with or without YAP overexpression. The data are shown as the mean ± SD from three biological replicates (including IB). Data in B–E, G–I, K, L were analyzed using a one-way ANOVA test. **P < 0.01, NS nonsignificant.
YAP boosts the transcription of 6PGL
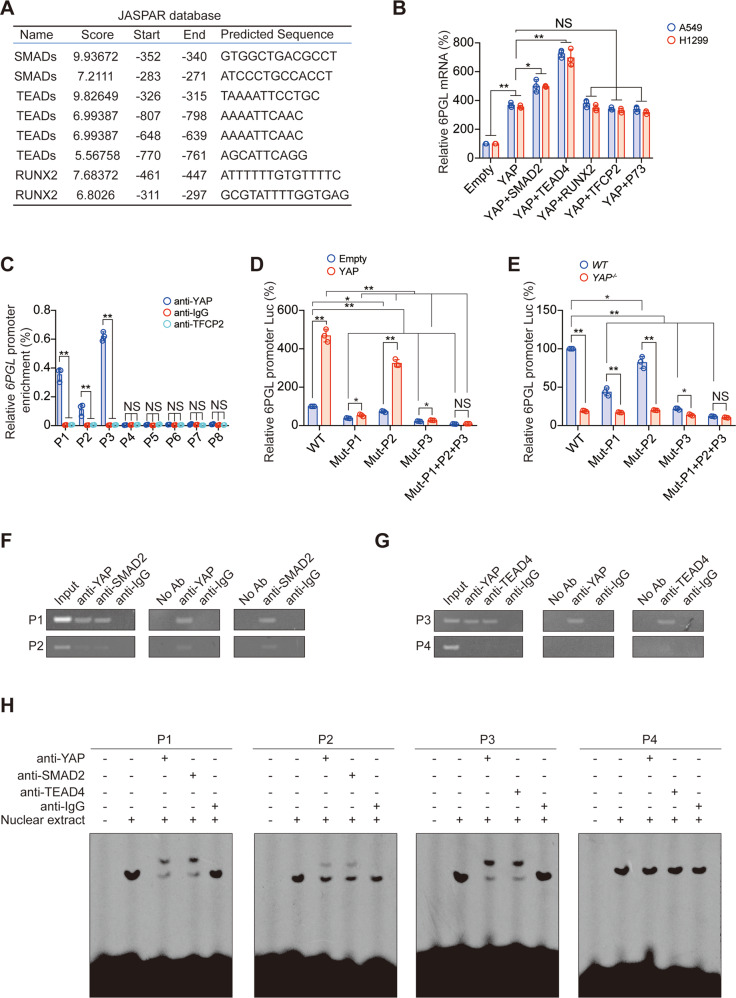
YAP usually acts as a transcription cofactor, promoting the binding of transcription factors to the promoter region of target genes, thereby promoting the transcription of target genes [32]. We used the JASPAR database to predict whether common YAP-associated transcription factors (including SMADs, TEADs, RUNX2, TFCP2, P73) could bind to the promoter region of 6PGL. The data revealed possible binding sites for SMADs, TEADs, and RUNX2 on the 6PGL promoter (Fig. 5A). In addition, SMAD2 and TEAD4 had an additional effect on the elevation of 6PGL mRNA level by YAP, while TFCP2, P73, and RUNX2 had no similar effect (Fig. 5B). Chromatin immunoprecipitation (ChIP) experiments suggested that YAP could bind to two SMADs-related peaks (P1 and P2) and one TEADs-related (P3) peak, but could not bind to the other five peaks (Fig. 5C). The mutation of P1, P2, or P3 significantly inhibited the activity of the 6PGL promoter and the promotion effect of YAP on the 6PGL promoter, and simultaneous mutations of P1–P3 had an additional effect compared to the single mutations of them (Fig. 5D, E). We also performed Re-ChIP and electrophoretic mobility shift assay (EMSA) experiments to reveal that YAP and SMAD2 bound to P1 and P2 simultaneously, while YAP and TEAD4 bound to P3 simultaneously (Fig. 5F–H). In addition, we observed that simultaneous knockout of SMADs and TEADs had additional effects on inhibiting the mRNA and promoter activity of 6PGL compared to the single knockout of one of them, and this inhibitory function could not be reversed by YAP overexpression (Fig. S5A–C). However, when YAP was knocked out, 6PGL mRNA level and promoter activity were significantly reduced and could not be rescued by overexpression of TEAD4 and SMAD2 (Fig. S5D, E). Therefore, these data suggested that the promoter of 6PGL was affected by both SMAD2 and TEAD4 in a YAP dependent-manner.
Fig. 5. SMAD and TEAD stimulate 6PGL transcription in a YAP-dependent manner.
A Predicting 6PGL promoter binding YAP-associated transcription factors by JASPAR database. B 6PGL mRNA was measured in A549 and H1299 cells with indicated plasmids overexpressed. C The enrichments of YAP at indicated regions of the 6PGL promoter were calculated as the percentage of Input chromosomal DNA via ChIP in A549 cells. Anti-IgG and anti-TFCP2 were used as parallel control. D, E Luciferase activity was analyzed in A549 cells co-expressing indicated 6PGL promoter–reporter with or without overexpressing (D) or knocking out YAP (E). F, G Co-occupancies of YAP and SMAD2 (F) and YAP and TEAD4 (G) were measured by ChIP and Re-ChIP experiments in A549 cells using indicated antibodies. H Nuclear extracts from A549 cells were incubated with indicated 6PGL promoter probes with or without the presence of indicated antibodies. The DNA–protein interactions were measured using EMSA. The data are shown as the mean ± SD from three biological replicates (including IB). Data in B–E were analyzed using a one-way ANOVA test. **P < 0.01, *P < 0.05, NS nonsignificant.
