Abstract
Seven Newcastle disease (ND) virus (NDV) isolates which were recovered from ND outbreaks in chicken and pigeon flocks in China and Taiwan between 1996 and 2000 were genotypically and pathotypically characterized. By phylogenetic analysis of the fusion protein genes, isolates Ch-A7/96, Ch/98-3, Ch/99, Ch/2000, and TW/2000 were placed into two novel subgenotypes, VIIc and VIId. Isolate Ch/98-1 was grouped into subgenotype VIb, while Ch-W6/96 was proven to be a mixture of isolates Ch-A7/96 and Ch/98-1. These isolates were pathotyped as viscerotropic velogenic for Ch/98-3, Ch/99, Ch/2000, and TW/2000; neurotropic velogenic for Ch-A7/96; and mesogenic for Ch/98-1. Three separate, comparative, genetic analyses of the F genes, including genetic distance measurement, phylogenetic tree analysis, and residue substitution analysis, were performed with our isolates and selected NDV strains from GenBank. Results showed that the close genetic similarity provided evidence for the epidemiological linkage between the outbreaks in China and Taiwan and that the 1990s outbreaks in Asia, the Middle East, Africa, and Europe constituted the fourth panzootic of ND. In combination with epidemiological analysis, an evolutionary model of the NDV strains, representative of the direction of transmission within the NDV strains, was proposed, and epidemiology of NDV transmission was evaluated with emphasis on molecular aspects. Finally, a cross-protective experiment indicated that at least one strain (Ch-A7/96) among our NDV isolates was an antigenic variant, responsible for recent outbreaks of ND in vaccinated chicken flocks.
Newcastle disease (ND) is one of the most serious diseases of poultry caused by ND virus (NDV), the only member of the avian paramyxovirus type 1 serotype (3). NDV belongs to the Rubulavirus genus of the family Paramyxoviridae and has a negative-sense, single-stranded-RNA genome that consists of approximately 15 kbp. The genome encodes six major polypeptides in the 5′-to-3′ direction, including RNA-directed RNA polymerase, hemagglutinin-neuraminidase, fusion protein, matrix protein, phosphoprotein, and nucleoprotein (15). The pathogenicity of any newly isolated strain can be assessed by determining the intracerebral pathogenicity index (ICPI) and the mean death time (MDT). By pathotyping, NDV strains were classified into the highly pathogenic (velogenic), intermediate or moderately pathogenic (mesogenic), and lowly pathogenic (lentogenic) categories. Some lentogenic strains of NDV are avirulent, whereas velogenic forms were further classified as viscerotropic velogenic (VV) and neurotropic velogenic (NV) types based on clinical manifestation and lesions (3). The primary molecular determinant for NDV pathogenicity is the amino acids of the F protein cleavage site (10, 16, 18).
Three panzootics of ND have occurred since the disease was first recognized (1, 3). By restriction site mapping and sequence analysis of the F gene, NDV strains were divided into eight genotypes (6, 11, 13). Among these, at least three genotypes (II, III, and IV) were involved in the first panzootic; genotypes V and VI were considered to be responsible for the second and third panzootics. In addition, it was indicated that the severe outbreaks in western Europe (13), South Africa and southern Europe (11), and Taiwan (29) in the 1990s were caused by prevalent genotype VII. Genotype I consists of the avirulent strains of NDV, whereas genotype VIII appears to be endemic to South Africa during the past few decades (11).
In the past 2 decades, due to a strict vaccination program, occurrences of ND were mild and sporadic and caused few deaths in chicken flocks in China. The sporadic cases, showing no typical clinical and pathological manifestation of the disease, were named “atypical ND” (28). Since 1996, however, outbreaks of ND have continuously occurred in the vaccinated chicken flocks of China. In Taiwan, four major outbreaks of ND have occurred since the first identification of ND in 1935 (29, 30). In this paper, seven NDV isolates recovered from outbreaks between 1996 and 2000 in China and Taiwan are characterized biologically and molecularly and the epidemiology of ND is evaluated on the basis of molecular analysis of the F protein gene.
MATERIALS AND METHODS
Source of virus.
Seven NDV isolates were collected between August 1996 and June 2000 in China and Taiwan (Table 1). These isolates were collected from outbreaks of ND in chicken and pigeon flocks. All chicken flocks from which viruses were isolated had been vaccinated with the NDV vaccine (LaSota/46) at least once depending on the age when the ND outbreaks occurred. The isolates were designated according to location of origin and year of isolation; Ch and TW are abbreviations for China and Taiwan, respectively. Ch-W6/96 was isolated from a racing-pigeon flock that suffered an outbreak of ND with 71.4% mortality shortly after an international race in 1996. Two months later, ND outbreaks occurred in chicken flocks in the same area where Ch-A7/96 was isolated from an outbreak with a 97% mortality rate. Ch/98-1 was considered typical of the virus responsible for the outbreaks in Chinese pigeon flocks in 1998, whereas Ch/98-3, Ch/99, and Ch/2000 were considered typical of the viruses responsible for outbreaks in Chinese chicken flocks between 1998 and 2000. TW/2000 was isolated from an outbreak in Taiwanese chicken flocks with a 80% mortality rate in 2000. It was noted that the affected chicken flocks of Taiwan had been vaccinated with LaSota/46 vaccine (live or killed) six times for age ranging from days 1 to 120 before the outbreak at day 160. Ch-F48E9/44 was biologically characterized (Table 2) as a virulent standard strain isolated from Beijing in 1944.
TABLE 1.
Pathogenicity and phylogenetic analysis of the NDV isolates from China and Taiwan
| Isolate | Host
|
MDT | ICPI | Pathotypeb | Cleavage site (112RRQKRF117)c | Genotype | Unique residue substitution at:
|
Accession no. of F gene | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | Vaca | 4 (K) | 71 (K) | 200 (I) | 255 (V) | 402 (A) | 489 (D) | 520d (V)e | |||||||
| Ch-W6/96f | Pigeon | − | 54 | 1.88 | NV | ||||||||||
| Ch-A7/96 | Fowl | + | 54 | 1.89 | NV | K—————g | VIIc | T | —g | L | Ij | — | — | — | AY 028995 |
| Ch/98-1 | Pigeon | − | 80 | 1.48 | M | K————— | VIb | T | — | L | I | — | — | Ik | AF 358785 |
| Ch/98-3 | Fowl | + | 48 | 1.89 | VV | —————— | VIId | T | Ri | L | I | T | E | G | AF 364835 |
| Ch/99 | Fowl | + | 58 | 1.84 | VV | —————— | VIId | T | R | L | I | T | E | G | AF 358787 |
| Ch/2000 | Fowl | + | 60 | 1.83 | VV | —————— | VIId | T | R | L | I | T | E | G | AF 358788 |
| TW/2000 | Fowl | + | 48 | 1.89 | VV | —————— | VIId | Rh | — | — | I | — | — | — | AF 358786 |
Vac, Vaccination. Column refers to immunity status of chicken flocks when outbreaks of ND occurred. −, not vaccinated; +, vaccinated.
