Abstract
Specific oligonucleotide probes were developed to identify medically important fungi that display yeast-like morphology in vivo. Universal fungal primers ITS1 and ITS4, directed to the conserved regions of ribosomal DNA, were used to amplify DNA from Histoplasma capsulatum, Blastomyces dermatitidis, Coccidioides immitis, Paracoccidioides brasiliensis, Penicillium marneffei, Sporothrix schenckii, Cryptococcus neoformans, five Candida species, and Pneumocystis carinii. Specific oligonucleotide probes to identify these fungi, as well as a probe to detect all dimorphic, systemic pathogens, were developed. PCR amplicons were detected colorimetrically in an enzyme immunoassay format. The dimorphic probe hybridized with DNA from H. capsulatum, B. dermatitidis, C. immitis, P. brasiliensis, and P. marneffei but not with DNA from nondimorphic fungi. Specific probes for H. capsulatum, B. dermatitidis, C. immitis, P. brasiliensis, P. marneffei, S. schenckii, C. neoformans, and P. carinii hybridized with homologous but not heterologous DNA. Minor cross-reactivity was observed for the B. dermititidis probe used against C. immitis DNA and for the H. capsulatum probe used against Candida albicans DNA. However, the C. immitis probe did not cross-react with B. dermititidis DNA, nor did the dimorphic probe hybridize with C. albicans DNA. Therefore, these fungi could be differentiated by a process of elimination. In conclusion, probes developed to yeast-like pathogens were found to be highly specific and should prove to be useful in differentiating these organisms in the clinical setting.
The incidence of disease caused by pathogenic and opportunistic fungi has been increasing over the past decade (1, 2, 4, 25, 34). Such increases are primarily the result of the human immunodeficiency virus epidemic and advances in modern medicine that maintain or prolong the lives of severely ill patients (12, 25, 34). Moreover, the true burden of fungal disease is most certainly underestimated because of the insensitivity of present diagnostic methods (8, 18, 25). Diagnosis of fungal infections is typically made by isolation of the infecting organism in culture, by serologic assays, or through histopathologic examination of tissue (13, 27). Histopathologic diagnosis is advantageous because it is more rapid than culture. Pathogenic fungi may require 2 to 3 weeks or longer to grow (39, 40). A positive culture may also represent colonization rather than true invasion, especially when opportunistic organisms are isolated (7, 16). Furthermore, an infectious etiology may not be suspected at the time of biopsy and the tissue is often placed in fixative, making culture impossible. Histopathologic diagnosis can also be more rapid than serology. Serologic tests on a single serum sample to detect circulating antifungal antibodies may be inconclusive (especially in immunosuppressed patients). The acquisition of paired acute- and convalescent-phase sera, which is necessary for definitive serologic diagnosis, requires an additional 3 to 4 weeks before convalescent-phase serum can be obtained (27). Therefore, histopathologic examination of tissue sections may be the most rapid or only way in which to diagnose invasive fungal disease.
Histopathologic diagnosis of fungal infections is typically made through morphological criteria using reagents that preferentially stain fungal structures (6). However, fungi may present with atypical morphological features, making definitive histopathologic identification difficult (19). Other methods for diagnosis are then required. Presently, the immunofluorescence assay (either direct or indirect) using a specific antibody is the primary method available for in situ identification (18, 31). However, polyclonal antibodies to detect specific fungi are in limited supply and are generally not available commercially. Generating specific polyclonal antibodies to replace decreasing supplies is time consuming, and the replacement antibodies are often not of the same avidity or specificity as the original. Monoclonal antibody preparations can provide a reproducible supply of reagent that is usually highly specific but may be less sensitive than the corresponding polyclonal antibodies (8). The relatively recent development of automated DNA synthesis has allowed production of molecular probes with consistently defined properties that may result in increased test sensitivity, specificity, and reproducibility.
Past research in the molecular identification of fungi has typically concentrated on a single species or genus of fungus (3, 9, 14, 21, 22, 26, 29, 30, 32, 33, 37) or has used methods that may be cumbersome for the routine diagnostic laboratory to perform (35, 36, 38). The present study employed a modification of a PCR-enzyme immunoassay (PCR-EIA) method (10, 11) for amplification and differentiation of the major fungal pathogens that possess a yeast-like morphology in vivo. This method allows amplification and specific identification of DNA from all fungi, using a convenient method that requires little specialized equipment. Fungal DNA is amplified using universal fungal primers (ITS1 and ITS4) directed towards the rRNA gene. The rRNA gene is a common target for molecular identification of fungi because this area of the genome contains both unique and conserved regions (41). PCR amplification using the ITS1 and ITS4 primers produces an approximately 600-bp amplicon which contains conserved regions among fungi, including the sequence for ITS3 (41). The hybridization EIA then colorimetrically detects amplicons using biotinylated ITS3 and specific oligonucleotide probes directed to rDNA regions unique for each fungus. The combination of universal fungal PCR amplification and colorimetric detection creates a PCR-EIA that can potentially detect and specifically identify DNA from any fungus. Whereas the ultimate goal of this research is to identify fungi in tissue, the specificity of these probes was first tested using the PCR-EIA format and DNA isolated from cultured fungal isolates. This paper describes the development of specific oligonucleotide probes to identify fungal pathogens that possess yeast-like morphology in vivo.
MATERIALS AND METHODS
DNA isolation.
One loopful of yeast-phase Blastomyces dermatitidis (strain 4478, KL-1 [ATCC 26198], or A2 [ATCC 60916]), was inoculated into 10 ml of brain heart infusion broth (BD, Sparks, Md.) in a 50-μl Erlenmeyer flask and was incubated at 37°C on a rotary shaker (140 rpm) for 48 to 72 h. The suspension was then transferred to a 30-ml Oak Ridge centrifuge tube (Nalge, Rochester, N.Y.) and was centrifuged for 3 min at 2,000 × g. Genomic DNA was extracted and purified using a commercial kit (PureGene Yeast and Gram Positive DNA Isolation Kit; Gentra Systems Inc., Minneapolis, Minn.) following the manufacturer's protocol.