YAP ISGylation is essential for its regulation of 6PGL
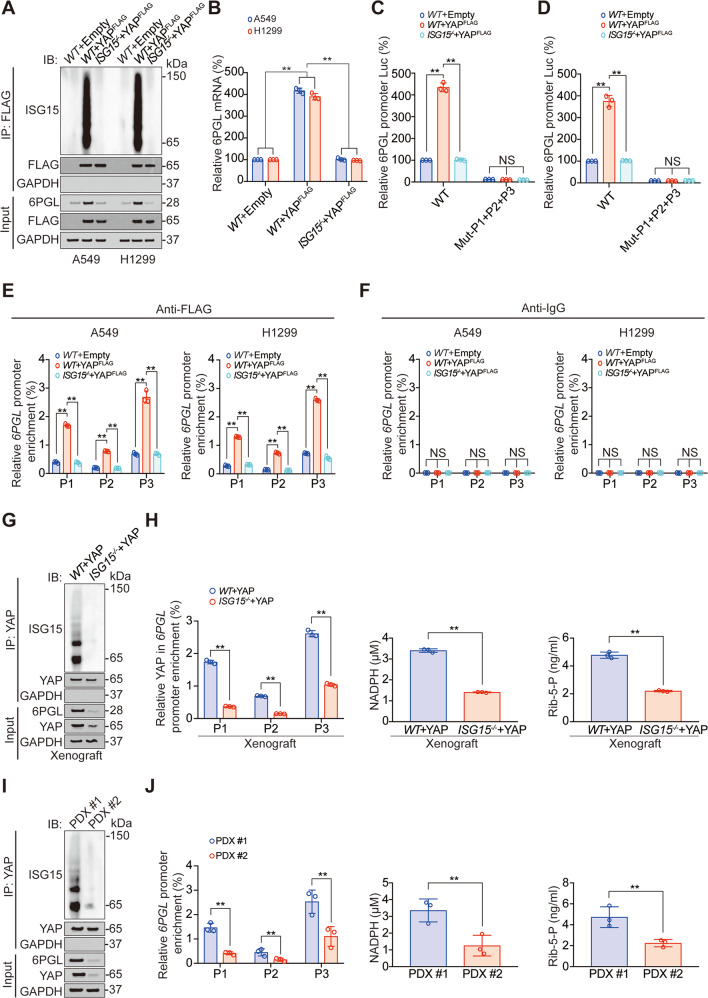
Since YAP ISGylation is essential for its pro-transcription function (Fig. 2), we subsequently investigated whether the stimulating role of YAP on 6PGL transcription depends on its ISGylation. We observed that knockout of ISG15 reversed YAP-induced 6PGL elevation in protein and mRNA level (Fig. 6A, B), as well as promoter activity (Fig. 6C, D). However, simultaneous mutations of P1–P3 abolished 6PGL promoter activity, and 6PGL promoter activity was no longer regulated by ISG15 (Fig. 6C, D). Moreover, YAP bindings to P1–P3 of the 6PGL promoter were also held back by ISG15 knockout (Fig. 6E, F). We used the previously described CDX and PDX models in Fig. 3A–I, and found in CDX models, ISG knockout significantly inhibited 6PGL expression, YAP bindings to P1–P3 of 6PGL promoter, NADPH, and Rib-5-P level (Fig. 6G, H). Similarly, in PDX#1 model (high YAP ISGylated model), 6PGL expression, YAP bindings to P1–P3 of 6PGL promoter, NADPH and Rib-5-P level were significantly higher than PDX#2 model (low YAP ISGylated model) (Fig. 6I, J). We also found in previously reported 20 primary cell lines in Fig. 3J–L, YAP ISGylation level was positively associated with 6PGL expression, NADPH, and Rib-5-P level (Fig. S6A–C). Collectively, these data indicated YAP ISGylation was critical for YAP-induced 6PGL transcription.
Fig. 6. YAP ISGylation stimulates 6PGL transcription.
A Co-IP experiments analyzing YAP ISGylation in WT or ISG−/− A549 and H1299 cells with or without overexpressing YAP, and YAP and 6PGL expression in Input sample were analyzed by IB. The FLAG level in YAPFLAG overexpressed co-IP samples was adjusted to the same protein content. B 6PGL mRNA level was measured in WT or ISG−/− A549 and H1299 cells with or without overexpressing YAP. C, D Luciferase activity was analyzed in WT or ISG15−/− A549 (C) and H1299 (D) cells co-expressing indicated 6PGL promoter–reporter with or without overexpressing YAP. E, F The enrichments of YAP at indicated regions of 6PGL promoter were calculated as the percentage of Input chromosomal DNA via ChIP in WT or ISG−/− A549 and H1299 cells with or without overexpressing YAP (E). Anti-IgG was used as parallel control (F). G Co-IP experiments analyzing YAP ISGylation in tumor xenografts overexpressing YAP with or without ISG15 knockout, and YAP and 6PGL expression in Input sample were analyzed by IB. The YAP level in each co-IP sample was adjusted to the same protein content. H The enrichments of YAP at indicated regions of 6PGL promoter, NADPH, and Rib-5-P level were analyzed in the same tumor xenografts as those in Panel G. I Co-IP experiments analyzing YAP ISGylation in PDX#1 and PDX#2, and YAP and 6PGL expression in Input sample were analyzed by IB. The YAP level in each co-IP sample was adjusted to the same protein content. J The enrichments of YAP at indicated regions of 6PGL promoter, NADPH, and Rib-5-P level were analyzed in PDX#1 and PDX#2. The data are shown as the mean ± SD from three biological replicates (including IB). Data in B–F were analyzed using a one-way ANOVA test. Data in H and J were analyzed using a student’s t test. **P < 0.01, NS nonsignificant.