M, mesogen VV, viscerotropic velogen; NV, neurotropic velogen.
The 112RRQKRF117 motif around the F2/F1 cleavage site represents the majority sequence.
Positions of residues in deduced amino acid sequence of the F0 protein.
Consensus sequence was constructed from the F0 protein.
Sequence data are not shown because Ch-W6/96 is a mixture of Ch-A7/96 and Ch/98-1.
Genetic identity with the consensus is marked with a dash (—).
R-for-K substitution is not unique and is shared by some NDV strains listed in Table 3.
R-for-K substitution is shared by Q-GB 506/97 (listed in Table 2).
I-for-V substitution is shared by Ch/97-1 (listed in Table 2).
I-for-V substitution is not unique and is shared by some NDV strains listed in Table 2.
TABLE 2.
Data for NDV strains and sequences
| Strain (origin) | Host | Genotype | Pathotype | ICPI | Cleavage site (112RRQKRF117) | Accession no. of F gene |
|---|---|---|---|---|---|---|
| Ch/97-1 (Guangdong, China) | Goose | VIId | VV | NA | ——————c | AF192406 |
| TW/98-4 | Fowl | VIId | NAb | NA | —————— | AF083964 |
| CZ 3898/96 (Czech Republic) | Fowl | VIIc | VV | 1.69 | —————— | AF109883 |
| CH 62/96 (Switzerland) | Fowl | VIIc | VV | 1.69 | —————— | AF109880 |
| TW/84C | Fowl | VIIc | NA | NA | —————— | AF083965 |
| TW/95-9 | Fowl | VIIc | VV | 1.68 | ——— ——— | AF083966 |
| TW/96P | Fowl | VIIc | NA | NA | ——— ——— | AF083971 |
| DE R143/95 (Germany) | Fowl | VIIa | VV | 1.86 | ——— ——— | AF109881 |
| TW/Owl95 | Owl | VIIa | VV | NA | ——— ——— | AF164966 |
| TW/94P | Fowl | VIIa | NA | NA | ——— ——— | AF083961 |
| Taiwan 95 | Fowl | VIIa | VV | NA | ——— ——— | U62620 |
| ZA 360/95 (South Africa) | Ostrich | VIIb | NA | NA | ——— ——— | AF109876 |
| ZW 3422/95 (Zimbabwe) | Ostrich | VIIb | NA | NA | ——— ——— | AF109877 |
| AE 232/1/96 (United Arab Emirates) | Partridge | VIIb | NA | NA | K——R—— | AF109884 |
| WB1-94 (India) | Fowl | VIIb | NA | NA | ——— R—— | AJ249530 |
| UP1-93 (India) | Fowl | VIIb | NA | NA | ——— ——— | AJ249529 |
| NP1-93 (India) | Fowl | VIIb | NA | NA | ——— ——— | AJ249526 |
| ASTR/74 (Russia) | Fowl | VIa | NA | NA | ——— ——— | Y18728 |
| Ch-HT40 | Pigeon | VIb | NA | NA | K————— | AF210635 |
| GB 1168/84 (United Kingdom) | Pigeon | VIb | M | 0.85 | G————— | AF109885 |
| Q-GB 506/97 (United Kingdom) | Finch | VIc | VV | 1.65 | ———R—— | AF136773 |
| AF2240 (Malaysia) | Fowl | VIII | VV | NA | ——— ——— | AF048763 |
| Italien/45 (Italy) | Fowl | IV | VV | 1.85 | ——— R—— | M17710 |
| Herts 33 (United Kingdom) | Fowl | IV | VV | 1.99 | ——— R—— | M24702 |
| Texas (United States) | Fowl | IV | VV | NA | ——— R—— | M33855 |
| GROZN/47 (Russia) | Fowl | IV | NA | NA | ——— R—— | AJ243387 |
| POK/70 (Russia) | Fowl | IV | NA | NA | ——— R—— | AJ243388 |
| SIMF/64 (Russia) | Fowl | IV | NA | NA | ———R—— | AJ243390 |
| Ch-PNDV1 | Pigeon | II | NA | NA | G——G—L | AF079324 |
| Clone 30a | II | L | 0.29 | G——G—L | Y18898 | |
| LaSota/46 (United States) | Fowl | II | L | 0.40 | G—— G—L | AF077761 |
| TN1-87 (India) | Fowl | II | NA | NA | G—— A—L | AJ249528 |
| Connecticut/9-12-60 (Canada) | Fowl | II | NA | NA | G——G—L | AF206617 |
| Beaudette C/45 (United States) | Fowl | II | M | 1.45 | ——— ——— | X04719 |
| Texas GB/48 (United States) | Fowl | II | NV | 1.74 | ——— ——— | M23407 |
| R2B (India) | Fowl | II | M | NA | ——— ——— | AJ249527 |
| Fuller (India) | Fowl | II | NA | NA | G——G—L | AJ249516 |
| D26/76 (Japan) | Duck | I | L | 0 | GK—G—L | M24692 |
| V4(Queensland)/66 | Fowl | I | L | 0.39 | GK—G—L | AF217084 |
| Ulster 2C/67 (Northern Ireland) | Fowl | I | L | 0 | GK—G—L | D00243 |
| AUS Victoria/32 | Fowl | III | NV | 1.66 | ——— ——— | M21881 |
| Miyadera/51 (Japan) | Fowl | III | VV | NA | ——— R—— | M18456 |
| Ch-F48E9/44 | Fowl | III | VV | 1.89 | ———R—— | AF079172 |
| Ch-DB3 | Fowl | III | NA | NA | ———R—— | AF079322 |
| TW/69 | Fowl | III | NA | NA | ——— ——— | AF083959 |
| TW/95-3 | Fowl | III | VV | 1.68 | ——— ——— | AF083970 |
From plaque cloning of LaSota/46.
NA, not available.
Genetic identity with the consensus is marked with dashes (—).
Virus isolation and biological characterization.
