Abstract
The administration of toxin-specific therapy in snake envenoming is predicated on improved diagnostic techniques capable of detecting specific venom toxins. Various serological tests have been used in detecting snakebite envenoming. Comparatively, enzyme-linked immunosorbent assay (ELISA) has been shown to offer a wider practical application. We report an inhibition ELISA for detecting three-finger toxin (3FTx) proteins in venoms of African spitting cobras. The optimized assay detected 3FTxs in N. ashei (including other Naja sp.) venoms, spiked samples, and venom-challenged mice samples. In venoms of Naja sp., the assay showed inhibition, implying the detection of 3FTxs, but showed little or no inhibition in non-Naja sp. In mice-spiked samples, one-way ANOVA results showed that the observed inhibition was not statistically significant between spiked samples and negative control (p-value = 0.164). Similarly, the observed differences in inhibition between venom-challenged and negative control samples were not statistically significant (p-value = 0.9109). At an LOD of 0.01 µg/mL, the assay was able to confirm the presence of 3FTxs in the samples. Our results show a proof of concept for the use of an inhibition ELISA model as a tool for detecting 3FTxs in the venoms of African spitting cobra snakes.
Keywords: inhibition ELISA, three-finger toxins, cytotoxic, snakebite envenoming, diagnostics
1. Introduction
Snakebite envenoming constitutes a major public health challenge in developing countries. It contributes significantly to cases of morbidity and mortality, particularly in poorly developed regions including sub-Saharan Africa and Asia [1]. The World Health Organization estimates that about 5.4 million people globally are bitten by snakes every year [2]. Of this, nearly 2.7 million people are believed to be envenomed, resulting in between 81,000 to 138,000 deaths [1]. Although likely grossly underreported, close to 15,000 snakebites occur each year in Kenya [3]. The African spitting cobra species comprises of various snake species including Naja nigricollis, Naja mossambica, Naja katiensis, Naja pallida, and others. Naja ashei belongs to the African spitting cobras, which are distributed widely across sub-Saharan Africa. N. ashei, also called the large brown spitting cobra was in 2007 considered a distinct species and classified accordingly [4]. As is the case with the other African spitting cobras, the venom of N. ashei is known to induce mainly cytotoxic effects caused by the predominant presence of 3FTx proteins and phospholipases A2 [5,6]. N. ashei is a medically important snake species in Kenya, Ethiopia, Uganda, Somalia, and Tanzania, and thus, is implicated in snakebites and associated morbidity, mortality, and disability [7].
Proteomic analysis of the venom of spitting cobras shows that the venoms are made up largely of three-finger toxins (3FTxs), accounting for about 67–80%. For instance, 3FTx proteins were found to constitute 73.3% of the total venom load of N. nigricollis [8]. Similarly, an analysis of the venom proteome of N. ashei revealed that 3FTx proteins accounted for 60–80% [5,6]. Three-finger toxins are non-enzyme-based polypeptides present in the venoms of elapids and colubrids [9,10], and are very rare in the venoms of vipers [11]. Three-finger toxins present in some elapid venoms are cytotoxic and are thus capable of inducing tissue necrosis [9]. They also include cytolytic cardiotoxins, which are mainly involved in the formation of ion pores [12].
Among other things, improved diagnostic methods are crucial in discriminating between various venoms [13]. With advancements in diagnostics capable of detecting specific toxins, implicating species in snakebite envenoming could potentially be deduced, allowing the administration of toxin-specific treatments as well as other species-specific bites [12,14]. Various serological tests are available for detecting snake venoms including agglutination assays, enzyme-linked immunosorbent assays (ELISA), florescence immunoassay, and others [15]. However, ELISA has been shown to offer superior practical application relative to the other tests [15,16,17] and has been employed clinically in the diagnosis of snakebites and studying envenoming-associated syndromes [15,16,17,18,19]. Inspite of this, ELISAs are not readily deployed in the field owing to constraints of time and requirements for specialized equipment and reagents [17,20,21], albeit the technique is currently being used for clinical purposes [22]. Even more promising is the fact that ELISA is the basis for developing snake venom detection kits, such as the CSL-SVDK, which is considered the only commercially available device for detecting snakebite envenoming [23]. In this study, we describe an inhibition ELISA for detecting 3FTx proteins, the most abundant toxin present in the venoms of African spitting cobras. The assay was tested for its ability to discriminate 3FTxs-containing and non-3FTxs-containing venoms. An inhibition ELISA is a variant of ELISA where an antigen-containing sample and primary antibody are incubated for specific binding. Following the incubation process, the resulting antigen-antibody complex is transferred to a plate coated with a standard antigen, resulting in binding between the coated antigen and free primary antibody. Due to the initial binding between the antigen-containing sample and primary antibody, there is a reduction in the reaction (reduced absorbance), which is inversely proportional to the analyte concentration in the sample [24].
An inhibition ELISA assay was developed, optimized, and evaluated for the detection of 3FTx proteins in African spitting cobras. The assay was able to detect 3FTxs in both spiked samples and venom-challenged mice samples.
2. Results
2.1. Optimization of ELISA Parameters
Following optimization of ELISA parameters, the highest signal-noise ratio of the conjugate was obtained at a dilution of 1:8000. Also, significant levels of detection were attained at 1 µg/mL of the coating antigen concentration. A two-tailed t-test analysis of substrate sensitivity showed that OPD produced significantly higher ODs compared to TMB (p-value = 0.0207). Consequently, these parameters were considered optimal and thus adopted for subsequent assays.
2.2. Determination of ELISA Cut-Off Point
The specificity of the assay was evaluated using homologous (same genera) and heterologous (different genera) venom samples. At all the inhibitor concentrations tested, a percent inhibition of greater than 18% was observed across all homologous venoms, with the pattern more closely related between N. ashei and N. nigricollis (18.80 and 21.00% respectively). On the other hand, a% inhibition less than 16% was observed among the heterologous samples across all the inhibitor concentrations tested (Table 1). On the basis of the specificity of the assay for both homologous and heterologous samples, a percent inhibition of greater than 30% was set as the identification criteria for the presence of the analyte of interest in homologous venom samples [24].
Table 1.
