Abstract
Stenotrophomonas maltophilia and Achromobacter (Alcaligenes) xylosoxidans have been increasingly recognized as a cause of respiratory tract colonization in cystic fibrosis (CF). Although both organisms have been associated with progressive deterioration of pulmonary function, demonstration of causality is lacking. To examine the molecular epidemiology of S. maltophilia and A. xylosoxidans in CF, isolates from patients monitored for up to 2 years were fingerprinted using a PCR-based randomly amplified polymorphic DNA (RAPD-PCR) method. Sixty-one of 69 CF centers screened had 183 S. maltophilia culture-positive patients, and 46 centers had 92 A. xylosoxidans-positive patients. At least one isolate from each patient was genotyped, and patients with ≥10 positive cultures (12 S. maltophilia cultures, 15 A. xylosoxidans cultures) had serial isolates genotyped. In addition, centers with multiple culture-positive patients were examined for evidence of shared clones. There were no instances of shared genotypes among different CF centers. Some patients demonstrated isolates with a single genotype throughout the observation period, and others had intervening or sequential genotypes. At the six centers with multiple S. maltophilia culture-positive patients and the seven centers with multiple A. xylosoxidans-positive patients, there were three and five instances of shared genotypes, respectively. The majority of shared isolates were from pairs who were siblings or otherwise epidemiologically linked. These findings suggest RAPD-PCR typing can distinguish unique CF isolates of S. maltophilia and A. xylosoxidans, person-to-person transmission may occur, there are not a small number of clones infecting CF airways, and patients with long-term colonization may either have a persistent organism or may acquire additional organisms over time.
Stenotrophomonas maltophilia and Achromobacter (Alcaligenes) xylosoxidans are aerobic, nonfermentative, gram-negative bacilli that are found in a wide variety of aquatic, soil, and rhizosphere environments. Both organisms have been isolated from hospital sources, and they have been increasingly recognized as nosocomial pathogens, particularly for immunocompromised patients (7, 13, 14, 19, 22, 31, 38, 41). Recent evidence suggests that they may also be emerging pathogens in cystic fibrosis (CF) patients (1, 4, 6, 9, 12, 28).
The prevalence of S. maltophilia in the respiratory tract of patients with CF has increased in recent years, with some clinics reporting rates in excess of 30% (1, 9). Interestingly, the source of the majority of S. maltophilia strains colonizing the respiratory tracts of CF patients cannot be linked to previously identified nosocomial sources, suggesting multiple, independent acquisitions from a variety of environmental sources (10). The data on A. xylosoxidans in CF is less complete, but the prevalence in CF patients may be as high as 8.7% (4). There are no studies identifying the source of A. xylosoxidans in CF patients, although nebulizers and respiratory therapy equipment have been implicated in nosocomial respiratory tract infections in non-CF patients (7).
Lung infection with gram-negative organisms is an important cause of morbidity and mortality in CF. Two of the most important CF pathogens, Pseudomonas aeruginosa and Burkholderia cepacia, are persistently isolated from CF sputum. However, very different epidemiological scenarios exist for B. cepacia and P. aeruginosa. Cross-infection with P. aeruginosa within a CF center is only rarely seen (26, 37), whereas epidemic spread of some strains of B. cepacia has been clearly demonstrated (17, 24, 25, 39).
The epidemiology of S. maltophilia and A. xylosoxidans in patients with CF is not well understood. To date, no genotyping study has analyzed isolates of these organisms from multiple patients at different CF centers in order to determine whether these isolates are closely related or unique. It is also unknown whether patients are persistently colonized, with the organism escaping detection on certain cultures, or whether a cycle of acquisition and clearing is occurring. It is important to investigate these questions, because clinicians report individual patients who exhibit deterioration of pulmonary function associated with isolation of these organisms from CF sputum. Establishing the role of S. maltophilia and A. xylosoxidans in CF lung infections could have significant treatment implications, because these organisms are often highly resistant to various antibiotics, including β-lactams, quinolones, aminoglycosides, and carbapenems (16, 18, 27, 33) that are commonly used for the management of CF lung infections. The increase in prevalence of S. maltophilia in the lungs of CF patients has been associated with the extensive use of antipseudomonal antibiotics for early treatment of P. aeruginosa colonization and for suppression of chronic P. aeruginosa respiratory tract infections (11). Parallel data are unavailable for A. xylosoxidans. Molecular typing may contribute to our knowledge of the epidemiology of these infections in CF, thus allowing the development of strategies to prevent their acquisition.