Except for TW/2000, all the viruses were isolated by two passages using embryonated, specific-pathogen-free (SPF) eggs. TW/2000 was isolated on the tissue culture of chicken kidney (first passage) and was cloned by using the virus plaque technique (second passage). Pathotyping of the isolates initially involved virus inoculation of 10-day-old, embryonated, SPF eggs to determine the MDT of the embryos, and further testing entailed inoculation of 1-day-old, SPF chicks to determine the ICPI values. Additionally, the intracloacal inoculation pathogenicity test was used to distinguish VV NDV from NV viruses in the SPF chickens. All the tests were performed using standard procedure (2).
Viral RNA extraction and RT-PCR.
A total of 20 ml of allantoic fluid or tissue culture supernatant was ultracentrifuged, and viral RNA was isolated from the pellet by using the RNAgents total RNA isolation system (Promega, Medison, Wis.) according to the manufacturer's instructions. The degenerate primers for reverse transcriptase PCR (RT-PCR) of the F gene, 5′GCGTCGACATGGGCYCYARAYCTTCTACCA3′ as forward primer and 5′TAGTCGACTCABATTYTTGTAGTGGCYCTCATCT3′ as reverse primer, were designed according to alignment of the F gene sequences from GenBank. The entire coding region of the F gene, a fragment of 1,662 bp (nucleotides ([nt] 47 to 1708) containing the variable region and cleavage site sequence (26), was amplified by RT-PCR (31) using Expand RT and the Expand high-fidelity PCR system (Boehringer Mannheim, Mannheim, Germany).
Sequencing of PCR products.
The recovered PCR products were directly sequenced (20) with Taq polymerase (Perkin-Elmer, Norwalk, Conn.). Firstly, the primers designed for PCR amplification were used for sequencing in opposite orientation. Thereafter, primers were designed based on the previous F gene sequences for the next sequencing. Ten picomoles of primer and 0.2 μg of double-strand DNA template were used in each sequencing reaction.
Phylogenetic analysis.
Alignment and phylogenetic analysis of the nucleotide sequences and the deduced amino acid sequences of F gene was performed with the Clustal V method of DNAStar software (12) (MegAlign, version 1.03, 1993). This method uses a multiple-alignment algorithm and the unweighted-pair group method with arithmetic mean algorithm to derive a preliminary phylogeny. The final phylogeny (dendrogram) was produced by the neighbor-joining method (23). The 100 replications were bootstrapped to construct a consensus phylogenetic tree.
Strains and accession numbers used for molecular analysis.
A total of 91 NDV strains, including our six isolates, were molecularly analyzed. Their F gene sequences, containing nt 47 to 420, 329 to 582, or 47 to 1708, were obtained from GenBank. The NDV strains studied and their accession numbers are listed in Table 1. The 85 strains that are compared (Table 2) (11, 13, 21, 22) were chosen to give a fair representation based on geographic distribution, year of isolation, and phylogenetic position. Among the sequences, nt 47 to 420 of TW/Owl95, Indian strains, and Ch-HT40 lack 24, 150, and 158 bp, respectively, at their 5′ end. That of TW/Owl95 lacks 15 bp at its 3′ end. The strains whose entire coding region of the F gene is available were chosen for analysis of the three regions.
Cross-protective test.
Two NDV strains, field isolate Ch-A7/96 and vaccine strain LaSota/46, were used to prepare monovalent oil-emulsion vaccines as described before (24). Selected 2-week-old, SPF chicks were vaccinated with either Ch-A7/96 or LaSota/46 oil-emulsion vaccine by the subcutaneous route, while the control group was injected with phosphate-buffered saline. Four weeks later, the birds were challenged by the eyedrop and intra-nasal routes, with Ch-A7/96 to determine cross-protection between Ch-A7/96 and LaSota/46. Ch-F48E9/44 was used as the challenge control to determine the efficiency of LaSota/46 oil-emulsion vaccine. The birds were observed for signs of disease for 2 weeks postchallenge.
RESULTS
Pathogenicity of isolates.
Initial biological characterization of the NDV isolates included MDT and ICPI determinations (Table 1). All viruses were virulent with MDT in embryonated chicken eggs being less than 60 h except for Ch/98-1, an isolate from pigeon that exhibited MDT within 80 h. The ICPI values ranged from 1.83 to 1.89 for the highly virulent NDV isolates and 1.48 for the moderately virulent isolate Ch/98-1. Intracloacal inoculation of chickens revealed that, among the highly virulent isolates, Ch/98-3, Ch/99, Ch/2000, and TW/2000 produced viscerotropic lesions, while Ch-W6/96 and Ch-A7/96 produced neurotropic lesions. The pigeon isolate Ch/98-1, which is a moderately virulent strain, did not produce apparent signs and lesions in chickens.
Phylogenetic analysis.
To assess the genetic relatedness among the NDV strains, phylogenetic analyses were performed with 3′ portions (nt 47 to 420) of the 76 F genes, 48 F gene subsequences (nt 329 to 582), and the entire coding regions of the 26 F genes (nt 47 to 1708). Results are shown in Fig. 1 and 2.
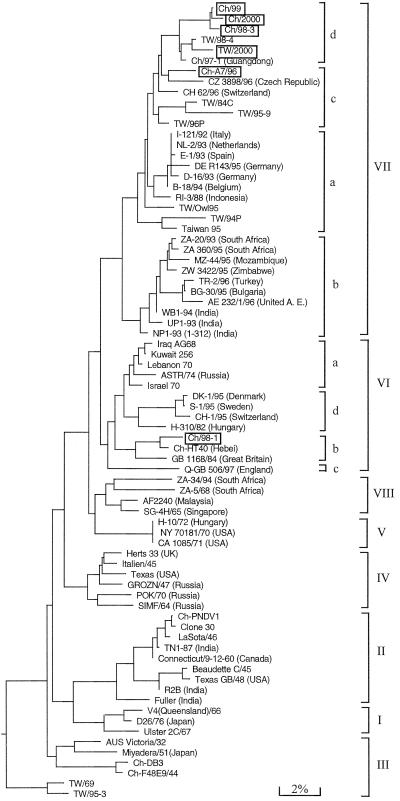
FIG. 1.
Phylogenetic tree of NDV strains based on nucleotide sequences from a portion (nt 47 to 420) of the F gene. Strains and their accession numbers are shown in Tables 1 and 2. Accession numbers of NDV strains in subgenotypes VIIa (except Taiwanese strains), VIa, and VId are given in reference 13; those of NDV strains ZA-5/68, ZA-20/93, ZA-34/94, MZ-44/95, BG-30/95, TR-2/96, and SG-4H/65 are given in reference 11. The strains characterized here are boxed. A. E., Arab Emirates.