Percent inhibition of 3FTxs induced by N. ashei, N. nigricollis, N. haje, B. arietans, and D. polylepsis venoms.
| Inhibitor Concentration (µg/mL) |
Naja ashei | Naja nigricollis | Naja haje | Bitis arietans | Dendroapsis polylepsis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| OD | % Inhibition | OD | % Inhibition | OD | % Inhibition | OD | % Inhibition | OD | % Inhibition | |
| 6.000 | 0.094 | 76.90 | 0.085 | 77.68 | 0.088 | 79.75 | 0.474 | 7.06 | 0.458 | 8.77 |
| 2.000 | 0.118 | 71.13 | 0.087 | 77.01 | 0.091 | 79.17 | 0.464 | 9.02 | 0.439 | 12.46 |
| 0.667 | 0.174 | 57.25 | 0.098 | 74.24 | 0.098 | 77.45 | 0.454 | 11.08 | 0.458 | 8.67 |
| 0.222 | 0.259 | 36.49 | 0.125 | 66.97 | 0.113 | 73.99 | 0.467 | 8.53 | 0.465 | 7.28 |
| 0.074 | 0.295 | 27.64 | 0.177 | 53.37 | 0.131 | 69.85 | 0.471 | 7.65 | 0.445 | 11.27 |
| 0.025 | 0.319 | 21.62 | 0.252 | 33.42 | 0.182 | 58.11 | 0.447 | 12.45 | 0.434 | 13.46 |
| 0.008 | 0.331 | 18.80 | 0.299 | 21.00 | 0.260 | 40.28 | 0.447 | 12.35 | 0.424 | 15.55 |
| NAC | 0.407 | 0.379 | 0.435 | 0.510 | 0.502 | |||||
Sensitivity of the inhibition ELISA was also determined using crude N. ashei venom at various concentrations. Negative controls made up of pre-immune mice serum samples were also analyzed. With a mean and standard deviation (of the negative control) of 0.835 and 0.207 respectively, the limit of detection (LOD) of the assay was determined to be approximately 0.01 µg/mL (Table 2). Although the lowest concentration in the assay was 0.04 µg/mL, the determined OD was 1.249 which fell between 0.04 µg/mL and the ‘No Antigen Control’ (NAC), hence the 0.01 µg/mL. This implies that at a minimum concentration of 0.01 µg/mL, the inhibition ELISA assay was capable of discriminating between positive and negative samples. Similarly, at the same cut-off, the assay was able to confirm the presence or otherwise of 3FTxs in a venom sample. It should be noted that familiarity with inhibition ELISA coupled with ‘NAC’ sample type may give rise to variations in cut-offs. Nevertheless, ODs of positive and negative control samples should be consistently different. As happens with inhibition ELISAs, lower ODs imply high levels of the analyte of interest, thus more competitive binding and reduced signal (OD) and the vice versa.
Table 2.
Percent inhibition for determination of assay sensitivity.
| % Inhibition for Determining Sensitivity of Inhibition ELISA | ||||
|---|---|---|---|---|
| N. ashei Venom | Negative Controls (n = 12) | |||
| Conc (µg/mL) | Replicate 1 | Replicate 2 | ||
| 27.00 | 76.69 | 78.68 | 49.60 | 49.70 |
| 9.00 | 75.28 | 77.22 | 47.71 | 48.91 |
| 3.00 | 69.62 | 76.04 | 46.52 | 45.97 |
| 1.00 | 64.39 | 70.54 | 26.80 | 20.83 |
| 0.33 | 54.18 | 56.96 | 25.09 | 25.82 |
| 0.11 | 45.02 | 44.95 | 20.46 | 21.92 |
| 0.04 | 34.44 | 38.55 | - | - |
2.3. Evaluation of the Developed Prototype
2.3.1. Inhibition ELISA for Detecting Three-Finger Toxins in Crude N. ashei and Other Venoms
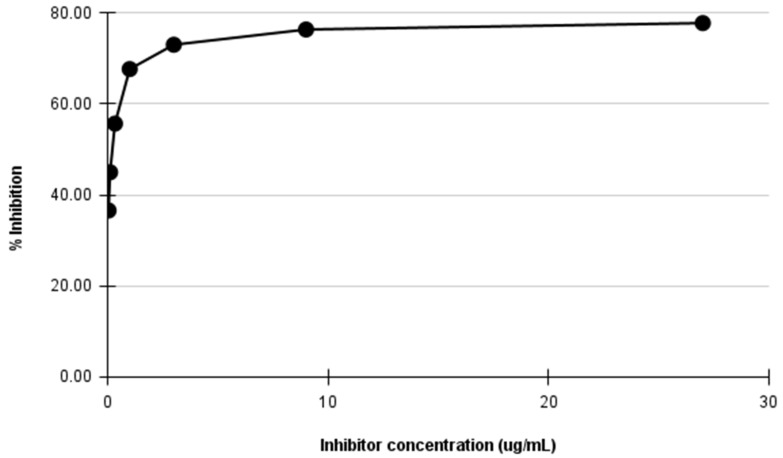
From the six-point three-fold dilution of the sample antigen, the percent inhibition was observed to be highest (77.70%) at 27.00 µg/mL and lowest (36.56%) at 0.04 µg/mL of the antigen, respectively (Figure 1). In terms of absorbance, the ODs at these two concentrations of the sample antigen were 0.380 and 1.081, respectively. In contrast, the ‘No Antigen Control’ recorded an OD of 1.704 (Table 3). The results indicate that the 3FTxs present in the crude N. ashei venom were inhibited across all the concentrations tested when compared with the NAC. The inhibition is reflected in the reduced signals (ODs) particularly in wells containing high concentration of the antigen. Suffice to say, in these wells, there were more 3FTxs antigen, resulting in more binding between the primary antibody and the antigen, hence the high percent inhibition.
Figure 1.
Inhibition ELISA curve showing % inhibition of the coated antigen by the sample-containing antigen at various concentrations of crude N. ashei venom (inhibitor). Sample antigen was diluted 3-fold from an initial concentration of 27 µg/mL to a final concentration of 0.04 µg/mL and run in duplicates.
Table 3.
