Abstract
Of 3,052 Staphylococcus aureus strains collected by the European SENTRY surveillance study, 35 were found to be nonsusceptible to quinupristin-dalfopristin (MIC of ≥2 mg/liter). These isolates originated from four hospitals in France and one in Spain. In isolates from two Parisian hospitals exhibiting the same SmaI macrorestriction pattern, streptogramin resistance was based on vatA and vgbA. One isolate from a hospital in Lyon and 22 from a hospital in Lille were of the vatB vgaB streptogramin A resistance genotype and possessed ermA and/or ermC. As deduced from the loss of either streptogramin A or streptogramin B resistance determinants in particular isolates, resistance to quinupristin-dalfopristin requires mechanisms conferring resistance to both compounds. The SmaI macrorestriction patterns of strains from hospitals in Lille and Lyon were different; however, similarity analysis suggested a relatedness of 20 methicillin-resistant S. aureus strains from the Lille hospital, a finding confirmed by PCR typing based on three different genomic polymorphisms. These groups of isolates were found to be hetero-glycopeptide-intermediate susceptible S. aureus. Information about the failure of glycopeptide chemotherapy has not been available.
Methicillin-resistant Staphylococcus aureus (MRSA) has become an important nosocomial pathogen in many countries. Glycopeptides have been the drugs of choice for treating MRSA infections. However, with the emergence of intermediate susceptibility to glycopeptides, first reported from sporadic cases in many countries (12, 20) and more recently also from an outbreak of infections (11), other treatment alternatives such as quinupristin-dalfopristin and linezolid have become important. Quinupristin-dalfopristin is a combination of streptogramin B and A compounds with a synergistic activity against most gram-positive bacteria (8, 14).
Resistance to the B component is mediated by methylation of 23S rRNA which confers macrolide-lincosamide-streptogramin B (MLSB) resistance when the erm genes are expressed constitutively (7). A second mechanism is hydrolysis of the drug by a lactonase encoded by the vgbA and the vgbB genes, respectively (1, 6). Two kinds of mechanisms are known to mediate resistance to the A compound: inactivation of the drug by acetylation and efflux. In staphylococci, three acetyltransferase genes, vatA, vatB, and vatC (3, 4, 6) and two genes for efflux pumps (ABC porters), vgaA and vgaB (2, 5), have been described. These streptogramin A and B resistance genes are often located on the same plasmids (6).
In this study we describe quinupristin-dalfopristin-resistant S. aureus strains collected during the course of the European SENTRY surveillance study in European countries (18). These strains were characterized in our laboratory with regard to streptogramin resistance genes, phenotypic resistance to other antibiotics, and genotype.
MATERIALS AND METHODS
Bacterial isolates.
A total of 35 isolates of S. aureus exhibiting insensitivity to quinupristin-dalfopristin originated from the strain culture collection of the European SENTRY study (18). S. aureus ES1767 (vatA vgaA vgbA), ES1768 (vatB vgaB), and ES1877 (vatC vgbB) were kindly provided by N. El Solh, Paris, France; S. aureus Mu 50 (13) was provided by K. Hiramatsu, Tokyo, Japan.
Antimicrobial susceptibility testing.
Isolates were tested by broth microdilution assay performed according to the NCCLS standard (16) and by using Isosensitest broth from Oxoid. The prepared panels were immediately stored at −20°C. S. aureus ATCC 25923 served as a control for the activity of the antibiotics tested and strains ES1767, ES1768, and ES1877 as controls for the expression of resistance to streptogramin compounds. The antibiotics tested included penicillin G, oxacillin, gentamicin, erythromycin, clindamycin, oxytetracycline, ciprofloxacin, moxifloxacin, sulfonamide-trimethoprim, fusidic acid, rifampin, quinupristin-dalfopristin (Synercid), quinupristin, dalfopristin, vancomycin, teicoplanin, and linezolid. Breakpoints were according to NCCLS standards (16), except for those antibacterials not included, such as phosphomycin (resistance, ≥128 mg/liter), fusidic acid (≥4 mg/liter), and linezolid (≥8 mg/liter).
Genotyping and PCR.