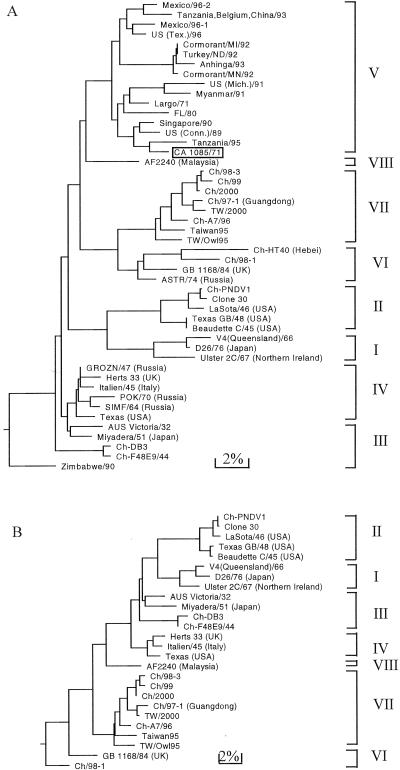
FIG. 2.
Phylogenetic trees of the nucleotide sequences of NDV strains from nt 329 to 582 (tree A) and 47 to 1708 (tree B) of the F gene. Zimbabwe/90 and strains from genotype V are listed in tree A. See details in references 21 and 22. Other strains and their accession numbers are listed in Tables 1 and 2. The boxed strain CA 1085/71 was selected from genotype V (Fig. 1) for analyses of tree A to determine the phylogenetic positions of the U.S. isolates.
The 76 NDV strains were separated into eight genotypes (I to VIII) by phylogenetic analysis of the 3′ portions (Fig. 1). Two novel subgenotypes, VIIc and VIId, were found in genotype VII, which consists of the most recent enzootic strains from China and Taiwan as well as of CH 62/96 and CZ 3898/96 from Switzerland and the Czech Republic, respectively. Our fowl isolates recovered during the outbreaks between 1998 and 2000, which include Ch/98-3, Ch/99, Ch/2000, and TW/2000, were grouped into subgenotype VIId, while fowl isolate Ch-A7/96, recovered in 1996, was placed in subgenotype VIIc (Table 1). The DNA sequence of pigeon isolate Ch-W6/96 exhibited overlapping profiles at all the different bases between fowl isolate Ch-A7/96 and pigeon isolate Ch/98-1 (data not shown), indicating that Ch-W6/96 was a mixture of Ch-A7/96 and Ch/98-1 isolates. Our pigeon isolate Ch/98-1, together with another two pigeon isolates, Ch-HT40 and GB 1168/84, was grouped under subgenotype VIb.
Similar results were obtained when the other two regions (nt 329 to 582 and 47 to 1708) of the F gene were used to analyze phylogenetic relatedness among the NDV strains, although the branch orders were somewhat different (Fig. 2). The NDV strains are displayed in the three trees (Fig. 1 and 2) as belonging to the same genotypes and subgenotypes, showing that each of the three regions in the F gene can be used to determine phylogenetic relatedness among NDV strains. The three trees share a very similar topology, indicating that no recombinant events occurred between F genes to generate strains of different lineage. By construction of the phylogenetic tree (Fig. 2A), the NDV strains from the outbreaks caused by exotic birds in the United States from 1971 to 1996 were grouped as genotype V, except for Zimbabwe/90, which belonged to genotype III.
The genetic similarities among the viruses from genotypes and subgenotypes, as well as the ranges of similarities, are presented in Table 3. For genotypes, the median similarities clustered in the range of 91.2 to 97.6%, while they were in the range of 94.9 to 98.4% for subgenotypes. Apparently, the intragenotype and intrasubgenotype genetic similarities were greater than the intergenotype and intersubgenotype similarities, except for genotype III (88.8 to 94.7%) and subgenotypes VIIa (91.7 to 100%) and VIIc (89.6 to 95.5%), in which some strains from Taiwan (Table 3) were responsible for the high divergences.
TABLE 3.
Genotype- and subgenotype-specific residue substitutions in deduced F0 protein sequences
| Genotype and subgenotype | Medium % of similaritya | Range of similarity (%)a | Consensus residue and its position in deduced F0 protein sequences
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 52 (I) | 69 (M) | 82 (E) | 93 (T) | 101 (R) | 104 (G) | 107 (S) | 118 (I) | 121 (I) | 124 (S) | 176 (A) | 192 (N) | 195 (Q) | 272 (N) | 288 (T) | 314 (F) | 341 (T) | 396 (M) | 482 (E) | |||
| I | 96.5 | (94.9–98.1) | —j | — | — | — | — | Ec | Td | — | — | G | — | K | — | — | — | — | — | — | — |
| II | 97.6 | (95.0–99.7) | — | L | Db | — | — | E | T | — | — | G | — | K | — | — | — | — | — | — | — |
| III | 91.2 | (88.8–94.7) | — | — | — | — | — | E | T | — | — | — | — | — | — | — | — | — | — | — | — |
| IV | 97.3 | (94.4–97.9) | — | — | — | — | — | E | T | — | — | — | — | — | — | — | — | — | — | — | — |
| V | (100) | — | — | — | — | — | — | T | Ve | — | — | — | — | — | — | — | — | — | — | — | |
| VI | 94.6 | (90.9–99.5) | |||||||||||||||||||
| a | 98.4 | (96.8–99.5) | — | — | — | — | — | — | T | — | — | — | — | — | — | — | — | — | — | — | — |
| ce | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| d | 98.4 | (94.7–99.7) | — | — | — | S | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| b | 95.4 | (93.0–97.7) | — | — | — | — | — | — | — | — | — | — | — | — | R | — | N | — | — | Ii | A |
| VIII | 91.2 | (90.4–92.0) | — | — | — | — | — | — | T | — | — | — | — | — | R | Y | N | — | Sh | — | A |
| VII | 92.1 | (86.6–99.2) | |||||||||||||||||||
| b | 97.4 | (95.7–99.2) | — | — | — | — | — | — | — | — | Vf | — | g | ||||||||
| a | 98.7 | (91.7–100) | — | — | — | — | K | — | — | — | V | — | S | R | Y | N | — | S | I | A | |
| c | 94.9 | (89.6–95.5) | — | — | — | — | K | — | — | — | V | — | S | R | Y | N | — | S | I | A | |
| d | 96.3 | (95.5–99.2) | V | — | — | — | K | — | — | — | V | — | S | R | Y | N | Y | S | I | A | |
The median genetic distance between the strain on top of the genotype or subgenotype and other strains in the same group is used as the median percentage of sequence similarity.