Clinical association among YAP ISGylation, YAP, and 6PGL level
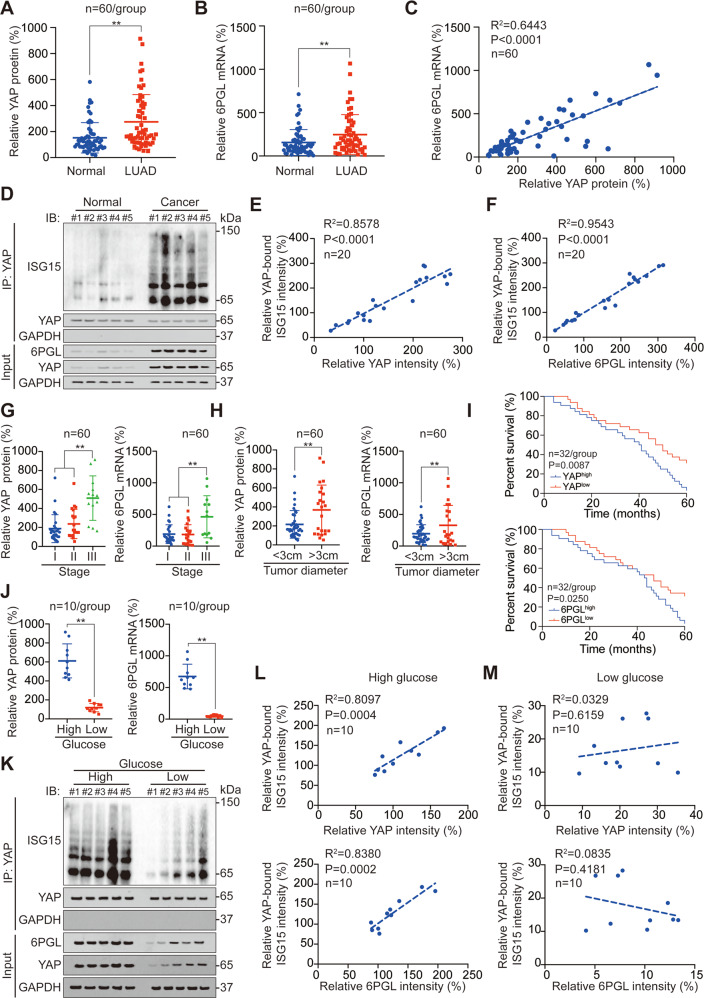
Subsequently, we analyzed the clinical associations of the factors in this study. After analyzing 60 paired tissues, we found that YAP protein (by ELISA) and 6PGL mRNA levels were significantly higher in LUAD tissues compared to adjacent normal tissues, and a positive correlation between 6PGL mRNA and YAP protein in LUAD tissues was also revealed (Fig. 7A–C). We also analyzed YAP and 6PGL protein as well as YAP ISGylation level in tissues using IB and found that they were all elevated in LUAD tissues compared to their adjacent normal tissues (Fig. 7D, Fig. S7A–C). Positive associations between YAP ISGylation and YAP protein level, as well as 6PGL protein level, were also revealed (Fig. 7E, F). Moreover, YAP protein and 6PGL mRNA levels were upregulated in stage III LUAD tissues compared to stage I and II LUAD tissues (Fig. 7G), as well as in >3 cm diameter LUAD tissues compared to <3 cm LUAD tissues (Fig. 7H). We also found that high expression of YAP protein and 6PGL mRNA were associated with poor LUAD prognosis (Fig. 7I). As a histological marker, YAP protein has an area under the receiver operating characteristic curve (AUC-ROC) of 0.714, a sensitivity of 71.7%, and a specificity of 65.0%. The AUC-ROC of 6PGL mRNA as a histological marker was 0.629, sensitivity was 62.3%, and specificity was 61.7% (Fig. S7D). In addition, neither YAP and 6PGL protein or mRNA could be detected in serum (Fig. S7E).
Fig. 7. Clinical association among YAP ISGylation, YAP, and 6PGL.
A YAP protein level in LUAD and adjacent normal tissues as measured by ELISA. B 6PGL mRNA level in LUAD and adjacent normal tissues as measured by qPCR. C Association between YAP protein and 6PGL mRNA level. D Co-IP experiments analyzing YAP ISGylation in LUAD and adjacent normal tissues, and YAP and 6PGL expression in Input sample were analyzed by IB. The YAP level in each co-IP sample was adjusted to the same protein content. E, F Association between YAP bound ISG15 intensity and YAP intensity (E), as well as YAP, bound ISG15 intensity and 6PGL intensity (F) in LUAD tissues. G, H YAP protein and 6PGL mRNA level in stage I–III LUAD samples (G), <3 cm and >3 cm diameter LUAD samples (H). I Survival analysis for LUAD patients with high or low YAP protein and 6PGL mRNA level. J YAP protein and 6PGL mRNA level in high and low glucose LUAD samples. K Co-IP experiments analyzing YAP ISGylation in high and low glucose LUAD tissues, and YAP and 6PGL expression in Input sample were analyzed by IB. The YAP level in each co-IP samples was adjusted to the same protein content. L, M Association between YAP bound ISG15 intensity and YAP intensity, as well as YAP, bound ISG15 intensity and 6PGL intensity in high (L) or low (M) glucose LUAD tissues. The data are shown as the mean ± SD from three biological replicates (including IB). Data in A, B, H, I were analyzed using a student’s t test. Data in C, E, F, L, M were analyzed using the Spearman rank correlation analysis. Data in G were analyzed using a one-way ANOVA test. Data in I were analyzed using a log-rank test. **P < 0.01.