SmaI macrorestriction patterns were obtained as described previously (24). The CHEF-III apparatus from Bio-Rad (Munich, Germany) was used with the following conditions: a charge of 6 V/cm, pulse times of 5 to 15 s for 7 h and 15 to 60 s for 19 h, an angle of 120°, and a temperature of 14°C. Image processing and cluster analysis for similarity were performed according to the method of Claus et al. (10): this program converts band positions into molecular masses by use of the patterns of S. aureus 8325 as an internal standard. Molecular mass patterns are analyzed for similarity by means of a matching algorithm. PCR typing by means of amplimer patterns for the 16S-23S rRNA gene spacer, the DNA stretches flanked by the transposon Tn916 attachment region and Shine-Dalgarno sequence (tar916-shida), and the ERIC-2 sequence were performed as described previously (24).
The following PCR primers were used for the detection of streptogramin resistance genes: the acetyltransferase gene vatA was amplified with primers vatA1 (5′-CAA TGA CCA TGG ACC TGA TC) and vatA2 (5′-AGC ATT TCG ATA TCT CC), vatB was amplified with primers vatB1 (5′-CCT GAT CCA AAT AGC ATA TAT CC) and vatB2 (5′-CTA AAT CAG AGC TAC AAA GTG), and vatC was amplified with primers vatC1 (5′-TGG CAA AAT CAG CAA GG) and vatC2 (5′-TCG TCT CTA TCT CTA GGT CC), the genes encoding efflux pumps vgaA were amplified with primers vgaA1 (5′-AGT GGT GGT GAA GTA ACA CG) and vgaA2 (5′-CTT GTC TCC TCC GCG AAT AC), and those encoding vgaB were amplified with primers vgaB1 (5′-TCT CTC AAT TAG AAG AAC C) and vgaB2 (5′-TTA TCT ATT CGT GTT TCC) (except for the primers for vgaB [3, 4, 6]). The streptogramin B resistance determinants vgbA and vgbB were amplified using primers vgbA1 (5′-CCA GAT TCA GCA CCC TAC G) and vgbA2 (5′-CCC CCC ATT CAG TAA ACC), as well as vgbB1 (5′-CAG CAG TCT AGA TCA GAG TGG) and vgbB2 (5′-CAT ACG GAT CCA TCT TTT CC) (1, 6). For erm genes the primers were used as described previously (9, 19). The PCR mixture consisted of ca. 20 ng of DNA, 100 pmol of each primer, a 200 μM concentration of each of the deoxyribonucleotides, and 2.5 U of the Replithern polymerase (Biozym, Markoldendorf, Germany) with an appropriate buffer. Initial denaturation at 94°C for 5 min was followed by 30 cycles of amplification with 94°C for 1 min, annealing at 50 to 58°C for 1 min, and extension at 72°C for 1 min (except for the last cycle with an extension step of 4 min).
Screening test for detection of GISA.
Brain heart infusion (BHI) agar plates containing 6 mg of vancomycin per liter were inoculated as described elsewhere (22). S. aureus ATCC 25923 was used as a control for the activity of vancomycin, and strain S. aureus Mu 50 was used as a control for the expression of the glycopeptide-intermediate susceptible S. aureus (GISA) phenotype. GISA strains are strains for which vancomycin MICs are 4 to 8 mg/liter (22).
Population analysis for GISA.
This was performed on BHI agar as described previously (13). Each experiment was performed in triplicate.
RESULTS
Of 3,052 S. aureus strains isolated during the European SENTRY surveillance study in 1997 and 1998 from 24 European hospitals, 32 isolates exhibited MICs of quinupristin-dalfopristin of ≥2 mg/liter: for 6 isolates, the MIC was 2 mg/liter, for 2 isolates it was 4 mg/liter, and for 24 isolates it was 8 mg/liter. The results of characterization are shown in Table 1.
TABLE 1.
