Abstract
The non-mevalonate or also called MEP pathway is an essential route for the biosynthesis of isoprenoid precursors in most bacteria and in microorganisms belonging to the Apicomplexa phylum, such as the parasite responsible for malaria. The absence of this pathway in mammalians makes it an interesting target for the discovery of novel anti-infectives. As last enzyme of this pathway, IspH is an oxygen sensitive [4Fe-4S] metalloenzyme that catalyzes 2H+/2e− reductions and a water elimination by involving non-conventional bioinorganic and bioorganometallic intermediates. After a detailed description of the discovery of the [4Fe-4S] cluster of IspH, this review focuses on the IspH mechanism discussing the results that have been obtained in the last decades using an approach combining chemistry, enzymology, crystallography, spectroscopies, and docking calculations. Considering the interesting druggability of this enzyme, a section about the inhibitors of IspH discovered up to now is reported as well. The presented results constitute a useful and rational help to inaugurate the design and development of new potential chemotherapeutics against pathogenic organisms.
Keywords: MEP pathway, antibiotics, IspH, LytB, [4Fe-4S] cluster, reductive dehydroxylation, bioorganometallic intermediate, inhibitors
1. IspH, an Enzyme Involved in the Biosynthesis of Isoprenoids
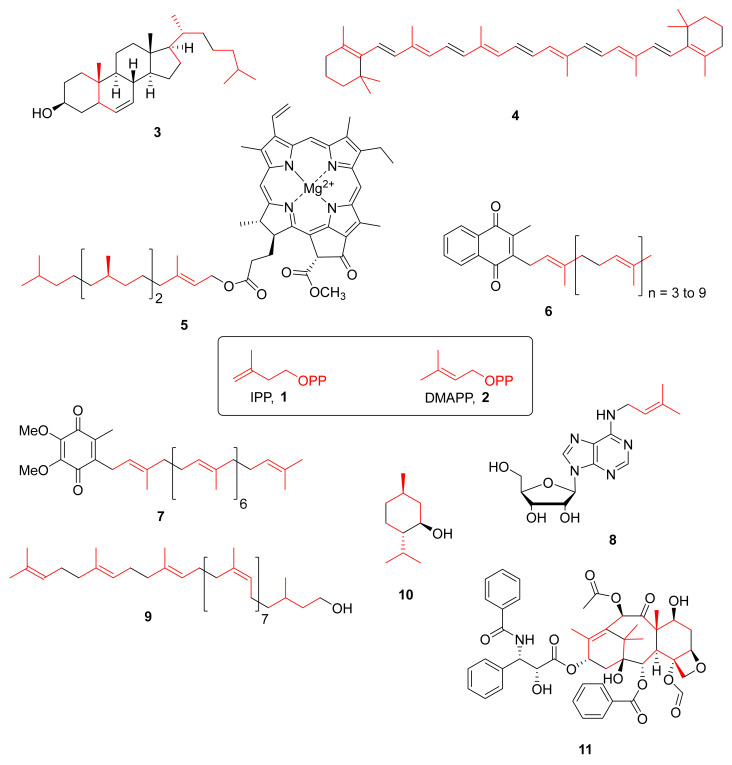
Isoprenoids, also known as terpenoids, constitute one of the largest classes of natural products with over 50,000 molecules. All terpenoids are derived from a common five-carbon precursor unit and are biosynthesized by the addition of one or more molecules of isopentenyl diphosphate (IPP, 1) to its isomer dimethylallyl diphosphate (DMAPP, 2) (Figure 1) [1]. The great structural diversity of isoprenoids is caused by different numbers of isoprene units, cyclization and oxidation reactions [2].
Figure 1.
Some examples of isoprenoids and their biological precursors. The common five-carbon precursors are highlighted in red.
Isoprenoids are present in all living organisms and are involved in numerous important biological processes. For instance, in most eukaryotes, sterols as for example cholesterol 3 play the role of membrane stabilizers, and in vertebrates, they play the role of precursors of steroid hormones and bile acids. In phototrophic organisms, carotenoids (β-carotene, 4) and chlorophylls (chlorophyll a, 5) are essential for the conversion of light into chemical energy. Menaquinone 6 or ubiquinone 7 are used for electron transport in cellular respiration. Isopentenyladenosine 8 is a modification found in some tRNAs in bacteria and eukaryotes. Bactoprenol 9 is a crucial isoprenoid for bacteria as it is responsible for the transport of the biosynthetic sugar precursors of peptidoglycan, an essential constituent of the bacterial envelop.
From early on, humans took advantage of isoprenoids as odor compounds in fragrance or as essential oils with one famous representative being menthol 10. More recently, taxol 11 was shown to be an efficient drug to treat some cancers.
Since the late 1950s, the mevalonate pathway was the only known metabolic pathway for the synthesis of IPP 1 and DMAPP 2 [3]. Importantly, this pathway is present in most eukaryotes including humans. One of its enzymes (HMGCoA reductase) is the target of drugs called statins that are given to patients suffering from hypercholesterolemia.
In the mid-1990s, an alternative pathway called the methylerythritol 4-phosphate (MEP) pathway was identified in bacteria, apicomplexan parasites and plant plastids for the biosynthesis of IPP and DMAPP [4,5,6].
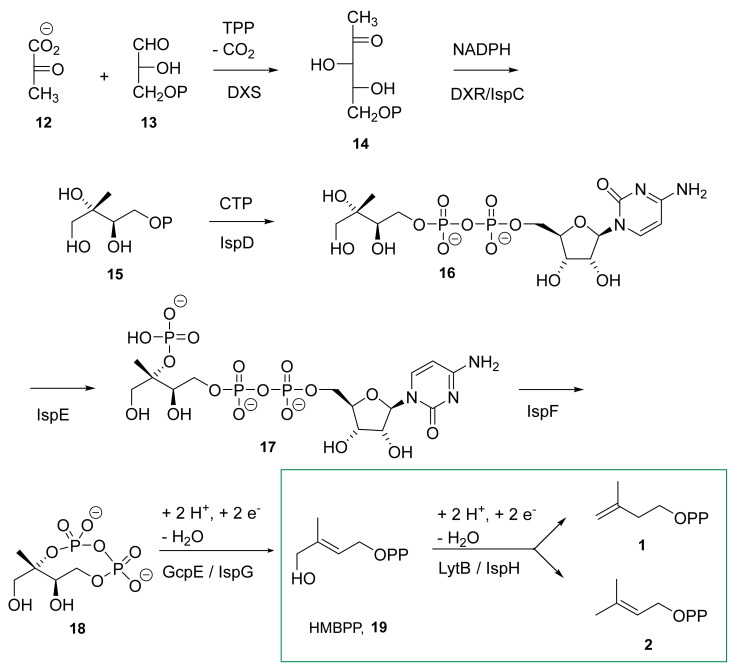
The MEP pathway (Scheme 1) starts with the condensation of pyruvate 12 and glyceraldehyde 3-phosphate 13 to form 1-deoxy-D-xylulose-5-phosphate (DXP, 14) catalyzed by the enzyme DXS. DXP is then converted into 2-C-methyl-D-erythritol-4-phosphate (MEP, 15) by IspC (also called DXR). A sequence of three enzyme-catalyzed steps (IspD, IspE, and IspF) further converts MEP 15 into 2-C-methyl-D-erythritol-2,4-cyclodiphosphate (MEcPP, 18) via cytidine diphosphate intermediates (4-diphosphocytidyl-2C-methyl-D-erythritol-2-phosphate 16 and 4-diphosphocytidyl-2C-methyl-D-erythritol-2-phosphate 17). IspG/GcpE catalyzes the penultimate reaction by reducing and opening the cyclic diphosphate intermediate 18 to form (E)-4-hydroxy-3-methylbut-2-en-1-yl diphosphate (HMBPP, 19).
Scheme 1.
Methylerythritol phosphate pathway. The reaction catalyzed by IspH is framed.
In the last step of this biosynthetic route, IspH, also called LytB (EC.1.17.7.4), catalyzes the conversion of HMBPP 19 into a mixture of IPP 1 and DMAPP 2. IspH, the subject of this review, catalyzes a two-electron reduction and OH-group elimination by using an oxygen sensitive [4Fe-4S]2+ center.
2. Occurrence of IspH
IspH as an enzyme of the MEP pathway occurs in most bacteria, plant chloroplasts, green algae, and apicomplexan but is absent in humans [7].
Interestingly, IspH is present in numerous disease-causing microbes including those posing a major challenge for new drug development. Indeed, antibiotic resistance is a growing concern as some infections have already become impossible to treat due to resistance. The worldwide emergence of carbapenemase-producing bacteria represents a health menace as carbapenems are often the last option for the treatment of patients infected by these bacteria [8]. Therefore, in 2017, the WHO published for the first time a list of antibiotic-resistant priority pathogens to guide research and development of new antibiotics as strains that cannot be fought by any antibiotic are emerging worldwide [8,9]. Importantly, 9 out of 12 of the classified bacteria contain an IspH enzyme (Table 1) highlighting its potential as a new target for antibiotic development.
Table 1.
Occurrence of IspH in the bacteria classified as priority by the WHO.
| Priority | Bacteria | IspH |
|---|---|---|
| critical | Acinetobacter baumannii, carbapenem-resistant | + |
| Pseudomonas aeruginosa, carbapenem-resistant | + | |
| Enterobacteriaceae, carbapenem-resistant, ESBL-producing | + | |
| high | Enterococcus faecium, vancomycin-resistant | − a |
| Staphylococcus aureus, methicillin-resistant, vancomycin-intermediate and resistant | − | |
| Helicobacter pylori, clarithromycin-resistant | + | |
| Campylobacter spp., fluoroquinolone-resistant | + | |
| Salmonellae, fluoroquinolone-resistant | + | |
| Neisseria gonorrhoeae, cephalosporin-resistant, fluoroquinolone-resistant | + | |
| medium | Streptococcus pneumoniae, penicillin-non-susceptible | − |
| Haemophilus influenzae, ampicillin-resistant | + | |
| Shigella spp., fluoroquinolone-resistant | + |
a partial IspH protein with incomplete catalytic site; rely on MVA pathway [14].
Mycobacterium tuberculosis, the bacteria responsible for tuberculosis is another health threat. The WHO estimated that 1.8 million people worldwide died from tuberculosis each year with 250,000 cases from antibiotic resistant M. tuberculosis [8]. Interestingly, M. tuberculosis synthesizes its isoprenoids according to the MEP pathway and therefore relies on a functional IspH for survival. Actually, it was reported that M. tuberculosis contained two IspH homologs called LytB1 and LytB2 with both showing activity when expressed in E. coli [10]. However, it was shown that LytB2 was essential for M. tuberculosis’ viability as LytB1 was unable to complement for the loss of LytB2 [11].
Malaria is transmitted to humans through bites from female mosquitoes infected by the apicomplexan Plasmodium falciparum parasite and is causing nearly half a million deaths worldwide each year, mostly children less than 5 years of age [12]. In the context of fighting this life-threatening disease, fosmidomycin, an inhibitor of the P. falciparum enzyme DXR, the second enzyme of the MEP pathway, is under clinical trial as an antimalarial agent in combination with piperaquine [13], making the MEP pathway an attractive target for drug development. As an enzyme of the MEP pathway, IspH is not only present in P. falciparum but also in Toxoplasma gondii, the parasite responsible for toxoplasmosis.
Since the discovery of IspH two decades ago, an intense research activity has been devoted to the understanding of its mechanism with the aim of developing unprecedented therapeutics with new modes of action that are urgently needed to combat infectious diseases.
3. The Discovery of the IspH Metalloenzyme
3.1. The Lytb Gene
The lytB gene was first described in Escherichia coli by the group of Ishiguro in 1993 as a gene involved in penicillin resistance [15] and was further reported to be present in other bacteria [16]. The group of Gantt revealed that lytB was an essential gene for a Synechocystis strain (cyanobacterium) as its disruption was lethal and that the growth of this mutant could only be restored by the addition of the alcohol analogs of IPP and DMAPP (isopentenol and dimethylallyl alcohol) in the media. Furthermore, the overexpression of this Synechocystis gene in an E. coli mutant engineered to produce carotenoids led to increased accumulation of these isoprenoids in E. coli highlighting the role of lytB in isoprenoid biosynthesis and most probably in the MEP pathway [17]. LytB was further shown to be an essential gene for E. coli and to be involved in the MEP pathway [18,19]. The group of Jomaa reported that an E. coli mutant harboring a deletion of the lytB gene and engineered for the utilization of exogenous mevalonate (to rescue the cells and allow the synthesis of crucial isoprenoids when the endogenous MEP pathway is disrupted), accumulates an immunogenic compound stimulating the proliferation of Vγ9/Vδ2 T cells. This compound was identified as HMBPP 19 after its isolation followed by a complete characterization using NMR spectroscopy and mass spectrometry, affording the first evidence that HMBPP was the substrate of the lytB gene product [20].
