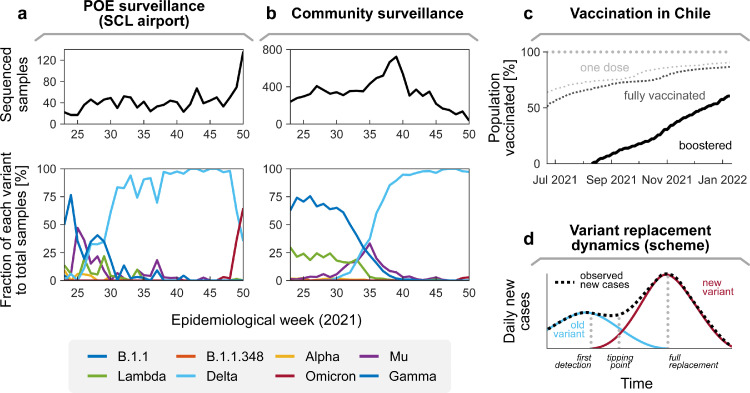
The emergence of SARS-CoV-2 variants with enhanced transmissibility and partial immune escape (also known as Variants of Concern, VOC) has characterised the second year of the COVID-19 pandemic. Genomic surveillance allows identifying hidden, fast-spreading lineages and VOCs before they become a public health threat. The Chilean Public Health Institute (Instituto de Salud Pública, ISP) has led SARS-CoV-2 genomic surveillance initiatives in Chile. Using sequencing data, the ISP has signalled the introduction of VOCs and characterised their transmissibility and mutational signatures, contributing to a better understanding of the local features of the pandemic.1
As of Dec 17 2021, the newly emerged Omicron VOC had quickly spread to 89 countries, including Chile.2 By Dec 21, the ISP had identified 119 samples as Omicron. Of these, 118 correspond to arrivals at the SCL airport and one to community transmission (Figure 1a, b). Although Omicron's epidemiological features remain under study, current reports suggest two worrisome facts: First, Omicron has an extensive escape from both natural and vaccine-elicited neutralisation, thus increasing the chances of reinfection and breakthrough infections. Specifically, Omicron's immune escape would reduce vaccine effectiveness against symptomatic disease to 35% and 73% for 2x freshly vaccinated and boosted individuals, respectively, considering mRNA vaccines (Pfizer-BioNTech's BNT162b2).3 Moreover, this protection against infection seems to wane within a few months and the escape rate is much higher for a heterologous BNT162b2 booster in individuals fully vaccinated with Sinovac's inactivated virus vaccine, CoronaVac.4,5 Although as of Jan 9, 2022 86.6% of the Chilean population is fully vaccinated and 60.5% has got a booster shot6 (Figure 1c), most of the administered doses correspond to CoronaVac (55.3%) followed by BNT162b2 (36.4%, typically booster),7 thus risking another wave.4 Second, initially and in the absence of new NPIs, Omicron reached a doubling time of 1.5-3 days,8 thus spreading faster than Delta VOC. However, this is better explained by Omicron's extensive immune escape rather than an inherent increase in the basic transmissibility.9 Therefore, more stringent NPIs can considerably reduce the transmission. Even though protection against severe course seems to remain for Omicron infections, high incidence among the population endangers the correct function of critical infrastructure, as large fractions of the workforce would require tests and quarantine. Therefore, it is fundamental for informed decision-making to determine the circulating variants and properties through genomic surveillance.
Figure 1.
SARS-CoV-2 genomic surveillance in Chile. (a) Surveillance at Points of Entry (POEs) has provided an early alert for the onset of community transmission of introduced variants. For example, the Lambda, Gamma, Mu, and Delta variants were markedly present in the samples collected at POEs at a given time and shortly after had a substantial contribution to community transmission (b). (c) As of 9 Jan 2022, 86.6% of the Chilean population is fully vaccinated and 60.5% has received a booster jab.6 Despite the high vaccine coverage, the booster scheme is typically heterologous; 55.3% of the administered vaccines are inactivated virus vaccines (Sinovac's CoronaVac) and 36.4% mRNA-based (Pfizer-BioNTech's BNT162b2).7 Early reports suggest that vaccine-elicited neutralisation against Omicron might be lower in these settings.4 (d) Scheme: In the absence of genomic surveillance, declining trends on a previously dominant variant can mask the early stages of the exponential spread of another with increased transmissibility. In such a situation, policymakers would notice the replacement only after the tipping point in case numbers, which is typically too late to prevent a major surge.