Mold-phase cultures of Sporothrix schenckii (ATCC 58251) and Penicillium marneffei (strains ATCC 64101, ATCC 58950, and JH05 [gift of William Merz, Johns Hopkins Medical School, Baltimore, Md.]) were grown in 50 ml of Sabouraud dextrose broth (Difco) in 250-ml Erlenmeyer flasks and were incubated at 25°C on a rotary shaker for 5 days. Growth was harvested by vacuum filtration through sterile filter paper, and the cellular mat was washed three times with sterile, distilled H2O by filtration. The cellular mat was then removed from the filter and placed into a sterile petri plate which was then sealed around the edges with Parafilm (American Can, Neenah, Wis.) and was frozen at −20°C until used.
DNA was extracted by grinding the cellular mats with a mortar and pestle in the presence of liquid nitrogen. Just before use, a portion of the frozen cellular mat, approximately equal in size to a quarter, was removed from the petri plate with sterile forceps and was placed into an ice-cold, sterile mortar (diameter, 6 in.). Liquid nitrogen was added to cover the mat and was added as needed to keep the mat frozen during grinding. The fungal mat was ground into a fine powder with a sterile pestle. Fungal DNA was then extracted and purified using serial proteinase K and RNase treatments followed by phenol extraction and ethanol precipitation by conventional methods (24).
Other DNA was kindly provided as a gift from the following persons: Histoplasma capsulatum (strains G186B [ATCC 26030], Down's, FLs-1, and B293 [var. duboisii]), Brent Lasker, Centers for Disease Control and Prevention (CDC), Atlanta, Ga.; Coccidioides immitis (strains C635 and C735), Garry Cole, Medical College of Ohio, Toledo, Ohio; Paracoccidioides brasiliensis (strains 265, Pb18, rh, and soil [soil isolate from Venezuela]), Maria Jose Soares Mendes Giannini, Faculdade de Ciencias Farmaceuticas, Universidade Estadual Paulista, Araraquara, Brazil, and Juan McEwen, Corporacion Para Investigaciones Biologicas, Medellin, Colombia; Cryptococcus neoformans (strains 9759-MU-1 [serotype A], BIH409 [serotype B], K24066TAN [serotype C], and 9375 [serotype D]) and all Candida species DNA (Candida albicans [strain B311], Candida glabrata [CDC Y-65], Candida krusei [CDC 259-75], Candida tropicalis [CDC 38], and Candida parapsilosis [ATCC 22019]), Cheryl Elie, CDC; and Pneumocystis carinii (rat isolate), Charles Beard, CDC.
Primers and probes.
All primers and probes were synthesized by β-cyanoethyl phosphoramidite chemistry using a 394 or expedite automated DNA synthesizer (PE Applied Biosystems, Foster City, Calif.). ITS3, a universal fungal sequence located in the 5.8S region of the rRNA gene and contained within the region amplified by ITS1 and ITS4 primers (23, 41), was biotinylated at the 5′ end by incorporating dimethyoxytrityl-biotin-carbon-6-phosphoramidite during its synthesis. This biotinylated probe (ITS3-B) was then purified by reverse-phase liquid chromatography. Digoxigenin-labeled probes were synthesized with a 5′-terminal amine group using 5′ Amino-Modifier C6 (Glen Research, Sterling, Va.), mixed with a 10-fold-molar excess of digoxigenin-3-O-methylcarbonyl-ɛ-aminocaproic acid N-hydroxysuccinimide ester (Roche Molecular Biochemicals, Indianapolis, Ind.) in 0.1 M sodium carbonate buffer, pH 9.0, and incubated at ambient temperature overnight. The digoxigenin-labeled probes were then purified by reverse-phase high-pressure liquid chromatography (5). Sequences and locations in the rRNA gene of these primers and probes are depicted in Table 1 and Fig. 1, respectively. All primers and probes were synthesized by the CDC Biotechnology Core Facility.
TABLE 1.
Sequences of oligonucleotide primers and probes
| Oligonucleotide probe or primer | Sequence (5′ to 3′) | Source or reference | Oligonucleotide labeling |
|---|---|---|---|
| PCR primers | |||
| ITS1 | TCCGTAGGTGAACCTGCGG | 31 | Universal forward primer |
| ITS4 | TCCTCCGCTTATTGATATGC | 31 | Universal reverse primer |
| Probes | |||
| ITS3-B | GCATCGATGAAGAACGCAGC | 31 | 5′-Biotin-labeled universal capture probe |
| Dm | GGACGTGCCCGAAATGCAGTGGCGG | U18363a | 5′-Digoxigenin-labeled probe for all endemic dimorphic fungi |
| Hc | ACCATCTCAACCTCCTTTTTCACACCAGG | U18363 | 5′-Digoxigenin-labeled probe for H. capsulatum |
| Bd | GGTCTTCGGGCCGGTCTCCCC | U18364 | 5′-Digoxigenin-labeled probe for B. dermatitidis |
| Ci | CTCTTTTTTTTATTATATCC | U18360 | 5′-Digoxigenin-labeled probe for C. immitis |
| Pb | CACTCATGGACCCCGG | AF322389 | 5′-Digoxigenin-labeled probe for P. brasiliensis |
| Ss | GACGCGCAGCTCTTTTTA | AF117945 | 5′-Digoxigenin-labeled probe for S. schenckii |
| Pm | GGGTTGGTCACCACCATA | L37406 | 5′-Digoxigenin-labeled probe for P. marneffei |
| Cn | CCTATGGGGTAGTCTTCGG | L14068 | 5′-Digoxigenin-labeled probe for C. neoformans |
| Pc | GTAGTAGGGTTAATTCAATT | L27658 | 5′-Digoxigenin-labeled probe for P. carinii |
GenBank sequence accession number.
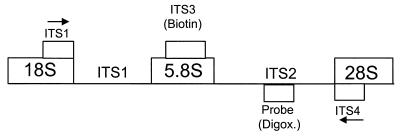
FIG. 1.
