Abstract
We previously observed a marked diversity of rotavirus strains and a high prevalence of the uncommon serotype G9 in a small survey of rotavirus strains collected from six centers in India. In the present study, we characterized a larger collection of strains from children hospitalized with severe diarrhea in seven Indian cities between 1996 and 1998. A total of 287 strains were G and P genotyped by reverse transcription-PCR, and some were further characterized by electropherotyping and subgrouping. Of the four strains common globally, three were found in only 43% of samples (P[8], G1, 15%; P[4], G2, 22%; P[8], G4, 6%), whereas G9 strains made up 17% of the total. Three different G9 strains were present: a P[8], G9 strain, which displayed the long electropherotype and subgroup II VP6 specificity, and two P[6], G9 strains, one with the long electropherotype and subgroup II specificity and the other with the short electropherotype and subgroup I specificity. Marked diversity was observed among strains collected from different cities and collected over time. Of the 253 strains that were fully typed, 54 (21%) had a mixed G or P genotype. Serotype G2 strains were detected more often in infections caused by single strains than in mixed infections (P < 0.05), whereas serotype G1 strains were found more often in mixed infections than in infections caused by single strains (P < 0.05). The diversity of rotavirus strains and the high prevalence of mixed infections confirm trends reported earlier and help to better characterize the strains of rotavirus circulating in India. Vaccines under development should clearly target G9 strains, and G9 should be included as one of the common global serotypes.
Group A rotavirus (RV) is the leading cause of gastroenteritis worldwide (17) and is the cause of significant childhood morbidity and mortality among children younger than 5 years of age in India. Of the approximately 600,000 annual deaths due to RV worldwide, more than 150,000 occur in India (V. Jain and U. Parashar, unpublished data, 2000), making the development and testing of an effective RV vaccine a particular priority in India.
In 1998, an RV vaccine composed of rhesus-human reassortant viruses that targeted the four globally common RV G serotypes was licensed for use in the United States. This vaccine was designed to provide serotype-specific immunity to the four most common RV strains circulating worldwide, and the vaccine demonstrated an efficacy of 80 to 100% for severe and dehydrating diarrhea caused by these common strains (16, 24). Subsequent work, however, showed that the distribution of RV strains varied greatly between geographic regions; and in some countries, such as Brazil and India (11, 21, 26), novel strains not commonly seen elsewhere were very prevalent.
Our initial pilot studies characterizing 133 RV strains from India made three interesting observations: first, serotype G9, previously not considered a common strain worldwide, was the most common serotype, with 17% of strains surveyed being of serotype G9 (7, 21). Second, in some Indian cities, as many as eight different strains were circulating at the same time, a diversity of strains far greater than that seen in most other countries. Third, the prevalence of mixed infections with two or more different RV strains was high (11%). This was one of the first reports demonstrating that type G9 could be a common cause of diarrhea in infants since its discovery in the United States and Japan in the mid-1980s (3; N. Ikegami, N., K. Akatani, T. Hosaka, and H. Ushijima, Abstr., VII. Int. Congr. Virol., p. 113, 1987). Since these initial reports from India (6, 21), the G9 serotype has been found in a variety of studies (for example, in Bangladesh, Malawi, the United Kingdom, and the United States) (4, 5, 13, 15, 27). In the United States, 4% of strains detected in a multicenter surveillance study were serotype G9, and serotype G9 strains were sometimes the predominant strains (i.e., 17 to 48% of isolates collected in several cities) (13, 22). Mixed infections are also common in other developing countries (e.g., Brazil and Bangladesh) where they have been sought (19, 26, 27). In addition, previous studies in India have demonstrated that neonatal genotype P[6] strains are also commonly found among pediatric patients (21) and can be present in association with many different G serotypes.
These initial observations in India, made from only 63 RV specimens, similar observations made in Delhi, India, by Husain and coworkers (14) led us to undertake a larger, longer-term study of the RV strains in circulation in India with the specific aim of confirming and extending our initial findings. This information will be critical for understanding the background of RV strains before assessing the potential efficacy of any vaccine predicated upon elicitation of serotype-specific immunity. We report here the results of a 3-year study of hospital inpatients (7, 21) in seven cities in India where RV was isolated and characterized for their G and P genotypes by using reverse transcription (RT)-PCR.