3.2. The IspH Catalyzed Reaction
A very elegant in vivo approach was used by Rohdich, Eisenreich, and collaborators to identify the reaction catalyzed by the lytB gene product. An E. coli strain overexpressing the xylB gene coding for D-xylulose kinase, known to phosphorylate 1-deoxy-D-xylulose (DX) in DXP, and all the genes of the MEP pathway including lytB was constructed. After feeding of [U-13C6]DX to this strain followed by 13C-NMR analysis of the bacterial crude extract, it was shown that this E. coli mutant accumulated [U-13C6]IPP and [U-13C6]DMAPP in a 5:1 ratio [21]. When lytB was absent in the construction, the mutant accumulated [U-13C6]HMBPP from [U-13C6]DX [22]. Together, these results indicate that the lytB gene product, further called LytB or IspH, catalyzes the conversion of HMBPP into IPP and DMAPP in a mixture of 5:1.
E. coli IspH has a mass of about 36 kDa [21]. First activity assays performed with purified E. coli IspH displayed no activity whereas the cell-free extract of E. coli expressing the recombinant IspH was active suggesting the involvement of auxiliary enzymes [23]. Purified E. coli IspH was further shown to convert HMBPP into IPP and DMAPP with an activity of 3 nmol min−1 mg−1 when assayed under anaerobic conditions in the presence of a reduction system such as photoreduced deazaflavin or NADPH/flavodoxin/flavodoxin reductase [24]. An approximatively 3000 times higher activity was reported by Jomaa and collaborators for recombinant IspH from the thermophilic eubacteria Aquifex aeolicus when purified in a glove-box under anaerobic conditions and using reduced methyl viologen as reduction system [25]. The authors pointed out the fact that the enzyme’s activity could not be maintained after contact with oxygen indicative of the enzyme’s oxygen sensitivity.
3.3. IspH, a [4Fe-4S] Metalloenzyme
The nature of the iron-sulfur cluster of the IspH enzyme has been the subject of debate for about a decade. The cysteine residue is a common ligand for iron centers of which many examples are known in literature [26]. Rohdich et al. assumed the presence of such a cluster due to three conserved cysteines which they found in ten different species containing the IspH protein [23]. The iron-sulfur cluster of IspH was first experimentally evidenced in the UV visible spectrum as a broad absorption maximum at a wavelength of 420 nm [25]. By comparison with spectroscopic data of other holoenzymes such as IspG [27], which is the enzyme prior to IspH in the MEP pathway, a [4Fe-4S]2+ cluster was proposed [24].
The early propositions concerning the [4Fe-4S]2+ cluster were backed up by Rohmer and collaborators conducting EPR spectroscopic measurements after reconstitution of the Fe/S cluster by incubation of the apoenzyme with FeCl3, Na2S, and dithiothreitol under anaerobic conditions [28]. [4Fe-4S]2+ itself does not show any signals on the EPR spectrum due to its diamagnetic nature. Upon reduction of the chemically reconstituted enzyme with dithionite, however, a paramagnetic species appeared having g values of 1.92(1) and 2.03(7). By analogy to earlier studies of iron-sulfur clusters measured by EPR spectroscopy, the microwave power half-saturation P1/2 was used as means to distinguish between a [2Fe-2S]+ and a [4Fe-4S]+ species [29,30]. With a rather high P1/2 value of 153 mW, a [4Fe-4S]+ cluster was proposed [28]. Iron species that could be detected on the enzyme purified under aerobic conditions were attributed to [3Fe-4S]+ (g = 2.02) and to ferric ions (g = 4.29) and exhibited signals already known from literature [31]. The discussion about the nature of the iron-sulfur cluster continued owing to new results from Rohdich et al. [32]. In their work, recombinant E. coli IspH protein was expressed in the presence of an isc operon, enabling the assembly of the full iron-sulfur cluster in vivo, and purified under anaerobic conditions. They found the same absorption maximum in the UV visible spectrum at about 410 nm as had been found before [28], but with a lower absorption coefficient (here: ε = 11,800 M−1 cm−1, compared to ε = 18,750 M−1 cm−1 in the previous study) suggesting a lower concentration of the iron-sulfur cluster. The corresponding EPR spectrum showed an anisotropic signal with g-values at 2.032(3) and 2.003(3). Its symmetry led them to the conclusion that a [3Fe-4S]+ was the active species as they found a high catalytic rate of 550 nmol mg−1 min−1 when using NADPH/flavodoxin reductase/flavodoxin as reduction system.
In order to solve this controversy, two groups independently investigated E. coli IspH purified under anaerobic conditions using Mössbauer spectroscopy [33,34]. Irrespective of its magnetic state, Mössbauer spectroscopy offers the possibility to gain structural information about the iron-sulfur cluster [35]. The analyses of the Mössbauer spectra indicated concordantly the structure of a [4Fe-4S]2+ cluster with a unique fourth iron site. The spectra could be simulated by the sum of three components with a ratio of 2:1:1 as shown in Table 2 and indicated an unusual [4Fe-4S]2+ center [33]. Two iron centers share one electron with an oxidation state of +2.5 each as derived from their isomer shift of δ = 0.42 mm s−1 and exhibit a tetrahedral coordination sphere with four sulfur ligands; besides the three sulfide ions, a cysteine residue is coordinated to each site as confirmed by X-ray structure analysis [36,37]. The two remaining iron sites consist of a high-spin Fe3+ ion and a high-spin Fe2+ ion, respectively [33]. The latter was assumed to have a coordination sphere with five or six ligands, most likely three sulfur and three nitrogen or oxygen ligands. Schünemann and collaborators further used Mössbauer spectroscopy to unequivocally access the nature of the Fe/S cluster of IspH existing in vivo in E. coli [33]. The authors recorded Mössbauer spectra of E. coli cells overexpressing IspH and of a WT strain, both strains grown under the same conditions. The difference of the spectra obtained for each type of cells could be simulated using the Mössbauer parameters obtained for pure 57Fe IspH. This proves that IspH assembles a [4Fe-4S]2+ cluster in vivo that is maintained when the protein is purified under anaerobic conditions. More details about the coordination sphere on this unique fourth iron atom were revealed by nuclear resonance vibrational spectroscopy (NRVS) on substrate-free IspH by the groups of Schünemann and Seemann [38]. NRVS is an advanced Mössbauer technique using synchrotron based radiation that is sensitive to vibrational modes involving iron [39]. By comparison of the experimental spectrum with DFT calculations obtained on an IspH structure harboring an incomplete Fe/S cluster (PDB: 3F7T) in which they inserted the missing iron, the authors concluded that the unique iron site was in a hexa-coordinated form with three water molecules. Together, these results highlight that IspH harbors an unusual [4Fe-4S] cluster linked to the three conserved cysteines with a unique iron site of oxidation state +2 that is linked to three sulfides of the cluster and to three water molecules.
Table 2.
Results of Mössbauer and NRVS spectroscopy on the [4Fe-4S]2+ cluster [33,38]. The Mössbauer spectra were recorded at T = 77 K and B = 0 T. The ΔEQ and δ values of the HMBPP-bound IspH are found in parentheses.
| ΔEQ [mm s−1] | δ [mm s−1] | Oxidation State and Coordination | |
|---|---|---|---|
| Component 1 (50%) | 1.21 (1.33) | 0.42 (0.42) | Tetrahedrally coordinated Fe2.5+, mixed valence iron pairs, 3 S2− and 1 Cys ligand |
| Component 2 (25%) | 0.89 (0.92) | 0.37 (0.38) | Tetrahedrally coordinated high-spin Fe3+, 3 S2− and 1 Cys ligand |
| Component 3 (25%) | 1.97 (1.00) | 0.89 (0.64) | Hexa-coordinated high-spin Fe2+, 3 S2− and 3 H2O (tetrahedrally coordinated, 3 S2− and OHHMBPP) |
The fact that the [4Fe-4S] cluster was the active species in the IspH enzyme was further confirmed by Duin et al. [40]. They could show that the enzyme’s activity was linearly dependent on the content of its cofactor.
It is now firmly established that IspH is an oxygen sensitive [4Fe-4S]2+ enzyme that catalyzes a reductive dehydroxylation and that the electrons needed for this reduction are provided in vivo by a biological system such as NADPH/flavodoxin reductase/flavodoxin in the case of E. coli or NADPH/ferredoxin reductase/ferredoxin in the case of P. falciparum [41].
4. The Mechanism of the Reductive Dehydroxylation
4.1. Protein Conformation in the Crystalline State
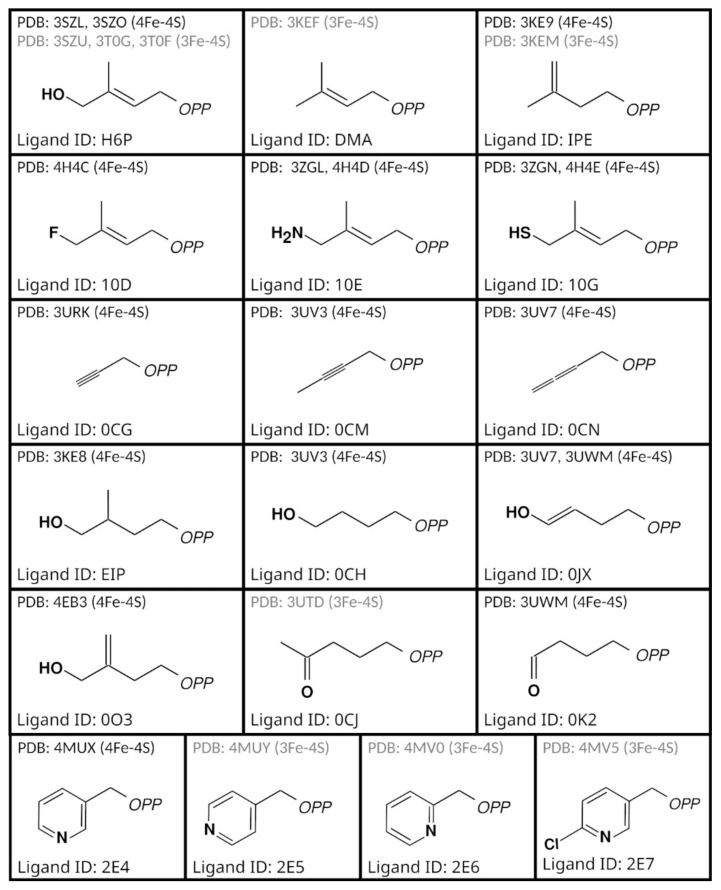
Crystal structures can give hints for the elucidation of the mechanism, but the propositions are usually confirmed by studies in solution. During the last two decades, 30 IspH crystallographic structures were deposited into the PDB database (Figure 2, Table 3). IspH structures from three different organisms are available. The first structure solved at the end of 2008 by Ermler and collaborators was the one from Aquifex aeolicus, a Gram-negative hyperthermophilic bacteria [36]. In 2009, Gräwert et al. released the first IspH structure of E. coli [42]. The last organism for which the crystallographic structure of IspH has been described is the one from the apicomplexan parasite P. falciparum [43]. For A. aeolicus and E. coli the full-length protein was crystallized whereas for P. falciparum the 217 first amino acids were not included in the construct used for the production and the crystallization of the enzyme.
Figure 2.
Structures of IspH deposited in the PDB database with their corresponding ligand.
Table 3.