Diagram of hybridization sites of primers and probes. Hybridization sites for the ITS1 and ITS4 primers are in the phylogenetically conserved 18S and 28S rDNA regions, and arrows designate the direction of amplification (ITS1, forward primer; ITS4, reverse primer). ITS3 (biotin) represents the biotinylated, universal fungal probe which binds in the phylogenetically conserved, 5.8S rDNA region. Probe (Digox.) represents digoxigenin-labeled, microbe-specific probes which bind to the less highly conserved ITS2 region.
Microbe-specific probes.
DNA sequences of the ITS2 region of the fungal rRNA gene were obtained from GenBank (Table 1). Those fungi that did not have sequences available in GenBank (P. brasiliensis, S. schenckii, and P. marneffei) were sequenced using a dye terminator cycle sequencing kit (ABI PRISM; Applied Biosystems, Perkin-Elmer, Foster City, Calif.), and sequences have since been deposited with GenBank by our laboratory or by others (accession numbers: S. schenckii, AF117945; P. brasiliensis, AF322389; and P. marneffei, L37406). Briefly, primary DNA amplifications were conducted using ITS1 and ITS4 as primers. The DNA was purified using QIAquick Spin Columns (Qiagen Corp., Chatsworth, Calif.) and was eluted with 50 μl of heat-sterilized Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, pH 8.0). Sequencing was performed in both the forward and reverse directions. The reaction mix (20 μl) containing 9.5 μl of terminator premix, 2 μl (1 ng) of DNA template, 1 μl of primer (either a forward or reverse primer, 3.2 pmol), and 7.5 μl of heat-sterilized, distilled H2O was placed into a preheated (96°C) Perkin-Elmer 9600 thermal cycler for 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. The PCR product was then purified before sequencing using CentriSep spin columns (Princeton Separations, Inc., Adelphia, N.J.). DNA was then vacuum dried, resuspended in 6 μl of formamide-EDTA (5 μl deionized formamide plus 1 μl of 50 mM EDTA, pH 8.0), and denatured for 2 min at 90°C before subjection to sequencing using an automated capillary DNA sequencer (model 373; ABI Systems, Bethesda, Md.).
Sequences were aligned and a comparison was performed to determine unique sequences that could be used for the development of specific digoxigenin-labeled oligonucleotide probes. The initial screen for specificity of the probe sequences was performed using basic local alignment search tool (BLAST) software (GCG, Madison, Wis.). Probe sequences determined to be unique were then synthesized and digoxigenin labeled as described above.
PCR conditions.
The PCR mix consisted of 10 mM Tris-HCl buffer containing 50 mM KCl, pH 8.0 (Roche), 1.5 mM MgCl2 (Roche), 0.2 mM deoxynucleoside triphosphate (TaKaRa Shuzo Co. Ltd., Otsu, Shiga, Japan), and 1.25 U of Taq polymerase (TaKaRa Shuzo). Primers ITS1 and ITS4 were added to a final concentration of 0.2 mM each. Template DNA was added at a final concentration of 1 ng per 50 μl of reaction mix. For each experiment, at least one reaction tube received water in place of template DNA as a negative control. Amplification was performed in a Model 9600 thermocycler (Perkin-Elmer, Emeryville, Calif.). Initial denaturation of template DNA was achieved by heating at 95°C for 5 min. This was followed by 30 cycles of 30 s at 95°C, 30 s at 58°C, and 1 min at 72°C. A final extension step was conducted for 10 min at 72°C. Appropriate controls were included and PCR contamination precautions were followed (11, 20).
Agarose gel electrophoresis.
Successful PCR amplification was confirmed by visualization of PCR amplicons after agarose gel electrophoresis. Gels consisted of 1% agarose LE (Boehringer Mannheim, Indianapolis, Ind.) and 1% NuSieve GTG agarose (FMC Bioproducts, Rockland, Maine) or 2% Metaphore agar (FMC Bioproducts) dissolved in Tris-borate-EDTA buffer (0.1 M Tris, 0.09 M boric acid, 0.001 M EDTA, pH 8.4). Five microliters of the PCR amplicons was combined with 1 μl of tracking dye (Roche) and was then added to each well of the agarose gel. Electrophoresis was conducted at 70 to 80 V for 45 to 60 min. The gel was stained with ethidium bromide for 30 min and washed in deionized water for 30 min before examination on a UV transilluminator.
EIA.
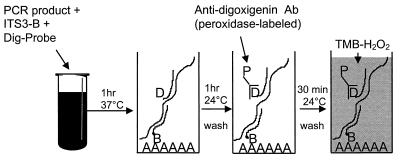
EIA identification of PCR products was performed as previously described (10, 11), with minor modifications (Fig. 2). Briefly, tubes containing 10 μl of heat-denatured (5 min at 95°C) PCR amplicons were placed on ice, and 200 μl of hybridization buffer (4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], pH 7.0, 0.02 M HEPES, 0.002 M EDTA, and 0.15% Tween 20) containing 10 ng of ITS3-B and 10 ng of a digoxigenin-labeled specific probe was added. Samples were mixed and incubated at 37°C for 1 h. One hundred microliters of the mixture was added in duplicate to wells of a strepavidin-coated, 96-well, microtiter plate (Roche) and was incubated at ambient temperature for 1 h on a microtiter plate shaker (∼350 rpm; Labline Instruments, Melrose Park, Ill.). Microtiter plates were washed six times with 0.01 M phosphate-buffered saline, pH 7.2 (GibcoBRL, Life Technologies, Grand Island, N.Y.), containing 0.05% Tween 20 (Sigma Chemical Co., St. Louis, Mo.) (PBST) before addition of 100 μl of a 1:1,000 dilution of horseradish peroxidase-labeled, anti-digoxigenin antibody (150 U/ml; Roche) per well. Plate contents were incubated for 1 h at ambient temperature with shaking and were then washed six times with PBST. 3,3′,5,5′-Tetramethylbenzidine (TMB)-H2O2 substrate (Kirkegaard & Perry, Gaithersburg, Md.) was then added to the wells, and the color reaction was allowed to develop at ambient temperature for 15 min. The optical density of each well was immediately read at a wavelength of 650 nm in a UVMax microtiter plate reader (Molecular Devices, Sunnyvale, Calif.). The optical density of the duplicate wells was averaged and used in the analysis of the results. The optical density results were then converted to an EIA index (EI), which was calculated by dividing the optical density of the wells which had received test DNA by the optical density of the PCR water control as follows: optical density of test DNA/optical density of water blank = EI.