Origin, grouping according to genotypes, resistance phenotypes, and particular resistance genes in quinupristin-dalfopristin-resistant S. aureusa
| Origin (hospital) | SmaI pattern | No. of isolates | Resistance phenotype | MIC (mg/liter)
|
Streptogramin resistance genes | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| VAN | TEC | LNZ | Q-D | QUI | DAL | |||||
| Paris (Salpetriene) | A | 1 | PEN, OXA, CIP, MXF, Q-D | 0.5 | 0.5 | 1.0 | 8 | 16 | 32 | vat(A), vgb(A) |
| Paris (St. Joseph) | A | 1 | PEN, OXA, GEN, ERY, CLI, OTE, SXT, CIP, MXF, RIF | 0.5 | 0.5 | 1.0 | 2 | 32 | 2 | |
| A | 1 | PEN, OXA, GEN, ERY, CLI, OTE, SXT, CIP, MXF, RIF, Q-D | 0.5 | 0.5 | 1.0 | 8 | 64 | 32 | vat(A), vgb(A) | |
| Lyon | B | 1 | PEN, OXA, PHO, GEN, ERY, CLI, OTE, SXT, CIP, MXF, RIF, Q-D | 0.5 | 0.5 | 0.5 | 8 | 64 | 32 | vat(B), vga(B) |
| Lille | C | 2 | PEN, OXA, GEN, ERY, CLI, OTE, CIP, MXF, SXT, RIF, FUS, Q-D | 2.0 | 2.0 | 0.5 | 8 | 64 | 32 | vat(B), vgaB |
| D | 6 | PEN, OXA, GEN, ERY, CLI, OTE, CIP, MXF, SXT, RIF, FUS, Q-D | 2.0 | 2.0 | 0.5 | 8 | 64 | 32 | vat(B), vga(B) | |
| D1 | 1 | PEN, OXA, GEN, ERY, CLI, OTE, CIP, MXF, SXT, RIF, FUS, Q-D | 2.0 | 2.0 | 0.5 | 8 | 64 | 32 | vat(B), vga(B) | |
| D2 | 1 | PEN, OXA, GEN, ERY, CLI, OTE, CIP, MXF, SXT, RIF, FUS, Q-D | 2.0 | 2.0 | 0.5 | 8 | 64 | 32 | vat(B), vga(B) | |
| E | 7 | PEN, OXA, GEN, ERY, CLI, OTE, CIP, MXF, SXT, RIF, FUS, Q-D | 2.0 | 2.0 | 0.5 | 8 | 64 | 32 | vat(B), vga(B) | |
| E1 | 1 | PEN, OXA, GEN, ERY, CLI, OTE, CIP, MXF, SXT, RIF, FUS, Q-D | 2.0 | 2.0 | 0.5 | 8 | 64 | 32 | vat(B), vga(B) | |
| F | 2 | PEN, OXA, GEN, ERY, CLI, CIP, MXF, Q-D | 2.0 | 2.0 | 0.5 | 8 | 64 | 32 | vat(B), vga(B) | |
| G | 1 | PEN, OXA, GEN, ERY, CLI, OTE, CIP, MXF, SXT, FUS, RIF, Q-D | 2.0 | 2.0 | 0.5 | 8 | 64 | 32 | vat(B), vga(B) | |
| G | 2 | PEN, OXA, GEN, OTE, CIP, MXF, SXT, FUS, RIF | 2.0 | 2.0 | 0.5 | 2 | 8 | 32 | vat(B), vga(B) | |
| G | 2 | PEN, OXA, GEN, ERY-ind., OTE, CIP, MXF, SXT, FUS, RIF | 2.0 | 2.0 | 0.5 | 2 | 8 | 32 | vat(B), vga(B) | |
| H | 2 | PEN, OXA, ERY, CLI, OTE, CIP, MXF, Q-D | 0.5 | 0.5 | 0.5 | 2 | 8 | 32 | vat(B), vga(B) | |
| I | 1 | ERY, Q-D | 0.5 | 0.5 | 0.5 | 4 | 16 | 16 | vat(B), vga(B) | |
| Seville | J | 1 | PEN, ERY, CLI, CIP, MXF, Q-D | 0.5 | 0.5 | 0.5 | 4 | 32 | 32 | |
PEN, penicillin G; OXA, oxacillin; CIP, ciprofloxacin; MXF, moxifloxacin; Q-D, quinupristin-dalfopristin (Synercid); GEN, gentamicin; ERY, erythromycin; ERY-ind., inducible erythromycin resistance; CLI, clindamycin; OTE, oxytetracycline; SXT, trimethoprim-sulfamethoxazole; RIF, rifampin; FUS, fusidic acid; VAN, vancomycin; TEC, teicoplanin; LNZ, linezolid; QUI, quinupristin; DAL, dalfopristin.