To examine the epidemiology of S. maltophilia and A. xylosoxidans we used random amplified polymorphic DNA PCR (RAPD-PCR). This technique utilizes a single, arbitrary oligonucleotide primer selected for its ability to discriminate among epidemiologically distinct isolates. This random primer anneals to multiple sites in the genome, resulting in a reproducible banding pattern. RAPD-PCR has previously been shown to be discriminatory for typing bacterial isolates from the lungs of patients with CF, including P. aeruginosa and B. cepacia (25, 26). Several authors have reported its utility in the typing of nosocomial outbreaks of S. maltophilia (21). Both enterobacterial repetitive intergenic consensus (ERIC) PCR and repetitive extragenic palindromic PCR have been used to type a small outbreak of A. xylosoxidans from the lungs of children with and without CF (12). ERIC PCR and repetitive extragenic palindromic PCR have also been compared with pulsed-field gel electrophoresis for the typing of nosocomial outbreaks and CF isolates of A. xylosoxidans (23, 28).
The isolates for this study were obtained from a collection of bacterial isolates from patients enrolled in paired clinical trials of inhaled tobramycin (6, 32). This collection of sequential isolates from a large number of CF patients from many centers across the United States offered the unique opportunity to investigate the molecular epidemiology of these emerging pathogens. The overall objectives of the study were (i) to determine whether specific clones of S. maltophilia and A. xylosoxidans could be identified at difference CF centers across the United States, (ii) to examine whether individual patients within a CF center shared common genotypes, and (iii) to identify patterns of organism acquisition, namely, do patients persistently have the same isolate or are different ones acquired and lost over time?
MATERIALS AND METHODS
Bacterial isolates and microbiological methods.
The clinical trials of inhaled tobramycin (6, 32) enrolled 520 patients at 69 CF centers in the United States. All gram-negative isolates from sputum and oropharyngeal cultures obtained during these trials were saved and identified using standard techniques, including the use of a biochemical panel for the identification of non-P. aeruginosa, non-lactose-fermenting gram-negative bacilli (36). Following identification, isolates were catalogued and frozen at −80°C in 0.5 ml of sterile skim milk. Isolates identified as S. maltophilia or A. xylosoxidans were recovered from frozen stocks and grown on Luria (L) agar with 24 to 48 h of incubation at 37°C for use in the present study.
Isolation of genomic DNA
A single colony was inoculated into 2 ml of L broth in a 20-ml glass tube and grown overnight in a shaking incubator at 37°C. After harvest by centrifugation, the bacterial pellet was resuspended in 50 mM glucose–25 mM EDTA–10 mM Tris-Cl, pH 8.0. Genomic DNA was isolated by a modified alkaline lysis preparation, which included an overnight digestion with pronase to degrade extracellular nucleases. Other than this modification, this was a standard preparation that used ammonium acetate and chloroform, to remove proteins and polysaccharides, and isopropanol, to precipitate genomic DNA (35). The resulting DNA pellet was resuspended in H2O containing RNase and quantified by A260. All preparations were frozen at −80°C until use.
RAPD typing
The RAPD primer sequences were provided by Eshwar Mahenthiralingam, University of British Columbia, Vancouver, Canada, and were as follows: for primer 270, 5′ TGCGCGCGGG 3′; for primer 272, 5′ AGCGGGCCAA 3′. Both primers were used to produce discriminatory polymorphisms from CF isolates of P. aeruginosa and B. cepacia, organisms with a G+C content similar to that of S. maltophilia and A. xylosoxidans (25, 26). Thus, they were screened for their utility with 12 S. maltophilia and 13 A. xylosoxidans isolates each from a different CF center. This confirmed the ability of the primers to produce discriminatory polymorphisms with these organisms. Primer 270 was initially used to type the isolates in this study; primer 272 was used for confirmation of identity in strains with similar patterns using primer 270.