FIG. 2.
Diagram of PCR-EIA procedure. PCR product is heat denatured and incubated with both a biotinylated capture probe (ITS3-B) and a microbe-specific digoxigenin-labeled probe (Dig-Probe) before addition to the wells of a streptavidin-coated (AAAAAA) microtiter plate. Probe hybridization is then detected using a peroxidase-labeled (P), anti-digoxigenin antibody (Ab) and a colorimetric substrate-hydrogen peroxide mixture (TMB-H2O2). Specifics are described in Materials and Methods.
Statistical analysis.
Student's t test was used to determine differences between the mean EIs of probe hybridization to homologous DNA and those of probe hybridization to heterologous DNA. Differences were considered significant when P was less than or equal to 0.05.
RESULTS
Confirmation of DNA amplification.
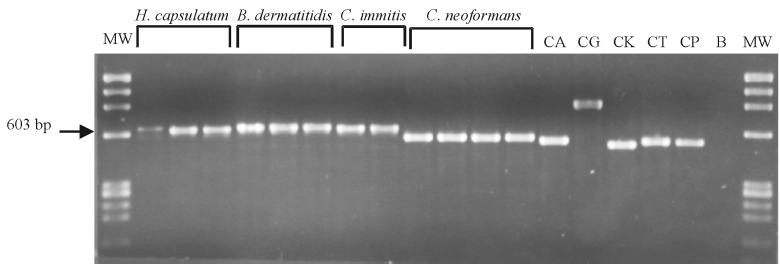
To verify that the specific DNA target was appropriately amplified and was of the expected size, the PCR amplicons were subjected to agarose gel electrophoresis and bands were visualized after ethidium bromide staining. The amplification of the rRNA gene using the ITS1 and ITS4 primers resulted in an approximately 600-bp-long amplicon for all fungi tested. The molecular sizes of amplicons were especially similar among the systemic, dimorphic fungi (Fig. 3). The greatest differences in amplicon size were observed among the five Candida species tested and were particularly pronounced for C. glabrata and C. krusei compared to all other Candida species (Fig. 3). However, specific identification of the fungi using amplicon size alone was not possible and is not generally recommended (28). Therefore, probes were designed to specifically identify each fungus using a modification of an EIA method previously described (10, 11).
FIG. 3.
Agarose gel of amplified products from yeast-like fungi. Lane abbreviations (left to right): MW, molecular weight markers (HaeIII digest of ΦX174 plasmid; Roche); H. capsulatum, DNA amplified from H. capsulatum strains B293, Down's, and Fls-1; B. dermatitidis, DNA amplified from B. dermatitidis strains 4478, KL-1, and A2; C. immitis, DNA amplified from C. immitis strains C635 and C735; C. neoformans, DNA amplified from C. neoformans strains 9759-MU-1 (serotype A), BIH409 (serotype B), K24066TAN (serotype C), and 9375 (serotype D); CA, CG, CK, CT, and CP, DNA amplified from C. albicans (strain B311), C. glabrata (CDC Y-65), C. krusei (CDC 259-75), C. tropicalis (strain CDC 38), and C. parapsilosis (ATCC 22019), respectively; B, water blank (negative control).
Probe specificity.
Digoxigenin-labeled probes directed to the ITS2 region of rDNA were designed to specifically detect PCR amplicons from the most medically important yeast-like fungi. In addition to the microbe-specific probes, a probe was also designed as a primary screening probe with which to identify only the systemic, dimorphic fungal pathogens. The specificity of these probes was confirmed using the PCR-EIA method in a checkerboard pattern (Table 2). The dimorphic screening probe (Dm) successfully hybridized with PCR amplicons from all strains of the major systemic, dimorphic fungi tested (H. capsulatum, B. dermatitidis, C. immitis, P. brasiliensis, and P. marneffei) but not with DNA from any strain of the other yeast-like fungi (S. schenckii, C. neoformans, Candida species, and P. carinii).
TABLE 2.
Specificity of oligonucleotide probes to DNA from yeast-like fungid
| Probea | Mean EI ± SE (n) using DNA from:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| H. capsulatum | B. dermatitidis | C. immitis | P. brasiliensis | P. marneffei | S. schenckii | C. neoformansb | P. carinii | Candida speciesc | |
| Dm | 11.0 ± 1.1 (34) | 9.4 ± 1.4 (24) | 16.9 ± 2.2 (14) | 13.9 ± 1.1 (21) | 3.0 ± 0.5 (31) | 0 | 0 | 0 | 0 |
| Hc | 15.8 ± 1.4 (37) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Bd | 0 | 11.9 ± 2.0 (20) | 4.3 ± 0.8 (10) | 0 | 0 | 0 | 0 | 0 | 0 |
| Ci | 0 | 0 | 21.9 ± 3.2 (13) | 0 | 0 | 0 | 0 | 0 | 0 |
| Pb | 0 | 0 | 0 | 10.8 ± 0.8 (21) | 0 | 0 | 0 | 0 | 0 |
| Pm | 0 | 0 | 0 | 0 | 7.6 ± 0.6 (36) | 0 | 0 | 0 | 0 |
| Ss | 0 | 0 | 0 | 0 | 0 | 23.6 ± 4.3 (12) | 0 | 0 | 0 |
| Cn | 0 | 0 | 0 | 0 | 0 | 0 | 42.7 ± 2.7 (31) | 0 | 0 |
| Pc | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.2 ± 2.7 (10) | 0 |
See Table 1 for definition of abbreviations.