Although genomic surveillance provides a snapshot of the circulating variants and their mutational signatures, its reliability depends on the quality of the sampling protocol. In the following, we summarise official recommendations for genomic surveillance10 into four simple rules to be considered for a robust sampling protocol for SARS-CoV-2 genomic surveillance: the CARD rules:
-
1.
Coordinated: Efforts from different entities should be centrally directed and harmonised to avoid selection biases, ensure representativeness, optimise resource allocation, and standardise sampling protocols. This entity should also ensure the transparency of sampling protocols, which should be reported along with genome information to GISAID.
-
2.
Adaptive: Sequencing intensity should be increased to provide better resolution when a new variant is signalled at POEs and detected in community transmission and decreased to its baseline rate when variant dominance is ascertained. By doing so, policymakers can act preventively (cf. Figure 1d).
-
3.
Representative: Sampling protocols should capture spatial heterogeneities, aim at different ethnographic groups, account for potential collider biases (e.g., as sampling only symptomatic, or unvaccinated individuals, or from the same contagion chain), and be proportional to the current incidence to maintain representativeness. In countries like Chile, where urban centres are sparsely distributed, representativeness must be assured on a regional basis.
-
4.
Differential: Samples collected at (i) points of entry (POEs) and (ii) surveillance centres should be dealt with and analysed separately, as they serve different purposes. Once novel threats are detected at POEs, community sampling should be increased so that early replacement dynamics between variants are accurately captured.
Since we already detected Omicron in community transmission, Chilean policymakers should dynamically adapt the stringency of non-pharmaceutical interventions to prevent its spread while offering booster mRNA vaccines to larger fractions of the population.11 In this stage, genomic surveillance is crucial for preparedness and policymakers should not only aim to surf the wave but also learn from it. Altogether, we have greater chances of succeeding if we play our CARDs right.
Declaration of interests
All authors declare no competing interests.
Acknowledgments
Contributions
All authors contributed equally.
Acknowledgements
We thank the molecular genetics laboratory (ISP Chile) for their hard work on genomic surveillance, completing more than 16.000 SARS-CoV-2 genomes to date. We gratefully acknowledge support from ANID Chile (projects PIA FB0001 and COVID0557).
References
- 1.Oróstica KY, Contreras S, Mohr SB, et al. Mutational signatures and transmissibility of SARS-CoV-2 Gamma and Lambda variants. arXiv preprint arXiv:2108.10018. 2021.
- 2.Viana R., Moyo S., Amoako D.G., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022 doi: 10.1038/d41586-021-03832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cele S., Jackson L., Khoury D.S., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021 doi: 10.1038/d41586-021-03824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pérez-Then E, Lucas C, Monteiro VS, et al. Immunogenicity of heterologous BNT162b2 booster in fully vaccinated individuals with CoronaVac against SARS-CoV-2 variants Delta and Omicron: the Dominican Republic Experience. medRxiv. 2021.
- 5.Lu L., Mok B.W., Chen L., et al. Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis. 2021:ciab1041. doi: 10.1093/cid/ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathieu E., Ritchie H., Ortiz-Ospina E., et al. A global database of COVID-19 vaccinations. Nat Hum Behav. 2021:1–7. doi: 10.1038/s41562-021-01122-8. [DOI] [PubMed] [Google Scholar]
- 7.168° informe epidemiológico, informe incidencia y gravedad de Casos covid-19 según antecedentes de Vacunación e informe Estrategia Nacional de Tta [Internet]. Departamento de Estadisticas e Información de Salud. [cited 2022Jan13]. Available from: https://deis.minsal.cl/
- 8.Torjesen I. Covid restrictions tighten as Omicron cases double every two to three days. BMJ. 2021;375:n3051. doi: 10.1136/bmj.n3051. [DOI] [PubMed] [Google Scholar]
- 9.Lyngse FP, Mortensen LH, Denwood MJ, et al. SARS-CoV-2 Omicron VOC transmission in Danish households. medRxiv. 2021 Dec.
- 10.World Health Organization. Guidance for surveillance of SARS-CoV-2 variants. Interim guidance. [Internet] 9 August 2021. Available from: https://www.who.int/publications-detail-redirect/WHO_2019-nCoV_surveillance_variants
- 11.Bauer S., Contreras S., Dehning J., et al. Relaxing restrictions at the pace of vaccination increases freedom and guards against further COVID-19 waves. PLoS Comput Biol. 2021;17(9) doi: 10.1371/journal.pcbi.1009288. [DOI] [PMC free article] [PubMed] [Google Scholar]