MATERIALS AND METHODS
Study sites.
A nationwide RV surveillance system consisting of six cities in India had been established previously (21). The stool samples analyzed in the present study came from inpatients younger than 5 years of age who sought treatment for severe diarrhea at hospitals in Bhopal, New Delhi, Davengere, Lucknow, Nagpur, and Shimla from 1996 to 1998. Hyderabad was added to the study in 1998. Patient information collected at the time of treatment included hospital and study identification numbers, age, and dates of treatment. Stool specimens were also collected at the time of treatment. Clinical data, such as severity of disease, symptoms, and history of prior RV infection, were not collected. Demographic and socioeconomic data were also not collected.
Detection of RV in stool specimens.
A total of 1,502 stool specimens from diarrheal patients were collected, frozen at −20°C, and shipped to the All India Institute of Medical Sciences, New Delhi, for testing. RV detection was performed with the Premier Rotaclone EIA kit (Meridian Diagnostics, Cincinnati, Ohio) according to the manufacturer's instructions, except that 15% stool suspensions made in 50 mM Tris-HCl (pH 7.5) were used. A total of 313 samples that tested positive for RV by the enzyme immunoassay were included in the study.
RNA extraction.
Rotavirus RNA was extracted from fecal suspensions by a modified version of the glass powder method reported previously (10). Briefly, 15% stool suspensions were first extracted with Freon and were then incubated with 6 M guanidine isothiocyanate (GITC) for 10 min at 65°C. Ten microliters of RNAMatrix glass powder (Bio 101, Inc., Vista, Calif.) was used to bind and pellet the RNA. The pellets were then washed with 4 M GITC in 50 mM Tris-HCl (pH 7.5) and RNA wash solution (Bio 101, Inc.). They were then air dried and resuspended in diethyl pyrocarbonate-treated H2O.
G and P typing by RT-PCR.
RV RNA was analyzed to determine both the VP7 and the VP4 genotypes by a modified protocol similar to those described previously (6, 10). For VP7 genotyping, RV RNA was subjected to heminested RT-PCR with primers 9con1 and 9con2, followed by 30 cycles of PCR with a cocktail of seminested primers, primers 9T1-1, 9T1-2, 9T-3P, 9T-4, and 9T-9B (each at a concentration of 20 μM). For VP4 genotyping, all procedures were identical to those described above, except that RT and the first amplification were done with primers con2 and con3 and the second amplification was done with con3 and VP4-genotyping multiplex primers 1T-1, 2T-1, 3T-1, 4T-1, and 5T-1. The samples were then resolved on 2% agarose gels to determine the G and P types. This combined typing scheme was designed to detect VP7 genotypes G1, G2, G3, G4, and G9 as well as VP4 genotypes P[4], P[6], P[8], P[9], and P[10]. All samples for which the RV VP4 genotype was not determined by this method were then subjected to RT-PCR with the ND2 primer, designed to detect P[11] strains.
Electropherotyping.
The electropherotype patterns of selected G2 and G9 strains were determined by subjecting RV RNA to electrophoresis on 10% polyacrylamide gels and silver staining.
VP6 subgroup determination.
A VP6-subgrouping enzyme-linked immunosorbent assay was performed as described previously (25) with monoclonal antibodies to subgroup I (monoclonal antibody 255-60) and subgroup II (monoclonal antibody 631-9) (12).
RESULTS
G and P typing.
RV RNA was extracted from 287 of the 313 RV-positive specimens and subjected to G and P genotyping by RT-PCR (Table 1). Of the RVs in 287 samples, RVs in 253 (88%) could be fully typed, while RVs in 34 (12%) samples had a nontypeable VP7 gene or VP4 gene, or both. The four globally common serotypes—P[8], G1; P[4], G2; P[8], G3; and P[8], G4—together accounted for only 43% of strains tested. Serotype G9 strains, associated with a variety of VP4 genotypes, accounted for a combined 17% of the total. In the collection, P[4], G2 strains were the most common (22%), followed by P[8], G1 (15%), P[6], G2 (10%), and P[6], G9 (9%). Among the G types, G2 was the most common serotype (34%), followed by G1 (18%) and G9 (17%). Among the P types, P[8] (31%) was the most common, followed by P[6] (24%) and P[4] (22%). Altogether, 11 distinct strains (excluding those from mixed infections) were detected in the survey (Table 1). Of note, the neonatal genotype P[11] strain was found at more than one surveillance site, albeit at a low overall prevalence (3%), confirming that these strains previously believed to cause asymptomatic infections (8) may potentially cause symptomatic infections in older children.