RAPD-PCR mixtures (25 μl) were optimized for both organisms and contained 100 ng of genomic DNA, 0.45 μM primer, 2.5 U of polymerase, and 200 μM (each) deoxynucleoside triphosphate. The reaction mixtures were amplified using an MJ Research PTC-100 thermocycler and the following conditions: (i) 1 cycle of 15 min at 95°C; (ii) 4 cycles with 1 cycle consisting of 5 min at 94°C, 5 min at 36°C, and 5 min at 72°C; and (iii) 30 cycles with 1 cycle consisting of 1 min at 94°C, 1 min at 36°C, and 1 min at 72°C, followed by a final extension step at 72°C for 10 min.
RAPD products (one-third of each reaction mixture) were separated by electrophoresis in 1.5% agarose gels. Molecular size standards, a positive control consisting of either S. maltophilia or A. xylosoxidans DNA previously amplified using primer 270, and a negative control which contained all reaction components except template DNA, were included on all gels. The gels were stained with ethidium bromide and photographed using a digital camera. RAPD fingerprints were analyzed visually with the molecular size standards used to correct for gel-to-gel migration variation. Polymorphisms that differed by two or more bands were considered distinct. All polymorphisms that had fewer than three bands or were the same using the two-band difference criterion were repeated with primer 272.
RESULTS
S. maltophilia isolates.
There were a total of 183 S. maltophilia culture-positive patients, with a range of 0 to 21 isolates per patient. Seventy-seven patients had a single isolation of the organism over a 2-year period. Of these 77, 15 were from throat swabs (19%) and 62 were from sputum cultures. Of the S. maltophilia-positive cultures overall, the percentage of isolates from throat swabs was similar (16%).
Sixty-one of the 69 CF centers in the study had patients with respiratory cultures positive for S. maltophilia. There were 16 centers with a single culture-positive patient and 10 sites with five or more culture-positive patients. From 55 centers, a single isolate from each patient was amplified. Isolates from the additional six sites, each with five or more culture-positive patients, were examined in greater detail with multiple isolates from each patient typed. A total of 309 isolates from 168 patients were examined.
A. xylosoxidans isolates.
The respiratory tract cultures from 92 patients grew A. xylosoxidans. Forty-five patients had a single isolation of the organism; five were from throat swabs (11%), compared with 9% of isolates from throat swabs in the A. xylosoxidans positive specimens, overall.
Forty-six of the study centers were found to have A. xylosoxidans culture-positive patients. There were 12 centers with only a single patient whose cultures grew A. xylosoxidans and three centers that had five or more culture-positive patients. From 33 of the 46 sites, a single isolate from each patient with A. xylosoxidans was amplified, resulting in discriminatory polymorphisms. The remaining 13 CF centers were investigated in more detail, either because they had a large number of patients with the organism or because a single patient had multiple isolates. A total of 290 isolates from 92 patients were examined.
Reproducibility of RAPD analysis.
Primers 270 and 272 gave reproducible polymorphisms suitable for strain differentiation, ranging from 1 to 14 bands over an approximate size range of 200 to 3,000 bp. To demonstrate genotype stability, three unique isolates of each organism from different centers were passaged consecutively on L agar five times, with RAPD-PCR performed after each passage. Both sets of organisms showed stable genotypes with both primers following each passage, as shown by identical polymorphisms.
Molecular epidemiology.
To investigate the genetic relatedness among isolates from patients at different CF centers, at least one isolate from each patient at each clinical site with culture-positive patients was examined by RAPD-PCR. For neither organism were there instances of shared genotypes among different centers.