Crystallization conditions and structural data of the reported IspH.
| PDB ID | Release Date | Source Organism | Crystallization Method (Vapor difusion) |
Crystal Growth Procedure | pH | Space Group | Res. (Å) | Cluster | Ligand ID | Ref |
|---|---|---|---|---|---|---|---|---|---|---|
| 3DNF | 30 December 2008 | A. aeolicus | Hanging drop | 0.1 M Tris-Hcl, 10% PEG 8000 | 8.0 | P 1 21 1 | 1.65 | Fe3 S4 | - | [36] |
| 4N7B | 20 November 2013 | P. falciparum | Sitting drop | 0.1 M Na citrate, 0.1 M (NH4)2SO4, 30% PEG 4000 | 5.6 | P 31 2 1 | 2.20 | Fe3 S4 | SO4 | [43] |
| 3F7T | 7 July 2009 | E. coli | Hanging drop | 1.6 M Potassium Phosphate | 8.0 | P 32 2 1 | 1.80 | Fe3 S4 | POP | [42] |
| 3KEF | 12 January 2010 | E. coli | Sitting drop | 0.1 M Bis-Tris, 0.2 M Li2SO4, 25% PEG 3350 | 6.5 | P 21 21 21 | 1.70 | Fe3 S4 | DMA | [37] |
| 3KEL | 1.80 | Fe3 S4 | POP | |||||||
| 3KE8 | 1.70 | Fe4 S4 | EIP | |||||||
| 3KE9 | 1.90 | Fe4 S4 | IPE | |||||||
| 3KEM | 7.0 | C 1 2 1 | 2.00 | Fe3 S4 | IPE | |||||
| 3T0G | 30 November 2011 | E. coli | Hanging drop | 0.1 M Bis-Tris, 0.2 M Li2SO4, 25% PEG3350 | 6.5 | P 21 21 21 | 2.10 | Fe3 S4 | H6P | [45] |
| 3T0F | 1.90 | Fe3 S4 | H6P | |||||||
| 3SZU | 1.40 | Fe3 S4 | H6P | |||||||
| 3SZL | 1.60 | Fe4 S4 | H6P | |||||||
| 3SZO | 1.60 | Fe4 S4 | H6P | |||||||
| 3URK | 5 September 2012 | E. coli | Sitting drop | 0.1 M Bis-Tris, 0.2 M (NH4)2SO4, 25% PEG3350 | 6.5 | P 21 21 21 | 1.50 | Fe4 S4 | 0CG | [46] |
| 3UTC | 1.90 | Fe4 S4 | 0JX | |||||||
| 3UTD | 1.70 | Fe3 S4 | OCJ | |||||||
| 3UV3 | 1.60 | Fe4 S4 | 0CM | |||||||
| 3UV6 | 1.70 | Fe4 S4 | 0CH | |||||||
| 3UV7 | 1.60 | Fe4 S4 | 0CN | |||||||
| 3UWM | 1.80 | Fe4 S4 | 0K2/0JX | |||||||
| 3ZGL | 9 January 2013 | E. coli | Hanging drop | 0.1M Bis-Tris, 0.2 M Li2SO4, 24% PEG 3350 | 6.5 | C 1 2 1 | 1.68 | Fe4 S4 | 10E | [47] |
| 3ZGN | 1.95 | Fe4 S4 | 10G | |||||||
| 4H4C | 23 January 2013 | E. coli | Sitting drop | 0.1 M Bis-Tris, 0.2 M (NH4)2SO4, 25% PEG3350 | 6.5 | P 21 21 21 | 1.80 | Fe4 S4 | 10D | [48] |
| 4H4D | 1.35 | Fe4 S4 | 10E | |||||||
| 4H4E | 1.70 | Fe4 S4 | 10G | |||||||
| 4EB3 | 6 February 2013 | E. coli | Sitting drop | 0.1 M Bis-Tris, 0.2 M (NH4)2SO4, 25% PEG3350 | 6.5 | P 21 21 21 | 1.90 | Fe4 S4 | 0O3 | [49] |
| 4MUY | 11 June 2014 | E. coli | Sitting drop | 0.1 M Bis-Tris, 0.2 M (NH4)2SO4, 25% PEG3350 | 6.5 | P 21 21 21 | 1.80 | Fe3 S4 | 2E5 | [50] |
| 4MV0 | 1.90 | Fe3 S4 | 2E6 | |||||||
| 4MV5 | 18 June 2014 | 1.90 | Fe3 S4 | 2E7 | ||||||
| 4MUX | 1.70 | Fe4 S4 | 2E4 |
The unprecedented fold of all IspH enzymes looks like a three-leaf clover and is composed of three structurally similar domains that do not display detectable similarity [42]. Each domain comprises a central parallel beta sheet containing four beta strands surrounded by three or four alpha helices. The active site is buried in a hydrophobic cavity embedded between the three domains. The [4Fe-4S] cluster, required for enzyme activity [28], is bound to the protein via three highly conserved cysteines [42]. Each domain contributes to the cluster coordination since these cysteines are located at the N-terminus of the first alpha helix of each of the three domains. E. coli and P. falciparum display an additional extended C-terminal loop containing a short beta sheet positioned above the active site pocket to protect the cluster from the surrounding solvent. In A. aeolicus, the dimerization of the protein is proposed to achieve the same role [44]. However, it should be noted that the crystal structure of the thermophilic enzyme does not reveal evidence of dimerization [36].
Among the 30 IspH structures solved only the one of A. aeolicus was obtained without a ligand into the binding pocket. This structure contained an incomplete [3Fe-4S] cluster. It should be mentioned that no crystal structure of substrate-free IspH in its [4Fe-4S] form has been reported to date. The inability to obtain such a structure is most probably due to the instability of the apical iron linked to three water molecules [38], which dissociates during the crystallization process. Comparison of this empty structure with the others reveals a conformational change. Indeed, upon substrate binding, a motion of the third domain of IspH results in the closing of the active site [42], and from the P. falciparum structure [43] it appears that a single sulfate ion bound in the substrate cavity is sufficient to induce the closing of the active site.
4.2. First Step: Binding of the Hydroxyl Group of HMBPP to the Iron-Sulfur Cluster
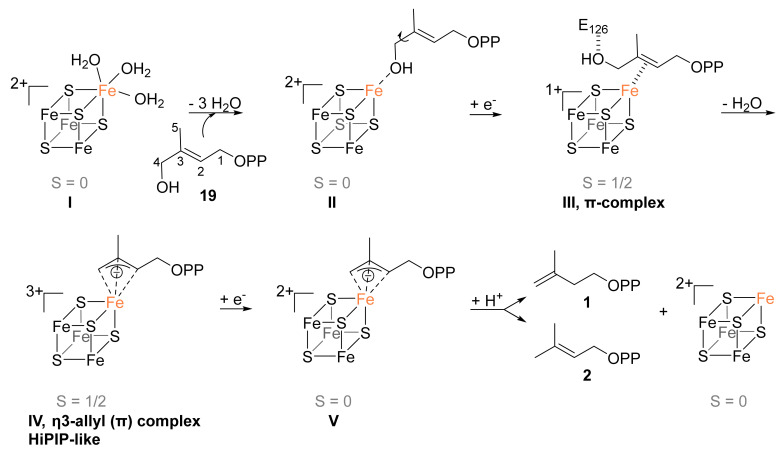
The proposed mechanistic course of the IspH enzyme is depicted in Scheme 2. Formally, the mechanism of IspH involves the elimination of water and the transfer of two electrons as well as two protons.
Scheme 2.
Proposition for the mechanism of the IspH catalyzed reductive dehydroxylation.
The active site of this reaction includes the [4Fe-4S] cluster that has already been described in detail in Section 3.3. Synthetic models for such 3:1 site-differentiated clusters were recently provided by the Suess group and it was shown that the unique fourth iron site could easily undergo ligand exchange [51] while the other ligands of the cluster were cysteine thiolates that are soft bases making stronger coordination bonds. It was proposed that the coordination geometry and the exchange at the unique iron site was controlled by steric effects provided by the ligands of the adjacent iron sites. Therefore, the opening of the coordination sphere of the apical iron required for its hexacoordination in IspH might be controlled by steric effects provided by the position of the cysteine ligands. Furthermore, broken-symmetry density functional theory (BS-DFT) studies performed on IspH indicated that the Fe-S bonds of the unique fourth iron site could be distorted which led to greater flexibility in ligand binding [52]. The three water molecules are good leaving groups contrary to the deprotonated cysteines exhibiting strong coordination bonds, suggesting a role of the apical iron of IspH in catalysis.
Docking studies of HMBPP using the A. aeolicus IspH structure in its [3Fe-4S]+ form in which the missing iron was computationally added revealed that the OH of HMBPP could bind to the fourth iron [36]. This observation was first experimentally confirmed using Mössbauer spectroscopy by adding the substrate HMBPP to 57Fe-LytB in solution [33]. A change in the isomer shift from δ3 = 0.89 mm s−1 to δ3′ = 0.64 mm s−1 of the component related to the unique fourth iron site of the cluster is observed in the presence of HMBPP, confirming the binding of the substrate to this iron site. The quadrupole splitting of this iron is also lowered from ΔEQ,3 = 1.97 mm s−1 to ΔEQ,3′ = 1.00 mm s−1 which indicates a change in the geometry of this iron ion. Upon substrate binding, the three water molecules of I are released and instead, the hydroxyl group of HMBPP is bound to the unique fourth iron site which is now tetrahedrally coordinated (intermediate II).
E. coli wild type or IspH mutants have been further crystallized with various ligands such as substrate, substrate analogs, products, or inhibitors (Figure 2, Table 3) [37,45,46,47,48,49,50]. In 2010, Groll, Bacher, and collaborators reported the first E. coli IspH structure with an intact [4Fe-4S] cluster as a complex with a substrate molecule (HMBPP; PDB ligand ID: H6P) [37]. (The ligand reported in the PDB file (3KE8) is not the substrate but an analog with the PDB ligand ID EIP, Figure 2, Table 3). The binding of HMBPP does not induce any significant backbone differences compared to the previously described structures. The diphosphate moiety inserts into a polar pocket composed of the side chains H41, 74, and 124, S225 and 269, T168, N227, and Q166 and is engaged in an extensive hydrogen bonding network. The substrate adopts a hairpin-like conformation to allow its C4 oxygen to form a coordination bond with the apical iron of the [4Fe-4S] cluster. The structures of the complex with each of the two reaction products (DMAPP and IPP) are also available [37]. Here again, the two molecules retain the same binding mode and their hairpin conformations but they are no longer bound to the apical iron.
The X-ray analysis of IspH in complex with its substrate confirmed that the hydroxyl moiety of HMBPP or its deprotonated form (the alkoxide) is bound to the unique fourth iron atom. With regards to the question of protonation, Blachly, Noodleman, and co-workers gave a plausible answer. By applying BS-DFT calculations, the protonated OH-form of HMBPP yields Mössbauer values comparable to the simulated values from experimental spectra [53]. Indeed, when HMBPP is bound to the apical iron as an alcohol, the HO-Fe distance is elongated with shortening of the distances of this iron and the S2− giving to the apical iron more ferrous character as revealed by Mössbauer spectroscopy.
Together these results show that the first step in the mechanism of IspH is the binding of the OH of the substrate to the unique iron site accompanied by the release of three water molecules. It is not clear what triggers the departure of the three water molecules but conformational change of the protein upon binding of the HMBPP substrate might generate movement of the cysteine ligands leading to reduction of the space around the apical iron and therefore favoring tetrahedral coordination geometry. Similar steric effects were reported in models [51]. Furthermore, the hydrogen bonding network revealed in the IspH structure in complex with the substrate will also increase the stability of this complex compared to the substrate-free enzyme harboring loosely bound water molecules.
4.3. Second Step: Reduction of the [4Fe-4S]2+ Cluster and Rotation of the CH2OH Group
Systematic studies by Liu et al. with different organic redox mediators along with the reducing agent dithionite revealed that the enzyme’s activity was dependent on the redox potential of these redox mediators [34]. The enzyme was most efficient when using a system of dithionite and a mediator with a redox potential of −450 mV vs. NHE while no detectable activity was found for redox mediators bearing redox potentials above −250 mV vs. NHE or below −720 mV vs. NHE. By using this chemical system, the authors reported an activity of 31.6 µmol min −1 mg −1 which is much higher than the activity of about 800 nmol min −1 mg −1 that is now frequently described for E. coli IspH using the natural NADPH/flavodoxin reductase/flavodoxin as reduction system [32,33].
After binding of the substrate to the apical iron atom, a rotation of the CH2OH group has been proposed upon reduction of the [4Fe-4S]2+ cluster to form intermediate III. The first who observed this rotation to be a part of the catalytic course were Oldfield et al. who conducted docking calculations of the substrate with reduced IspH [54]. After molecular mechanics optimization in silico, the hydroxyl group did not bind to the iron-sulfur cluster anymore but pointed towards the opposite direction as is depicted in Figure 3.
Figure 3.
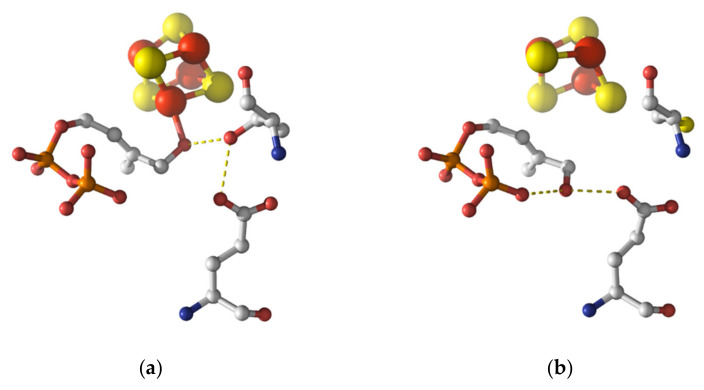
Crystallographic structures depicting the rotation of the hydroxymethyl group [45]. (a) The IspH WT was incubated with the substrate HMBPP. The full iron-sulfur cluster is present and the CH2OH group is coordinated to the apical iron, stabilized by T167. (b) The IspH T167C mutant was incubated with the substrate HMBPP. The CH2OH group has rotated about almost 180° and is now stabilized by the diphosphate group as well as the glutamate E126. The crystal structure only shows a [3Fe-4S] cluster.
Intermediate III was then observed experimentally by Groll and collaborators who provided crystal structures of E. coli IspH enzyme with bound HMBPP [45]. The authors compared the structures before and after irradiation with X-rays as synchrotron irradiation is known to reduce some metalloenzymes [55,56]. Based on the measured electron density of the crystal, two HMBPP conformers were suggested: the early intermediate in which the OH was bound to the apical iron and a new conformer with the CH2OH group partially rotated away from the apical iron. The hydroxyl group was now stabilized intramolecularly by the pyrophosphate group as well as by the amino acid residue E126 of IspH, which in turn formed a hydrogen bond with T167. However, the interpretation was still ambiguous as the initial binding of the hydroxyl group to the iron-sulfur cluster could also be detected.