TABLE 1.
G and P types of RV strains in the multicenter study
| Genotype | No. (%) of strainsa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G9 | G Mxb | G NTc | Total | |
| P[4] | 62 (22)d | —e | — | — | 1f | 63 (22) | ||
| P[6] | — | 29 (10) | 2 | 2 | 26 (9) | 2g | 8 (3) | 69 (24) |
| P[8] | 44 (15)d | 1 | — | 16 (6)d | 14 (5) | 1 (4)h | 2 | 88 (31) |
| P[11] | — | 4 (1) | 11 | 5 (2)i | 10 (3) | |||
| P[Mx] | 5 (2)j | 6 (2)k | 7 (2)l | 5 (2)m | 10 (3)n | 33 (11) | ||
| P[NT] | 2 | 1 | 3 (1) | 4 (1) | 2 | 12 (4) | 24 (8) | |
| Total | 51 (18) | 99 (34) | 2 | 32 (11) | 50 (17) | 31 (11) | 22 (8) | 287 |
Values indicate number of strains (percentage of total).
G Mx, mixed infection.
NT, nontypeable.
Globally common strain.
—, strain presumed to be present as part of mixed infection on the basis of the RT-PCR result.
P[4], G2 + G3.
P[6], G2 + 9; P[6], G3 + 4.
Of 11 P[8], G mixed strains, 2 were G1 + G2, 1 was G1 + G3, 3 were G1 + G4, 3 were G1 + G9, 1 was G2 + G4, and 1 was G4 + G9.
Of five P[11], G mixed strains, four were G1 + G9 and one was G1 + G4.
P[6+8], G1.
Of six P mixed, G2 strains, five were P[4] + [P6] and one was P[4] + P[8].
Of seven P mixed, G4 strains, five were P[6+8] and one was P[4] + P[8].
Of five P mixed, G9 strains, three were P[4] + P[6] and two were P[6] + P[8].
Of 10 strains, 7 were P[4] + P[8], G1 + G2; one was P[4] + P[6] + P[8], G1 + G2; 1 was P[6] + P[8], G1 + G2; and 1 was P[6] + P[8], G4 + G9.
Mixed RV infections.
Mixed infections with RVs of different G or P genotypes were detected in 21% of the 253 fecal specimens whose RV strains were fully typed (Table 1). Mixed VP7 genotypes were most often associated with P[8] strains; mixed VP4 genotypes were most frequently observed with mixed VP7 types (Table 1).
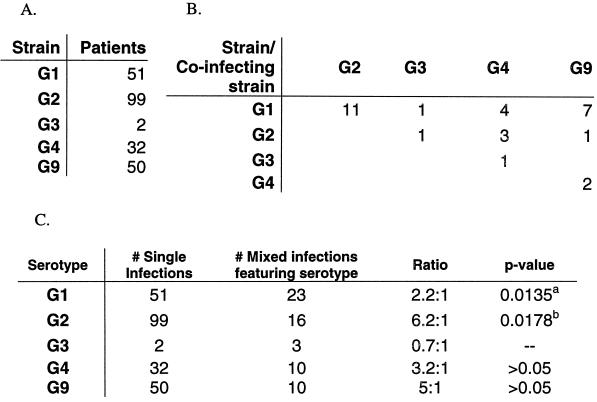
By diagramming the mixed VP7 genotypes of all strains (Fig. 1), we observed that serotype G2 strains were most often present alone as the sole infecting strain, with a ratio of 6.2:1 for single versus mixed infections (P ≥ 0.02). Other strains did not display a significant association.
FIG. 1.