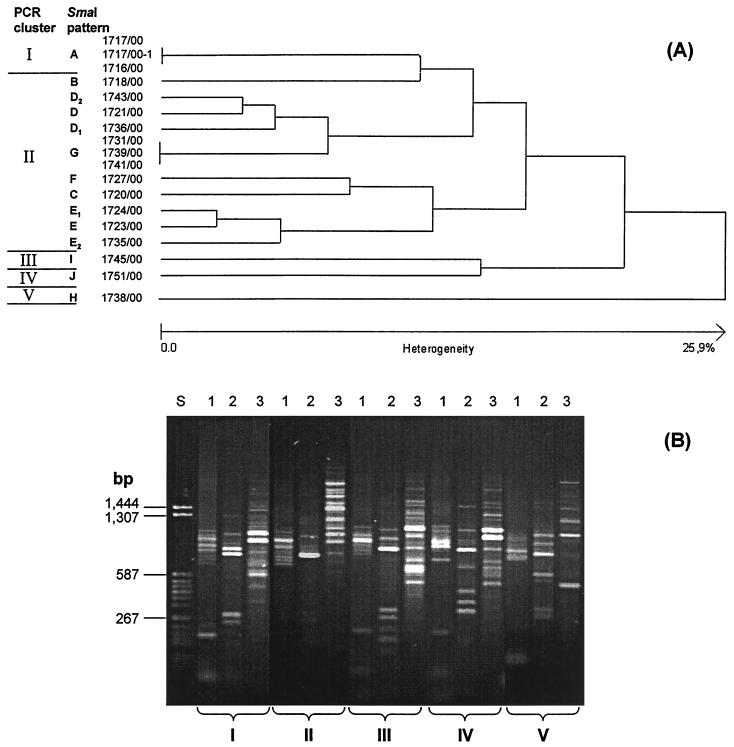
All 32 isolates have been typed by means of SmaI macrorestriction patterns separated by pulsed-field gel electrophoresis. Ten different patterns (A to J) could be discriminated when a difference in four and more fragments was used as a criterion (as described in reference 21).
Analysis of these patterns for similarity (Fig. 1A) leads to different lineages, which indicate but not necessarily document clonal relatedness. Therefore, three additional genomic polymorphisms were assessed by PCR (see Materials and Methods). PCR typing revealed five different, unrelated clusters (clonal groups I to V): for isolates with SmaI pattern A (PCR pattern I), I (PCR pattern III), J (PCR pattern IV), and H (PCR pattern V). Isolates with SmaI patterns B to G belonged to a group of ancestral relatedness because they all exhibit PCR pattern II (Fig. 1B).
FIG. 1.
Dendrogram of similarity of SmaI macrorestriction patterns of quinupristin-dalfopristin-resistant S. aureus strains in relation to patterns of PCR typing. (A) Similarity dendrogram. (B) PCR typing patterns found in quinupristin-dalfopristin-resistant S. aureus. Lanes: 1, rRNA gene spacer; 2, tar916-shida; 3, ERIC-2; S, molecular mass standard.
The two isolates of SmaI pattern A were MRSA and originated from two Parisian hospitals. Both carried vatA and vgbA. It is of particular interest that the isolate from the Salpetriene Hospital lacked macrolide-lincosamide resistance and did not possess any of the tested erm(A/B/C) genes. The isolate from St. Joseph Hospital exhibited a broader resistance phenotype, including erythromycin-clindamycin resistance encoded by the ermC determinant. A spontaneous in vitro derivative of this isolate (1717/00–1) detected by lacking hemolysis on blood agar obviously was determined to be only intermediately susceptible to quinupristin-dalfopristin and has lost vatA and vgbA.
Isolates of SmaI patterns B to G (PCR typing cluster II) are MRSA strains and originated from a hospital in Lille. These strains carried the vatB and vgaB streptogramin A resistance genes. Isolates belonging to patterns B, C, D, E, and F possessed ermA and partly also ermC. Of particular interest were the five isolates exhibiting pattern G. Isolate 1741/00 harbored ermC and was found to be constitutively resistant to erythromycin and lincomycin and also resistant to quinupristin-dalfopristin (MIC, 8 mg/liter). Two isolates (1739/00 as an example in Fig. 1A) also possessed ermC but exhibited an inducible MLSB resistance, as demonstrated by disk diffusion (reduction of the inhibition zone of lincomycin by a neighboring erythromycin disk). The MIC of quinupristin-dalfopristin was 2 mg/liter. Two isolates (1731/00 as an example in Fig. 1A) had no erm determinant, were sensitive to erythromycin and to lincomycin, and exhibited an MIC of 2 mg/liter of quinupristin-dalfopristin.