D-for-E substitution is shared by TW/69 and TW/95-3 in genotype III.
Residue E is retained in AE 232/1/96 of genotype VIIb.
Residue T is not retained in ZA-5/68 of genotype VIII or in TW/69 and TW/95-3 of genotype III.
Only one value was obtained, so no median or range is given.
e V-for-I substitution is shared by ZA-5/68 grouped in genotype VIII.
V-for-I substitution in genotype VII is shared by TW/69 and TW/95-3 in genotype III and AF2240 in genotype VIII.
No complete sequence of the F genes is available.
S-for-T substitution is shared by GB 1168/84 in genotype VIb.
I-for-M substitution is shared by Ch-F48E9/44 in genotype III.
—, genetic identity with the consensus.
Unique or genotype-specific residue substitutions.
The predicted amino acid sequences of the F genes listed in Fig. 1 (residues 1 to 125), 2A (95 to 179) and 2B (residues 126 to 553) exhibit a high degree of identity in pairwise comparisons (data not shown). Cluster, subgenotype, and genotype-specific residue substitutions in the deduced sequences are listed in Tables 1 and 3.
As shown in Table 1, our isolates, especially the ones from China, shared some unique amino acid substitutions despite being grouped in different genotypes or subgenotypes. Chinese isolates, including VIIc virus Ch-A7/96, VIId viruses Ch/98-3, Ch/99, and Ch/2000, and VIb virus Ch/98-1, shared unique T4-for-K and L200-for-I substitutions. These isolates, together with the Taiwanese isolate TW/2000 (Table 1) and a Chinese goose isolate, Ch/97-1 (Table 2), shared a unique I255-for-V substitution. Furthermore, Ch/98-3, Ch/99, and Ch/2000, among the Chinese isolates, produced four unique amino acid substitutions at residues 71 (R-for-K), 402 (T-for-A), 489 (E-for-D), and 520 (G-for-V). These geography-specific substitutions, not related to genotype and subgenotype, were unique among the NDV strains compared. However, the 5 potential N-linked glycosylation sites and 10 cysteine residues (7, 14) were absolutely conserved in the six isolates.
As shown in Table 3, many genotype- and subgenotype-specific residue substitutions were found in comparison with viruses from the ancient genotypes I to IV. They include V52-for-I and Y314-for-F substitutions for VIId; K101-for-R substitution for VIIa, -c, and -d; Y272-for-N and S341-for-T substitutions for VII and VIII; I396-for-M substitution for VIb and VII; R195-for-Q, N288-for-T, and A482-for-E substitutions for VIb, VII, and VIII; and V118-for-I substitution for genotype V. These lineage-specific amino acids constitute the molecular basis of genotyping NDV strains in Fig. 1 and 2.
By comparison of the deduced amino acid sequences, the NDV strains having unique genetic features were found (Table 3) in some geographic areas. In Southeast Asia, TW/69 and TW/95-3 (genotype III) shared some genetic features of the genotype II viruses (D82-for-E substitution) and the VII viruses (V121-for-I substitution) but did not have the genotype-III-specific T107-for-S substitution. Similarly, AF2240 (genotype VIII) shared the characteristic V121-for-I substitution of the VII viruses. In the Middle East, TR-2/96 (VIIb) did not contain the V121-for-I substitution of genotype VII, while AE 232/1/96 (VIIb) retained the specific residue (E) of genotypes I to IV; GB 1168/84 (VIb) shared VII- and VIII-specific S341-for-T substitution. In South Africa, genotype VIII virus ZA-5/68 did not contain the specific residue of the genotypes I to IV, VIa, and VIII (T107) but shared the V-specific V118-for-I substitution.
Proteolytic cleavage site of F0 protein.
Cleavage site motifs of the 76 NDV strains listed in Fig. 1 are compared (Tables 1 and 2) (11, 13). Of the cleavage site motifs of the 67 virulent strains that were compared, 49 showed 112RRQKR/F117, 13 showed 112RRQRR/F117, 3 showed 112KRQKR/F117, 1 showed 112KRQRR/F117 (AE 232/1/96), and 1 showed 112GRQKR/F117 (GB 1168/84). The cleavage sites of our isolates showed motifs 112RRQKR/F117 and 112KRQKR/F117 (Table 1). Of the cleavage site motifs of the nine low-virulence and avirulent strains (Table 2), five showed 112GRQGR/L117, three showed 112GKQGR/L117, and one showed 112GRQAR/L117 (TN1-87). Results showed that the virulent NDV strains exhibit sequence motif 112R(K)-R-Q-K(R)-R/F117, while the low-virulence and avirulent NDV strains exhibit sequence motif 112G-R(K)-Q-G(A)-R/L117.
Of the genotype II viruses, although LaSota/46 shared 94.9 and 95.2% similarity with Beaudette C/45 and Texas GB/48, respectively (data not shown), their cleavage sites (112GRQGR/L117 and 112RRQKR/F117) were very different. Similarly, although Beaudette C/45 shared 99.2% similarity and an identical cleavage site (112RRQKR/F117) with Texas GB/48, the virulence of the two strains was very different, with ICPI values of 1.45 and 1.74. This showed that no necessary correlation between the phylogenetic relatedness and the virulence exists among NDV strains.
Neutralizing epitopes and antigenic variants.
At least seven neutralizing epitopes, positioned at residues 72, 74, 75, 78, 79, 157 to 171, and 343 of the F protein, have been identified (17, 25, 32). Our analysis of the amino acid sequences showed some neutralizing epitope variants. They include POK/70 and SIMF/64 in genotype IV (G72-for-D substitution), ZW 3422/95 (V75-for-A substitution) and ZA-20/93 (R78-for-K substitution) in subgenotype VIIb, TW/94P and Taiwan 95 in the subgenotype VIIa and four genotype VIII viruses (T/P79-for-A substitution), and V4(Queensland)/66 in the genotype I and some of the genotype V viruses (N170-for-D substitution). Most of these variants are distributed geographically in Southeast Asia and southern Africa despite their genotyping. However, these epitopes are highly conserved among most strains, including our isolates from China and Taiwan.
Epidemiological linkage of NDV transmission between China and Taiwan.
As shown in Fig. 1, TW/2000 was closely related to the isolate TW/98-4 (97.9% similarity), which is very similar to the goose isolate Ch/97-1 (98.4% similarity), which in turn is highly virulent in geese and chickens (Table 2). These results, in conjunction with the analysis of the unique residue substitutions from Table 1, showed that TW/98-4 was introduced into China from Taiwan before 1997. By producing a unique I255-for-V substitution, TW/98-4 became Ch/97-1 and caused the outbreaks of ND in goose and chicken flocks. Thereafter, the Ch/97-1 evolved rapidly by producing multiple, unique amino acid substitutions (Table 1), becoming the Ch/98-3, Ch/99, and Ch/2000 isolates that caused the outbreaks in Chinese chicken flocks between 1998 and 2000.