Independently, Dickschat and collaborators conducted feeding experiments with deuterated isotopologues of the precursor of the DOX pathway, 1-deoxy-D-xylulose [57]. These in vivo experiments were conducted on the bacteria Streptomyces avermitilis that are known to produce the isoprenoid pentalenene for which the biosynthetic route was deciphered. After conversion, the configuration of the IPP and DMAPP products was determined by tracing back the course of the deuterium atoms in pentalenene. Given that only one of the two possible deuterated IPP analogs was formed, a rotation of the CH2OH group seemed likely to be involved in the formation of the required intermediate.
More evidence for this rotation was given by HYSCORE experiments on the E. coli IspH E126Q mutant incubated with 17O-labeled HMBPP ([4-17O]HMBPP) [49]. In previous experiments, this mutant exhibited only 0.3% activity compared to the wild type [45]. It was proposed that E126 played a role in the proton transfer step, so mutation to glutamine might hinder the subsequent protonation needed for water elimination. After reduction of the [4Fe-4S]2+ cluster of IspH, an intermediate species was detected with a very low 17O hyperfine coupling constant (Aiso = 1 MHz) compared to those of direct Fe-O bonds [49]. This led the authors to the conclusion that the substrate’s oxygen atom must have had a greater distance to the apical iron atom, most likely as a result of a rotation of the CH2OH group. The same authors had previously suggested that based on 13C-ENDOR experiments using the inactive E126A IspH mutant and [U-13C6]HMBPP a π- or π/σ-complex between the reduced cluster and the C2-C3 double bond of the substrate was formed. A similar complex had already been confirmed for a nitrogenase FeMo cofactor mutant in complex with allyl alcohol or ethylene [58,59]. The g-values of the IspH E126Q as well as the IspH E126A mutant were comparable with those of the Fe alkenyl complexes [49,54].
Similarly to the in vivo experiments, deuterated substrates were later prepared for more straightforward in vitro experiments with IspH [60]. HMBPP was deuterated on the C4-position to yield the two stereoisomers (S)-[4-2H1]HMBPP and (R)-[4-2H1]HMBPP, respectively. After incubation of each substrate with IspH in the presence of its naturally occurring reducing system, the configuration of the deuterium atoms on the products was determined by NMR spectroscopy. The S- and the R-configured substrates both yield [4-2H1]DMAPP without any stereocenters. However, the (S)-configured substrate only yields (E)-[4-2H1]IPP whereas the (R)-configured substrate only yields (Z)-[4-2H1]IPP. With regard to these results, Seemann and coworkers concluded that after reduction of the iron-sulfur cluster the CH2OH group rotated by almost 180° compared to the initial binding of the hydroxyl moiety to the apical iron atom as suggested from the previous structural data [45].
The driving force of this rotation may be explained by the fact that the apical ferrous ion becomes even softer upon reduction of the [4Fe-4S]2+ cluster [60]. According to the HSAB concept [61], this iron is more prone to form a complex with the soft alkene than with the harder hydroxyl group. The flexibility of the [4Fe-4S] cluster has been described in great detail by Holm et al. [62] and the reduced form of the cluster exhibits a greater core volume and distortions facilitating the ligand exchange. An η2-ring conformation of HMBPP was suggested by Blachly, Noodleman and coworkers [52]. The rotated CH2OH-group may be stabilized by forming intramolecular hydrogen bonds with the diphosphate moiety which was found to be the energetically most favorable configuration according to BS-DFT calculations when the [4Fe-4S] is reduced by one electron. Another possibility could be the formation of an η2-trans state where the CH2OH group is stabilized by the nearby E126. When the [4Fe-4S] is reduced by one electron both η2 configurations were showed to be lower in energy than the configuration having the hydroxyl of the substrate bound to the apical iron.
An alternative pathway where rotation of the CH2OH-group occurs before the reduction has also been proposed based on computed reduction potentials obtained from BS-DFT calculations [52].
4.4. Third Step: Protonation of the Hydroxyl Group and Elimination of Water
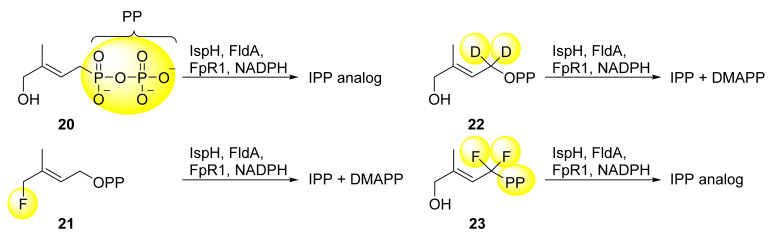
Mechanistic studies with substrate analogs were conducted by Liu et al. in order to get insights into the presumed C-O bond cleavage of the substrate’s hydroxyl group [63]. Replacing the diphosphate group at the C1 atom with a pyrophosphonate group (see Scheme 3, compound 20) still leads to the IPP analog after incubation with IspH and the naturally occurring reducing system as confirmed by 1H NMR spectroscopy. Thus, C-O bond cleavage at the C1 site of HMBPP can be excluded despite the fact that diphosphate is a better leaving group than the hydroxyl group. When incubating the fluorine analog 21 with the system mentioned above both IPP and DMAPP are obtained. Hence, the mechanism is thought to be analogous to that for HMBPP. As the fluorine atom exhibits a high electronegativity, only a heterolytic cleavage of the C-F bond comes into consideration which is assumed for the C4-O bond as well. Furthermore, isotopic labeling of the C1 hydrogen atoms with deuterium (compound 22) or substitution of hydrogen with fluorine (compound 23) still allows some turnover of the substrate, and at least one of the two products (the IPP analog in case of compound 23) are synthesized [64]. Therefore, it is unlikely that the C1 atom is deprotonated or otherwise directly involved in the mechanistic pathway.
Scheme 3.
Substrate analogs of HMBPP used for the study of C-O bond cleavage [63,64]. Discrepancies from the natural substrate HMBPP are highlighted in yellow.
As mentioned earlier, a possible proton source for the formation of water after hydroxyl elimination may be the glutamate residue E126. It lies in proximity to the hydroxyl group [45] and upon mutation to glutamine, aspartate, or alanine, the respective mutants E126Q, E126D, and E126A exhibit almost no activity [37,42,54]. The proton may be transferred to this glutamate residue by the adjacent histidine residue H124. It was shown that the respective mutants alanine (H124A) or phenylalanine (H124F) had a very low activity also [40,54]. Quantum mechanistics/molecular mechanistics (QM/MM) investigations support evidence that the dehydroxylation is triggered by a proton transfer from protonated E126 to the hydroxyl group of HMBPP mediated by a conserved water molecule [65].
4.5. Fourth Step: Formation of a HiPIP-like Cluster
The next intermediate (IV) was first observed by Duin et al. by cw EPR spectroscopy [40]. It was trapped by incubation of one electron-reduced IspH with its substrate without further electron source for complete turnover. The resulting paramagnetic species had g-values of 2.173, 2.013, and 1.997, comparable to those of ferredoxin:thioredoxin reductase that contains a [4Fe-4S]3+ cluster [66]. The authors proposed that the intermediate in the IspH catalyzed reaction was linked to [4Fe-4S]3+, but further interpretation of the data remained obscure.
Oldfield et al. clarified the nature of this intermediate. By freeze-quenching a reaction mixture containing IspH, dithionite as the only reducing agent, and the substrate, the reaction was slowed down and the trapped intermediate IV could be analyzed by EPR spectroscopy. Eventually, they measured the same intermediate as did Duin et al. [49]. The g-tensor was found to be g = [2.171, 2.010, 1.994] and was also similar to that found in other high-potential iron-sulfur proteins (HiPIP) [67]. The authors confirmed the presence of a [4Fe-4S]3+ cluster, which implicates that formally a two-electron transfer from the cluster to the substrate and the elimination of the hydroxyl group would occur.
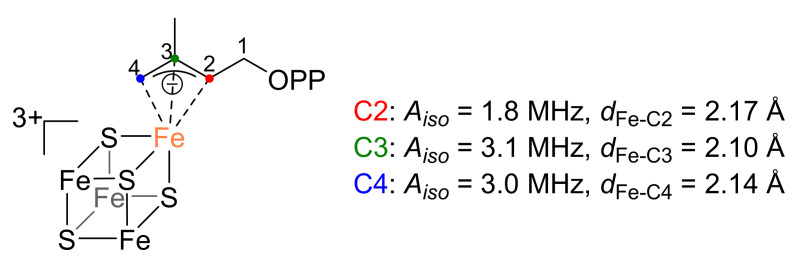
Support for this [4Fe-4S]3+-intermediate was provided by the same group after HYSCORE experiments on one-electron reduced IspH incubated with HMBPP [68]. In these experiments, the electron source for the second electron transfer was again removed before incubation with the substrate, thus a possible reaction intermediate could be trapped. For the HYSCORE measurements, the oxygen of the substrate’s hydroxyl group had been isotopically labeled with 17O. As no 17O hyperfine coupling was detected, it was suggested that in the intermediate under discussion the hydroxyl group had already been eliminated. Furthermore, in another experiment one of the respective C2, C3, and C4 atoms of HMBPP had been labeled with 13C. In this way, possible hyperfine couplings to the unique fourth Fe species of the cluster could be measured. The 13C labeling led to hyperfine coupling constants of Aiso = 1.8 MHz (C2), Aiso = 3.1 MHz (C3) and Aiso = 3.0 MHz (C4) (Figure 4). By DFT calculations, Fe-C distances of 2.17, 2.10, and 2.14 Å for the respective C2, C3, and C4 atoms were estimated in silico. As simulated Aiso values were in the range of the experimental values, the proposed distances were taken as a good approximation and suggest an allyl anion that forms an η3-complex with the [4Fe-4S]3+ cluster. The Fe-C distances of 2.6–2.7 Å measured previously in the crystal structure of IspH in complex with the “converted substrate” reported by Gräwert et al. presumably represent this same intermediate [37]. The HiPIP-like cluster in complex with the delocalized π-system is considered as one of the rare examples of a bioorganometallic intermediate found in nature.
Figure 4.
Schematic representation of the HiPIP-like η3-allyl (π/σ) complex. The hyperfine coupling constants Aiso obtained after HYSCORE measurements of 13C-labelled HMBPP as well as the iron-carbon distances dFe-C estimated by DFT calculations are given for the C2, C3, and C4 atom [68].
The formation of the η3-complex occurs after two-electron transfer from the cluster to the substrate and the elimination of the hydroxyl group. It is still unclear if the OH leaves the substrate as OH or as water after protonation by E126 and if the two-electron transfers are concomitant with the water formation.
4.6. Fifth Step: Protonation and Formation of the Two Products IPP and DMAPP
IspH catalyzes the reductive dehydroxylation of HMBPP into IPP and DMAPP with a ratio of about 4–6:1 [23]. This ratio is different from the equilibrium provided by isopentenyl diphosphate isomerase that favors the thermodynamically more stable DMAPP with a ratio of 3:7 (IPP:DMAPP) [24]. Obviously, protonation on the C2 atom of the allyl anion complex yields IPP while on the C4 atom, it yields DMAPP. The source of proton is still a matter of debate and several hypotheses for the proton mediator were done.
The proton source was identified early by Rohmer and co-workers without knowledge of the IspH-catalyzed mechanism [69,70] using the bacterium Zymomonas mobilis that due to its peculiar metabolic pathways (metabolization of glucose according to the Entner-Doudoroff pathway and no conversion of pyruvate into glyceraldehyde phosphate) is able to synthesize the two possible NAD(P)2H isotopomers from [1-2H]glucose. After feeding Z. mobilis with [1-2H]glucose, two pentacyclic triterpenes of the hopanoid series were analyzed by 1H- and 13C-NMR spectroscopy. These hopanoids exhibited deuterium atoms on definite positions of the carbon skeleton that could be traced back to the C2 and C4 positions of IPP and DMAPP. While the deuterium atom on the C4 position could be explained by an earlier reduction step in the MEP pathway catalyzed by DXR, the deuterium atom on the C2 position revealed the signature of a further reduction step in the formation of IPP and DMAPP that could not be explained at that time. The authors also observed that the deuterium was introduced in the pro-S position of IPP. Later on, the authors concluded that the deuterium was introduced by IspH and that NADPH was the proton donor for this catalytic step at least for this bacterium [28,71]. Feeding experiments of different methylerythritol isotopomers in E. coli [69] or of labelled deoxyxylulose in E. coli and in several plant cells [71] by Arigoni and co-workers and by the groups of Rohmer and Bach shed light on the introduction of a deuterium on C2 of IPP in the pro-S position [71,72,73,74].