Since YAP is a positive regulator for glycolysis, PPP, and HBP in stimulating glucose metabolism (Figs. 4–6 [14, 15]), we further analyzed YAP and 6PGL level as well as YAP ISGylation in different blood glucose level tissues. We observed that YAP protein, 6PGL mRNA, and YAP ISGylation were all significantly elevated in high glucose LUAD tissues compared to low glucose LUAD tissues (Fig. 7J, K, Fig. S7F). In addition, YAP ISGylation level was significantly associated with YAP and 6PGL levels in high glucose LUAD tissues (Fig. 7L), whereas in low glucose LUAD tissues, no significant correlations were uncovered among YAP ISGylation, YAP, and 6PGL level (Fig. 7M). We also found that although blood glucose was significantly elevated in stage III LUAD patients compared to stage I and II patients (Fig. S7G), no significant correlation was found between blood glucose and LUAD stage (Fig. S7H). These data suggested that YAP ISGylation, YAP, and 6PGL level were positively associated with each other, especially in high glucose LUAD tissues.
Discussion
YAP is regulated by a variety of post-translational modifications. It is best known that YAP is phosphorylated at Ser127 (Ser, serine) and then inhibited [20]. Our Lab first reported the YAP O-GlcNAcylation, which inhibits YAP phosphorylation, promotes YAP to enter the nucleus, and activates downstream transcription [15]. Here, we observed that YAP could undergo a kind of ubiquitination-like modification, ISGylation. In this study, we identified the possible YAP ISGylation site as K497. This ISGylation inhibited ubiquitination of YAP at the same site, further inhibited YAP proteasome degradation, and promoted downstream transcription. These findings further enrich the theory that YAP is regulated by post-translational modification.
Initially, it is thought that YAP is mainly regulated by the Hippo pathway [33]. However, recently, some regulations of YAP are found to be independent of upstream Hippo signaling. For example, enhanced YAP O-GlcNAcylation stimulates YAP to enter the nucleus and promote downstream transcription [15, 34]. Moreover, CREB is identified by our Lab as a transcription factor up-regulating YAP mRNA level [35]. In addition, erb-b2 receptor tyrosine kinase 2 (ERBB2) is also identified as an upstream transcription factor of YAP [36]. The YAP ISGylation was also independent of the Hippo pathway and enhanced the stability of YAP by directly inhibiting its ubiquitination. All the above studies indicate that in addition to the Hippo pathway, YAP is also regulated by many other ways, which are also crucial to the regulation of YAP [3, 37].
YAP is a crucial regulator of glucose metabolism. As for glycolysis, YAP can directly promote the transcription of phosphoglycerate mutase 1 (PGAM1) or promote the transcription of pyruvate kinase-2 (PKM-2) through hypoxia-inducible factor 1 subunit alpha (HIF-1α) to activate glycolysis [14, 38]. Moreover, YAP stimulates HBP by activating the transcription of two key enzymes, nudix hydrolase 9 (NUDT9) and solute carrier family 5 member 3 (SLC5A3) [15]. In this study, we observed that YAP was also a stimulator for PPP. Without regulating other enzymes, YAP stimulated the transcription of 6PGL through SMAD2 and TEAD4. The influence of transcription factors on the target gene promoter is very complex, and one promoter can be affected by multiple transcription factors at the same time [39, 40]. In our study, it was found that the transcription of 6PGL could not be activated by overexpression of TEAD4 and SMAD2 after knocking out YAP. Therefore, we speculated that if we want to inhibit the transcription of an oncogene in clinical treatment, it is not feasible to inhibit the direct transcription factors, because the oncogene will be activated by multiple transcription factors at the same time, while suppressing transcription cofactors like YAP can achieve the purpose of simultaneously inhibiting multiple transcription factors.
YAP and high glucose have mutually promoting feedback effects, YAP can generate a large number of intermediate products needed for tumor proliferation through glycolysis, PPP, and HBP, while high glucose can activate YAP such as promoting its O-GlcNAcylation and interaction with TEADs [14, 15, 38, 41]. In this study, we found that YAP protein and 6PGL mRNA had certain diagnostic effects as tissue markers, but their optimal diagnostic specificities and sensitivities were all less than 75%, and they could not be detected in blood without trauma, so they were of little significance for early diagnosis (Figure S7D-E). Factors downstream of YAP, such as membrane protein melanoma cell adhesion molecule, chaperonin containing t-complex 1 subunit 3, circRNA104075 which are blood detectable indicators, can indeed play a certain role in the early diagnosis of tumors [42–44]. However, due to the high expression of YAP in a variety of tumors, the diagnostic specificity of YAP-related indicators is somewhat deficient [1, 45, 46], and their sensitivity in the diagnosis of LUAD is not as good as that of low-dose spiral CT at present [47]. Therefore, we believed that it may be more meaningful to further explore the role of YAP in prognostic monitoring and as an individual therapeutic target, or to integrate YAP-related markers into a multi-marker panel. In this study, we further demonstrated that YAP is highly expressed and ISGylated in patients with hyperglycemic cancer. Therefore, targeting YAP and its ISGylation for hyperglycemic tumors may achieve promising therapeutic effects. This is not only limited to LUAD discussed in this study, but may also include liver, colorectal, breast, pancreatic cancer, and other tumors that are more closely related to hyperglycemia [48]. Therefore, further research on YAP promoting tumors through glucose metabolism is needed, which is conducive to the proposal of individualized tumor diagnosis and treatment.