The 1996 outbreaks in Chinese chicken flocks due to Ch-A7/96 were caused by sickness in the pigeon flocks after an international pigeon race. This shows that Ch-A7/96, a strain similar to CZ 3898/96 from the Czech Republic (Fig. 1), is not the direct origin of our subgenotype VIId viruses (Ch/98-3, Ch/99, Ch/2000, and TW/2000). Also, the Ch/98-1 (VIb virus) originated from the 1996 outbreaks after the pigeon race and shared a common ancestor with Ch-HT40 (97.7% similarity).
Evolution and molecular epidemiology of NDV strains.
Based on the residue substitutions (Table 3), the phylogenetic relatedness (Fig. 1 and 2), and epidemiological data (Table 2) of the NDV strains, we proposed an evolutionary model of NDV strains throughout the world and evaluated the epidemiology of ND transmission.
The viruses of genotype I to IV are the most ancient strains. Among them, genotype III and genotype IV viruses became genotype I viruses by producing G124-for-S and K192-for-N substitutions, which further became genotype II viruses by producing L69-for-M and D82-for-E substitutions. The outbreaks caused by viruses of genotypes I to IV constituted the first panzootic of ND. A dramatic change, G104-for-E substitution, caused evolution of genotypes III and IV into the subgenotype VIa. By producing a V118-for-I substitution, subgenotype VIa was changed into genotype V. At the same time, genotype VIII viruses also evolved from the VIa viruses by producing five amino acid substitutions at the conserved region of the F protein (residues 195 to 482) but retained an ancient residue, T107. The outbreaks caused by viruses from genotypes V, VIa, and VIII constituted the second panzootic of ND. Subgenotype VIa was changed to VIc by production of a crucial S107-for-T substitution and further became VId by production of an S93-for-T substitution. Thereafter, the evolution from subgenotype VIc to VIb occurred by production of four amino acid changes at the conserved region (residues 195 to 482). The outbreaks caused by viruses from subgenotypes VIb, VIc, and VId constituted the third panzootic of ND.
Subgenotype VIIb evolved from VIb via production of a VII-specific V121-for-I substitution and then changed to subgenotypes VIIa and VIIc by producing a K101-for-R substitution and became VIId by producing additional V52-for-I and Y314-for-F substitutions. To date, genotype VII viruses have caused many outbreaks of ND around the world (Fig. 1). They include the outbreaks caused by subgenotypes VIIc and -d in China between 1996 and 2000; subgenotypes VIIa, -c, and -d in Taiwan between 1984 and 2000; subgenotype VIIa in western Europe between 1992 and 1996; subgenotype VIIb in southern Africa between 1990 and 1995; and subgenotypes VIIb and -c in the Middle East, northern and eastern Europe, and India between 1993 and 1996. Based on our phylogenetic analysis (Fig. 1), the outbreaks in the United Kingdom and northern Europe between 1996 and 1997 (4) were caused by subgenotype VIIb viruses. Thus, genotype VII viruses, which appeared in Taiwan and Indonesia in the 1980s and caused outbreaks in Europe, southern Africa, the Middle East, and Asia in the 1990s, constitute the fourth panzootic of ND. The VIId viruses atop the phylogenetic tree (Fig. 1), from China and Taiwan, represent the most newly emerging NDV strains. Since the fourth panzootic caused by the VII viruses, cocirculation of the virulent viruses grouped in III, V, VIb, -c, and -d, and VIII has been occurring in some countries (Fig. 1 and 2A; Table 2).
Thus, amino acid substitution analysis of the deduced F protein sequences (Table 3) was consistent with the phylogenetic analysis of the 5′ terminal F genes (Fig. 1) in defining evolutionary and epidemiological linkage among the NDV strains. However, the phylogenetic analyses of other regions (nt 329 to 582 and 47 to 1708) of the F genes (Fig. 2) could not specify the branch order that exhibits the evolutionary direction of NDV strains.
Cross-protection.
In view of recent outbreaks of ND in vaccinated chicken flocks of China and Taiwan and the genetic features of the isolates, we conducted a laboratory investigation to determine whether there was evidence of cross-protection between vaccine strain LaSota/46 and field strain Ch-A7/96. Results showed that the oil-emulsion vaccine of Ch-A7/96 protected all chicks against morbidity and death from challenge with Ch-A7/96 and Ch-F48E9/44. However, the vaccine strain LaSota/46 reduced overall clinical protection to 40% when challenged with Ch-A7/96, although it protected 9 out of the 10 chicks challenged with Ch-F48E9/44. No control chicks were protected after challenge. These results showed that Ch-A7/96 was responsible for the outbreaks of ND in China during 1996 as an immune response-escaping antigenic variant.
DISCUSSION
In this study, seven NDV isolates recovered from China and Taiwan between 1996 and 2000 were genotypically and pathotypically characterized. Among them, the highly virulent VIIc and VIId viruses, representative of the most newly emerging NDV strains, were responsible for outbreaks of ND in vaccinated chicken flocks. Furthermore, a cross-protective experiment showed that the VIIc virus Ch-A7/96 is an immuneresponse-escaping antigenic variant. By comparative genetic analyses, epidemiological linkage of NDV transmission between China and Taiwan and of the fourth panzootic of ND in the 1990s was determined. On the basis of establishing an evolutionary model of the NDV strains, the epidemiology of NDV infection was evaluated.
It was known that ND viruses exist as a single serotype based on the neutralizing test and cross-protective analysis (3). Nonetheless, all the chicken-derived strains reported here were isolated from vaccinated flocks. Phylogenetic analysis (Fig. 1) and unique residue substitution analysis (Table 1) suggested that these isolates might be antigenic variants. To investigate the possibility of vaccination failure caused by antigenic variation, a cross-protective experiment was performed. Results revealed that LaSota/46-vaccinated chickens were only partially protected against challenge with Ch-A7/96, indicating that Ch-A7/96 indeed was an antigenic variant that escapes the protective immune response of the LaSota/46 vaccine strain.