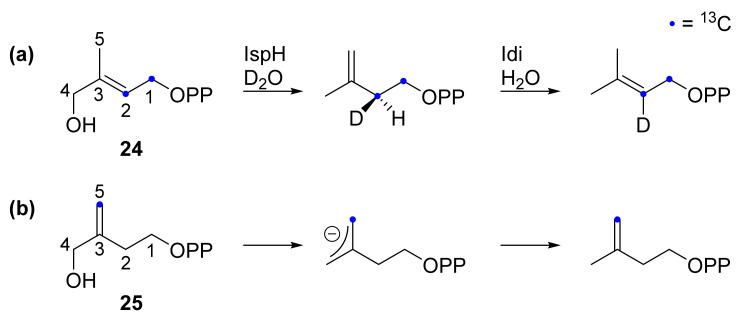
The stereochemical course of the IPP formation was further investigated by Rohdich, Eisenreich et al. using enzymology on IspH [75]. The authors conducted 13C-NMR spectroscopy on isotopically labeled HMBPP (24) after incubation with IspH in D2O followed by incubation with isopentenyl diphosphate isomerase in H2O (Scheme 4a). The E. coli IPP isomerase is known to eliminate the pro-R-proton of IPP. The resulting DMAPP exhibits a deuterium atom on the C2 atom which thus corresponds to the pro-S-proton of IPP. Hence, it is proposed that protonation occurs only from the si face of the allyl anion. Based on the crystallographic structure, the diphosphate moiety of the substrate may serve as proton mediator on this site as it is in close vicinity to the C2 and C4 atoms [37,42].
Scheme 4.
(a) Stereochemical course of deuterium and hydrogen after incubation with IspH in D2O and isopentenyl diphosphate isomerase in H2O [75]. (b) Substrate analog 25 used by Liu et al. and the product formed in the IspH catalyzed reaction [76].
In line with these assumptions, the product formation of the 13C isotopically labeled substrate analog 25 was investigated and it was found that only the IPP analog was formed [76] showing that the protonation was sterically restricted to the C4 atom. The authors explained this observation by the higher distance between the C5 of 25 and its diphosphate once 25 was in the active site of IspH that would prevent the diphosphate to protonate C5.
5. IspH Inhibitors
Since the first breakthrough into the IspH mechanism, it has been readily exploited for the generation of potent inhibitors to develop unprecedented and safe therapeutics to fight against multiresistant bacteria and the malaria parasite Plasmodium falciparum.
Most of the compounds which have been classified as IspH inhibitors interact with the apical iron of the [4Fe-4S] cluster through different mechanisms. The group of Hirsch demonstrated that the active site and the neighboring pockets of IspH are druggable and, moreover, molecules larger than the natural substrate might be able to access this spacious site [77]. All the inhibitors studied so far can be divided into four classes: substrate analogs, pyridine diphosphates, alkyne derivatives, and non-diphosphate compounds.
5.1. Analogs of the HMBPP Substrate as Inhibitors
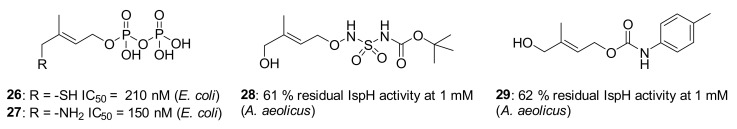
The first step in the IspH mechanism is the binding of the hydroxyl group of the substrate to the apical iron (Fe2+) site of the [4Fe-4S]2+ cluster. Replacement of this OH by a softer ligand such as a thiol was envisioned. Furthermore, substrate analogs not able to undergo the OH elimination step such as amino analogs were considered. In this context, Poulter, Seemann, and collaborators conceived and tested two HMBPP analogs where a thiol or an amino group replaces the hydroxyl group of the substrate (Figure 5) [78,79]. A complete kinetic investigation under anaerobic conditions revealed that the thiol analog 26 and the amino analog 27 were extremely potent inhibitors of E. coli IspH displaying competitive mode of inhibition with Ki values in the nanomolar range (Ki = 24 nM for 26 and Ki = 54 nM for 27). Furthermore, 27 presented a slow binding inhibition mode with the slow step being the formation of the enzyme-inhibitor complex. This might be explained by the fact that the amino group undergoes a deprotonation at physiological pH to bind to the apical iron, therefore, the authors assumed that the slow step was the deprotonation of the amine [78]. Previous Mössbauer spectroscopy studies showed a transition from octahedral to tetrahedral geometry for the apical iron when adding 26 or 27 to the enzyme, as it was already observed when HMBPP is in complex with IspH [79]. This finding validates that these inhibitors are bound via their amino and thiol group to the unique iron of the [4Fe-4S] cluster. The same binding mode was observed in the crystal structures of E. coli IspH in complex with these two potent inhibitors (AMBPP 27; PDB ligand ID: 10E and TMBPP 26; PDB ligand ID: 10G), confirming that 27 is bound to the apical iron of the [4Fe-4S]2+ cluster by its amino group and 26 is bound to this iron by its thiol group [47,48].
Figure 5.
Substrate analogs as inhibitors of IspH.
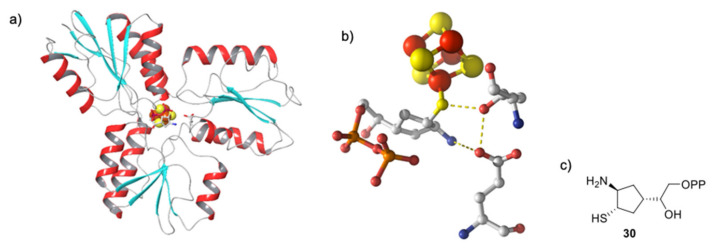
Nuclear resonance vibrational spectroscopy (NRVS) data were further recorded by Schünemann and collaborators for IspH bound to either of these inhibitors [38]. The measured NRVS data and those calculated based on the X-ray structure reported by Span et al. [48] showed reasonable agreement and the deviation was attributed to the presence of different IspH conformations in protein crystals and in solution. These differences were better explained once Borel and collaborators reported new structures of E. coli IspH in complex with these inhibitors in which the authors ascertained that the apical iron was present with an occupancy of 100% [47] as 35–45% of IspH molecules in the previously reported [48] E. coli IspH structure in complex with the amino analog 27 seemed to have an incomplete cluster. Furthermore, a detailed analysis of the E. coli IspH:26 complex revealed that 26 bound in each monomer adopted two different conformations [47]: a classical one, superimposed on HMBPP already described, and one in which the C4 atom of 26 was pulled away from the diphosphate group and E126. The fact that IspH can accommodate more than one substrate conformer was proposed to be related to the selectivity of the protonation and consequently to the genesis of the two products. The presence of two conformers was further exploited to generate in silico putative IspH inhibitors with enhanced affinity by mimicking the space occupied by both conformers by a cyclopentyl group linked to a thiol and a diphosphate moiety [47]. Interestingly, the best virtual inhibitor candidate (30) that arose from these in silico evolution experiments is displayed in Figure 6 and revealed binding of the thiol moiety to the apical iron of the [4Fe-4S]2+ cluster and formation of a salt bridge between an amino group and E126. However, no data of the potency of inhibition of 30 on IspH was reported.
Figure 6.
In silico docking result of the IspH inhibitor candidate 30 designed based on the structure of E. coli IspH:26 (PDB ID: 3ZGN) and showing the best docking score. (a) Docking result of 30 binding to IspH; (b) active site; (c) chemical structure of 30.
Two more substrate analogs have been designed by the group of Van Calenbergh and the group of Jomaa by replacing the diphosphate of HMBPP with a N-acyl-N′-oxy sulfamate function leading to 28 and a carbamate moiety leading to 29 (Figure 5). These compounds, tested at 1 mM concentration, showed a very low inhibition of IspH from A. aeolicus [80]. The poor inhibition potency of both molecules might be related to the fact that they lack the diphosphate group which is involved in important interactions with different histidine residues, such as His42 and His124 that are highly conserved in IspH of several species [36].
5.2. Pyridine Diphosphate as Inhibitors
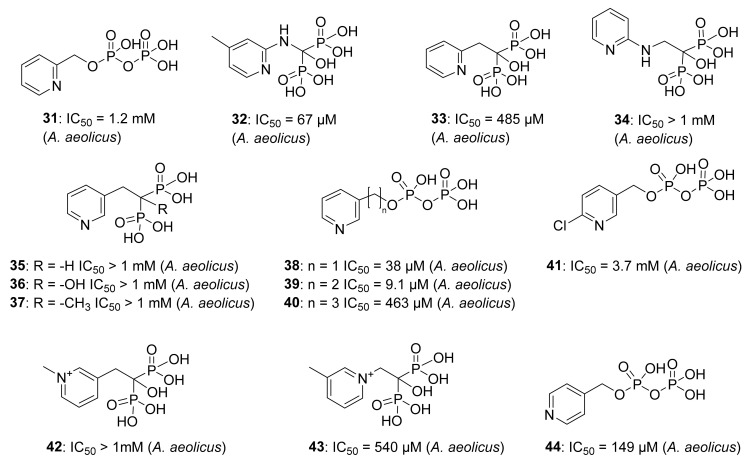
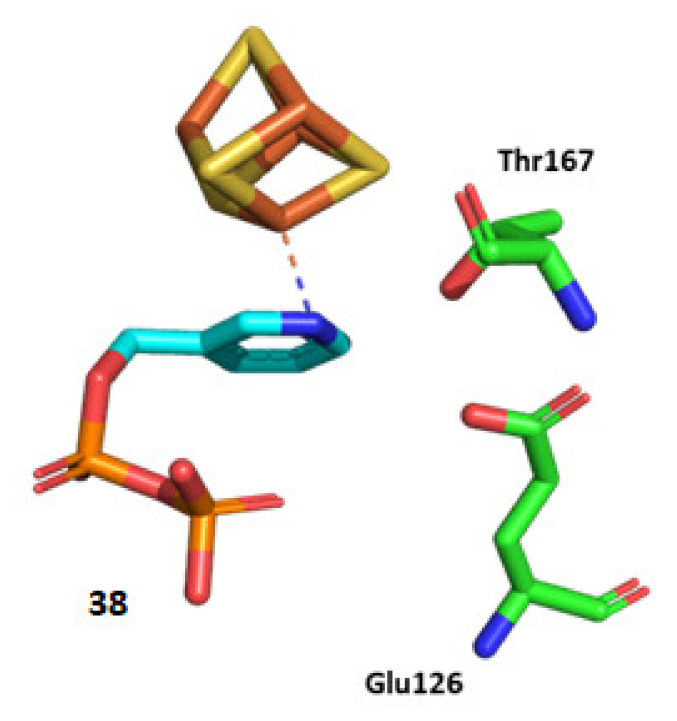
Wang et al. studied a set of pyridine derivatives substituted in ortho, meta, and para (31–44) (Figure 7), as potential inhibitors of IspH from A. aeolicus [81]. In some compounds, the diphosphate has been replaced by the bioisostere bisphosphonate, which is more stable to phosphatase hydrolysis in vivo [82]. 14N and 15N HYSCORE investigations by Oldfield and co-workers demonstrate that inhibitor 38 binds to the apical iron probably via a η1 bonding between the nitrogen and the [4Fe-4S] cluster, whereas no 14N hyperfine interactions were observed with ortho and para pyridinyl analogs [83]. Furthermore, crystallographic studies reported by Span, Groll, Oldfield, and collaborators confirmed that the pyridine is close to the apical iron (2.3–2.4 Å), and the authors observed a continuous electron density between the nitrogen of the pyridine derivative and the apical iron. According to these observations and quantum chemical calculations, Span et al. provided evidence that the binding occurs through a η2 coordination [50]. Moreover, the authors explored the orientation that the pyridine ring adopts in relation to the [4Fe-4S] cluster. X-ray crystallographic studies on E. coli IspH demonstrated that the nitrogen of the pyridine points in a direction likely represented in Figure 8. This orientation is presumably the result of the interaction of 38 with two pockets of the catalytic site, a lipophilic one characterized by four Val15, Val43, Val73, Val99 residues, and a hydrophilic site composed of Glu126, Thr167, Thr168, Thr200, and Asn227 residues [50].
Figure 7.
Compounds 31–44, tested as inhibitors of IspH from A. aeolicus.
Figure 8.
Structural representation of Ec-IspH in complex with compound 38 (PDB code 4MUX) [50]. The amino acids are shown as sticks, as well as the [4Fe-4S] cluster and the ligands.
Considering the insights acquired about the pyridine binding mode, it seems clearer why 38 and 39 showed the best inhibitory potency in this class. For example, 41 is not an inhibitor probably due to electron-withdrawing effect of the chlorine in ortho-position, which reduces the capacity of the nitrogen as electron donor Lewis-base. For similar reasons, the cation pyridinium analogs 42 and 43 did not show any biological activity.