Conclusions
YAP ISGylation is critical for maintaining its stability and further activation of PPP. Targeting ISGylated YAP might be promising for hyperglycemia cancer treatment.
Material and methods
Cell culture
Established A549 and H1299 cell lines were purchased from Fuheng Biotechnology (Shanghai, China). All the cell lines were validated by short tandem repeat analysis. Patient-derived primary LUAD cells were established from LUAD tissues as previously described [49]. Briefly, fresh tissues less than 1.0 cm3 without necrosis were immediately rinsed 3 times with cold Dulbecco’s Phosphate Buffered saline and then re-suspended in Dulbecco’s Modified Eagle Medium (DMEM) containing collagenase I (2 mg/ mL, Solarbio, Shanghai) at 37 °C for 4 h. After being rinsed with DMEM 3 times, cells were cultured at 37 °C, 5% CO2 condition. For monolayer culture, cells were cultured in DMEM containing 10% fetal bovine serum and 1% penicillin/streptomycin. For 3D spheroid culture, basement membrane extract (BME) (Trevigen, Gaithersburg, MD, USA) was seeded in a 96-well plate at 50 μl/well and pre-warmed at 37 °C for 0.5 h. Then, cells were seeded on top of the plate coated with BME at a density of 105 cells per well. Images were captured using a microscope [49].
Mouse experiments and tissue samples
For a generation of CDX mouse models, A549 cells (initial 5 × 106, including control, YAP overexpression, ISG15 knockout with YAP overexpression cells) were subcutaneously injected into 6-week-old athymic nude mice (Jiesijie, Shanghai, China). For a generation of PDX mouse models, fresh LUAD specimens (high YAP and ISGylation level as PDX#1, low YAP, and ISGylation level as PDX#2) in a size of 2–3 mm3 were implanted into six-week-old athymic nude mice. The third generation of PDX-bearing mice was used for further analysis [49]. The tumor volume was calculated as 0.5 × L × W2 (L indicating length while W indicating width). All the tissue and serum specimens were recruited in Shanghai Chest Hospital (Shanghai, China) (mean age ± SD, 63.91 ± 11.27 years; male: female ratio, 1.17:1) were recruited in Shanghai Chest Hospital (Shanghai, China) from March 2015 to December 2020. Informed written consents were obtained from all patients.
Regents and plasmids
For regents, CHX (Sigma, St Louis, MO, USA), MG132 (MedChemExpress, Monmouth Junction, NJ, USA), 3-MA (MedChemExpress), CHQ (MedChemExpress), Bort (MedChemExpress), and IFNα (Sigma) were used to treat cells. For plasmids, ISG15 and 6PGL expression plasmids were bought from Origene (Beijing, China). LentiCRISPR v2 based constructs were used for knockout ISG15, UbCH8, HERC5, 6PGL, SMADs, TEADs. pGL4.21 vector was used to construct a 6PGL promoter-luciferase vector. YAPK280R-HA, YAPK321R-HA, YAPK497R-HA, 6PGLMut-P1, 6PGLMut-P2, 6PGLMut-P3, and 6PGLMut-P1+P2+P3 mutant plasmids were constructed using overlapping PCR. YAP, ATG5, PSMB5, and βTrCP knockout constructs, YAPWT-HA, YAPFLAG, SMAD2, TEAD4, RUNX2, TFCP2, P73, pUAS-Luc/TEAD-Gal4 plasmids were acquired from previous studies [15, 31, 43, 49, 50]. The primers are listed in Supplementary Table 1.
Immunofluorescence (IF), immunohistochemistry (IHC), immunoblotting (IB), and enzyme-linked immunosorbent assay (ELISA)
IF and IHC were performed according to the conventional protocols. The primary antibodies used for IF were anti-YAP (Abcam, Hong Kong, China #ab52771) and anti-βTrCP (Abcam, #ab233638). The primary antibodies used for IHC were: anti-YAP (Santa Cruz Biotechnology, Santa Cruz, CA, USA, #sc-101199) and anti-ISG15 (Abcam, #ab233071). For IB, the proteins were resolved on SDS-PAGE gels according to the conventional protocols. The primary antibodies used were anti-ISG15 (Abcam, #ab233071), anti-YAP (Abcam, #ab52771 and Santa Cruz, #sc-101199), anti-GAPDH (CST, #5174 and #51332), anti-Ub (Abcam, #ab7780 and #ab7254), anti-PSMB5 (Abcam, #ab167341), anti-βTrCP (Abcam, #ab71753 and #ab233638), anti-YAPO241 (developed by Biolynx, Hangzhou, China), anti-YAPP127 (Abcam, #ab76252), anti-YAPP397 (CST, Boston, MA, USA, #13619), anti-HA (Abcam, #ab9110 and #ab18181), anti-TEAD4 (Abcam, #ab197589 and #ab58310), anti-6PGL (Abcam, #ab229872), anti-FLAG (CST, #8146 and #2368), anti-UbCH8 (Abcam, #ab177485), anti-HERC5 (Invitrogen, Carbsland, CA, USA, #703675), anti-ATG5 (Abcam, #ab221604), anti-LATS1 (Abcam, #ab243656), anti-CK1 (Abcam, #ab270997 and #ab115293), anti-SMAD2 (Abcam, #ab40855), anti-alpha fetoprotein (AFP, Abcam, #ab284388) and anti-albumin (Alb, Abcam, #ab207327). ELISA kits (Yingxin, Shanghai, China) were used to measure the concentration of YAP protein and Rib-5-P.