Inhibition of 3FTxs by sample antigen in crude N. ashei venom.
| Inhibitor Concentration (µg/mL) | OD (492 nm) | % Inhibition |
|---|---|---|
| 27.00 | 0.380 | 77.70 |
| 9.00 | 0.404 | 76.29 |
| 3.00 | 0.461 | 72.95 |
| 1.00 | 0.552 | 67.61 |
| 0.33 | 0.756 | 55.63 |
| 0.11 | 0.938 | 44.95 |
| 0.04 | 1.081 | 36.56 |
| No antigen control (NAC) | 1.704 | - |
The results of the inhibition ELISA were used to confirm, first of all, the presence of three-finger toxins in the crude venoms assayed and secondly, to determine the % inhibition caused by the different sample antigens present in the crude venoms at the different concentrations tested. The ANOVA summary (Table 4) shows that there was a significant difference (p-value < 0.0001) within the sample antigen-induced inhibition in terms of the means. Nearly 72% of the variation in inhibition caused by the sample antigen therein in the crude venoms is explained by the analysis (R-squared = 0.7185).
Table 4.
One-way ANOVA results a.
| F | 19.14 |
| p value | <0.0001 |
| p value summary | **** |
| Significant diff. among means (p < 0.05)? | Yes |
| R squared | 0.7185 |
a ANOVA summary table showing whether or not inhibition induced by the sample-containing homologous (N. ashei, N. nigricollis, and N. haje) and heterologous (B. arietans and D. polylepsis) venom samples are statistically significant. At a p-value of <0.0001, the differences were found to be significantly different. This experiment was carried out to determine how specific the assay is for homologous samples, while being non-specific for heterologous samples.
From the post hoc results (Table 5), it can be observed that there was a significant difference in the inhibition caused by the 3FTx-containing sample antigens between all three Naja species and their non-Naja counterparts, with the difference being particularly significant between N. nigricollis vs. B. arietans, N. nigricollis vs. D. polylepsis, N. haje vs. B. arietans, and N. haje vs. D. polylepsis (p-value < 0.0001 in all cases). Also, there was no significant difference in the 3FTx-induced inhibition between all three spitting cobra species. The multiple comparisons test also show that the sample antigens-induced inhibition (if any) between B. arietans vs. D. polylepsis was not significant (p-value = 0.9999).
Table 5.
Multiple comparisons b.
| Tukey’s Multiple Comparisons Test | Adjusted p Value | Significant? |
|---|---|---|
| N. ashei vs. N. nigricollis | 0.5409 | No |
| N. ashei vs. N. haje | 0.0649 | No |
| N. ashei vs. B. arietans | 0.0034 | Yes |
| N. ashei vs. D. polylepsis | 0.0051 | Yes |
| N. nigricollis vs. N. haje | 0.7312 | No |
| N. nigricollis vs. B. arietans | <0.0001 | Yes |
| N. nigricollis vs. D. polylepsis | <0.0001 | Yes |
| N. haje vs. B. arietans | <0.0001 | Yes |
| N. haje vs. D. polylepsis | <0.0001 | Yes |
| B. arietans vs. D. polylepsis | 0.9999 | No |
b Tukey posthoc multiple comparisons test was carried out to determine where the differences in sample-induced inhibition existed following one-way ANOVA analysis. Venom samples were compared against one another in order to determine if the differences were statistically significant or not.
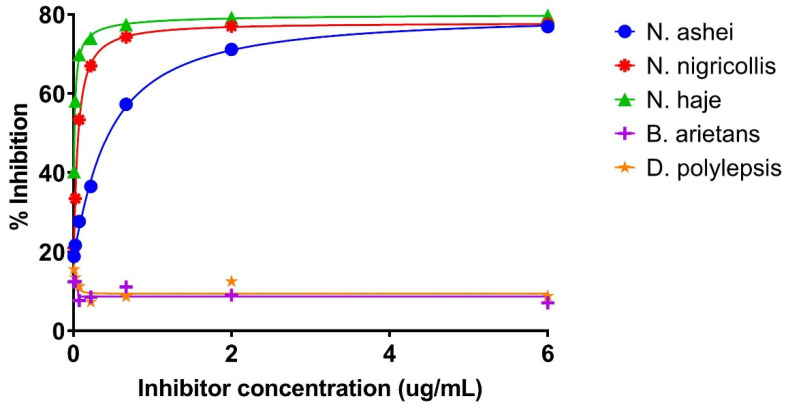
It was also be observed that at high inhibitor concentrations (Figure 2), the ODs of all three Naja species were low. The ODs, however, increased with decreasing inhibitor concentrations. As low as 0.008 µg/mL inhibitor concentration, the sample antigens induced varying levels of inhibition (18.80, 21.00, and 40.28% for N. ashei, N. nigricollis, and N. haje, respectively). Conversely, increasing inhibitor concentrations resulted in an increase in the percent inhibition for the same species.
Figure 2.
Inhibition ELISA curves showing % inhibition of the coated antigen (pure 3FTx fraction) by sample antigens from crude N. ashei, N. nigricollis, N. haje, B. arietans, and D. polylepsis venoms at different concentrations. The different sample antigen-containing venoms were diluted 3-fold from an initial concentration of 6 µg/mL to a final concentration of 0.008 µg/mL. Samples were then analyzed in duplicates at each concentration.
For all the venoms tested, the ‘No Antigen Control’ (NAC) produced the highest OD values, an indication of inhibition when compared with wells containing different inhibitor concentrations. ODs obtained for B. arietans and D. polylepsis suggest that there was no or minimal inhibition mainly because the venoms either contain little or none of the antigen of interest. As such, the ODs are comparable to those of the NAC (Table 1).