Rates of mixed rotavirus infections in Indian patients. (A) Numbers of patients infected with a single RV strain of one of the various VP7 genotypes. (B) Numbers of patients infected with two of more genotypes of RV strains. (C) Strains with statistically significant associations with either single or mixed infections are indicated by a P value of <0.05, as determined by a t test (with the Mantel-Haenszel correction). Superscript letters: a, G1 strains significantly more likely to be present in mixed infections than in single infections; b, G2 strains significantly more likely to be present in single infections than in mixed infections.
Geographic variation.
A comparison of the strains from the seven cities from which strains were collected for this study showed highly variable numbers of circulating strains (Table 2). Only 2 to 3 strains were present in Nagpur, Shimla, and Davengere, while in New Delhi, 10 distinct strains circulated over the 3-year study period. Larger sample sizes and a possibly longer study period would both allow a more thorough examination of this geographic diversity.
TABLE 2.
Geographic distribution of RV strains
| City | No. of strains
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total tested | Globally common genotypes
|
Uncommon and neonatal serotypes
|
Mixed Infections
|
Other strains | NFTa | |||||||||
| P[8], G1 | P[4], G2 | P[8], G4 | P[6], G2 | P[6], G3 | P[6], G4 | P[6], G9 | P[8], G9 | P[4] or P[8], G1 or G2 | P[6] or P[8], G4 or G9 | Other mixtures | ||||
| Bhopal | 46 | 6 | 7 | 3 | 1 | 4 | 4 | 2b | 1 | 16c | 2 | |||
| New Delhi | 75 | 7 | 27 | 2 | 2 | 2 | 2 | 1 | 8 | 1 | 8d | 2e | 13 | |
| Davengere | 17 | 3 | 10 | 2f | 2 | |||||||||
| Hyderabad | 31 | 8 | 13 | 4 | 2g | 1 (p[8], G2) | 3 | |||||||
| Lucknow | 76 | 20 | 12 | 11 | 2 | 1 | 1 | 14h | 3 (p[11], G4) | 12 | ||||
| Nagpur | 5 | 3 | 2 | |||||||||||
| Shimla | 37 | 24 | 9 | 1 | 3 | |||||||||
| Totals | 287 | 44 | 62 | 16 | 29 | 2 | 2 | 26 | 14 | 8 | 1 | 42 | 6 | 35 |
NFT, not fully typed: either the G type, or the P type, or both, were nontypeable.
One was P[4] + P[6] + P[8].
Three strains each were P[6] + P[8], G1; P[4] + P[6], G2; and P[4] + P[6], G9. Two strains were P[6] + P[8], G4. One strain each was P[8], G1 + G3; P[6], G2 + G9; P[6], G3 + G4; P[8], G4 + G9; and P[6] + P[8], G9.
One strain each was P[6] + P[8], G1 + G2; P[4], G2 + G3; and P[6] + P[8]. Two strains were P[8], G1 + G2. Three strains were P[6] + P[8], G4.
P[11], G4; P[9], G4.
P[6] + P[8], G1.
P[4] + P[6], G2; P[4] + P[8], G2.
One strain each was P[11], G1 + G4; P[4] + P[6], G2; P[8], G2 + G4; and P[6] + P[8], G4. Three strain each were P[8], G1 + G4 and P[8], G1 + G9. Four strains were P[11], G1 + G9.
Electropherotype analysis and subgrouping.
After all strains were P and G genotyped, a high level of strain diversity was apparent. To characterize other genotypes of these strains, we electropherotyped several selected strains by polyacrylamide gel electrophoresis and silver staining and subgrouped the same set (Table 3). All P[4], G2 and P[6], G2 strains tested had a short electropherotype and all P[8], G9 strains displayed a long electropherotype. The 16 P[6], G9 strains included 12 with long patterns and 4 with short patterns, indicating further diversity within this group. Subgrouping, which determines the origin of the gene encoding VP6, the inner capsid protein, indicated that all P[6], G9 strains with short electropherotypes had a subgroup I VP6 antigen, while P[6], G9 strains with long electropherotypes had a subgroup II VP6 antigen (Table 3). All P[8], G9 strains possessed a subgroup II VP6 antigen.
TABLE 3.
Electropherotype analysis of selected strains
S, short; L, long.
Nine strains were subgroup I and three were not subgroupable.
Three strains were subgroup II and one was not subgroupable.
Thirteen strains were subgroup II and one was not subgroupable.