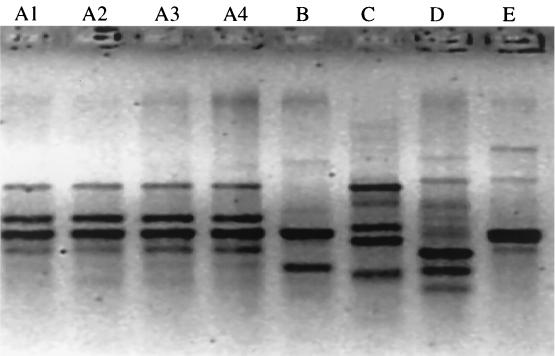
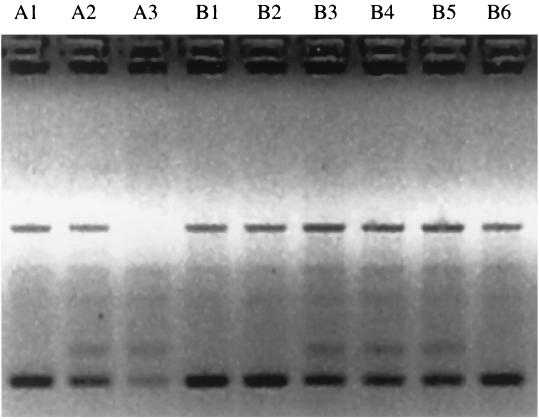
To investigate the genetic relatedness among isolates from patients at a single site, those CF centers with multiple culture-positive patients were examined in more depth, examining the majority of isolates from those centers (Fig. 1). Six clinical sites with five or more S. maltophilia culture-positive patients were examined. However, because there were only three such sites for A. xylosoxidans, sites with four or more culture-positive patients were examined. Of the six sites with five or more S. maltophilia-positive patients, three had patients with shared genotypes and three had patients with unique genotypes. Of the pairs with shared genotypes, one was a sibling pair and the other two were unrelated. Of the seven centers with multiple A. xylosoxidans-positive patients, five sites had patient pairs with shared genotypes. Of these, two pairs were siblings, one pair was friends who were frequently hospitalized at the same time, and two were epidemiologically unrelated. An example of shared genotypes of A. xylosoxidans in siblings is shown in Fig. 2.
FIG. 1.
A. xylosoxidans polymorphisms from five patients (A, B, C, D, and E) from the same CF center. The polymorphisms were generated using RAPD primer 270. Sequential isolates from patient A are listed as A1 through A4.
FIG. 2.
Identical A. xylosoxidans polymorphisms from two siblings (A and B). The polymorphisms shown here were generated using RAPD primer 270 and are arranged longitudinally. The genotypes were identical with primer 272. Sequential isolates from patient A are in lanes A1 through A3; those for patient B are in lanes B1 through B6. The DNA from lane A3 did not produce optimal amplification of polymorphisms.
Persistence of colonizing isolates.
In an attempt to determine whether CF patients acquired a single isolate that they kept for years or were periodically recolonized, serial isolates on a subpopulation of patients who were culture-positive for each organism were examined. Patients with ten or more isolates collected over a maximum of 2 years were targeted to determine the genotypic pattern of these serial isolates.
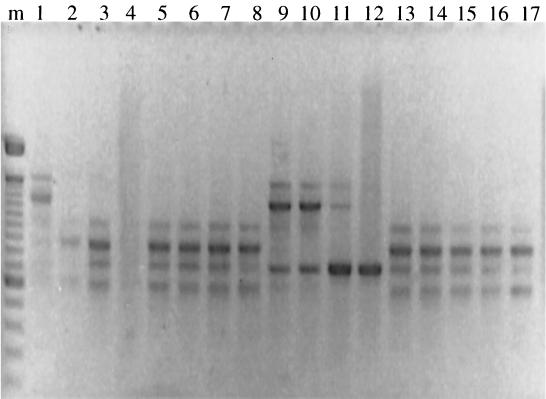
There were 15 patients with ≥10 S. maltophilia isolates collected longitudinally; isolates from 12 of them were genotyped. Because of loss during archiving or failure to prime, not all isolates could be examined for all 12 patients. The number of potential isolates for each individual patient ranged from 10 to 21, and the actual number of isolates genotyped ranged from 8 to 17. Five of the 12 patients had a single genotype identified. In the other seven only two genotypes were identified per patient. If genotypes are designated A, B, C, etc. in the order of appearance, the pattern in four patients was ABA and those in one patient each were AB, ABAB (Fig. 3), and ABABA. In patients in whom throat isolates were examined at some visits because of the patient's inability to expectorate, the genotypes always correlated with a sputum isolate at a previous or subsequent visit.
FIG. 3.