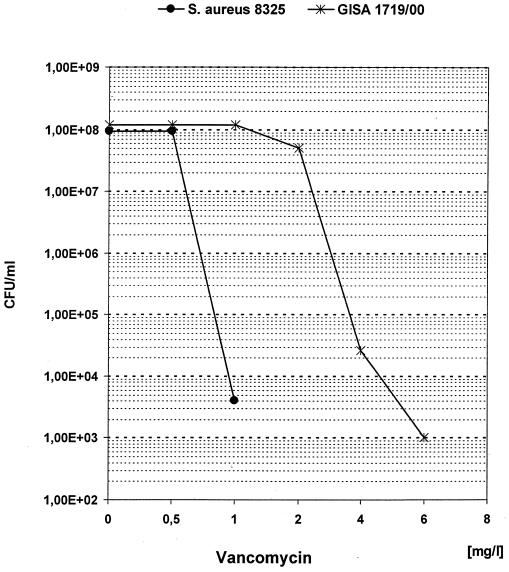
All isolates of PCR cluster II exhibited an elevated MIC (2 to 4 mg/liter) of vancomycin and teicoplanin and grew on screening plates for assessing the GISA phenotype. When we performed a population analysis, these isolates were determined to be hetero-GISA, as shown for strain 1719/00 as a representative in Fig. 2. For the antimicrobials checked, only MICs of phosphomycin and linezolid were in the sensitive range.
FIG. 2.
Population analysis of GISA 1719/00 and S. aureus 8325.
The two MRSA strains with SmaI pattern H (PCR cluster V) from Lille possessed vatB and vgaB, as well as ermC, but exhibited an MIC of quinupristin-dalfopristin of only 2 mg/liter. Another isolate from Lille which was not an MRSA (phenotypically sensitive, no mecA demonstrated) and belonged to a quite separate genotype (SmaI pattern I, PCR cluster III) had an MIC of quinupristin-dalfopristin of 4 mg/liter but also possessed vatB, vgaB, and ermC. One isolate from a hospital in Seville (1751/00) exhibited an MIC of 4 mg/liter of quinupristin-dalfopristin and resistance to both compounds when tested separately. It possessed ermC (constitutively expressed), but none of the known determinants conferring resistance to streptogramin A could be detected.
DISCUSSION
Strain collection by the European SENTRY study in 1997 and 1998 was performed before quinupristin-dalfopristin was licensed in Europe. All but one streptogramin-resistant S. aureus strain originated from three French hospitals. This is most probably due to selective pressure by the use of pristinamycins in France, although the consumption volume for this antibiotic has not been substantial. In the resistant strains, streptogramin A resistance genes have been detected as already described for S. aureus and other staphylococci from this country (15). The results from this study further show that different mechanisms of resistance to both compounds are essential for quinupristin-dalfopristin resistance in S. aureus. As evident from the SmaI pattern A strain originating from the Salpetriene Hospital, S. aureus can achieve quinupristin-dalfopristin resistance by the vatA-encoded acetyltransferase (inactivation of streptogramin A) and the vgbA-encoded lactonase (inactivation of streptogramin B). In other isolates resistance to the streptogramin A-streptogramin B combination is based on acetylation and efflux for streptogramin A resistance and erm-encoded 23S rRNA methylases for streptogramin B resistance. The loss of resistance either to streptogramin A or to streptogramin B leads to only intermediate susceptibility, as concluded from the data for isolates of SmaI patterns A (1717/00–1) and G (1731/00).
A cluster of related MRSA strains possessing vatB and vgaB originated from two hospitals, namely, in Lyon and Lille, with a broader dissemination in the latter one. The vatB and vgaB determinants have also been found in two S. aureus strains from the hospital in Lille; these strains exhibited other genotypical typing patterns, which suggests a dissemination between different strains of the same hospital. In both of the SmaI pattern H (PCR cluster V) isolates, the MIC of quinupristin-dalfopristin is only 2 mg/liter. Although they are fully resistant to erythromycin and lincomycin, the MIC of quinupristin was only 8 mg/liter.