Temporal analysis of RV strains.
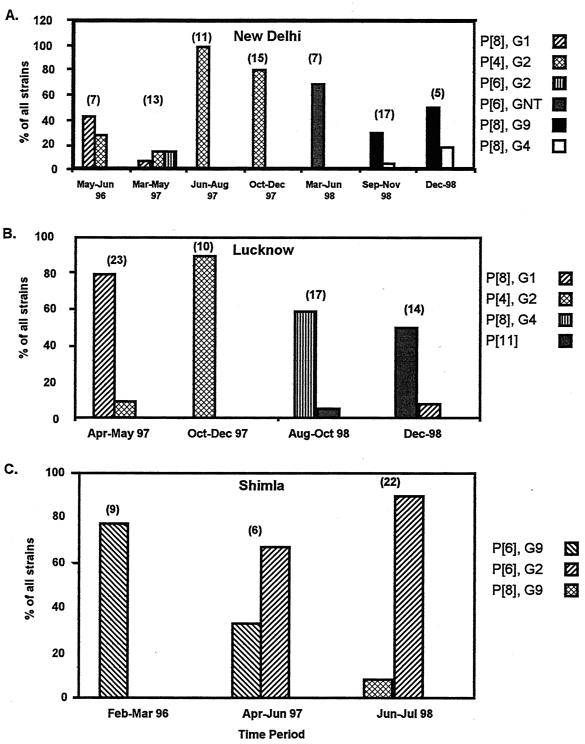
The G and P genotypes of the predominant strains from different cities demonstrated regular but unpredictable temporal shifts (Fig. 2). In Fig. 2 the predominant as well as the second most common strains for each city are shown as percentages of the total number of strains within a given period. Although the periods were not uniform, patterns of emergence and disappearance were evident. For example, in New Delhi (Fig. 2A), P[8], G1 strains began as the predominant strain and then waned as P[4], G2 strains became the most prevalent. Later in the study period, P[8], G9 and P[8], G4 strains accounted for the highest percentage of the total. In Lucknow (Fig. 2B), each of the four periods charted displayed a different dominant strain. In Shimla (Fig. 2C), P[6], G9 strains were observed to decrease concomitantly with an increasing share of P[6], G2 strains.
FIG. 2.
Temporal analysis of shifting rotavirus strains in New Delhi (A), Lucknow (B), and Shimla (C). The predominant and the second most common strains are shown as a percentage of the total number of strains for each period indicated. The total number of strains in each period is indicated above the columns.
DISCUSSION
The results presented here confirm and extend the results of preliminary studies in India that documented the marked diversity of RV strains in circulation, a high prevalence of types G9 and P[6], the presence of unusual strains (e.g., P[11], G4) not found in the United States and other developed countries (6, 7, 14, 18, 21). Furthermore, the present study confirms the previously noted high prevalence of mixed RV infections among hospital inpatients (7, 21). Within the group of serotype G9 strains, the diversity observed in the present study was greater than that observed in our previous study: both P[6] and P[8] VP4 genotypes were detected, and both long- and short-electropherotype strains were found among the P[6], G9 strains. In our previous study (21), all P[6], G9 strains had a long electropherotype and subgroup II specificity. The present results are reminiscent of those of some other recent studies documenting the elevated prevalence of G9 strains, in that P[6], G9 strains with short electropherotypes and subgroup I antigens were common and one to five different G9 reassortants were detected (4, 13, 15, 23)]. These results support a growing body of evidence suggesting that novel G9 strains distinct from prototype strains isolated in the mid-1980s (3; Ikegami et al., Abstr. VII Int. Congr. Virol.) are now emerging globally.
The epidemiologic significance of the results of the present study is twofold. First, given that present RV vaccines are aimed at eliciting serotype-specific immunity to RV, our results indicate that a successful vaccine against RV in India will need to elicit immunity to unusual G serotypes such as G9. Whether the presently licensed RRV-TV vaccine can elicit protective immunity to serotype G9 RV remains to be demonstrated conclusively. Second, the high prevalence of mixed infections may be intimately related to the observed diversity of strains. Patients harboring multiple RVs may offer a unique environment for the reassortment of RV genes, leading to the generation of novel strains and the maintenance of the observed diversity. Separately, the high prevalence of mixed infections may be explained by greater environmental contamination with RV, coupled with greater contact between children and the environment around them. Whether better sanitation and hygienic standards will lessen the prevalence of mixed infections remains to be demonstrated.