Includes DNA from serotypes A, B, C, and D.
Candida species included C. albicans, C. glabrata, C. krusei, C. tropicalis, and C. parapsilosis; excludes results for Hc probe against C. albicans; mean EI ± SE = 6.5 ± 1.0 (n = 10).
Mean EI ± standard error (n) for all heterologous DNA tested with the following probes: Dm 1.13 ± 0.04 (n = 89); Hc, 1.2 ± 0.3 (n = 118); Bd, 1.41 ± 0.12 (n = 103); Ci, 1.0 ± 0.01 (n = 96); Pb, 1.01 ± 0.02 (n = 88); Pm, 0.99 ± 0.02 (n = 98); Ss, 0.99 ± 0.01 (n = 103); Cn, 0.98 ± 0.01 (n = 95); and Pc, 0.98 ± 0.01 (n = 96). All probes significantly hybridized to homologous DNA but not heterologous DNA at P < 0.001 except for Pc (P < 0.05) and Bd versus C. immitis DNA (P < 0.01). A value of 0 was assigned for all EI values less than 1.75 for ease of presentation.
Microbe-specific probes, designed to detect only DNA amplified from their homologous fungus, were tested against PCR amplicons from all strains of both homologous as well as heterologous yeast-like fungi. The results in Table 2 demonstrate that the microbe-specific probes hybridized with DNA from homologous fungi and not with DNA from heterologous fungi (P < 0.001 or P < 0.05) with minor exceptions. There was some reactivity of the B. dermatitidis probe observed when it was tested against C. immitis DNA. However, the hybridization signal for the B. dermatitidis probe tested against B. dermatitidis DNA was statistically greater than for C. immitis DNA (11.9 ± 2.0 versus 4.3 ± 0.8; P < 0.01). In addition, the reverse (i.e., the C. immitis probe tested against B. dermititidis DNA) was negative and could be used to differentiate the two fungi by a process of elimination. There was also a hybridization signal observed for the H. capsulatum probe reacted with DNA from C. albicans (15.8 ± 1.4 versus 6.5 ± 1.0; P < 0.001), but no signal was observed for any of the other Candida species tested using this probe. The dimorphic probe, however, did not hybridize with C. albicans DNA, and the C. albicans probe did not hybridize with H. capsulatum DNA (10, 11; data not shown). Therefore, analyzing results obtained using these probes would eliminate any doubt regarding the identity of the organism from which the DNA was derived.
Confirmation of probe specificity using multiple strains of homologous and heterologous fungi.
To further analyze each probe's capacity to hybridize with only DNA from homologous fungi, DNA from multiple strains of each of the systemic, dimorphic fungi was tested in the PCR-EIA (Table 3). The probe designed to identify all systemic, dimorphic fungi hybridized with DNA from all strains of H. capsulatum, B. dermatitidis, C. immitis, and P. brasiliensis tested. In addition, the probes specific for individual dimorphic fungi hybridized only to DNA isolated from homologous fungi but not to DNA isolated from heterologous fungi (Table 3). The minor hybridization signal observed for the B. dermatitidis probe tested against C. immitis DNA was similar for both strains of C. immitis tested.
TABLE 3.
Reactivity of oligonucleotide probes to dimorphic pathogens against DNA from multiple strains of homologous and heterologous dimorphic fungic
| Probeb | Mean EI ± SE (n) using DNA froma:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
H. capsulatum isolates
|
B. dermatitidis isolates
|
C. immitis isolates
|
P. brasiliensis isolates
|
|||||||||
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 1 | 2 | 1 | 2 | 3 | |
| Dm | 13.7 ± 2.8 (8) | 8.8 ± 1.8 (7) | 10.3 ± 2.0 (11) | 11.4 ± 2.8 (8) | 9.2 ± 1.8 (9) | 12.5 ± 3.3 (9) | 4.9 ± 0.8 (6) | 14.8 ± 1.8 (6) | 18.5 ± 3.6 (8) | 13.3 ± 2.2 (7) | 14.1 ± 2.3 (7) | 14.3 ± 2.0 (6) |
| Hc | 17.3 ± 3.8 (9) | 15.1 ± 2.3 (6) | 15.8 ± 2.6 (10) | 14.7 ± 2.6 (9) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Bd | 0 | 0 | 0 | 0 | 9.7 ± 2.4 (6) | 15.4 ± 3.5 (9) | 8.3 ± 3.3 (5) | 4.7 ± 1.2 (5) | 3.8 ± 1.2 (5) | 0 | 0 | 0 |
| Ci | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 23.8 ± 4.2 (6) | 20.2 ± 4.9 (7) | 0 | 0 | 0 |
| Pb | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9.4 ± 1.5 (7) | 12.3 ± 1.4 (7) | 12.0 ± 0.4 (6) |
Isolates used: H. capsulatum 1, G186B; 2, B293; 3, Down's; and 4, FLs-1; B. dermatitidis 1, 4478; 2, A2; and 3, KL-1; C. immitis 1, C634; and 2, C735; and P. brasiliensis 1, Pb18; 2, rh; and 3, soil.
See Table 1 for definition of abbreviations.
Mean EI ± standard error (n) for heterologous DNA tested with the following probes: Hc, 1.2 ± 0.05 (n = 42); Bd, 1.74 ± 0.27 (n = 44); Bd without Ci DNA, 0.99 ± 0.02 (n = 34); Ci, 1.0 ± 0.02 (n = 42); and Pb, 0.99 ± 0.03 (n = 33). All probes significantly hybridized to homologous DNA but not to heterologous, dimorphic DNA from areas of endeminity at P < 0.001. A value of 0 was assigned for all EI values less than 1.75 for ease of presentation.
Sensitivity of probes using PCR-EIA.