The molecular determinants responsible for the antigenic variant remain to be determined. Since the neutralizing epitopes (32), the cysteine residues, and N-linked glycosylation sites (7) of the F0 protein were conserved and since no deletions and insertions (data not shown) and recombinations occurred, the accumulation of point mutations that induces amino acid substitutions should be responsible for the production of the antigenic variant. Thus, a unique I255-for-V substitution, shared by Ch-A7/96 and other strains from China and Taiwan (Table 1) and a goose isolate, Ch/97-1 (Table 2), may have been responsible for the reduction of cross-protection between Ch-A7/96 and LaSota/46. The other unique substitutions presented in Table 1 and the genotype- and subgenotype-specific substitutions presented in Table 3 might also have contributed to the molecular basis of this antigenic variant.
Evolution of NDV, depending on the accumulation of point mutations that induce amino acid substitutions, is a slow process (26). However, it proceeded rapidly after the second panzootic that started in the late 1960s, when vaccination was widely used. Based on our analysis, the recent Chinese isolates originated from Taiwan. However, the evolutionary rate of the Chinese isolates was higher than that of Taiwan's, according to the unique amino acid substitutions (Table 1). Immune pressure of the host may contribute to the difference in evolutionary rates. In China a strict vaccination program against ND has been performed for 20 years, but in Taiwan, most of the chicken flocks had not been vaccinated when the outbreaks occurred in 1995 (30). These results suggested that selective immune pressure of the host enhanced the evolutionary process of NDV, which was rapid over the last 30 years compared to that of the previous 40 years.
Our comparative results (Table 2) signified that the differences in virulence among NDV strains mainly correlate with the number and arrangement of the basic amino acid residues in the cleavage site (8, 16, 27). Direct evidence has been achieved by introducing three amino acid changes into the F0 cleavage site of LaSota/46. When the cleavage site 112GRQGR/L117 was changed to 112RRQRR/F117, the ICPI of the modified LaSota/46 rose from 0.00 to 1.28 (19). It appears that, under natural conditions, it is difficult to turn vaccine or naturally avirulent strains into virulent strains by producing three nucleotide mutations that induce three amino acid substitutions at the same time. Nonetheless, there was a case in Australia in 1998 (5), in which virulent isolates had 99% identity to an endemic, nonvirulent Australian strain of NDV isolated recently but were quite different genetically from overseas isolates of virulent NDV that had been sequenced.
However, neither the highly virulent and moderately virulent NDV strains nor the low-virulence and avirulent NDV strains can be distinguished by the cleavage site motifs (Table 2). This indicated that the virulence of NDV strains can be qualified rather than quantified by the analysis of cleavage site motifs and that pathogenicity tests such as MDT and ICPI cannot be replaced by analysis of the cleavage site to precisely characterize virulence and pathogenicity of NDV strains. HN protein precursors also play a role in the virulence of NDVs (9, 16), due to which the cleavage site motifs of the F0 proteins cannot be used to precisely characterize the virulence of NDV strains. In addition, the phylogenetic relationship among NDV strains does not correlate well with cleavage site sequences and virulence (Fig. 1 and Table 2).
By comparative analysis of the amino acid sequences, we found that some NDV strains have unique genetic features (Table 3) characteristic of geographic areas. They include TW/69, TW/95-3, and AF2240 in Southeast Asia; GB 1168/84, AE 232/1/96, and TR-2/96 in the Middle East; and ZA-5/68 in South Africa. To have a counterpart, we also found that most of the neutralizing epitope variants had originated in Southeast Asia and southern Africa. These unusual strains suggested that NDVs had a long history of evolution in Southeast Asia, the Middle East, and South Africa and that these geographic areas were the original places of transmission and panzootic of ND. Epidemiological data (1) have indicated that the three panzootics of ND originated in Southeast Asia (panzootic I) and the Middle East (panzootics II and III). Since genotype VII viruses first appeared in Taiwan (VIIa and -c) and Indonesia (VIIb) in the 1980s and then caused the fourth panzootic of ND in the 1990s, we conclude that the fourth panzootic originated in Southeast Asia.
Analysis of viral nucleotide sequence distances, inferred phylogenetic relationship, and substitutions of deduced amino acid sequences played a central role in the investigation of this cluster of ND outbreaks and the epidemiological analysis of NDV infection. Genetic analysis is also proving to be valuable in tracking the spread and origin of NDVs in countries. This investigation demonstrates that detailed analysis of NDV genetic variation is a powerful tool for understanding the epidemiology of NDV transmission.
ACKNOWLEDGMENT
These investigations were supported by the National Science and Technology Board of Singapore.
REFERENCES
- 1.Alexander D J. Historical aspects. In: Alexander D J, editor. Newcastle disease. Boston, Mass: Kluwer Academic Publishers; 1988. pp. 1–10. [Google Scholar]
- 2.Alexander D J. Newcastle disease. In: Purchase H G, Arp L H, Domermuth C H, Pearson J E, editors. A laboratory manual for the isolation and identification of avian pathogens. 3rd ed. Kennett Square, Pa: American Association of Avian Pathologists, Inc.; 1989. pp. 114–120. [Google Scholar]
- 3.Alexander D J. Newcastle disease and avian paramyxovirus infections. In: Calnek B W, Barnes H J, Beard C W, McDougald L R, editors. Diseases of poultry. 10th ed. Ames, Iowa: Iowa State University Press; 1997. pp. 541–569. [Google Scholar]
- 4.Alexander D J, Banks J, Collins M S, Manvell R J, Frost K M, Speidel E C, Aldous E W. Antigenic and genetic characterization of Newcastle disease viruses isolated from outbreaks in domestic fowl and turkeys in Great Britain during 1997. Vet Rec. 1999;145:417–421. doi: 10.1136/vr.145.15.417. [DOI] [PubMed] [Google Scholar]
- 5.Anonymous. Outbreak of virulent Newcastle disease. Anim Health Surveill Q. 1998;3(3):1–3. [Google Scholar]