Apart from the overall diversity of strains, a marked geographic variance within India was also observed. The predominant RV strain in cities varied from year to year and from city to city, despite the sometimes close distances between them. Both temporal and geographic variances were readily evident from our study, underscoring the need for a comprehensive vaccine that will provide immunity despite these constantly changing and unpredictable conditions.
An interesting finding of the present study was that, compared with nonserotype G2 strains, serotype G2 strains were found more frequently in infections caused by single RV serotype strains than in mixed infections, an association that was statistically significant (P < 0.05). One explanation for this observation is that serotype G2 RV strains are more pathogenic and can cause disease alone, while other, less pathogenic strains may require the assistance of a coinfecting strain to elicit symptomatic disease. If serotype G2 infections are more severe, it is conceivable that the prevalence of serotype G2 found here is an overestimate relative to the prevalence of all G2 infections in the study area due to increased clinic visits associated with more severe infection.
The increased pathogenicity of serotype G2 strains has been noted before. Previous studies of the clinical severity of RV infections stemming from strains of different serotypes conducted in the United States, Bangladesh, and Italy concluded that serotype G2 strains are responsible for more severe disease (1, 2). In addition, during an outbreak of very severe RV diarrhea in Brazilian children and adults caused by a serotype G2 strain, it was suggested that it may have been due to serotype G2 infections in an immunologically naïve population, although the serotype prevalence in the community before that outbreak had not been studied (25). Other examinations of serotype and its relation to disease severity, however, have been limited by small sample sizes and have not supported this hypothesis (9, 20, 28). In the present study, when serotype G1 strains were similarly examined, they were found to be significantly associated with mixed infections, displaying a trend opposite that noted for serotype G2 strains.
Serotype G2 rotaviruses represent a genogroup quite distinct from the Wa genogroup, which includes the other common serotypes, serotypes G1, G3, G4, and G9. If immunity is mediated by several cross-reactive epitopes, it seems likely that first infections with a strain of genogroup Wa would offer the best protection against the other strains of this genogroup that generally share their VP4, VP6, and NSP4 antigens, all of which have been associated in the past with viral virulence and immune protection. The serotype G2 RV strains would then remain the odd strain and might be expected to behave differently. Although these findings may suggest differences in the relative pathogenicities of RV strains, larger sample sizes and a more thorough assessment of clinical severity must be undertaken to explore this issue.
These observations of RV strain diversity in India define a profile characteristic of a developing country and quite distinct from what we have observed in the United States and other developed countries. In the United States, the four or five common strains still represent >90% of the strains in circulation, mixed infections are rare (<2%), and in any single city, three or fewer strains are usually in circulation at any time. The RV strain diversity profile for a developing country has been observed in other studies of RV strain diversity such as those in Brazil and Bangladesh and in other areas where PCR diagnostics have been intensively applied to examine nontypeable strains. These contrasts may reflect differences in the predominant mode of virus transmission. It is possible, for example, that a mixed infection might result from exposure to a larger and more diverse inoculum of RV in water or food from a fecally contaminated environment. These modes of transmission, thought to be more common in developing country settings, may underlie the higher level of strain diversity observed. These differences hold clues to the differences in RV epidemiology as well as the possible efficacies of future vaccines.
The epidemiology of RV in India remains in constant flux. With the introduction of an RV vaccine being a major health priority, future studies should continue to document the high degree of diversity and the unusual and changing nature of RV strains in India. Vaccine strategies, furthermore, will necessarily have to account for and target this marked diversity of strains.