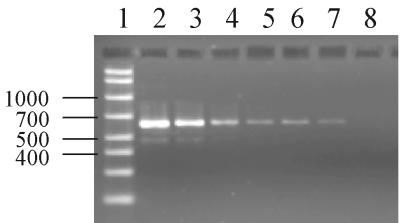
To assess the limit of sensitivity of the PCR-EIA method, compared to that for detection of amplicons by agarose gel electrophoresis, H. capsulatum (Down's strain) DNA was serially diluted prior to PCR amplification and was then assessed by both agarose gel electrophoresis and PCR-EIA. Agarose gel electrophoresis and ethidium bromide staining allowed detection of amplicons at a concentration as low as 16 pg per reaction (Fig. 4; Table 4). In contrast, as little as 3.2 pg of DNA per reaction could be detected by PCR-EIA (Table 4).
FIG. 4.
Agarose gel of titrated H. capsulatum DNA (Down's strain) amplified by PCR. Lane 1, molecular size markers in base pairs (AmpliSize molecular ruler; Bio-Rad, Hercules, Calif.). The number of picograms of DNA per reaction for lanes 2 to 8 was 20,000, 10,000, 2,000, 400, 80, 16, and 3.2, respectively. Limit of sensitivity of agarose gel electrophoresis and ethidium bromide staining, 16 pg per reaction.
TABLE 4.
Evaluation of the sensitivity of PCR-EIA
| DNA concna | EIb | Reading in agarose gelc |
|---|---|---|
| 10,000 | 53.3 | + |
| 2,000 | 50.4 | + |
| 400 | 38.2 | + |
| 80 | 13.9 | + |
| 16 | 5.3 | + |
| 3.2 | 2.2 | − |
| 0.64 | 1.5 | − |
| 0.128 | 1.2 | − |
| 0.0256 | 1.1 | − |
Concentration is given in picograms per reaction.
EI using H. capsulatum (Down's strain) DNA.
+, visually positive band in gel after ethidium bromide staining.
DISCUSSION
This paper describes the development of a PCR-EIA that could amplify and identify DNA sequences from the rRNA gene of fungi that have yeast-like morphology in vivo. By use of universal fungal primers and a biotinylated universal probe, all fungal DNA was amplified and bound to strepavidin-coated microtiter plate wells. Identification of the fungi from which the DNA was isolated was then confirmed by microbe-specific oligonucleotide probes. The probes designed in this study could specifically identify DNA isolated from fungi that display yeast-like morphology in vivo. Our laboratory has had comparable success using a similar method for the identification of Candida species (10, 11).
The sequential use of universal fungal primers for PCR amplification and microbe-specific probes to identify fungi has several advantages over methods used by others. The focus of most researchers has been to develop methods for the amplification and identification of a single species or genus of a particular fungal organism (3, 14, 21, 26, 29, 30, 32, 33, 37). This was accomplished in some cases through the use of single-copy gene targets, such as the ERG11 gene for Candida species (26, 30) and the URA5 gene for C. neoformans (37), or by use of microbe-specific primers for the amplification and identification of a single infecting pathogen (3, 29, 37). In the present study, universal fungal primers directed to the highly conserved ITS1 and ITS4 regions of ribosomal DNA allowed amplification of all fungal DNA rather than that from only a single organism. A complete array of different fungi could be identified following a single PCR amplification and the application of specific probes. The rRNA gene was chosen as an amplification target, not only because it contains binding sites for universal fungal primers but because the chromosome on which this gene is located contains approximately 100 gene copies (23) that serve as a “preamplification” step to increase amplicon yield and test sensitivity. Therefore, the use of universal primers and a multiple-copy gene target has greater utility and sensitivity for the identification of fungi in clinically diverse specimens.
The EIA format described in this study also has advantages over methods used by others for amplicon detection. Some investigators detected amplicons produced by microbe-specific primers after electrophoresis in agarose gels and ethidium bromide staining. The presence of a band was considered a positive result for those using specific primers (3, 14, 21, 29, 33). However, specific identification of fungi using amplicon size alone is not generally recommended (28) since different fungi may produce similarly sized amplicons, as was noted in the present study. Alternatively, the presence of a unique banding pattern after restriction enzyme digestion of the PCR product was used for species identification (26). Although the use of restriction enzymes is rapid and provides increased specificity compared to gel electrophoresis, results may be difficult to reproduce and can be expensive. Often, two or more enzymes may be required for adequate specificity (26). In addition, each enzyme may need different conditions for optimal restriction activity (24). Others developed specific probes to obtain a final identification of the organism using time-consuming and labor-intensive Southern blot or slot blot methods (9, 30, 32, 35, 36, 37). The slot blot method uses membranes that are stripped and reprobed sequentially each time that a probe for a different organism is to be tested (17, 35, 36). In contrast, the EIA is very rapid (3 h) and simple to perform, and unlike the slot blot method, all probes can be tested simultaneously.
A unique method using universal primers was developed by Turenne et al. (38) for determining the exact size of amplified DNA using an automated fluorescent capillary electrophoresis system. However, it was difficult to conclusively differentiate some fungi from others using this method because the size of the amplicons produced was similar if not identical for more than one fungus. In addition, the cost of the necessary equipment would make it difficult to use this assay in smaller laboratories.
The development of the probes described in our study should not only provide a means to identify fungi in culture but should also aid in the histologic identification of fungi in clinical specimens. Application of these probes to fungi in tissue sections will allow the differentiation of truly invasive organisms from simple colonizers. Further testing of larger numbers of organisms from pure culture and application to clinical specimens are planned.
Whereas the organisms under study have unique characteristics that often allow histologic identification, atypical tissue forms may resemble other fungi (15, 19). Therefore, methods other than physical characteristics are needed to confirm identification. Multiple techniques may be employed to identify fungi in tissue using molecular probes. First, fungal DNA can be extracted from the tissue and PCR-EIA can be performed using methods similar to those described in this paper. Second, the use of these probes in an in situ hybridization procedure would allow for the localization of fungal DNA directly in the tissue. Finally, the combination of these two procedures, where the target DNA is amplified and probes are hybridized in situ, may be employed. None of these methods should be considered mutually exclusive. When fungi are found in large numbers in tissue, the faster, more versatile DNA extraction and PCR-EIA method may be most useful. In instances where there are very few fungal elements present in the tissue, in situ hybridization may be more useful because very little DNA would be present and might get “lost” when one tries to extract it from the tissue. Keeping the DNA localized and “bringing the probe to it” may prove more advantageous. However, the universal aspects of the PCR-EIA would be lost since a single probe would have to be selected and run individually on each slide.