S. maltophilia polymorphisms from a single patient. The polymorphisms are arranged longitudinally. The pattern seen here is ABAB (lanes 1, 2 through 8, 9 through 12, and 13 through 17, respectively). The DNA from lanes 4 and 12 did not produce optimal amplification of polymorphisms. Molecular size markers were run in lane m.
There were twenty patients with ≥10 A. xylosoxidans isolates collected longitudinally; isolates from 15 of them were examined in depth. The number of potential isolates per patient ranged from 13 to 24, and the number of isolates genotyped ranged from 13 to 19. Thirteen of the fifteen patients had a single genotype identified. The other two patients each had an ABA pattern, with a single intervening culture with a markedly different genotype and reversion to the original genotype in subsequent cultures.
DISCUSSION
An arbitrary primed PCR typing method was used to examine the molecular epidemiology of two organisms that have recently been described as potential pathogens in patients with CF, A. xylosoxidans and S. maltophilia. This study systematically examined the relationship between genotypes of isolates from patients within a given CF center as well as between patients at different CF centers. In addition, multiple serial isolates from patients who were culture positive for up to 2 years were genotyped in an attempt to determine the course of infection. An understanding of the epidemiology of these organisms may help us better understand their role in CF lung disease.
The results of the present study demonstrate that there are multiple, unique clones of both S. maltophilia and A. xylosoxidans that can colonize CF patients. A tropism of a small number of specific clones for the CF lung, such as has been demonstrated with B. cepacia (25, 39), was not identified in this population. The diversity of genotypes seen with RAPD-PCR in the present study is consistent with the findings of nosocomial typing studies with both of these organisms. It appears that, in general, the majority of patients have unique isolates, and only occasional small clusters of indistinguishable strains have been identified (2, 15, 21, 34, 40, 44). The present results are also consistent with several smaller studies of S. maltophilia and A. xylosoxidans isolated from patients with CF (12, 28, 43). In addition, Denton et al. (10) reported that S. maltophilia isolates from the respiratory tract of patients with CF possess a genotype which is significantly distinct from strains collected from other patients or from the environment.
The issue of patient-to-patient transmissibility of these organisms was not fully elucidated in the present study. Patients from a single center occasionally shared a genotypically identical organism, and in many of those cases there was an obvious epidemiological link. This finding was similar to results with S. maltophilia reported by Demko et al. (8), suggesting that some transmission between siblings occurs. They found 10 sibling pairs (out of 40) in which both acquired S. maltophilia, but in only 5 pairs were both siblings positive at the same time. Unfortunately, those organisms were not genotyped, so it is unknown whether they represented shared isolates or concurrent acquisition of distinct strains. Using ERIC PCR, Denton et al. (10) found that a total of 33 out of 41 CF patients were colonized with unique strains of S. maltophilia and four pairs of patients shared strains. However, further investigation found no evidence of patient-to-patient transmission.
The epidemiology of A. xylosoxidans in CF patients has been less well studied. In nosocomial outbreaks, some investigations have demonstrated person-to-person or common-source infection (12, 42), and others have found strains to be unrelated (2). However, two small studies at different pediatric centers suggested little evidence of cross-infection or a common-source outbreak in CF (12, 43).
Perhaps most interestingly, the present data showed that many patients intermittently carry more than one strain of S. maltophilia. This is more reminiscent of the epidemiologic picture seen with early P. aeruginosa infection, where sequential or intermittent genotypes are identified (5). The finding that S. maltophilia was intermittently isolated from CF patients is also supported by the study by Demko et al. (8). Their data suggested that the persistence of S. maltophilia varied greatly, with 50% of the S. maltophilia-positive patients examined having only one positive culture between 1982 and 1994. Twelve percent had up to three positive cultures, with intervening negative cultures, but unfortunately, genotyping was not done on the isolates in that study. These results suggest either separate episodes of acquisition or inadequate microbiology techniques to isolate or identify the organism on intervening cultures (4).
Epidemiologic studies of serial CF isolates of A. xylosoxidans have not been performed. The present study showed a higher percentage of colonized patients with multiple isolations of A. xylosoxidans (20 of 92 [22%]) compared with S. maltophilia (15 of 183 [8%]) and far fewer patients with more than a single genotype. This suggests that either this organism may be more persistent in CF patients than S. maltophilia and P. aeruginosa or that certain organisms may have a tropism for CF airways.