S. aureus only possessing constitutively expressed erm genes is still not resistant to quinupristin-dalfopristin (breakpoint of ≥4 mg/liter). As known from earlier studies (15), resistance to both streptogramins needs additonal streptogramin A resistance determinants. Prevention of further dissemination of streptogramin A resistance genes requires early detection in bacteriological routine. This might be problematic when quinupristin-dalfopristin is tested as a combination. Therefore, a separate testing of dalfopristin resistance is desirable as has already been suggested (4).
In one isolate from a hospital in Seville resistance to quinupristin-dalfopristin was found, although no streptogramin A resistance genes were detected. There are obviously further genes (mechanisms?) conferring streptogramin A resistance.
All but two of the quinupristin-dalfopristin-insensitive isolates are MRSA strains, and most of them are already resistant to other classes of antibiotics.
All isolates from PCR cluster II of the SmaI patterns C to G from the Lille hospital were determined to be hetero-GISA. They are only susceptible to phosphomycin and linezolid. We have no information about failure of glycopeptide chemotherapy in the patients affected. However, this has recently been described for infections with hetero-GISA also exhibiting an MIC of 2 mg/liter of vancomycin (23). Recently, an outbreak of MRSA strains which were also determined to be hetero-GISA has been reported from Boroussias Hospital in Paris (11), and GISA isolates have also been reported from two hospitals in Lyon (17). These isolates, however, have been found to be susceptible to quinupristin-dalfopristin.
PCR typing of the hetero-GISA by means of three independent DNA polymorphisms, with each of them covering a comparably small (PCR-detectable) region of the genome, revealed a genotypical relatedness, although these isolates clearly exhibited different SmaI macrorestriction patterns that are due to genomic rearrangements over the whole genome. This indicates that these particular MRSA strains were already endemic to the hospital in Lille for a longer period of time.
ACKNOWLEDGMENTS
The skillful technical assistance of B. Pasemann and E. Baier is greatly appreciated.
The SENTRY Antimicrobial Resistance Surveillance Program was supported by Bristol-Myers Squibb Pharmaceuticals.
REFERENCES
- 1.Allignet J, Loncle V, Mazodier P, El Solh N. Nucleotide sequence of a staphylococcal plasmid gene, vgb, encoding a hydrolase inactivating the B components of virginiamycin-like antibiotics. Plasmid. 1988;20:271–275. doi: 10.1016/0147-619x(88)90034-0. [DOI] [PubMed] [Google Scholar]
- 2.Allignet J, Loncle V, El Solh N. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene. 1992;117:45–51. doi: 10.1016/0378-1119(92)90488-b. [DOI] [PubMed] [Google Scholar]
- 3.Allignet J, Loncle V, Simenel C, Delepierre M, El Solh N. Sequence of a staphylococcal gene, vat, encoding an acetyltransferase inactivating the A-type compounds of virginiamycin-like antibiotics. Gene. 1993;130:91–98. doi: 10.1016/0378-1119(93)90350-c. [DOI] [PubMed] [Google Scholar]
- 4.Allignet J, El Solh N. Diversity among the gram-positive acetyltransferases inactivating streptogramin A and structurally related compounds and characterization of a new staphylococcal determinant, vatB. Antimicrob Agents Chemother. 1995;39:2027–2029. doi: 10.1128/aac.39.9.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allignet J, El Solh N. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transfer conferring resistance to streptogramin A and related compounds. Gene. 1997;202:133–138. doi: 10.1016/s0378-1119(97)00464-2. [DOI] [PubMed] [Google Scholar]
- 6.Allignet J, Liassine N, El Solh N. Characterization of a staphylococcal plasmid related to pUB110 and carrying two novel genes, vatC and vgbB, encoding resistance to streptogramins A and B and similar antibiotics. Antimicrob Agents Chemother. 1998;42:1794–1798. doi: 10.1128/aac.42.7.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arthur M, Brisson-Noel A, Courvalin P. Origin and evolution of genes specifying resistance to macrolides, lincosamides and streptogramin antibiotics: data and hypothesis. J Antimicrob Chemother. 1987;20:783–802. doi: 10.1093/jac/20.6.783. [DOI] [PubMed] [Google Scholar]