The advantage of the PCR-EIA method is its ability to amplify and detect small quantities of DNA. However, some tissue may contain very low quantities of DNA, below the level of sensitivity. The sensitivity of a PCR-based assay can be enhanced by various modifications of the technique. First, one may use nested PCR (28, 33). This employs a second set of primers internal to the original set of primers to reamplify the target DNA using the amplicons from the first PCR as a template for the second PCR. This method has been shown to enhance test sensitivity (28, 33). Extreme care must be taken, however, so as not to contaminate reagents or the laboratory with amplicons from the first amplification. Second, one may continue the PCR through more cycles, continuing the geometric increase of DNA amplified. New forms of Taq polymerase have been developed that have increased stability and accuracy throughout an increased number of PCR cycles. Preliminary results from our laboratory indicate that increasing the number of PCR cycles may increase the sensitivity of the PCR-EIA (data not shown).
In conclusion, oligonucleotide probes for yeast-like fungi have been developed and evaluated in a PCR-EIA format. These probes have been shown to be sensitive and specific and able to identify DNA obtained from fungi in pure culture. These characteristics, along with the rapid and convenient EIA detection format and its potential for automation, make it useful for applications in the clinical laboratory setting.
REFERENCES
- 1.Ampel N M, Mosley D G, England B, Vertz P D, Komatsu K, Hajjeh R A. Coccidioidomycosis in Arizona: increase in incidence from 1990 to 1995. Clin Infect Dis. 1998;27:1528–1530. doi: 10.1086/515044. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1990 to May 1999, issued June 1999. Am J Infect Control. 1999;27:520–532. doi: 10.1016/s0196-6553(99)70031-3. [DOI] [PubMed] [Google Scholar]
- 3.Aoki F H, Imai T, Tanaka R, Mikami Y, Taguchi H, Nishimura N F, Nishimura K, Miyaji M, Schreiber A Z, Branchini M L. New PCR primer pairs specific for Cryptococcus neoformans serotype A or B prepared on the basis of random amplified polymorphic DNA fingerprint pattern analyses. J Clin Microbiol. 1999;37:315–320. doi: 10.1128/jcm.37.2.315-320.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong G L, Conn L A, Pinner R W. Trends in infectious disease mortality in the United States during the 20th century. JAMA. 1999;281:61–66. doi: 10.1001/jama.281.1.61. [DOI] [PubMed] [Google Scholar]
- 5.Becker C R, Efcavitch J W, Heiner C R, Kaiser N F. Use of a reverse phase column for the HPLC purification of synthetic oligonucleotides. J Chromatogr. 1985;326:293–299. [Google Scholar]
- 6.Chandler F W, Watts J C. Pathologic diagnosis of fungal infections. Chicago, Ill: ASCP Press; 1987. [Google Scholar]
- 7.de Repentigny L. Serodiagnosis of candidiasis, aspergillosis, and cryptococcosis. Clin Infect Dis. 1992;14(Suppl. 1):S11–S22. doi: 10.1093/clinids/14.supplement_1.s11. [DOI] [PubMed] [Google Scholar]
- 8.de Repentigny L, Kaufman L, Cole G T, Kruse D, Latge J P, Matthews R C. Immunodiagnosis of invasive fungal infections. J Med Vet Mycol. 1994;32(Suppl. 1):239–252. doi: 10.1080/02681219480000871. [DOI] [PubMed] [Google Scholar]
- 9.Einsele H, Hebart H, Roller G, Loffler J, Rothenhofer I, Muller C A, Bowden R A, van Burik J, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elie C M, Lott T J, Reiss E, Morrison C J. Rapid identification of Candida species with species-specific DNA probes. J Clin Microbiol. 1998;36:3260–3265. doi: 10.1128/jcm.36.11.3260-3265.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita S, Lasker B A, Lott T J, Reiss E, Morrison C J. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J Clin Microbiol. 1995;33:962–967. doi: 10.1128/jcm.33.4.962-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajjeh R A. Disseminated histoplasmosis in persons infected with human immunodeficiency virus. Clin Infect Dis. 1995;21(Suppl. 1):S108–S110. doi: 10.1093/clinids/21.supplement_1.s108. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton A J. Serodiagnosis of histoplasmosis, paracoccidioidomycosis, and penicilliosis marneffei: current status and future trends. Med Mycol. 1998;36:351–364. [PubMed] [Google Scholar]
- 14.Imai T, Sano A, Mikami Y, Watanabe K, Aoki F H, Branchini M L M, Negroni R, Nishimura K, Miyaji M. A new PCR primer for the identification of Paracoccidioides brasiliensis based on rRNA sequences coding the internal transcribed spacers (ITS) and 5.8S regions. Med Mycol. 2000;38:323–326. doi: 10.1080/mmy.38.4.323.326. [DOI] [PubMed] [Google Scholar]
- 15.Jensen H E, Schonheyder H C, Hotchi M, Kaufman L. Diagnosis of systemic mycoses by specific immunohistochemical tests. APMIS. 1996;104:241–258. doi: 10.1111/j.1699-0463.1996.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 16.Jones J M. Laboratory diagnosis of invasive candidiasis. Clin Microbiol Rev. 1990;3:32–45. doi: 10.1128/cmr.3.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kappe R, Okeke C N, Fauser C, Maiwald M, Sonntag H G. Molecular probes for the detection of pathogenic fungi in the presence of human tissue. J Med Microbiol. 1998;47:811–820. doi: 10.1099/00222615-47-9-811. [DOI] [PubMed] [Google Scholar]