Co-immunoprecipitation (co-IP)
Co-immunoprecipitation (co-IP) was performed as described previously [31]. After cells were harvested, the lysates were mixed with 50 ul of protein A/G-magnetic beads (Novex, Oslo, Norway) and incubated at 4 °C overnight with the indicated antibodies cross-linked to protein A/G magnetic beads. The beads were washed three times with Western/IP lysis buffer (Beyotime, Haimen, China), suspended in SDS-PAGE loading buffer, and then detected by IB with relevant antibodies. The antibodies used for co-IP were: anti-YAP (Abcam, #ab52771 and Santa Cruz, #sc-101199), anti-βTrCP (Abcam, #ab71753), anti-HA (Abcam #ab18181), anti-FLAG (CST, #2368), anti-CK1 (Abcam, #ab270997), anti-TEAD4 (Abcam, #ab197589) and anti-LATS1 (Abcam, #ab234820).
Proteasome isolation
Proteasomes were isolated using the proteasome isolation kit (Sigma, #539176). Isolation was performed strictly in accordance with guidelines provided by the manufacturer. Affinity and control beads were used to isolate the proteasome and serve as a negative control.
Proximity ligation assay
PLA was performed using the Duolink In Situ Red Starter Kit (Sigma) as previously described [31]. Cells were seeded on glass coverslips in 24-well plates. On the second day, the cells were fixed with 4% PFA and blocked with a blocking buffer. Then, the cells were incubated overnight at 4 °C in suitable primary antibodies. The primary antibodies used were anti-YAP (Abcam, China #ab52771) and anti-βTrCP (Abcam, #ab233638).
Measurements of metabolites, luciferase activity, cell viability, caspase 3/7 activity, and anchorage-independent colony formation
NADPH (Sigma) concentration was measured using the appropriate kits according to the manufacturer’s instructions. Luciferase activities were measured using a dual-luciferase reagent (Promega, Madison, WI, USA). Cell viability was measured using a CCK8 kit (Beyotime). Caspase3/7 activity was determined using Caspase 3/7 Glo luciferase reagent (Promega). As for the anchorage-independent colony formation assay, LUAD cells were seeded in a 6-well plate containing 0.3% agarose in DMEM at a density of 6 × 103 cells per well. Two weeks later, the numbers of colonies were calculated under a microscope.
Quantitative RT-PCR (qPCR)
Total RNA was extracted using Trizol (Ambion, Carlsbad, CA, USA) and reverse-transcribed into complementary DNA using the PrimeScriptTM RT reagent kit (Takara, Dalian, China). The SYBR premix Ex Taq (Takara) kit was used for real-time qPCR. For the evaluation of the mRNA level of YAP and 6PGL in serum, semi-qPCR was performed. The PCR was terminated at cycle 29 and the products were visualized by agarose gel electrophoresis. The primers are listed in Supplementary Table 1.
ChIP and Re-ChIP
ChIP and Re-ChIP experiments were performed using the kits from Active Motif (Carlsbad, CA, USA) as previously described [31], and strictly in accordance with guidelines provided by the manufacturer. The primary antibodies used in ChIP and Re-ChIP experiments were: anti-YAP (CST, #14074), anti-TFCP2 (CST, #80784), anti-SMAD2 (Abcam, #ab40855), anti-TEAD4 (Abcam, #ab155244) and anti-IgG (CST, #3900). The primers are listed in Supplementary Table 1.
EMSA
EMSA was performed as described in the previous study [49]. The light shift kit (Pierce, Rockford, IL, USA) was used. Nuclear extracted proteins were prepared using the kit from Active Motif (Carlsbad) and incubated in the reaction buffer on ice followed by the addition of the biotin-labeled probes (synthesized and 5′ labeled by Sangon Inc., Shanghai, China). For supershift assays, antibodies against YAP (CST, #14074), TEAD4 (Abcam, #ab155244), and SMAD2 (Abcam, #ab40855) were added to the mixture before adding the probe. The probes used are listed in Supplementary Table 1.
Statistical analysis
Tests used in this study included student’s t test, one-way, two-way ANOVA log-rank, χ2 test, and the Spearman rank correlation analysis. A P < 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81872288 and 81902869), Project of Clinical Research Supporting System, Clinical Medicine First-class Discipline, Grant Support “Chen Guang” project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (18CG16), Shanghai Sailing Program (19YF1444800). Talent training plan of Shanghai Chest Hospital in 2020.
Author contributions
XX researched, analyzed data, and wrote the paper. XT constructed the plasmids and analyzed the data. CZ researched and analyzed the data. YM performed the mouse experiments and analyzed the data. YW, SQ, HW, JC, and LC researched and analyzed data. FS contributed to the discussions. YQ designed the study. XZ designed the study and wrote the paper.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethic declarations
The study (including animal experiments) was approved by the institutional ethics committee of Shanghai Chest Hospital.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yongxia Qiao, Email: yongxia.qiao@shsmu.edu.cn.