The association of S. maltophilia and A. xylosoxidans with CF has been documented for almost 2 decades (3, 20). However, the role of these organisms in CF pulmonary disease is unclear. Because both these organisms are highly antibiotic resistant and can cause significant disease in non-CF patients (19, 22, 23, 27, 29, 30), there is a suggestion that they have potential for pathogenicity in CF pathogens. In a retrospective study of 211 S. maltophilia culture-positive CF patients, Demko et al. (8) found that S. maltophilia-positive patients had a lower mean percent predicted forced expired volume in one s (48.1% vs. 57.2%) and a higher proportion of concurrent P. aeruginosa colonization (84% vs. 76%). However, 2-year mortality did not appear to be related to whether patients were ever S. maltophilia positive, nor did S. maltophilia acquisition have an obvious deleterious effect on pulmonary status over the same 2 years. Similar types of studies are lacking for A. xylosoxidans. Based upon current data, it is not possible to rule out the possibility that S. maltophilia and A. xylosoxidans cause short-term mortality or that, in patients with severe disease, the presence of these resistant organisms makes management more difficult.
While the present study has done much to elucidate the epidemiology of S. maltophilia and A. xylosoxidans in patients with CF, further studies will be required to ascertain the mode of acquisition and source of the organisms. And a definitive epidemiological study that correlates the presence of S. maltophilia or A. xylosoxidans with clinical outcomes in CF will be essential to determining the pathogenicity of this organism. Finally, the present study does not provide sufficient data to definitively state whether segregation of these patients would be a beneficial infection control measure. However, the increasing prevalence of resistant gram-negative pathogens in CF patients suggests the need for caution in dealing with any multiply resistant organism.
REFERENCES
- 1.Ballestero S, Virseda I, Escobar H, Suarez L, Baquero F. Stenotrophomonas maltophilia in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 1995;14:728–729. doi: 10.1007/BF01690887. [DOI] [PubMed] [Google Scholar]
- 2.Benaoudia F, Bingen E. Evidence for the genetic unrelatedness of nosocomial Alcaligenes xylosoxidans strains in a pediatric hospital. Infect Control Hosp Epidemiol. 1997;18:132–134. doi: 10.1086/647568. [DOI] [PubMed] [Google Scholar]
- 3.Blessing J, Walker J, Maybury B, Yeager A S, Lewiston N. Pseudomonas cepacia and Pseudomonas maltophilia in the cystic fibrosis patient. Am Rev Respir Dis. 1979;119:262. . (Abstract.) [Google Scholar]
- 4.Burns J L, Emerson J, Stapp J R, Yim D L, Krzewinski J, Louden L, Ramsey B W, Clausen C R. Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis. 1998;27:158–163. doi: 10.1086/514631. [DOI] [PubMed] [Google Scholar]
- 5.Burns J L, Gibson R L, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith A L, Ramsey B W. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis. 2001;183:444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 6.Burns J L, Van Dalfsen J M, Shawar R M, Otto K L, Garber R L, Quan J M, Montgomery A B, Albers G M, Ramsey B W, Smith A L. Effect of chronic intermittent administration of inhaled tobramycin on respiratory microbial flora in patients with cystic fibrosis. J Infect Dis. 1999;179:1190–1196. doi: 10.1086/314727. [DOI] [PubMed] [Google Scholar]