2.3.2. Inhibition ELISA for Detecting 3FTx Proteins in Spiked Samples
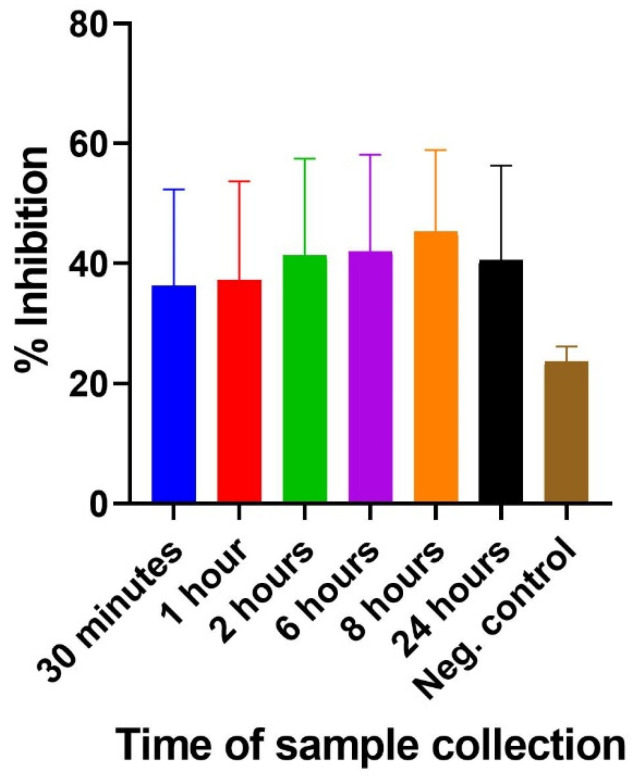
The utility of the inhibition ELISA was further tested for its capacity to detect 3FTxs in blood samples spiked with crude venom at different times. The spiked samples were assayed alongside previously collected pre-immune serum samples as control. An analysis of the results shows that there was inhibition of the coated antigen (purified 3FTxs) by the N. ashei 3FTxs-containing venom at all the dilution points. In contrast, the negative control showed no inhibition. The % inhibition was largely similar across the different sample collection times, but varied significantly relative to the negative control. One-way ANOVA results showed no significant difference among the means (p-value = 0.164) in terms of sample collection times. Similarly, no significant difference was found between the negative control and the other samples collected at the respective times. The results nonetheless demonstrate that the inhibition ELISA is capable of detecting 3FTxs in blood samples spiked with crude venom as depicted in the Figure 3 below.
Figure 3.
Comparison of % inhibition of coated antigen (3FTxs) by N. ashei venom spiked samples collected at different times (30 min–24 h).
2.3.3. Inhibition ELISA for Detecting 3FTxs in Mice Challenged with N. ashei Venom
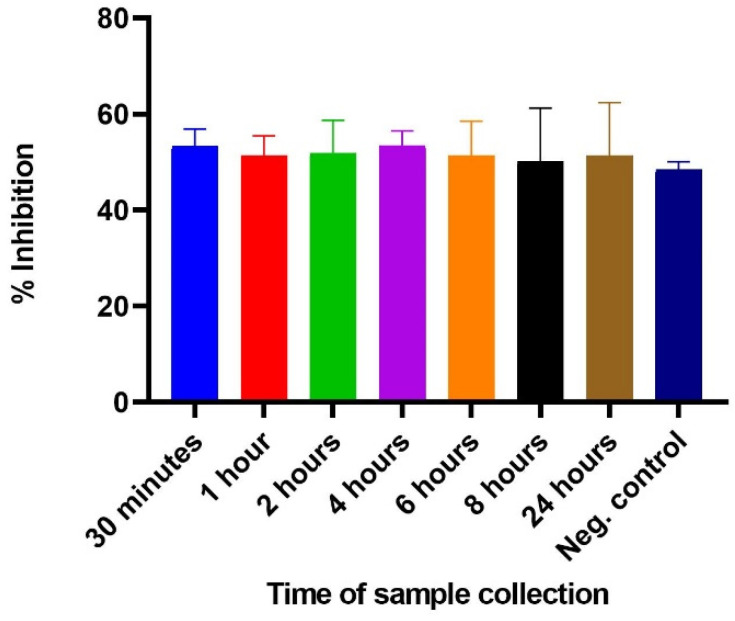
The potential use of the inhibition ELISA in snakebite envenoming was evaluated in mice challenged with crude N. ashei venom. The results demonstrate that 3FTxs were detected, evidenced in the observed inhibition in the sample-containing wells. Albeit varied inhibition between the samples and negative control were observed, the differences were not statistically significant (p-value = 0.9109) as determined by one-way ANOVA. Similarly, multiple comparison analysis showed that the inhibition between the negative control and the samples collected at different times were not significantly different. It is worthy to note that regardless of the differences, the assay was able to detect 3FTxs in the samples tested as shown in the observed inhibition (Figure 4).
Figure 4.
Comparison of % inhibition of coated antigen (3FTxs) by N. ashei venom in challenged mice with samples collected at different times (30 min–24 h).
3. Discussion
The determination of optimal concentrations for antigen coating, dilutions of primary and detection antibodies, and substrate sensitivity among others is crucial for the development of a successful assay [24]. In line with this, antigen coating concentration of 1 µg/mL was found to be optimal as previously reported by Laustsen and colleagues [25]. A 1:8000 dilution of the detection antibody was found to be more beneficial and represents a more cost-effective choice. Also, OPD, in addition to showing consistently higher absorbance readings was found to be statistically different from TMB, and thus, adopted for subsequent assays albeit Hosoda et al [26] and Goka and Farthing [27] had previously reported contradictory observations.
The results of the inhibition ELISA indicate that the 3FTxs present in the crude N. ashei venom were inhibited across all the concentrations tested when compared with the NAC. The inhibition is reflected in the reduced signals (ODs), particularly in samples containing high concentrations of the antigen. Suffice to say, in these wells, there were more 3FTxs antigen, resulting in more binding between the primary antibody and the 3FTxs antigen, hence the high percent inhibition. ODs obtained for B. arietans and D. polylepsis suggest that there was no or minimal inhibition mainly because the venoms either contain little or none of the antigen of interest. As such, the ODs are comparable to those of the NAC (Table 1). The case of B. arietans could be attributed to the fact that 3FTxs are thought to be uncommon in viper venoms [11]. D. polylepsis, although an elapid too, is reported to have 31.0% of its total protein content made up of 3FTxs toxins [28]. However, a majority of these toxins are neurotoxic (including muscarinic and short neurotoxins) and appear to lack the cytotoxic 3FTxs that characterize the Naja sp., hence the seeming absence of inhibition as observed earlier. Again, the preponderance of neurotoxic 3FTxs in the venom proteome of D. polylepsis is supported by Blanchet et al. [29]. Thus, the apparent lack of inhibition as observed in D. polylepsis may be attributed to the high level of neurotoxic 3FTx proteins as previously reported [28,29]. The relatively high and comparable inhibition observed in N. ashei, N. nigricollis, and N. haje may confirm two things; first, the similarity in the venom proteomes of the cobras (both spitting and non-spitting) and second, the venom proteomes of Naja sp. contain more 3FTxs. This finding is consistent with Hus et al. [5,6] and Petras et al. [8], who found that proteomic analysis of spitting cobra venoms shows that 3FTxs proteins make up the bulk. Hus and colleagues [5,6] found that 3FTxs accounted for 60–80% of the protein composition of N. ashei venom, while the same toxin was found to constitute 67–73% of N. nigricollis venom [8]. Thus, the inhibition patterns demonstrate that all three Naja species are made up of largely 3FTxs. Also, the similarities in the inhibition among the three Naja snakes is supported by Casassola et al. [30] who observed that similar biochemical, antigenic and toxinological characteristics are shared by snake venoms that belong to the same genera. Similarly, the venom of Hemachatus haemachatus, an African spitting cobra-like species found predominantly in Southern Africa was reported to contain 63.3% 3FTx proteins [31], although it is closely related to the Naja genus phylogenetically.