Xiao Zhang, Email: zhangxiao_sjtu@126.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41420-022-00842-8.
References
- 1.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Totaro A, Panciera T, Piccolo S. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 2018;20:888–99. doi: 10.1038/s41556-018-0142-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–10. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–27. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford S, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–33. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noguchi S, Saito A, Nagase T. YAP/TAZ signaling as a molecular link between fibrosis and cancer. Int J Mol Sci. 2018;19:3674. [DOI] [PMC free article] [PubMed]
- 8.Zhou B, Flodby P, Luo J, Castillo DR, Liu Y, Yu FX, et al. Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. J Clin Invest. 2018;128:970–84. doi: 10.1172/JCI90429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y, Zhang W, Han X, Li F, Wang X, Wang R, et al. YAP inhibits squamous transdifferentiation of Lkb1-deficient lung adenocarcinoma through ZEB2-dependent DNp63 repression. Nat Commun. 2014;5:4629. doi: 10.1038/ncomms5629. [DOI] [PubMed] [Google Scholar]
- 10.Chaib I, Karachaliou N, Pilotto S, Codony Servat J, Cai X, Li X, et al. Co-activation of STAT3 and YES-associated protein 1 (YAP1) pathway in EGFR-mutant NSCLC. J Natl Cancer Inst. 2017;109:djx014. [DOI] [PMC free article] [PubMed]
- 11.Liu Y, Ren H, Zhou Y, Shang L, Zhang Y, Yang F, et al. The hypoxia conditioned mesenchymal stem cells promote hepatocellular carcinoma progression through YAP mediated lipogenesis reprogramming. J Exp Clin Cancer Res. 2019;38:228. doi: 10.1186/s13046-019-1219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang CS, Stampouloglou E, Kingston NM, Zhang L, Monti S, Varelas X. Glutamine-utilizing transaminases are a metabolic vulnerability of TAZ/YAP-activated cancer cells. EMBO Rep. 2018;19:e43577. [DOI] [PMC free article] [PubMed]
- 13.Koo JH, Guan KL. Interplay between YAP/TAZ and metabolism. Cell Metab. 2018;28:196–206. doi: 10.1016/j.cmet.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Li Y, Ma Y, Yang L, Wang T, Meng X, et al. Yes-associated protein (YAP) binds to HIF-1alpha and sustains HIF-1alpha protein stability to promote hepatocellular carcinoma cell glycolysis under hypoxic stress. J Exp Clin Cancer Res. 2018;37:216. doi: 10.1186/s13046-018-0892-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Qiao Y, Wu Q, Chen Y, Zou S, Liu X, et al. The essential role of YAP O-GlcNAcylation in high-glucose-stimulated liver tumorigenesis. Nat Commun. 2017;8:15280. doi: 10.1038/ncomms15280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan F, Qian M, He Q, Zhu H, Yang B. The posttranslational modifications of Hippo-YAP pathway in cancer. Biochim Biophys Acta. 2020;1864:129397. doi: 10.1016/j.bbagen.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Lapi E, Di Agostino S, Donzelli S, Gal H, Domany E, Rechavi G, et al. PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol Cell. 2008;32:803–14. doi: 10.1016/j.molcel.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Fang L, Teng H, Wang Y, Liao G, Weng L, Li Y, et al. SET1A-mediated mono-methylation at K342 regulates YAP activation by blocking its nuclear export and promotes tumorigenesis. Cancer Cell. 2018;34:103–18. doi: 10.1016/j.ccell.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Hata S, Hirayama J, Kajiho H, Nakagawa K, Hata Y, Katada T, et al. A novel acetylation cycle of transcription co-activator Yes-associated protein that is downstream of Hippo pathway is triggered in response to SN2 alkylating agents. J Biol Chem. 2012;287:22089–98. doi: 10.1074/jbc.M111.334714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skaug B, Chen ZJ. Emerging role of ISG15 in antiviral immunity. Cell. 2010;143:187–90. doi: 10.1016/j.cell.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YJ, Kim ET, Kim YE, Lee MK, Kwon KM, Kim KI, et al. Consecutive inhibition of ISG15 expression and ISGylation by cytomegalovirus regulators. PLoS Pathog. 2016;12:e1005850. doi: 10.1371/journal.ppat.1005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu D, Zhang T, Xiao J, Zhu K, Wei R, Wu Z, et al. Modification of BECN1 by ISG15 plays a crucial role in autophagy regulation by type I IFN/interferon. Autophagy. 2015;11:617–28. doi: 10.1080/15548627.2015.1023982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okumura A, Pitha PM, Harty RN. ISG15 inhibits ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci USA. 2008;105:3974–9. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu H, Yan F, Yuan T, Qian M, Zhou T, Dai X, et al. USP10 promotes proliferation of hepatocellular carcinoma by deubiquitinating and stabilizing YAP/TAZ. Cancer Res. 2020;80:2204–16. doi: 10.1158/0008-5472.CAN-19-2388. [DOI] [PubMed] [Google Scholar]
- 27.Lee MK, Kim YJ, Kim YE, Han TH, Milbradt J, Marschall M, et al. Transmembrane protein pUL50 of human cytomegalovirus inhibits ISGylation by downregulating UBE1L. J Virol. 2018;92:e00462–18. [DOI] [PMC free article] [PubMed]