REFERENCES
- 1.Bern C, Unicomb L, Gentsch J R, Banul N, Yunus M, Sack R B, Glass R I. Rotavirus diarrhea in Bangladeshi children: correlation of disease severity with serotypes. J Clin Microbiol. 1992;30:3234–3238. doi: 10.1128/jcm.30.12.3234-3238.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cascio A, Vizzi E, Alaimo C, Arista S. Rotavirus gastroenteritis in Italian children: can severity of symptoms be related to the infecting virus? Clin InfectDis. 2001;32:1126–1132. doi: 10.1086/319744. [DOI] [PubMed] [Google Scholar]
- 3.Clark H F, Hoshino Y, Bell L M, Groff J, Hess G, Bachman P, Offit P A. Rotavirus isolate WI61 representing a presumptive new human serotype. J Clin Microbiol. 1987;25:1757–1762. doi: 10.1128/jcm.25.9.1757-1762.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cubitt W D, Steele A D, Iturriza M. Characterisation of rotaviruses from children treated at a London hospital during 1996: emergence of strains G9P2A[6] and G3P2A[6] J Med Virol. 2000;61:150–154. doi: 10.1002/(sici)1096-9071(200005)61:1<150::aid-jmv24>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 5.Cunliffe N A, Gondwe J S, Broadhead R L, Molyneux M E, Woods P A, Bresee J S, Glass R I, Gentsch J R, Hart C A. Rotavirus G and P types in children with acute diarrhea in Blantyre, Malawi, from 1997 to 1998: predominance of novel P[6]G8 strains. J Med Virol. 1999;57:308–312. [PubMed] [Google Scholar]
- 6.Das B K, Gentsch J R, Cicirello H G, Woods P A, Gupta A, Ramachandran M, Kumar R, Bhan M K, Glass R I. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994;32:1820–1822. doi: 10.1128/jcm.32.7.1820-1822.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das B K, Kumar R K, Bhan M K. Rotavirus gastroenteritis and vaccine development Indian. J Pediatr. 1998;65:S36–S44. [Google Scholar]
- 8.Flores J, Midthun K, Hoshino Y, Green K, Gorziglia M, Kapikian A Z, Chanock R M. Conservation of the fourth gene among rotaviruses recovered from asymptomatic newborn infants and its possible role in attenuation. J Virol. 1986;60:972–979. doi: 10.1128/jvi.60.3.972-979.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flores J, Taniguchi K, Green K, Perez-Schael I, Garcia D, Sears J, Urasawa S, Kapikian A Z. Relative frequencies of rotavirus serotypes 1, 2, 3, and 4 in Venezuelan infants with gastroenteritis. J Clin Microbiol. 1988;26:2092–2095. doi: 10.1128/jcm.26.10.2092-2095.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gentsch J R, Glass R I, Woods P, Gouvea V, Gorziglia M, Flores J, Das B K, Bhan M K. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouvea V, de Castro L, Timenetsky M D C, Greenberg H, Santos N. Rotavirus serotype G5 associated with diarrhea in Brazilian children. J Clin Microbiol. 1994;32:1408–1409. doi: 10.1128/jcm.32.5.1408-1409.1994. . (Erratum, 32:1834.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg H, McAuliffe V, Valdesuso J, Wyatt R, Flores J, Kalica A, Hoshino Y, Singh N. Serological analysis of the subgroup protein of rotavirus, using monoclonal antibodies. Infect Immun. 1983;39:91–99. doi: 10.1128/iai.39.1.91-99.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin D D, Kirkwood C D, Parashar U D, Woods P A, Bresee J S, Glass R I, Gentsch J R. Surveillance of rotavirus strains in the United States: identification of unusual strains. The National Rotavirus Strain Surveillance System collaborating laboratories. J Clin Microbiol. 2000;38:2784–2787. doi: 10.1128/jcm.38.7.2784-2787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Husain M, Seth P, Dar L, Broor S. Classification of rotavirus into G and P types with specimens from children with acute diarrhea in New Delhi, India. J Clin Microbiol. 1996;34:1592–1594. doi: 10.1128/jcm.34.6.1592-1594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iturriza-Gomara M, Cubitt D, Steele D, Green J, Brown D, Kang G, Desselberger U, Gray J. Characterisation of rotavirus G9 strains isolated in the UK between 1995 and 1998. J Med Virol. 2000;61:510–517. doi: 10.1002/1096-9071(200008)61:4<510::aid-jmv15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 16.Joensuu J, Koskenniemi E, Pang X L, Vesikari T. Randomised placebo-controlled trial of rhesus-human reassortant rotavirus vaccine for prevention of severe rotavirus gastroenteritis. Lancet. 1997;350:1205–1209. doi: 10.1016/S0140-6736(97)05118-0. [DOI] [PubMed] [Google Scholar]