A variety of methods are available for determining the analytical sensitivity of assays [32,33,34]. The determination of limit of detection of an assay among other parameters is required for establishing limits of identification. From the percent inhibition of the sensitivity assay, the LOD was determined to be 0.01 µg/mL. At this cut-off, the assay was able to confirm the presence of the analyte of interest in the samples tested. The detection of envenoming by our inhibition ELISA model is consistent with previously reported ELISA methods, although different LODs were found. An avidin-biotin ELISA kit by Dong and colleagues reported a detection limit of 0.2–1.6 ng/mL in venom samples from South Vietnam [17]. In aiding the management of snakebites in Taiwan, a sandwich ELISA approach was found to have a limit of quantification of 0.78 and 0.39 ng/mL for haemorrhagic and neurotoxic venoms, respectively [35]. Similarly, Shaikh and colleagues, in developing a sandwich ELISA assay for detecting venoms of common Indian snakes found the sensitivity of the assay to be 0.01 ng/mL [36]. Additionally, Watanabe et al. [37] and Selvanayagam [38] in their respective ELISAs for detecting snake venoms and toxins in mice serum and tissue homogenate, respectively, found sensitivities of 50 and 0.1 ng/mL. These studies, when considered in combination with our findings demonstrate capacity and utility of various ELISA methods to detect snake venom in different samples. Thus, albeit rarely used in snakebite envenoming, inhibition ELISA could prove useful in detecting cobra venoms.
In both blood samples spiked with crude venom and mice challenged with the same, the inhibition ELISA was able to detect the presence of 3FTx proteins, as demonstrated in the observed differences in % inhibition when compared to the negative control. This finding is significant to the extent that such an assay could be useful not only in discriminating between venoms [13], but also allow the identification of snakes implicated in envenoming and ultimately pave the way for the administration of toxin-specific therapies [12,14]. Thus, although preliminary, the inhibition ELISA may prove useful in snakebite envenoming, although it is rarely employed in this field.
4. Materials and Methods
4.1. Snake Venoms
Lyophilized crude N. ashei venom was obtained from snakes maintained at the Bioken snake farm in Watamu, Kenya. The venom was reconstituted in distilled water and stored at −20 °C until use. Three-finger toxin (3FTx) fraction was purified using size-exclusion chromatography as described elsewhere [39]. Venoms from Naja nigricollis, Naja haje, Bitis arietans, and Dendroaspis polylepis were obtained from Baringo Snake Park, Kenya, in liquid form.
4.2. Animals
Eight-week-old adult female Balb/c mice were challenged with 3FTxs (50 and 100 µg). Animals were sampled in February 2021 fortnightly in the tail vein for antibody screening for about 14 weeks. Blood samples were centrifuged to obtain serum, which were then stored at −20 °C until use, which antibodies were used as primary antibodies in all experiments. Animals were kept at the animal facility of the Kenya Medical Research Institute (KEMRI) and provided with food and water ad libitum.
4.3. Ethical and Institutional Approvals
Approval for the study was obtained from the KEMRI Center for Biotechnology Research and Development (CBRD), KEMRI’s Animal Care and Use Committee (ACUC), and KEMRI Scientific and Ethical Committee Unit (SERU) with protocol number KEMRI/SERU/CBRD/229/4340.
4.4. Quantification of Protein Content of Venoms
Protein content of all venoms was determined using the BCA kit in line with the manufacturer recommendations [40].
4.5. Optimization of ELISA Parameters
Goat anti-mouse HRP conjugated antibody, antigen coating concentration, and substrate sensitivity were optimized using an indirect ELISA method described by Islam and Jones [41].