- 28.Pincetic A, Kuang Z, Seo EJ, Leis J. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J Virol. 2010;84:4725–36. doi: 10.1128/JVI.02478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long X, Song K, Hu H, Tian Q, Wang W, Dong Q, et al. Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J Exp Clin Cancer Res. 2019;38:345. doi: 10.1186/s13046-019-1329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu C, Li L, Zhang Z, Bi M, Wang H, Su W, et al. A non-canonical role of YAP/TEAD is required for activation of estrogen-regulated enhancers in breast cancer. Mol Cell. 2019;75:791–806. doi: 10.1016/j.molcel.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Sun F, Qiao Y, Zheng W, Liu Y, Chen Y, et al. TFCP2 is required for YAP-dependent transcription to stimulate liver malignancy. Cell Rep. 2017;21:1227–39. doi: 10.1016/j.celrep.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 32.Vigneswaran K, Boyd NH, Oh SY, Lallani S, Boucher A, Neill SG, et al. YAP/TAZ transcriptional coactivators create therapeutic vulnerability to verteporfin in EGFR-mutant glioblastoma. Clin Cancer Res. 2021;27:1553–69. doi: 10.1158/1078-0432.CCR-20-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–92. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 34.Peng C, Zhu Y, Zhang W, Liao Q, Chen Y, Zhao X, et al. Regulation of the hippo-YAP pathway by glucose sensor O-GlcNAcylation. Mol Cell. 2017;68:591–604. doi: 10.1016/j.molcel.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Ma L, Weng W, Qiao Y, Zhang Y, He J, et al. Mutual interaction between YAP and CREB promotes tumorigenesis in liver cancer. Hepatology. 2013;58:1011–20. doi: 10.1002/hep.26420. [DOI] [PubMed] [Google Scholar]
- 36.Aharonov A, Shakked A, Umansky KB, Savidor A, Genzelinakh A, Kain D, et al. ERBB2 drives YAP activation and EMT-like processes during cardiac regeneration. Nat Cell Biol. 2020;22:1346–56. doi: 10.1038/s41556-020-00588-4. [DOI] [PubMed] [Google Scholar]
- 37.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen Y, Zhao S, Wang S, Pan X, Zhang Y, Xu J, et al. S1P/S1PR3 axis promotes aerobic glycolysis by YAP/c-MYC/PGAM1 axis in osteosarcoma. EBioMedicine. 2019;40:210–23. doi: 10.1016/j.ebiom.2018.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P, Nolan TM, Yin Y, Bassham DC. Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy. 2020;16:123–39. doi: 10.1080/15548627.2019.1598753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khan A, Shrestha A, Bhuyan K, Maiti IB, Dey N. Structural characterization of a novel full-length transcript promoter from Horseradish Latent Virus (HRLV) and its transcriptional regulation by multiple stress responsive transcription factors. Plant Mol Biol. 2018;96:179–96. doi: 10.1007/s11103-017-0693-6. [DOI] [PubMed] [Google Scholar]
- 41.Enzo E, Santinon G, Pocaterra A, Aragona M, Bresolin S, Forcato M, et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 2015;34:1349–70. doi: 10.15252/embj.201490379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Zhang X, Lin J, Chen Y, Qiao Y, Guo S, et al. CCT3 acts upstream of YAP and TFCP2 as a potential target and tumour biomarker in liver cancer. Cell Death Dis. 2019;10:644. doi: 10.1038/s41419-019-1894-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9:1091. doi: 10.1038/s41419-018-1132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Tang X, Weng W, Qiao Y, Lin J, Liu W, et al. The membrane protein melanoma cell adhesion molecule (MCAM) is a novel tumor marker that stimulates tumorigenesis in hepatocellular carcinoma. Oncogene. 2015;34:5781–95. doi: 10.1038/onc.2015.36. [DOI] [PubMed] [Google Scholar]
- 45.Dey A, Varelas X, Guan KL. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat Rev Drug Discov. 2020;19:480–94. doi: 10.1038/s41573-020-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zanconato F, Cordenonsi M, Piccolo S. YAP and TAZ: a signalling hub of the tumour microenvironment. Nat Rev Cancer. 2019;19:454–64. doi: 10.1038/s41568-019-0168-y. [DOI] [PubMed] [Google Scholar]
- 47.Sun YJ, Lou J, Xu QL, Xing YG, Zhao XZ, Zhao LL, et al. Comparison of clinical diagnostic value of spiral CT with different dose in patients with early-stage peripheral lung cancer. Clin Transl Oncol. 2021;23:1128–33. doi: 10.1007/s12094-020-02503-7. [DOI] [PubMed] [Google Scholar]
- 48.Hammer M, Storey S, Hershey DS, Brady VJ, Davis E, Mandolfo N, et al. Hyperglycemia and cancer: a state-of-the-science review. Oncol Nurs Forum. 2019;46:459–72. doi: 10.1188/19.ONF.459-472. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Yu K, Ma L, Qian Z, Tian X, Miao Y, et al. Endogenous glutamate determines ferroptosis sensitivity via ADCY10-dependent YAP suppression in lung adenocarcinoma. Theranostics. 2021;11:5650–74. doi: 10.7150/thno.55482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo S, Chen Y, Yang Y, Zhang X, Ma L, Xue X, et al. TRIB2 modulates proteasome function to reduce ubiquitin stability and protect liver cancer cells against oxidative stress. Cell Death Dis. 2021;12:42. doi: 10.1038/s41419-020-03299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.