- 18.Kaufman L, Kovacs J A, Reiss E. Clinical immunomycology. In: Rose N R, de Macario E C, Folds J D, Lane H C, Nakamura R M, editors. Manual of clinical laboratory immunology. 5th ed. Washington, D.C.: American Society for Microbiology; 1997. pp. 585–604. [Google Scholar]
- 19.Kaufman L, Valero G, Padhye A A. Misleading manifestations of Coccidioides immitis in vivo. J Clin Microbiol. 1998;36:3721–3723. doi: 10.1128/jcm.36.12.3721-3723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 21.LoBuglio K F, Taylor J W. Phylogeny and PCR identification of the human pathogenic fungus Penicillium marneffei. J Clin Microbiol. 1995;33:85–89. doi: 10.1128/jcm.33.1.85-89.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loffler J, Hebart H, Sepe S, Schumcher U, Klingebiel T, Einsele H. Detection of PCR-amplified fungal DNA by using a PCR-ELISA system. Med Mycol. 1998;36:275–279. doi: 10.1080/02681219880000441. [DOI] [PubMed] [Google Scholar]
- 23.Lott T J, Kuykendall R J, Reiss E. Nucleotide sequence analysis of the 5.8S rDNA and adjacent ITS2 region of Candida albicans and related species. Yeast. 1993;9:1199–1206. doi: 10.1002/yea.320091106. [DOI] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 25.McNeil M M, Nash S L, Hajjeh R A, Phelan M A, Conn L A, Plikaytis B D, Warnock D W. Trends in mortality due to invasive mycotic diseases in the United States, 1980–1997. Clin Infect Dis. 2001;33:641–647. doi: 10.1086/322606. [DOI] [PubMed] [Google Scholar]
- 26.Morace G, Sanguinetti M, Posteraro B, Lo Cascio G, Fadda G. Identification of various medically important Candida species in clinical specimens by PCR-restriction enzyme analysis. J Clin Microbiol. 1997;35:667–672. doi: 10.1128/jcm.35.3.667-672.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrison, C. J., and M. D. Lindsley. Serological approaches to the diagnosis of invasive fungal infections. In R. Calderone and R. Cihlar (ed.), Fungal pathogenesis: principles and practice. Marcel Dekker, Inc., New York, N.Y., in press.
- 28.Podzorski R P, Persing D H. Molecular detection and identification of microorganisms. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 130–157. [Google Scholar]
- 29.Poonwan N, Imai T, Mekha N, Yazawa K, Mikami Y, Ando A, Nagata Y. Genetic analysis of Histoplasma capsulatum strains isolated from clinical specimens in Thailand by a PCR-based random amplified polymorphic DNA method. J Clin Microbiol. 1998;36:3073–3076. doi: 10.1128/jcm.36.10.3073-3076.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posteraro B, Sanguinetti M, Masucci L, Romano L, Morace G, Fadda G. Reverse cross blot hybridization assay for rapid detection of PCR-amplified DNA from Candida species, Cryptococcus neoformans, and Saccharomyces cerevisiae in clinical samples. J Clin Microbiol. 2000;38:1609–1614. doi: 10.1128/jcm.38.4.1609-1614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powers C N. Diagnosis of infectious diseases: a cytopathologist's perspective. Clin Microbiol Rev. 1998;11:341–365. doi: 10.1128/cmr.11.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prariyachatigul C, Chaiprasert A, Meevootisom V, Pattanakitsakul S. Assessment of a PCR technique for the detection and identification of Cryptococcus neoformans. J Med Vet Mycol. 1996;34:251–258. doi: 10.1080/02681219680000431. [DOI] [PubMed] [Google Scholar]
- 33.Rappelli P, Are R, Casu G, Fiori P L, Cappuccinelli P, Aceti A. Development of a nested PCR for detection of Cryptococcus neoformans in cerebrospinal fluid. J Clin Microbiol. 1998;36:3438–3440. doi: 10.1128/jcm.36.11.3438-3440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rees J R, Pinner R W, Hajjeh R A, Brandt M E, Reingold A L. The epidemiological features of invasive mycotic infections in the San Francisco Bay area, 1992–1993: results of population-based laboratory active surveillance. Clin Infect Dis. 1998;27:1138–1147. [PubMed] [Google Scholar]
- 35.Sandhu G S, Aleff R A, Kline B C, da Silva Lacaz C. Molecular detection and identification of Paracoccidioides brasiliensis. J Clin Microbiol. 1997;35:1894–1896. doi: 10.1128/jcm.35.7.1894-1896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandhu G S, Kline B C, Stockman L, Roberts G D. Molecular probes for diagnosis of fungal infections. J Clin Microbiol. 1995;33:2913–2919. doi: 10.1128/jcm.33.11.2913-2919.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka K, Miyazaki T, Maesaki S, Mitsutake K, Kakeya H, Yamamoto Y, Yanagihara K, Hossain M A, Tashiro T, Kohno S. Detection of Cryptococcus neoformans gene in patients with pulmonary cryptococcosis. J Clin Microbiol. 1996;34:2826–2828. doi: 10.1128/jcm.34.11.2826-2828.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turenne C Y, Sanche S E, Hoban D J, Karlowsky J A, Kabani A M. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol. 1999;37:1846–1851. doi: 10.1128/jcm.37.6.1846-1851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheat L J. Diagnosis and management of histoplasmosis. Eur J Clin Microbiol Infect Dis. 1989;8:480–490. doi: 10.1007/BF01964063. [DOI] [PubMed] [Google Scholar]
- 40.Wheat L J, French M L V, Kohler R B, Zimmerman S E, Smith W R, Norton J A, Eitzen H E, Smith C D, Slama T G. The diagnostic laboratory tests for histoplasmosis: analysis of experience in a large urban outbreak. Ann Intern Med. 1982;97:680–685. doi: 10.7326/0003-4819-97-5-680. [DOI] [PubMed] [Google Scholar]
- 41.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]