4.6. Determination of Specificity and Sensitivity of Inhibition ELISA
Tests that are developed for identification purposes should be able to discriminate between closely related and other venoms. In line with this, the specificity and sensitivity of the assay were determined according to the methods described by Sharma et al [24] and Armbruster and Pry [32]. The specificity of the assay was assessed by the observed percent inhibition at various sample antigen concentrations (6–0.008 µg/mL) in three homologous (N. ashei, N. nigricollis, N. haje) and two heterologous (B. arietans and D. polylepsis) venom samples (Table 1). From the three-fold serial dilution of the samples, the percent inhibition was calculated across all the inhibitor concentrations. Inhibition patterns were then compared between the homologous and heterologous samples. Similarly, to determine sensitivity of the inhibition ELISA, crude N. ashei venom was analyzed from an initial concentration of 27 µg/mL to a final concentration of 0.04 µg/mL following a three-fold serial dilution (Table 5). Samples were analyzed in duplicates. At the same time, 12 negative control samples that consisted of pre-immune mice serum samples and sample buffer were analyzed. The negative controls (devoid of the analyte of interest) were used for determining the limit of detection. The mean, standard deviation, and percent inhibition were then calculated. The limit of detection (LOD) of the assay was thus determined according to the method described by Armbruster and Pry [32] using the formula below based on the signal-noise approach;
4.7. Inhibition ELISA for Detecting Three-Finger Toxins in Crude N. ashei Venom
The method described by Sharma and colleagues [24] with modifications was used. Wells of a 96-well maxisorp plate (NUNC, Denmark) were coated overnight at 4 °C with 100 µL of 1 µg/mL purified 3FTx toxin constituted in 0.1 M carbonate-bicarbonate buffer, pH 9.6. On the next day, 150 µL of the sample antigen (crude N. ashei venom) prepared to a final concentration of 27 µg/mL in blocking buffer (2% BSA + 0.05% PBST, pH 7.4) was added to row A of a separate plate (2). One hundred microliters (100 µL) of blocking buffer was added to rows B to H. A three-fold serial dilution was then performed by transferring 50 µL from row A to B and so on until G. The final 50 µL was discarded. Serum sample containing primary antibodies was prepared in blocking buffer at 1:1000 and 100 µL added to the duplicate wells of the same plate (including row H). The plate was then incubated at 37 °C for 60 min. The coating buffer in the earlier plate (1) was then aspirated and the plate washed three times with the washing/blocking buffer. The plate was then blocked with 200 µL of blocking buffer per well for 60 min at room temperature, following which washing was done as described previously. The content of the plate (2) containing the sample antigen and primary antibody was then transferred to the coated plate (1) and incubated at 37 °C for 60 min. The plate was washed again three times and 100 µL of goat-anti mouse IgG conjugated to horseradish peroxidase (American Qualex, San Clemente, CA, USA) added to each well. After a one-hour incubation at 37 °C, the washing step was repeated (with an extra wash) and 200 µL of 5 mg OPD substrate (Sigma Aldrich, St. Louis, MO, USA) added to the wells. The plate was again incubated for 60 mins at 37 °C for color development and then stopped with 50 µL per well of 3 M H2SO4. The plate was then read at 492 nm with a plate reader (Multiscan EX reader, Thermo Scientific, Massachusetts, MA, USA). Row H was considered the ‘No Antigen Control’ (NAC) and had its wells containing the blocking buffer and primary antibody without sample antigen. The NAC wells recorded the highest ODs. Subsequently, the percentage inhibition of the different wells containing different concentration of the sample antigen was determined against the absorbance of the NAC as per the formula below
Testing Inhibition ELISA for Detecting 3FTxs in Other Naja sp. and Non-Naja sp. Venoms
The inhibition ELISA described above was tested for its ability to discriminate between venoms containing 3FTxs and those without using crude venoms from two other Naja sp. (N. nigricollis and N. haje) and two non-Naja sp. (B. arietans and D. polylepsis). The inhibition ELISA protocol described in Section 4.7 was implemented, except that the sample antigen concentration for all venoms was prepared to a final concentration of 6 µg/mL in the diluent previously described. Thus, a six point three-fold serial dilution was performed until a final concentration of 0.008 µg/mL sample antigen was obtained. All washing, blocking, and incubation steps were carried out as previously described. The plate was finally read at 492 nm and the percentage inhibition per well determined.
4.8. Detection of Three-Finger Toxins in Spiked Samples
The inhibition ELISA described earlier was also tested for its capacity to detect 3FTxs proteins in blood samples spiked with crude N. ashei venom. To spike samples, blood samples were collected from mice and then mixed with crude venom. Whereas the components of the assay were similar to the one described earlier, the sample collection and preparation were slightly different. In this experiment, six different Swiss mice were bled through the tail vein at different times—30 min, 1 h, 2 h, 6 h, 8 h, and 24 h. Immediately after sample collection, blood samples were spiked with equal volume of reconstituted crude N. ashei venom. Samples were then stored at −20 °C until assayed as earlier described. Pre-immune samples were added as negative control. The % inhibition was determined taking into account the NAC and the individual OD values.
4.9. Detection of Three-Finger Toxins in N. ashei Venom-Challenged Mice
The applicability of the inhibition ELISA in snakebite envenoming diagnostics was tested in mice challenged with crude N. ashei venom. Post the challenge, mice were bled in the tail vein at 30 min, 1 h, 2 h, 4 h, 6 h, 8 h, and 24 h. Pre-immune samples were added as negative control. The samples were processed, subjected to the inhibition ELISA procedure and % inhibition determined based on the OD values.
4.10. Data Analysis
Data analysis was done using GraphPad version 8.4.3 software (GraphPad Software, San Diego, CA, USA) and Microsoft Excel 2013. Differences in group means was determined by one-way ANOVA. In comparing the means within the comparisons, the one-way ANOVA was proceeded by Tukey’s or Dunnett’s post hoc multiple comparison test. At p-value < 0.05 cut-off, all comparisons were considered statistically significant.
5. Conclusions
The study reports an inhibition ELISA model and its potential application in snakebite envenoming, especially in relation to cobra bites. Albeit not for diagnostic purposes, the inhibition ELISA described herein demonstrates the possibility of detecting three-finger toxins present in the venoms of cobra snakes. Venoms of all five snakes tested with the assay are considered ‘category 1’, constituting the greatest threat to public health [42]. The detection of 3FTxs proteins in venom-challenged mice samples may suggest that the inhibition ELISA assay could potentially be used in detecting the presence or otherwise of the same in human clinical samples. However, the utility of this assay in human envenoming, thus has to be adequately evaluated. Also, studies involving a wide range of venoms may be necessary to further test the sensitivity of the assay.
Acknowledgments
The authors acknowledge the support of Benson Mwangi and Omar Samuel of the Production Department, KEMRI.
Author Contributions
Conceptualization: E.Z.M., J.H.K., K.C.M. and J.K.G.; Three-finger toxin fractionation, A.B., K.K.H., V.P. and J.L.; Methodology, E.Z.M.; Software, E.Z.M.; Validation, J.H.K., K.C.M., J.K.G., A.B., K.K.H. and V.P.; Formal analysis, E.Z.M.; Investigation, E.Z.M., J.H.K., K.C.M. and J.K.G.; Writing—original draft preparation, E.Z.M.; Writing—review and editing, E.Z.M., J.H.K., K.C.M., J.K.G., A.B., K.K.H., V.P. and J.H.K.; Supervision, J.H.K., K.C.M. and J.K.G.; Project administration, E.Z.M. and J.H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported with funding from the Pan African University under the African Union scholarship and partial funding from the Africa-ai-Japan Project under JICA, 2018/02/X/NZ6/00840 research grant from National Science Center, Poland, APVV-17-0017 (Slovak Research and Development Agency Departmental Organization of the Ministry of Education, Science, Research and Sports of the Slovak Republic) and VEGA 1/0241/18 (Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic and the Slovak Academy of Sciences).