5.3. Alkyne Derivatives as Inhibitors
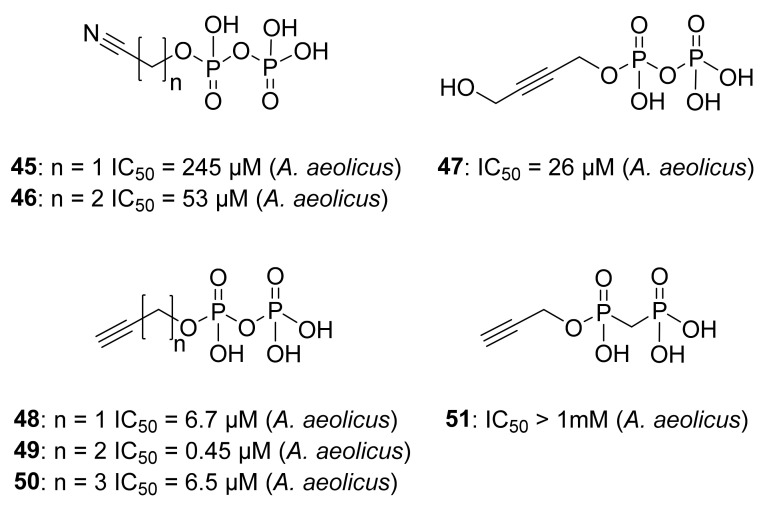
Wang et al. designed a series of alkyne diphosphates 45–50 and a phosphonate analog 51 (Figure 9), which were first screened by EPR spectroscopy. Furthermore, EPR investigations and specifically 13C-ENDOR results of reduced A. aeolicus IspH with [13C3]-48 revealed strong 13C hyperfine coupling interactions (6 MHz) [54], allowing to conclude that the acetylenic diphosphates bind through the alkyne fragment to the [4Fe-4S]+ cluster by π or π-σ interactions. The A. aeolicus IspH inhibition assays demonstrated that 49 is the best inhibitor of this category (IC50 = 0.45 µM). The substitution of the diphosphate with a phosphonate for 51 led to a dramatic drop in inhibition potency, which might be due to the fact that the diphosphate fits better in the conserved cationic pocket seen in previous crystallographic investigations [37]. Moreover, Span et al. examined the compounds 48–50 bound to the oxidized IspH from E. coli by Mössbauer spectroscopy [46]. Previous investigations [33] helped to easily interpret the results obtained by the authors, then stating that the apical iron undergoes a conversion from octahedral to tetrahedral geometry with a 3S1O coordination, likewise what happens with the natural substrate HMBPP. Co-crystallographic structures of 48 with oxidized IspH were in accordance with the unexpected Mössbauer spectroscopy results and demonstrated that the binding between the alkyne residue and the fourth iron is not direct, indeed a water molecule is in between [46]. This result was further validated using NRVS measurements of the IspH-48 complex [84]. NRVS studies of the free-substrate IspH had previously shown that the apical iron was bound to three sulfide ions of the cluster and to three molecules of water [38]. This finding was analog for the NRVS results of IspH in its [4Fe-4S]2+ state in presence of 48, where the fourth iron was in a tetrahedral geometry not linked to the acetylene residue but to a single molecule of water.
Figure 9.
Compounds 45–51, tested as inhibitors of IspH from A. aeolicus.
On the other hand, due to the impossibility of studying the oxidized IspH by EPR, the inhibitor 48 was examined by EPR in complex with the reduced IspH. The results obtained by O’Dowd et al. [84] revealed plausible π-interactions between the inhibitor 48 and the reduced [4Fe-4S] cluster.
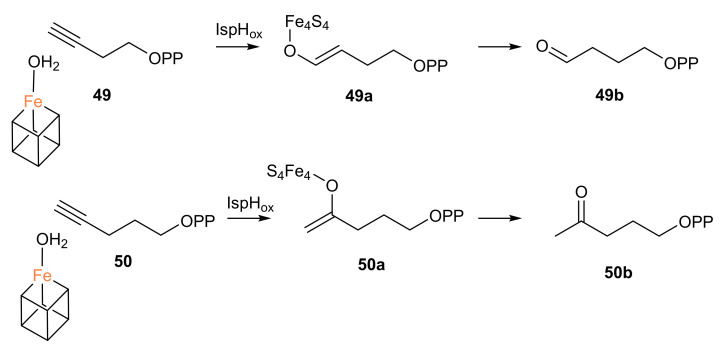
Surprisingly, X-ray structures of the best inhibitor 49 showed that IspH converts this compound to an aldehyde (Scheme 5), revealing two ligand conformations. One is an η1-O-enolate in complex with the apical iron, and the other one is a cyclic conformation of the aldehyde which binds to the Glu126 residue. Moreover, 50 was shown to react with the oxidized IspH, yielding in this case a ketone (Scheme 5). These unexpected products were also confirmed by 1H-NMR spectroscopy and electrospray ionization mass spectrometry (ESI-MS) [46].
Scheme 5.
Reactions of the inhibitors 49 and 50 catalyzed by the enzyme IspH in the oxidized state. The ligand bound to the apical iron could also be an OH.
5.4. Non-Diphosphate Inhibitors
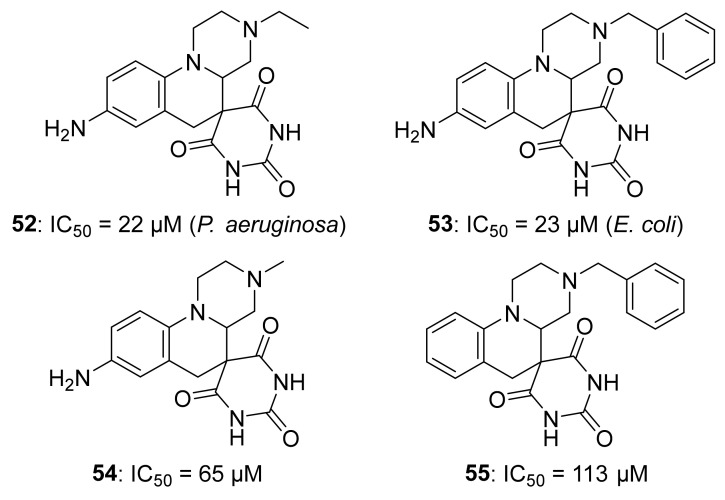
O’Dowd et al. used an in silico approach to screen a series of compounds from ZINC and NCI libraries and after having tested some hits for IspH inhibition, they identified 52 and 53 as two new IspH inhibitors (Figure 10) [84]. In both compounds, it is possible to see a common barbituric acid sub-structure which makes the hits more drug-like, similarly to antibacterial agents developed by Pharmacia Corporation [85]. The compound 52 resulted to be a good inhibitor of IspH from P. aeruginosa, with an IC50 = 22 µM. On the other hand, the analog 53 inhibits IspH from E. coli, showing as result an IC50 = 23 µM [84]. After docking experiments of 53 into the A. aeolicus IspH structure (PDB ID: 3DNF), the authors proposed a plausible binding mode where the barbiturate might chelate the apical iron of the [4Fe-4S]2+ cluster, similarly to how barbiturate enolates do with Zn2+ in other metalloenzymes [86,87]. However, EPR studies for 53 in complex with the reduced IspH from P. aeruginosa did show a very low signal intensity, probably due to a change of the redox potential upon ligand binding or to a different relaxation mode [84]. These compounds could be developed as potential drugs for several reasons: they solved the problem related to the lability of the diphosphate moiety present in other inhibitors in vivo [82]; they respect the Lipinski’s rule [88] and they are not PAINS compounds [89]. Although they are promising drug-like compounds, further studies to validate the binding mode should be realized.
Figure 10.
Compounds 52– 55 as inhibitors of IspH. The origin of IspH for inhibitions assays of 54 and 55 was not mentioned by the authors.
Potent IspH inhibitors have already been discovered with 26 and 27 showing the best inhibition properties. However, it should be noticed that no inhibitors having an activity on bacteria or apicomplexan parasites have been reported to date.
6. Conclusions
The oxygen sensitivity of IspH explains why the MEP pathway was overlooked for so many decades and not evidenced in the late 1950s when the mevalonate pathway was discovered by Bloch, Lynen, Cornforth. At that time, the researchers performed their enzymology experiments on the bench and did not have the equipment to work under anaerobic conditions nor the knowledge and spectroscopies needed for the characterization of metalloenzymes. Thanks to the advent of EPR, Mössbauer spectroscopy, and bioinorganic chemistry, it is now clear that IspH contains a peculiar [4Fe-4S]2+ cluster that is the active species and enables the electron transfer towards the substrate HMBPP in concert with a biological reduction system. Crystal structures, (labelled) substrate analogs synthesis and spectroscopic techniques permitted insights into the catalytic mechanism of IspH and led to the discovery of the first examples of bioorganometallic intermediates involving [4Fe-4S] centers. However, the complete mechanism of the IspH-catalyzed reaction is not fully understood. While it is now established that the first step is the binding of the substrate to the [4Fe-4S]2+ cluster, that a rotation of the OH of the substrate occurs upon reduction of the [4Fe-4S]2+ cluster and that the S = 1/2 η3-allyl-[4Fe-4S]3+ intermediate is formed, it is still unclear how the OH or water is removed and how the electron transfer occurs. As most investigations relied on paramagnetic species, no evidence of transient diamagnetic intermediates could be trapped. X-ray crystallography would be an elegant method to trap such intermediates. However, it suffers from the high lability of the apical iron and the obtained structures do not necessarily represent the intermediates present in solution. Obviously, Mössbauer spectroscopy will be a key player in the identification of such intermediates and more sensitive methods such as NRVS will be needed. Further investigations will also rely on the synthesis of new biomimetic models or other innovative molecular tools. The proton source involved in the formation of the two final products is still under discussion. Another question is the ratio of the products formed. What triggers the formation of about six times more of the less thermodynamically favored IPP compared to DMAPP? This raises also the question about the reduction systems. Why do some species use flavodoxin and others ferredoxin to transfer the electrons to the [4Fe-4S]2+ center of IspH? How are these electrons transferred in vivo? One puzzling question is the maintenance of an oxygen sensitive [4Fe-4S]2+ cluster in microorganisms living under aerobic conditions or in plant chloroplasts that produce O2 in phototropic conditions. Is the cluster continuously repaired? Are protecting enzymes involved? ErpA and NfuA proteins might be such candidates as they were shown to interact with IspH in E. coli, and NfuA was proposed to assist ErpA in Fe-S cluster delivery under oxidative stress [90].
Although some questions are still unanswered, most insights acquired about the mechanism of IspH were already exploited to discover promising inhibitors. However, none of these potent inhibitors were reported to be active on cells. Therefore, one of the big remaining challenges will also be the discovery of such molecules as they would represent good starting points to bring to the market innovative drugs with unprecedented modes of action that are urgently needed to fight against multidrug-resistant infectious diseases.
Acknowledgments
We are very grateful to our dear colleague and friend Jean-Luc Ferrer who sadly passed away in 2020. We would like to dedicate this manuscript to his memory and to his outstanding contribution in protein crystallography.