- 7.Cheron M, Abachin E, Guerot E, El-Bez M, Simonet M. Investigation of hospital-acquired infections due to Alcaligenes denitrificans subsp. xylosoxidans by DNA restriction fragment length polymorphism. J Clin Microbiol. 1994;32:1023–1026. doi: 10.1128/jcm.32.4.1023-1026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demko C A, Stern R C, Doershuk C F. Stenotrophomonas maltophilia in cystic fibrosis: incidence and prevalence. Pediatr Pulmonol. 1998;25:304–308. doi: 10.1002/(sici)1099-0496(199805)25:5<304::aid-ppul3>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 9.Denton M. Stenotrophomonas maltophilia: an emerging problem in cystic fibrosis patients. Rev Med Microbiol. 1997;8:15–19. [Google Scholar]
- 10.Denton M, Todd N J, Kerr K G, Hawkey P M, Littlewood J M. Molecular epidemiology of Stenotrophomonas maltophilia isolated from clinical specimens from patients with cystic fibrosis and associated environmental samples. J Clin Microbiol. 1998;36:1953–1958. doi: 10.1128/jcm.36.7.1953-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denton M, Todd N J, Littlewood J M. Role of antibiotics in the emergence of Stenotrophomonas maltophilia in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 1996;15:402–405. doi: 10.1007/BF01690098. [DOI] [PubMed] [Google Scholar]
- 12.Dunne W M, Jr, Maisch S. Epidemiological investigation of infections due to Alcaligenes species in children and patients with cystic fibrosis: use of repetitive-element-sequence polymerase chain reaction. Clin Infect Dis. 1995;20:836–841. doi: 10.1093/clinids/20.4.836. [DOI] [PubMed] [Google Scholar]
- 13.Flaherty J P, Garcia-Houchins S, Chudy R, Arnow P M. An outbreak of gram-negative bacteremia traced to contaminated o-rings in reprocessed dialyzers. Ann Intern Med. 1993;119:1072–1078. doi: 10.7326/0003-4819-119-11-199312010-00003. [DOI] [PubMed] [Google Scholar]
- 14.Garcia de Viedma D, Marin M, Cercenado E, Alonso R, Rodriguez-Creixems M, Bouza E. Evidence of nosocomial Stenotrophomonas maltophilia cross-infection in a neonatology unit analyzed by three molecular typing methods. Infect Control Hosp Epidemiol. 1999;20:816–820. doi: 10.1086/501590. [DOI] [PubMed] [Google Scholar]
- 15.Gerner-Smidt P, Bruun B, Arpi M, Schmidt J. Diversity of nosocomial Xanthomonas maltophilia (Stenotrophomonas) as determined by ribotyping. Eur J Clin Microbiol Infect Dis. 1995;14:137–140. doi: 10.1007/BF02111874. [DOI] [PubMed] [Google Scholar]
- 16.Glupczynski Y, Hansen W, Freney J, Yourassowsky E. In vitro susceptibility of Alcaligenes denitrificans subsp. xylosoxidans to 24 antimicrobial agents. Antimicrob Agents Chemother. 1988;32:276–278. doi: 10.1128/aac.32.2.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govan J R W, Brown P H, Maddison J, Doherty C J, Nelson J W, Dodd M, Greening A P, Webb A K. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet. 1993;342:15–19. doi: 10.1016/0140-6736(93)91881-l. [DOI] [PubMed] [Google Scholar]
- 18.Khadori N, Ruben A, Rosenbaum B, Roston K, Bodey G P. In vitro susceptibility of Xanthomonas maltophilia to newer antibiotics agents. Antimicrob Agents Chemother. 1990;34:1609–1610. doi: 10.1128/aac.34.8.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khadori N, Elting L, Wong E, Schable B, Bodey G P. Nosocomial infections due to Xanthomonas maltophilia (Pseudomonas maltophilia) in patients with cancer. Rev Infect Dis. 1990;12:997–1003. doi: 10.1093/clinids/12.6.997. [DOI] [PubMed] [Google Scholar]
- 20.Klinger J D, Thomassen M J. Occurrence and antimicrobial susceptibility of gram negative nonfermentative bacilli in cystic fibrosis patients. Diagn Microbiol Infect Dis. 1985;3:149–158. doi: 10.1016/0732-8893(85)90025-2. [DOI] [PubMed] [Google Scholar]