Institutional Review Board Statement
The study was approved by the Kenya Medical Research Institute (KEMRI) Scientific and Ethical Review Unit with protocol number KEMRI/SERU/CBRD/229/4340.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gutiérrez J.M., Calvete J., Habib A.G., Harrison R.A., Williams D., Warrell D.A. Snakebite envenoming. Nat. Rev. Dis. Primers. 2017;3:17063. doi: 10.1038/nrdp.2017.63. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization Snakebite Envenoming. [(accessed on 15 January 2022)];2021 Available online: https://www.who.int/news-room/fact-sheets/detail/snakebite-envenoming.
- 3.Ministry of Health . Guidelines for Prevention, Diagnosis and Management of Snakebite Envenoming in Kenya. Ministry of Health; Nairobi, Kenya: 2019. [Google Scholar]
- 4.Wüster W., Broadley D.G. Get an eyeful of this: A new species of giant spitting cobra from eastern and north-eastern Africa (Squamata: Serpentes: Elapidae: Naja) Zootaxa. 2007;1532:51–68. doi: 10.11646/zootaxa.1532.1.4. [DOI] [Google Scholar]
- 5.Hus K.K., Buczkowicz J., Petrilla V., Petrillová M., Łyskowski A., Legáth J., Bocian A. First Look at the Venom of Naja ashei. Molecules. 2018;23:609. doi: 10.3390/molecules23030609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hus K.K., Marczak Ł., Petrilla V., Petrillová M., Legáth J., Bocian A. Different Research Approaches in Unraveling the Venom Proteome of Naja ashei. Biomolecules. 2020;10:1282. doi: 10.3390/biom10091282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okumu M.O., Mbaria J.M., Gikunju J.K., Mbuthia P.G., Madadi V.O., Ochola F.O. Enzymatic activity and brine shrimp lethality of venom from the large brown spitting cobra (Naja ashei) and its neutralization by antivenom. BMC Res. Notes. 2020;13:325. doi: 10.1186/s13104-020-05167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petras D., Sanz L., Segura A., Herrera M., Villalta M., Solano D., Vargas M., León G., Warrell D.A., Theakston R.D.G., et al. Snake Venomics of African Spitting Cobras: Toxin Composition and Assessment of Congeneric Cross-Reactivity of the Pan-African EchiTAb-Plus-ICP Antivenom by Antivenomics and Neutralization Approaches. J. Proteome Res. 2011;10:1266–1280. doi: 10.1021/pr101040f. [DOI] [PubMed] [Google Scholar]
- 9.Bermúdez-Méndez E., Fuglsang-Madsen A., Føns S., Lomonte B., Gutiérrez J.M., Laustsen A.H. Innovative Immunization Strategies for Antivenom Development. Toxins. 2018;10:452. doi: 10.3390/toxins10110452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kini R.M., Doley R. Structure, function and evolution of three-finger toxins: Mini proteins with multiple targets. Toxicon. 2010;56:855–867. doi: 10.1016/j.toxicon.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Bernardoni J.L., de Sousa L., Wermelinger L.S., Lopes A.S., Prezoto B.C., Serrano S.M.T., Zingali R.B., Moura-Da-Silva A.M. Functional Variability of Snake Venom Metalloproteinases: Adaptive Advantages in Targeting Different Prey and Implications for Human Envenomation. PLoS ONE. 2014;9:e109651. doi: 10.1371/journal.pone.0109651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams H.F., Layfield H.J., Vallance T., Patel K., Bicknell A.B., Trim S.A., Vaiyapuri S. The Urgent Need to Develop Novel Strategies for the Diagnosis and Treatment of Snakebites. Toxins. 2019;11:363. doi: 10.3390/toxins11060363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams H.F., Vaiyapuri R., Gajjeraman P., Hutchinson G., Gibbins J.M., Bicknell A.B., Vaiyapuri S. Challenges in diagnosing and treating snakebites in a rural population of Tamil Nadu, India: The views of clinicians. Toxicon. 2017;130:44–46. doi: 10.1016/j.toxicon.2017.02.025. [DOI] [PubMed] [Google Scholar]
- 14.Bulfone T.C., Samuel S.P., Bickler P.E., Lewin M.R. Developing Small Molecule Therapeutics for the Initial and Adjunctive Treatment of Snakebite. J. Trop. Med. 2018;2018:4320175. doi: 10.1155/2018/4320175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selvanayagam Z., Gopalakrishnakone P. Tests for detection of snake venoms, toxins and venom antibodies: Review on recent trends (1987–1997) Toxicon. 1999;37:565–586. doi: 10.1016/S0041-0101(98)00203-7. [DOI] [PubMed] [Google Scholar]
- 16.Kulawickrama S., O’Leary M., Hodgson W., Brown S., Jacoby T., Davern K., Isbister G. Development of a sensitive enzyme immunoassay for measuring taipan venom in serum. Toxicon. 2010;55:1510–1518. doi: 10.1016/j.toxicon.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Van Dong L. Immunogenicity of venoms from four common snakes in the South of Vietnam and development of ELISA kit for venom detection. J. Immunol. Methods. 2003;282:13–31. doi: 10.1016/S0022-1759(03)00277-1. [DOI] [PubMed] [Google Scholar]
- 18.Morais V., Berasain P., Ifrán S., Carreira S., Tortorella M.N., Negrín A., Massaldi H. Humoral immune responses to venom and antivenom of patients bitten by Bothrops snakes. Toxicon. 2012;59:315–319. doi: 10.1016/j.toxicon.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Norris R.L., Pfalzgraf R.R., Laing G. Death following coral snake bite in the United States—First documented case (with ELISA confirmation of envenomation) in over 40 years. Toxicon. 2009;53:693–697. doi: 10.1016/j.toxicon.2009.01.032. [DOI] [PubMed] [Google Scholar]
- 20.Hung D.-Z., Liau M.-Y., Lin-Shiau S.-Y. The clinical significance of venom detection in patients of cobra snakebite. Toxicon. 2003;41:409–415. doi: 10.1016/S0041-0101(02)00336-7. [DOI] [PubMed] [Google Scholar]
- 21.Warrell D.A. Snake bite. Lancet. 2010;375:77–88. doi: 10.1016/S0140-6736(09)61754-2. [DOI] [PubMed] [Google Scholar]