- 8.Bouanchaud D H. In vitro and in vivo antibacterial activity of quinupristin/dalfopristin. J Antimicrob Chemother. 1997;39:15–21. doi: 10.1093/jac/39.suppl_1.15. [DOI] [PubMed] [Google Scholar]
- 9.Braulke C, Heuck D, Witte W. Ergebnisse der Tätigkeit des Nationalen Referenzzentrums für Staphylokokken im Jahr 1998. Bundesgesundheitsbl Gesundheitsforsch Gesundheitsschutz. 1999;42:499–506. [Google Scholar]
- 10.Claus H, Cuny C, Pasemann B, Witte W. A database system for fragment patterns of genomic DNA of Staphylococcus aureus. Zentbl Bakteriol. 1998;287:105–116. doi: 10.1016/s0934-8840(98)80154-0. [DOI] [PubMed] [Google Scholar]
- 11.Guerin F, Buu-Hoï A, Mainardi J-L, Kac G, Colardelle N, Vaupré S, Gutmann L, Podglajen I. Outbreak of methicillin-resistant Staphylococcus aureus with reduced susceptibility to glycopeptides in a Parisian hospital. J Clin Microbiol. 2000;38:2985–2988. doi: 10.1128/jcm.38.8.2985-2988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover F C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 14.Jones N J, Ballow C H, Biedenbach D J, Deinhart J A, Schentag J H. Antimicrobial activity of quinupristin/dalfopristin (RP 59500, Synercid) tested against over 28,000 recent clinical isolates from 200 medical centers in the United States and Canada. Diagn Microb Infect Dis. 1998;30:437–451. doi: 10.1016/s0732-8893(98)80002-3. [DOI] [PubMed] [Google Scholar]
- 15.Lina G, Quaglia A, Reverdy M-E, Leclercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother. 1999;43:1062–1066. doi: 10.1128/aac.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7–A4, 4th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17.Reverdy M-E, Jarraud S, Bobin-Dubreux S, Busel E, Girardo P, Lina G, Vandenesch F, Etienne J. Incidence of Staphylococcus aureus with reduced susceptibility to glycopeptides in two French hospitals. Clin Microbiol Infect. 2001;7:267–272. doi: 10.1046/j.1198-743x.2001.00256.x. [DOI] [PubMed] [Google Scholar]
- 18.Schmitz F-J, Verhoef J, Fluit A C The Sentry Participants Group. Prevalence of resistance to MLS antibiotics in 20 European university hospitals participating in the European SENTRY surveillance programme. J Antimicrob Chemother. 1999;43:783–792. doi: 10.1093/jac/43.6.783. [DOI] [PubMed] [Google Scholar]
- 19.Schmitz F-J, Sadurski R, Kray A, Boos M, Geisel R, Köhrer K, Verhoef J, Fluit A C. Prevalence of macrolide resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J Antimicrob Chemother. 2000;45:891–894. doi: 10.1093/jac/45.6.891. [DOI] [PubMed] [Google Scholar]
- 20.Sieradzki K, Roberts R B, Haber S W, Tomasz A. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N Engl J Med. 1999;340:517–523. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 21.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenover F C, Lancaster M V, Hill B C, Steward C D, Stocker S A, Hancock G A, O'Hara C M, Clark N C, Hiramatsu K. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J Clin Microbiol. 1998;36:1020–1027. doi: 10.1128/jcm.36.4.1020-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trakulsomboon S, Danchaivijitr S, Rongrungruang Y, Dhiraputra C, Susaemgrat W, Ito T, Hiramatsu K. First report of methicillin-resistant Staphylococcus aureus with reduced susceptibility to vancomycin in Thailand. J Clin Microbiol. 2001;39:591–595. doi: 10.1128/JCM.39.2.591-595.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witte W, Kresken M, Braulke C, Cuny C. Increasing incidence and widespread dissemination of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals in central Europe, with special reference to German hospitals. Clin Microbiol Infect. 1997;3:414–422. doi: 10.1111/j.1469-0691.1997.tb00277.x. [DOI] [PubMed] [Google Scholar]