- 6.Ballagi-Pordany A, Wehmann E, Herczeg J, Belak S, Lomniczi B. Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Arch Virol. 1996;141:243–261. doi: 10.1007/BF01718397. [DOI] [PubMed] [Google Scholar]
- 7.Chambers P, Millar N S, Emmerson P T. Nucleotide sequence of the gene encoding the fusion glycoprotein of Newcastle disease virus. J Gen Virol. 1986;67:2685–2694. doi: 10.1099/0022-1317-67-12-2685. [DOI] [PubMed] [Google Scholar]
- 8.Glickman R L, Syddall R J, Iorio R M, Sheehan J P, Bratt M A. Quantitative basic residue requirements in the cleavage-activation site of the fusion glycoprotein as a determinant of virulence for Newcastle disease virus. J Virol. 1988;62:354–356. doi: 10.1128/jvi.62.1.354-356.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorman J J, Nestorowicz A, Mitchell S J, Corino G L, Selleck P W. Characterization of the sites of proteolytic activation of Newcastle disease virus membrane glycoprotein precursors. J Biol Chem. 1988;263:12522–12531. [PubMed] [Google Scholar]
- 10.Gotoh B, Ohnishi Y, Inocencio N M, Esaki E, Nakayama K, Barr P J, Thomas G, Nagai Y. Mammalian subtilisin-related proteinases in cleavage activation of the paramyxovirus fusion glycoprotein: superiority of furin/PACE to PC2 or PC1/PC3. J Virol. 1992;66:6391–6397. doi: 10.1128/jvi.66.11.6391-6397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herczeg J, Wehmann E, Bragg R R, Travassos Dias P M, Hadjiev G, Werner O, Lomniczi B. Two novel genetic groups (VIIb and VIII) responsible for recent Newcastle disease outbreaks in Southern Africa, one (VIIb) of which reached Southern Europe. Arch Virol. 1999;144:2087–2099. doi: 10.1007/s007050050624. [DOI] [PubMed] [Google Scholar]
- 12.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 13.Lomniczi B, Wehmann E, Herczeg J, Ballagi-Pordany A, Kaleta E F, Werner O, Meulemans G, Jorgensen P H, Mante A P, Gielkens A L, Capua I, Damoser J. Newcastle disease outbreaks in recent years in western Europe were caused by an old (VI) and a novel genotype (VII) Arch Virol. 1998;143:49–64. doi: 10.1007/s007050050267. [DOI] [PubMed] [Google Scholar]
- 14.McGinnes L W, Morrison T G. Nucleotide sequence of the gene encoding the Newcastle disease virus fusion protein and comparisons of paramyxovirus fusion protein sequences. Virus Res. 1986;5:343–356. doi: 10.1016/0168-1702(86)90028-6. [DOI] [PubMed] [Google Scholar]
- 15.Millar N S, Emmerson P T. Molecular cloning and nucleotide sequencing of Newcastle disease virus. In: Alexander D J, editor. Newcastle disease virus. Boston, Mass: Kluwer Academic Publishers; 1988. pp. 79–97. [Google Scholar]
- 16.Nagai Y, Klenk H D, Rott R. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology. 1976;72:494–508. doi: 10.1016/0042-6822(76)90178-1. [DOI] [PubMed] [Google Scholar]
- 17.Neyt C, Geliebter J, Slaoui M, Morales D, Meulemans G, Burny A. Mutations located on both F1 and F2 subunits of the Newcastle disease virus fusion protein confer resistance to neutralization with monoclonal antibodies. J Virol. 1989;63:952–954. doi: 10.1128/jvi.63.2.952-954.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogasawara T, Gotoh B, Suzuki H, Asaka J, Shimokata K, Rott R, Nagai Y. Expression of factor X and its significance for the determination of paramyxovirus tropism in the chick embryo. EMBO J. 1992;11:467–472. doi: 10.1002/j.1460-2075.1992.tb05076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peeters B P, de Leeuw O S, Koch G, Gielkens A L. Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J Virol. 1999;73:5001–5009. doi: 10.1128/jvi.73.6.5001-5009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seal B S, King D J, Bennett J D. Characterization of Newcastle disease virus isolates by reverse transcription PCR coupled to direct nucleotide sequencing and development of sequence database for pathotype prediction and molecular epidemiological analysis. J Clin Microbiol. 1995;33:2624–2630. doi: 10.1128/jcm.33.10.2624-2630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seal B S, King D J, Locke D P, Senne D A, Jackwood M W. Phylogenetic relationships among highly virulent Newcastle disease virus isolates obtained from exotic birds and poultry from 1989 to 1996. J Clin Microbiol. 1998;36:1141–1145. doi: 10.1128/jcm.36.4.1141-1145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Stone H D. Efficacy of oil-emulsion vaccines prepared with pigeon paramyxovirus-1, Ulster, and La Sota Newcastle disease viruses. Avian Dis. 1989;33:157–162. [PubMed] [Google Scholar]
- 25.Toyoda T, Gotoh B, Sakaguchi T, Kida H, Nagai Y. Identification of amino acids relevant to three antigenic determinants on the fusion protein of Newcastle disease virus that are involved in fusion inhibition and neutralization. J Virol. 1988;62:4427–4430. doi: 10.1128/jvi.62.11.4427-4430.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toyoda T, Sakaguchi T, Hirota H, Gotoh B, Kuma K, Miyata T, Nagai Y. Newcastle disease virus evolution. II. Lack of gene recombination in generating virulent and avirulent strains. Virology. 1989;169:273–282. doi: 10.1016/0042-6822(89)90152-9. [DOI] [PubMed] [Google Scholar]
- 27.Toyoda T, Sakaguchi T, Imai K, Inocencio N M, Gotoh B, Hamaguchi M, Nagai Y. Structural comparison of the cleavage-activation site of the fusion glycoprotein between virulent and avirulent strains of Newcastle disease virus. Virology. 1987;158:242–247. doi: 10.1016/0042-6822(87)90261-3. [DOI] [PubMed] [Google Scholar]
- 28.Wei J, Shen Y, Xie S, Chu F, Wu Y. Atypical Newcastle disease. Chin J Anim Poult Infect Dis. 1998;20(Suppl.):31–38. [Google Scholar]
- 29.Yang C Y, Shieh H K, Lin Y L, Chang P C. Newcastle disease virus isolated from recent outbreaks in Taiwan phylogenetically related to viruses (genotype VII) from recent outbreaks in western Europe. Avian Dis. 1999;43:125–130. [PubMed] [Google Scholar]
- 30.Yang C Y, Chang P C, Hwang J M, Shieh H K. Nucleotide sequence and phylogenetic analysis of Newcastle disease virus isolates from recent outbreaks in Taiwan. Avian Dis. 1997;41:365–373. [PubMed] [Google Scholar]
- 31.Yu L, Liu W, Schnitzlein W M, Tripathy D N, Kwang J. Study of protection by recombinant fowlpox virus expressing C-terminal nucleocapsid protein of infectious bronchitis virus against challenge. Avian Dis. 2001;42:340–348. [PubMed] [Google Scholar]
- 32.Yusoff K, Nesbit M, McCartney H, Meulemans G, Alexander D J, Collins M S, Emmerson P T, Samson A C. Location of neutralizing epitopes on the fusion protein of Newcastle disease virus strain Beaudette C. J Gen Virol. 1989;70:3105–3109. doi: 10.1099/0022-1317-70-11-3105. [DOI] [PubMed] [Google Scholar]