- 22.Hung D.Z., Lin J.H., Mo J.F., Huang C.F., Liau M.Y. Rapid diagnosis of Naja atrasnakebites. Clin. Toxicol. 2014;52:187–191. doi: 10.3109/15563650.2014.887725. [DOI] [PubMed] [Google Scholar]
- 23.CSL Snake venom detection Kit (SVDK)-detection and identification of snake. Clin. Toxicol. 2017;55:1–19. [Google Scholar]
- 24.Sharma N., Hanif S., Upadhyay D., Chhikara M.K. Inhibition ELISA as a putative tool for the identification and quantification of meningococcal A and X polysaccharides at various stages of vaccine development. J. Immunol. Methods. 2019;473:112634. doi: 10.1016/j.jim.2019.112634. [DOI] [PubMed] [Google Scholar]
- 25.Laustsen A.H., Engmark M., Clouser C., Timberlake S., Vigneault F., Gutiérrez J.M., Lomonte B. Exploration of immunoglobulin transcriptomes from mice immunized with three-finger toxins and phospholipases A2 from the Central American coral snake, Micrurus nigrocinctus. PeerJ. 2017;5:e2924. doi: 10.7717/peerj.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosoda H., Takasaki W., Oe T., Tsukamoto R., Nambara T. A comparison of chromogenic substrates for horseradish peroxidase as a label in steroid enzyme immunoassay. Chem. Pharm. Bull. 1986;34:4177–4182. doi: 10.1248/cpb.34.4177. [DOI] [PubMed] [Google Scholar]
- 27.Goka A.K.J., Farthing M.J. The Use of 3, 3′, 5, 5′-Tetramethylbenzidine as a Peroxidase Substrate in Microplate Enzyme-Linked Immunosorbent Assay. J. Immunoass. 1987;8:29–41. doi: 10.1080/01971528708063053. [DOI] [PubMed] [Google Scholar]
- 28.Laustsen A.H., Lomonte B., Lohse B., Fernández J., Gutiérrez J.M. Unveiling the nature of black mamba (Dendroaspis polylepis) venom through venomics and antivenom immunoprofiling: Identification of key toxin targets for antivenom development. J. Proteom. 2015;119:126–142. doi: 10.1016/j.jprot.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Blanchet G., Collet G., Mourier G., Gilles N., Fruchart-Gaillard C., Marcon E., Servent D. Polypharmacology profiles and phylogenetic analysis of three-finger toxins from mamba venom: Case of aminergic toxins. Biochimie. 2014;103:109–117. doi: 10.1016/j.biochi.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Casasola A., Ramos-Cerrillo B., de Roodt A.R., Saucedo A.C., Chippaux J.-P., Alagón A., Stock R.P. Paraspecific neutralization of the venom of African species of cobra by an equine antiserum against Naja melanoleuca: A comparative study. Toxicon. 2009;53:602–608. doi: 10.1016/j.toxicon.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Sánchez A., Herrera M., Villalta M., Solano D., Segura Á., Lomonte B., Gutiérrez J.M., León G., Vargas M. Proteomic and toxinological characterization of the venom of the South African Ringhals cobra Hemachatus haemachatus. J. Proteom. 2018;181:104–117. doi: 10.1016/j.jprot.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Armbruster D.A., Pry T. Limit of Blank, Limit of Detection and Limit of Quantitation. Clin. Biochem. Rev. 2008;29((Suppl. S1)):S49–S52. [PMC free article] [PubMed] [Google Scholar]
- 33.Classen D.C., Morningstar J.M., Shanley J.D. Detection of antibody to murine cytomegalovirus by enzyme-linked immunosorbent and indirect immunofluorescence assays. J. Clin. Microbiol. 1987;25:600–604. doi: 10.1128/jcm.25.4.600-604.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lardeux F., Torrico G., Aliaga C. Calculation of the ELISA’s cut-off based on the change-point analysis method for detection of Trypanosoma cruzi infection in Bolivian dogs in the absence of controls. Memórias Inst. Oswaldo Cruz. 2016;111:501–504. doi: 10.1590/0074-02760160119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C.C., Yu J.S., Wang P.J., Hsiao Y.C., Liu C.H., Chen Y.C., Lai P.F., Hsu C.P., Fann W.C., Lin C.C. Development of sandwich ELISA and lateral flow strip assays for diagnosing clinically significant snakebite in Taiwan. PLOS Negl. Trop. Dis. 2018;12:e0007014. doi: 10.1371/journal.pntd.0007014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaikh I.K., Dixit P.P., Pawade B.S., Waykar I.G. Development of dot-ELISA for the detection of venoms of major Indian venomous snakes. Toxicon. 2017;139:66–73. doi: 10.1016/j.toxicon.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe M., Sakai A., Kouda T., Sawai Y. Detection of Agkistrodon b. blomho i venom in serum of mice by enzyme-linked immunosorbent assay (ELISA) Snake. 1988;20:25–29. [Google Scholar]
- 38.Selvanayagam Z. Ph.D. Thesis. University of Madras; Madras, India: 1995. Studies on Ehretia Buxifolia, a Herbal Antidote for Echis Carinatus Envenomation and Development of ELISA for Detection of Snake Venoms. [Google Scholar]
- 39.Dyba B., Barbasz A., Czy A., Hus K.K. Effects of 3FTx Protein Fraction from Naja ashei Venom on the Model and Native Membranes: Recognition and Implications for the Mechanisms of Toxicity. Molecules. 2021;26:2164. doi: 10.3390/molecules26082164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ThermoFisher Scientific Pierce BCA Protein Assay Kit User Guide. Illinois, USA. 2020. [(accessed on 20 March 2021)]. Available online: https://www.thermofisher.com/bca.
- 41.Islam M., Jones R. An enzyme-linked immunosorbent assay for measuring antibody titre against avian reovirus using a single dilution of serum. Avian Pathol. 1988;17:411–425. doi: 10.1080/03079458808436459. [DOI] [PubMed] [Google Scholar]
- 42.WHO . WHO Expert Committee on Biological Standardization. World Health Organization; Geneva, Switzerland: 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.