Author Contributions
Writing—original draft preparation, H.J., G.I.B., F.B., P.C., M.S.; writing—review and editing, H.J., G.I.B., F.B., P.C., M.S.; funding acquisition—F.B., M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 860816 and from the ‘Université franco-allemande’. This work of the Interdisciplinary Thematic Institute InnoVec, as part of the ITI program of the University of Strasbourg, CNRS and Inserm, was supported by IdEx Unistra (ANR-10-IDEX-0002), and by SFRI-STRAT’US project (ANR-20-SFRI-0012) under the framework of the French Investments for the Future Program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cane D.E., Barton D.H.R., Nakanishi K., Meth-Cohn O., editors. Comprehensive Natural Products Chemistry: Isoprenoids, Including Carotenoids and Steroids. Volume 2 Elsevier; Oxford, UK: 1999. [Google Scholar]
- 2.Sacchettini J.C., Poulter C.D. Creating isoprenoid diversity. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 3.Bloch K. Sterol molecule: Structure, biosynthesis, and function. Steroids. 1992;57:378–383. doi: 10.1016/0039-128X(92)90081-J. [DOI] [PubMed] [Google Scholar]
- 4.Rohmer M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 1999;16:565–574. doi: 10.1039/a709175c. [DOI] [PubMed] [Google Scholar]
- 5.Eisenreich W., Schwarz M., Cartayrade A., Arigoni D., Zenk M.H., Bacher A. The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem. Biol. 1998;5:R221–R233. doi: 10.1016/S1074-5521(98)90002-3. [DOI] [PubMed] [Google Scholar]
- 6.Lichtenthaler H.K. The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1999;50:47–65. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
- 7.Rohmer M., Grosdemange-Billiard C., Seemann M., Tritsch D. Isoprenoid biosynthesis as a novel target for antibacterial and antiparasitic drugs. Curr. Opin. Investig. Drugs. 2004;5:154–162. [PubMed] [Google Scholar]
- 8.Priorization of Pathogens to Guide Discovery, Research and Development of New Antibioitics for Drug-Resistant Bacterial Infections, Including Tuberculosis. World Health Organization; Geneva, Switzerland: 2017. Report No. WHO/EMP/IAU/2017.12. [Google Scholar]
- 9.Miethke M., Pieroni M., Weber T., Brönstrup M., Hammann P., Halby L., Arimondo P.B., Glaser P., Aigle B., Bode H.B., et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021;5:726–749. doi: 10.1038/s41570-021-00313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mann F.M., Xu M., Davenport E.K., Peters R.J. Functional characterization and evolution of the isotuberculosinol operon in Mycobacterium tuberculosis and related mycobacteria. Front. Microbiol. 2012;3:368. doi: 10.3389/fmicb.2012.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown A.C., Kokoczka R., Parish T. LytB1 and LytB2 of Mycobacterium tuberculosis are not genetically redundant. PLoS ONE. 2015;10:e0135638. doi: 10.1371/journal.pone.0135638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kafai N.M., Odom John A.R. Malaria in children. Infect. Dis. Clin. North Am. 2018;32:189–200. doi: 10.1016/j.idc.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mombo-Ngoma G., Remppis J., Sievers M., Zoleko Manego R., Endamne L., Kabwende L., Veletzky L., Nguyen T.T., Groger M., Lötsch F., et al. Efficacy and safety of fosmidomycin–piperaquine as nonartemisinin-based combination therapy for uncomplicated falciparum malaria: A single-arm, age de-escalation proof-of-concept study in Gabon. Clin. Infect. Dis. 2018;66:1823–1830. doi: 10.1093/cid/cix1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hedl M., Sutherlin A., Wilding E.I., Mazzulla M., McDevitt D., Lane P., Burgner J.W., Lehnbeuter K.R., Stauffacher C.V., Gwynn M.N., et al. Enterococcus faecalis acetoacetyl-coenzyme a thiolase/3-hydroxy-3-methylglutaryl-coenzyme a reductase, a dual-function protein of isopentenyl diphosphate biosynthesis. J. Bacteriol. 2002;184:2116–2122. doi: 10.1128/JB.184.8.2116-2122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gustafson C.E., Kaul S., Ishiguro E.E. Identification of the Escherichia coli lytB gene, which is involved in penicillin tolerance and control of the stringent response. J. Bacteriol. 1993;175:1203–1205. doi: 10.1128/jb.175.4.1203-1205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potter S., Yang X., Boulanger M.J., Ishiguro E.E. Occurrence of homologs of the Escherichia coli lytB gene in gram-negative bacterial species. J. Bacteriol. 1998;180:1959–1961. doi: 10.1128/JB.180.7.1959-1961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham F.X., Lafond T.P., Gantt E. Evidence of a role for LytB in the nonmevalonate pathway of isoprenoid biosynthesis. J. Bacteriol. 2000;182:5841–5848. doi: 10.1128/JB.182.20.5841-5848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altincicek B., Kollas A.-K., Sanderbrand S., Wiesner J., Hintz M., Beck E., Jomaa H. GcpE is involved in the 2-C-methyl-D-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. J. Bacteriol. 2001;183:2411–2416. doi: 10.1128/JB.183.8.2411-2416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McAteer S., Coulson A., McLennan N., Masters M. The lytB gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. J. Bacteriol. 2001;183:7403–7407. doi: 10.1128/JB.183.24.7403-7407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altincicek B., Kollas A.-K., Eberl M., Wiesner J., Sanderbrand S., Hintz M., Beck E., Jomaa H. LytB, a novel gene of the 2-C-methyl-D-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. FEBS Lett. 2001;499:37–40. doi: 10.1016/S0014-5793(01)02516-9. [DOI] [PubMed] [Google Scholar]
- 21.Rohdich F., Hecht S., Gartner K., Adam P., Krieger C., Amslinger S., Arigoni D., Bacher A., Eisenreich W. Studies on the nonmevalonate terpene biosynthetic pathway: Metabolic role of IspH (LytB) protein. Proc. Natl. Acad. Sci. USA. 2002;99:1158–1163. doi: 10.1073/pnas.032658999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hecht S., Eisenreich W., Adam P., Amslinger S., Kis K., Bacher A., Arigoni D., Rohdich F. Studies on the nonmevalonate pathway to terpenes: The role of the GcpE (IspG) protein. Proc. Natl. Acad. Sci. USA. 2001;98:14837–14842. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adam P., Hecht S., Eisenreich W., Kaiser J., Gräwert T., Arigoni D., Bacher A., Rohdich F. Biosynthesis of terpenes: Studies on 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase. Proc. Natl. Acad. Sci. USA. 2002;99:12108–12113. doi: 10.1073/pnas.182412599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohdich F., Zepeck F., Adam P., Hecht S., Kaiser J., Laupitz R., Amslinger S., Eisenreich W., Bacher A., Arigoni D. The deoxyxylulose phosphate pathway of isoprenoid biosynthesis: Studies on the mechanisms of the reactions catalyzed by IspG and IspH protein. Proc. Natl. Acad. Sci. USA. 2002;100:1586–1591. doi: 10.1073/pnas.0337742100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altincicek B., Duin E.C., Reichenberg A., Hedderich R., Kollas A.-K., Hintz M., Wagner S., Wiesner J., Beck E., Jomaa H. LytB protein catalyzes the terminal step of the 2-C-methyl-D-erythritol-4-phosphate pathway of isoprenoid biosynthesis. FEBS Lett. 2002;532:437–440. doi: 10.1016/S0014-5793(02)03726-2. [DOI] [PubMed] [Google Scholar]
- 26.Beinert H. Iron-sulfur proteins: Ancient structures, still full of surprises. J. Biol. Inorg. Chem. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- 27.Seemann M., Tse Sum Bui B., Wolff M., Tritsch D., Campos N., Boronat A., Marquet A., Rohmer M. Isoprenoid biosynthesis through the methylerythritol phosphate pathway: The (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase (GcpE) is a [4Fe–4S] protein. Angew. Chem. Int. Ed. 2002;41:4337–4339. doi: 10.1002/1521-3773(20021115)41:22<4337::AID-ANIE4337>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 28.Wolff M., Seemann M., Tse Sum Bui B., Frapart Y., Tritsch D., Estrabot A.G., Rodríguez-Concepción M., Boronat A., Marquet A., Rohmer M. Isoprenoid biosynthesis via the methylerythritol phosphate pathway: The (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase (lytB/IspH) from Escherichia coli is a [4Fe-4S] protein. FEBS Lett. 2003;541:115–120. doi: 10.1016/S0014-5793(03)00317-X. [DOI] [PubMed] [Google Scholar]
- 29.Rupp H., Rao K.K., Hall D.O., Cammack R. Electron spin relaxation of iron-sulphur proteins studied by microwave power saturation. Biochim. Biophys. Acta Protein Struct. 1978;537:255–269. doi: 10.1016/0005-2795(78)90509-3. [DOI] [PubMed] [Google Scholar]
- 30.Ollagnier S., Mulliez E., Schmidt P.P., Eliasson R., Gaillard J., Deronzier C., Bergman T., Gräslund A., Reichard P., Fontecave M. Activation of the anaerobic ribonucleotide reductase from Escherichia coli. J. Biol. Chem. 1997;272:24216–24223. doi: 10.1074/jbc.272.39.24216. [DOI] [PubMed] [Google Scholar]
- 31.Petrovich R.M., Ruzicka F.J., Reed G.H., Frey P.A. Characterization of iron-sulfur clusters in lysine 2,3-aminomutase by electron paramagnetic resonance spectroscopy. Biochemistry. 1992;31:10774–10781. doi: 10.1021/bi00159a019. [DOI] [PubMed] [Google Scholar]
- 32.Gräwert T., Kaiser J., Zepeck F., Laupitz R., Hecht S., Amslinger S., Schramek N., Schleicher E., Weber S., Haslbeck M., et al. IspH protein of Escherichia coli: Studies on iron−sulfur cluster implementation and catalysis. J. Am. Chem. Soc. 2004;126:12847–12855. doi: 10.1021/ja0471727. [DOI] [PubMed] [Google Scholar]
- 33.Seemann M., Janthawornpong K., Schweizer J., Böttger L.H., Janoschka A., Ahrens-Botzong A., Tambou E.N., Rotthaus O., Trautwein A.X., Rohmer M., et al. Isoprenoid biosynthesis via the MEP pathway: In vivo Mössbauer spectroscopy identifies a [4Fe-4S]2+ center with unusual coordination sphere in the LytB protein. J. Am. Chem. Soc. 2009;131:13184–13185. doi: 10.1021/ja9012408. [DOI] [PubMed] [Google Scholar]
- 34.Xiao Y., Chu L., Sanakis Y., Liu P. Revisiting the IspH catalytic system in the deoxyxylulose phosphate pathway: Achieving high activity. J. Am. Chem. Soc. 2009;131:9931–9933. doi: 10.1021/ja903778d. [DOI] [PubMed] [Google Scholar]
- 35.Schünemann V., Winkler H. Structure and dynamics of biomolecules studied by Mössbauer spectroscopy. Rep. Prog. Phys. 2000;63:263–353. doi: 10.1088/0034-4885/63/3/202. [DOI] [Google Scholar]
- 36.Rekittke I., Wiesner J., Röhrich R., Demmer U., Warkentin E., Xu W., Troschke K., Hintz M., No J.H., Duin E.C., et al. Structure of (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase, the terminal enzyme of the non-mevalonate pathway. J. Am. Chem. Soc. 2008;130:17206–17207. doi: 10.1021/ja806668q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gräwert T., Span I., Eisenreich W., Rohdich F., Eppinger J., Bacher A., Groll M. Probing the reaction mechanism of IspH protein by X-ray structure analysis. Proc. Natl. Acad. Sci. USA. 2010;107:1077–1081. doi: 10.1073/pnas.0913045107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faus I., Reinhard A., Rackwitz S., Wolny J.A., Schlage K., Wille H.-C., Chumakov A., Krasutsky S., Chaignon P., Poulter C.D., et al. Isoprenoid biosynthesis in pathogenic bacteria: Nuclear resonance vibrational spectroscopy provides insight into the unusual [4Fe-4S] cluster of the E. Coli LytB/IspH protein. Angew. Chem. Int. Ed. 2015;54:12584–12587. doi: 10.1002/anie.201502494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheidt W., Durbin S., Sage J. Nuclear resonance vibrational spectroscopy? NRVS. J. Inorg. Biochem. 2005;99:60–71. doi: 10.1016/j.jinorgbio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Xu W., Lees N.S., Hall D., Welideniya D., Hoffman B.M., Duin E.C. A closer look at the spectroscopic properties of possible reaction intermediates in wild-type and mutant (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase. Biochemistry. 2012;51:4835–4849. doi: 10.1021/bi3001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Röhrich R.C., Englert N., Troschke K., Reichenberg A., Hintz M., Seeber F., Balconi E., Aliverti A., Zanetti G., Köhler U., et al. Reconstitution of an apicoplast-localised electron transfer pathway involved in the isoprenoid biosynthesis of Plasmodium falciparum. FEBS Lett. 2005;579:6433–6438. doi: 10.1016/j.febslet.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 42.Gräwert T., Rohdich F., Span I., Bacher A., Eisenreich W., Eppinger J., Groll M. Structure of active IspH enzyme from Escherichia coli provides mechanistic insights into substrate reduction. Angew. Chem. Int. Ed. 2009;48:5756–5759. doi: 10.1002/anie.200900548. [DOI] [PubMed] [Google Scholar]
- 43.Rekittke I., Olkhova E., Wiesner J., Demmer U., Warkentin E., Jomaa H., Ermler U. Structure of the (E)-4-hydroxy-3-methyl-but-2-enyl-diphosphate reductase from Plasmodium falciparum. FEBS Lett. 2013;587:3968–3972. doi: 10.1016/j.febslet.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 44.Frank A., Groll M. The methylerythritol phosphate pathway to isoprenoids. Chem. Rev. 2017;117:5675–5703. doi: 10.1021/acs.chemrev.6b00537. [DOI] [PubMed] [Google Scholar]