- 17.Kapikian A Z, Chanock R M. Rotaviruses. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1657–1708. [Google Scholar]
- 18.Kelkar S D. Prevalence of human group A rotavirus serotypes in Pune, India (1990–1993) Indian J. Med Res. 1997;106:508–512. [PubMed] [Google Scholar]
- 19.Leite J P, Alfieri A A, Woods P A, Glass R I, Gentsch J R. Rotavirus G and P types circulating in Brazil: characterization by RT-PCR, probe hybridization, and sequence analysis. Arch Virol. 1996;141:2365–2374. doi: 10.1007/BF01718637. [DOI] [PubMed] [Google Scholar]
- 20.Pitson G A, Grimwood K, Coulson B S, Oberklaid F, Hewstone A S, Jack I, Bishop R F, Barnes G L. Comparison between children treated at home and those requiring hospital admission for rotavirus and other enteric pathogens associated with acute diarrhea in Melbourne, Australia. J Clin Microbiol. 1986;24:395–399. doi: 10.1128/jcm.24.3.395-399.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramachandran M, Das B K, Vij A, Kumar R, Bhambal S S, Kesari N, Rawat H, Bahl L, Thakur S, Woods P A, Glass R I, Bhan M K, Gentsch J R. Unusual diversity of human rotavirus G and P genotypes in India. J Clin Microbiol. 1996;34:436–439. doi: 10.1128/jcm.34.2.436-439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramachandran M, Gentsch J R, Parashar U D, Jin S, Woods P A, Holmes J L, Kirkwood C D, Bishop R F, Greenberg H B, Urasawa S, Gerna G, Coulson B S, Taniguchi K, Bresee J S, Glass R I. Detection and characterization of novel rotavirus strains in the United States. J Clin Microbiol. 1998;36:3223–3229. doi: 10.1128/jcm.36.11.3223-3229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramachandran M, Kirkwood C D, Unicomb L, Cunliffe N A, Ward R L, Bhan M K, Clark H F, Glass R I, Gentsch J R. Molecular characterization of serotype G9 rotavirus strains from a global collection. Virology. 2000;278:436–444. doi: 10.1006/viro.2000.0682. [DOI] [PubMed] [Google Scholar]
- 24.Rennels M B, Glass R I, Dennehy P H, Bernstein D I, Pichichero M E, Zito E T, Mack M E, Davidson B L, Kapikian A Z. Safety and efficacy of high-dose rhesus-human reassortant rotavirus vaccines—report of the National Multicenter Trial. United States Rotavirus Vaccine Efficacy Group. Pediatrics. 1996;97:7–13. [PubMed] [Google Scholar]
- 25.Taniguchi K, Urasawa T, Morita Y, Greenberg H B, Urasawa S. Direct serotyping of human rotavirus in stools by an enzyme-linked immunosorbent assay using serotype 1-, 2-, 3-, and 4-specific monoclonal antibodies to VP7. J Infect Dis. 1987;155:1159–1166. doi: 10.1093/infdis/155.6.1159. [DOI] [PubMed] [Google Scholar]
- 26.Timenetsky M D C, Santos N, Gouvea V. Survey of rotavirus G and P types associated with human gastroenteritis in Sao Paulo, Brazil, from 1986 to 1992. J Clin Microbiol. 1994;32:2622–2624. doi: 10.1128/jcm.32.10.2622-2624.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unicomb L E, Podder G, Gentsch J R, Woods P A, Hasan K Z, Faruque A S, Albert M J, Glass R I. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type G9 in 1995. J Clin Microbiol. 1999;37:1885–1891. doi: 10.1128/jcm.37.6.1885-1891.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yolken R H, Wyatt R G, Zissis G, Brandt C D, Rodriguez W J, Kim H W, Parrott R H, Urrutia J J, Mata L, Greenberg H B, Kapikian A Z, Chanock R M. Epidemiology of human rotavirus types 1 and 2 as studied by enzyme-linked immunosorbent assay. N Engl J Med. 1978;299:1156–1161. doi: 10.1056/NEJM197811232992103. [DOI] [PubMed] [Google Scholar]