- 45.Span I., Gräwert T., Bacher A., Eisenreich W., Groll M. Crystal structures of mutant IspH proteins reveal a rotation of the substrate’s hydroxymethyl group during catalysis. J. Mol. Biol. 2012;416:1–9. doi: 10.1016/j.jmb.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 46.Span I., Wang K., Wang W., Zhang Y., Bacher A., Eisenreich W., Li K., Schulz C., Oldfield E., Groll M. Discovery of acetylene hydratase activity of the iron–sulphur protein IspH. Nat. Commun. 2012;3:1042. doi: 10.1038/ncomms2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borel F., Barbier E., Krasutsky S., Janthawornpong K., Chaignon P., Poulter C.D., Ferrer J.-L., Seemann M. Further insight into crystal structures of Escherichia coli IspH/lytB in complex with two potent inhibitors of the MEP pathway: A starting point for rational design of new antimicrobials. ChemBioChem. 2017;18:2137–2144. doi: 10.1002/cbic.201700363. [DOI] [PubMed] [Google Scholar]
- 48.Span I., Wang K., Wang W., Jauch J., Eisenreich W., Bacher A., Oldfield E., Groll M. Structures of fluoro, amino, and thiol inhibitors bound to the [Fe4S4] protein IspH. Angew. Chem. Int. Ed. 2013;52:2118–2121. doi: 10.1002/anie.201208469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang W., Wang K., Span I., Jauch J., Bacher A., Groll M., Oldfield E. Are free radicals involved in IspH catalysis? An EPR and crystallographic investigation. J. Am. Chem. Soc. 2012;134:11225–11234. doi: 10.1021/ja303445z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Span I., Wang K., Eisenreich W., Bacher A., Zhang Y., Oldfield E., Groll M. Insights into the binding of pyridines to the iron–sulfur enzyme IspH. J. Am. Chem. Soc. 2014;136:7926–7932. doi: 10.1021/ja501127j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown A.C., Suess D.L.M. Controlling substrate binding to Fe4S4 clusters through remote steric effects. Inorg. Chem. 2019;58:5273–5280. doi: 10.1021/acs.inorgchem.9b00360. [DOI] [PubMed] [Google Scholar]
- 52.Blachly P.G., Sandala G.M., Giammona D.A., Bashford D., McCammon J.A., Noodleman L. Broken-symmetry DFT computations for the reaction pathway of IspH, an iron–sulfur enzyme in pathogenic bacteria. Inorg. Chem. 2015;54:6439–6461. doi: 10.1021/acs.inorgchem.5b00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blachly P.G., Sandala G.M., Giammona D.A., Liu T., Bashford D., McCammon J.A., Noodleman L. Use of broken-symmetry density functional theory to characterize the IspH oxidized state: Implications for IspH mechanism and inhibition. J. Chem. Theory Comput. 2014;10:3871–3884. doi: 10.1021/ct5005214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W., Wang K., Liu Y.-L., No J.-H., Li J., Nilges M.J., Oldfield E. Bioorganometallic mechanism of action, and inhibition, of IspH. Proc. Natl. Acad. Sci. USA. 2010;107:4522–4527. doi: 10.1073/pnas.0911087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuss J.O., Tsai C.-L., Ishida J.P., Tainer J.A. Emerging critical roles of Fe–S clusters in DNA replication and repair. Bioch. Biophys. Acta Mol. Cell Res. 2015;1853:1253–1271. doi: 10.1016/j.bbamcr.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ter Beek J., Parkash V., Bylund G.O., Osterman P., Sauer-Eriksson A.E., Johansson E. Structural evidence for an essential Fe–S cluster in the catalytic core domain of DNA polymerase ϵ. Nucleic Acids Res. 2019;47:5712–5722. doi: 10.1093/nar/gkz248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Citron C.A., Brock N.L., Rabe P., Dickschat J.S. The stereochemical course and mechanism of the IspH reaction. Angew. Chem. Int. Ed. 2012;51:4053–4057. doi: 10.1002/anie.201201110. [DOI] [PubMed] [Google Scholar]
- 58.Lee H.-I., Igarashi R.Y., Laryukhin M., Doan P.E., Dos Santos P.C., Dean D.R., Seefeldt L.C., Hoffman B.M. An organometallic intermediate during alkyne reduction by nitrogenase. J. Am. Chem. Soc. 2004;126:9563–9569. doi: 10.1021/ja048714n. [DOI] [PubMed] [Google Scholar]
- 59.Lee H.-I., Sørlie M., Christiansen J., Yang T.-C., Shao J., Dean D.R., Hales B.J., Hoffman B.M. Electron inventory, kinetic assignment (En), structure, and bonding of nitrogenase turnover intermediates with C2H2 and CO. J. Am. Chem. Soc. 2005;127:15880–15890. doi: 10.1021/ja054078x. [DOI] [PubMed] [Google Scholar]
- 60.Chaignon P., Petit B.E., Vincent B., Allouche L., Seemann M. Methylerythritol phosphate pathway: Enzymatic evidence for a rotation in the Lytb/IspH catalyzed reaction. Chem. Eur. J. 2020;26:1032–1036. doi: 10.1002/chem.201904676. [DOI] [PubMed] [Google Scholar]
- 61.Pearson R.G. Hard and soft acids and bases. J. Am. Chem. Soc. 1963;85:7. doi: 10.1021/ja00905a001. [DOI] [Google Scholar]
- 62.Rao P.V., Holm R.H. Synthetic analogues of the active sites of iron−sulfur proteins. Chem. Rev. 2004;104:527–559. doi: 10.1002/chin.200421274. [DOI] [PubMed] [Google Scholar]
- 63.Xiao Y., Zhao Z.K., Liu P. Mechanistic studies of IspH in the deoxyxylulose phosphate pathway: Heterolytic C-O bond cleavage at C4 position. J. Am. Chem. Soc. 2008;130:2164–2165. doi: 10.1021/ja710245d. [DOI] [PubMed] [Google Scholar]
- 64.Xiao Y., Liu P. IspH protein of the deoxyxylulose phosphate pathway: Mechanistic studies with C1 -deuterium-labeled substrate and fluorinated analogue. Angew. Chem. Int. Ed. 2008;47:9722–9725. doi: 10.1002/anie.200803452. [DOI] [PubMed] [Google Scholar]
- 65.Abdel-Azeim S., Jedidi A., Eppinger J., Cavallo L. Mechanistic insights into the reductive dehydroxylation pathway for the biosynthesis of isoprenoids promoted by the IspH enzyme. Chem. Sci. 2015;6:5643–5651. doi: 10.1039/C5SC01693B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walters E.M., Garcia-Serres R., Jameson G.N.L., Glauser D.A., Bourquin F., Manieri W., Schürmann P., Johnson M.K., Huynh B.H. Spectroscopic characterization of site-specific [Fe4S4] cluster chemistry in ferredoxin:thioredoxin reductase: Implications for the catalytic mechanism. J. Am. Chem. Soc. 2005;127:9612–9624. doi: 10.1021/ja051909q. [DOI] [PubMed] [Google Scholar]
- 67.Belinskii M. Spin coupling model for tetrameric iron clusters in ferredoxins. I. Theory, exchange levels, g-factors. Chem. Phys. 1993;172:189–211. doi: 10.1016/0301-0104(93)80116-Q. [DOI] [Google Scholar]
- 68.Li J., Wang K., Smirnova T.I., Khade R.L., Zhang Y., Oldfield E. Isoprenoid biosynthesis: Ferraoxetane or allyl anion mechanism for IspH catalysis? Angew. Chem. Int. Ed. 2013;52:6522–6525. doi: 10.1002/anie.201302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Charon L., Pale-Grosdemange C., Rohmer M. On the reduction steps in the mevalonate independent 2-C-methyi-D-erythritol 4-phosphate (MEP) pathway for isoprenoid biosynthesis in the bacterium Zymomonas Mobilis. Tetrahedron Lett. 1999;40:7231–7234. doi: 10.1016/S0040-4039(99)01531-2. [DOI] [Google Scholar]
- 70.Charon L., Hoeffler J.-F., Pale-Grosdemange C., Lois L.-M., Campos N., Boronat A., Rohmer M. Deuterium-labelled isotopomers of 2-C-methyl-D-erythritol as tools for the elucidation of the 2-C-methyl-D-erythritol 4-phosphate pathway for isoprenoid biosynthesis. Biochem. J. 2000;346:737–742. doi: 10.1042/bj3460737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoeffler J.-F., Hemmerlin A., Grosdemange-Billiard C., Bach T.J., Rohmer M. Isoprenoid biosynthesis in higher plants and in Escherichia coli: On the branching in the methylerythritol phosphate pathway and the independent biosynthesis of isopentenyl diphosphate and dimethylallyl diphosphate. Biochem. J. 2002;366:573–583. doi: 10.1042/bj20020337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giner J.-L., Jaun B., Arigoni D. Biosynthesis of isoprenoids in Escherichia coli: The fate of the 3-H and 4-H atoms of 1-deoxy-d-xylulose. Chem. Commun. 1998:1857–1858. doi: 10.1039/a804461i. [DOI] [Google Scholar]
- 73.Rieder C., Jaun B., Arigoni D. On the early steps of cineol biosynthesis in Eucalyptus globulus. Helv. Chim. Acta. 2000;83:2504–2513. doi: 10.1002/1522-2675(20000906)83:9<2504::AID-HLCA2504>3.0.CO;2-Z. [DOI] [Google Scholar]
- 74.Arigoni D., Eisenreich W., Latzel C., Sagner S., Radykewicz T., Zenk M.H., Bacher A. Dimethylallyl pyrophosphate is not the committed precursor of isopentenyl pyrophosphate during terpenoid biosynthesis from 1-deoxyxylulose in higher plants. Proc. Natl. Acad. Sci. USA. 1999;96:1309–1314. doi: 10.1073/pnas.96.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Laupitz R., Gräwert T., Rieder C., Zepeck F., Bacher A., Arigoni D., Rohdich F., Eisenreich W. Stereochemical studies on the making and unmaking of isopentenyl diphosphate in different biological systems. Chem. Biodivers. 2004;1:1367–1376. doi: 10.1002/cbdv.200490099. [DOI] [PubMed] [Google Scholar]
- 76.Chang W., Xiao Y., Liu H., Liu P. Mechanistic studies of an IspH-catalyzed reaction: Implications for substrate binding and protonation in the biosynthesis of isoprenoids. Angew. Chem. Int. Ed. 2011;50:12304–12307. doi: 10.1002/anie.201104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Masini T., Kroezen B.S., Hirsch A.K.H. Druggability of the enzymes of the non-mevalonate-pathway. Drug Discov. Today. 2013;18:1256–1262. doi: 10.1016/j.drudis.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 78.Janthawornpong K., Krasutsky S., Chaignon P., Rohmer M., Poulter C.D., Seemann M. Inhibition of IspH, a [4Fe–4S]2+ enzyme involved in the biosynthesis of isoprenoids via the methylerythritol phosphate pathway. J. Am. Chem. Soc. 2013;135:1816–1822. doi: 10.1021/ja309557s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahrens-Botzong A., Janthawornpong K., Wolny J.A., Tambou E.N., Rohmer M., Krasutsky S., Poulter C.D., Schünemann V., Seemann M. Biosynthesis of isoprene units: Mössbauer spectroscopy of substrate and inhibitor binding to the [4Fe-4S] cluster of the LytB/IspH enzyme. Angew. Chem. Int. Ed. 2011;50:11976–11979. doi: 10.1002/anie.201104562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Hoof S., Lacey C.J., Röhrich R.C., Wiesner J., Jomaa H., Van Calenbergh S. Synthesis of analogues of (E)-1-hydroxy-2-methylbut-2-enyl 4-diphosphate, an isoprenoid precursor and human Γδ T cell activator. J. Org. Chem. 2008;73:1365–1370. doi: 10.1021/jo701873t. [DOI] [PubMed] [Google Scholar]
- 81.Wang K., Wang W., No J.-H., Zhang Y., Zhang Y., Oldfield E. Inhibition of the Fe4S4 -cluster-containing protein IspH (lytB): Electron paramagnetic resonance, metallacycles, and mechanisms. J. Am. Chem. Soc. 2010;132:6719–6727. doi: 10.1021/ja909664j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elliott T.S., Slowey A., Ye Y., Conway S.J. The use of phosphate bioisosteres in medicinal chemistry and chemical biology. Med. Chem. Commun. 2012;3:735. doi: 10.1039/c2md20079a. [DOI] [Google Scholar]
- 83.Wang W., Li J., Wang K., Smirnova T.I., Oldfield E. Pyridine inhibitor binding to the 4Fe-4S protein, A. Aeolicus IspH (LytB): A HYSCORE Investigation. J. Am. Chem. Soc. 2011;133:6525–6528. doi: 10.1021/ja2008455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.O’Dowd B., Williams S., Wang H., No J.H., Rao G., Wang W., McCammon J.A., Cramer S.P., Oldfield E. Spectroscopic and computational investigations of ligand binding to IspH: Discovery of non-diphosphate inhibitors. ChemBioChem. 2017;18:914–920. doi: 10.1002/cbic.201700052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller A.A., Bundy G.L., Mott J.E., Skepner J.E., Boyle T.P., Harris D.W., Hromockyj A.E., Marotti K.R., Zurenko G.E., Munzner J.B., et al. Discovery and characterization of QPT-1, the progenitor of a new class of bacterial topoisomerase inhibitors. Antimicrob. Agents Chemother. 2008;52:2806–2812. doi: 10.1128/AAC.00247-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brandstetter H., Grams F., Glitz D., Lang A., Huber R., Bode W., Krell H.-W., Engh R.A. The 1.8-Å crystal structure of a matrix metalloproteinase 8-barbiturate inhibitor complex reveals a previously unobserved mechanism for collagenase substrate recognition. J. Biol. Chem. 2001;276:17405–17412. doi: 10.1074/jbc.M007475200. [DOI] [PubMed] [Google Scholar]
- 87.Dunten P., Kammlott U., Crowther R., Levin W., Foley L.H., Wang P., Palermo R. X-ray structure of a novel matrix metalloproteinase inhibitor complexed to stromelysin. Protein Sci. 2001;10:923–926. doi: 10.1110/ps.48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Del. Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 89.Baell J.B., Holloway G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 90.Py B., Gerez C., Huguenot A., Vidaud C., Fontecave M., Ollagnier de Choudens S., Barras F. The ErpA/NfuA complex builds an oxidation-resistant Fe-S cluster delivery pathway. J. Biol. Chem. 2018;293:7689–7702. doi: 10.1074/jbc.RA118.002160. [DOI] [PMC free article] [PubMed] [Google Scholar]