- 21.Laing F P Y, Ramotar K, Read R R, Alfieri N, Kureishi A, Henderson E A, Louie T J. Molecular epidemiology of Xanthomonas maltophilia colonization and infection in the hospital environment. J Clin Microbiol. 1995;33:513–518. doi: 10.1128/jcm.33.3.513-518.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legrand C, Anaissie E. Bacteremia due to Achromobacter xylosoxidans in patients with cancer. Clin Infect Dis. 1992;14:479–484. doi: 10.1093/clinids/14.2.479. [DOI] [PubMed] [Google Scholar]
- 23.Lin Y H, Liu P Y, Shi Z Y, Lau Y J, Hu B S. Comparison of polymerase chain reaction and pulsed-field gel electrophoresis for the epidemiological typing of Alcaligenes xylosoxidans subsp. xylosoxidans in a burn unit. Diagn Microbiol Infect Dis. 1997;28:173–178. doi: 10.1016/s0732-8893(97)00062-x. [DOI] [PubMed] [Google Scholar]
- 24.LiPuma J J, Mortensen J E, Dasen S E, Edlind T D, Schidlow D V, Burns J L, Stull T L. Ribotype analysis of Pseudomonas cepacia from cystic fibrosis centers. J Pediatr. 1988;113:859–862. doi: 10.1016/s0022-3476(88)80018-0. [DOI] [PubMed] [Google Scholar]
- 25.Mahenthiralingam E, Campbell M E, Henry D A, Speert D P. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by random amplified polymorphic DNA fingerprinting. J Clin Microbiol. 1996;34:2914–2920. doi: 10.1128/jcm.34.12.2914-2920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahenthiralingam E, Campbell M E, Foster J, Lam J S, Speert D P. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:1129–1135. doi: 10.1128/jcm.34.5.1129-1135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall W F, Keating M R, Anhalt J P, Steckelberg J M. Xanthomonas maltophilia: an emerging nosocomial pathogen. Mayo Clin Proc. 1989;64:1097–1104. doi: 10.1016/s0025-6196(12)64979-9. [DOI] [PubMed] [Google Scholar]
- 28.Moissenet D, Baculard A, Valcin M, Marchand V, Tournier G, Garbarg-Chenon A, Vu-Thien H. Colonization by Alcaligenes xylosoxidans in children with cystic fibrosis: a retrospective clinical study conducted by means of molecular epidemiological investigation. Clin Infect Dis. 1997;24:274–275. doi: 10.1093/clinids/24.2.274. [DOI] [PubMed] [Google Scholar]
- 29.Morrison A J, Hoffmann K K, Wenzel R P. Associated mortality and clinical characteristics of nosocomial Pseudomonas maltophilia in a university hospital. J Clin Microbiol. 1986;24:52–55. doi: 10.1128/jcm.24.1.52-55.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muder R R, Yu V L, Dummer J S, Vinson C, Lumish R M. Infections caused by Pseudomonas maltophilia: expanding clinical spectrum. Arch Intern Med. 1987;147:1672–1674. [PubMed] [Google Scholar]
- 31.Orr K, Gould F K, Sission P R, Lightfoot N F, Freeman R, Burdess D. Rapid inter-strain comparison by pyrolysis mass spectrometry in nosocomial infection with Xanthomonas maltophilia. J Hosp Infect. 1991;17:187–195. doi: 10.1016/0195-6701(91)90230-6. [DOI] [PubMed] [Google Scholar]
- 32.Ramsey B W, Pepe M S, Quan J M, Otto K L, Montgomery A B, Williams-Warren J, Vasiljev K M, Borowitz D, Bowman C M, Marshall B C, Marshall S, Smith A L. Intermittent administration of inhaled tobramycin in patients with cystic fibrosis. N Engl J Med. 1999;340:23–30. doi: 10.1056/NEJM199901073400104. [DOI] [PubMed] [Google Scholar]
- 33.Rolston K V I, Messer M. The in-vitro susceptibility of Alcaligenes denitrificans subsp. xylosoxidans to 40 antimicrobial agents. J Antimicrob Chemother. 1990;26:857–860. doi: 10.1093/jac/26.6.857. [DOI] [PubMed] [Google Scholar]
- 34.Sader H S, Pignatari A C, Frei R, Hollis R J, Jones R N. Pulsed-field gel electrophoresis of restriction-digested genomic DNA and antimicrobial susceptibility of Xanthomonas maltophilia strains from Brazil, Switzerland, and the USA J. Antimicrob Chemother. 1994;33:615–618. doi: 10.1093/jac/33.3.615. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. p. A1. [Google Scholar]
- 36.Shigei J. Test methods used in the identification of commonly isolated aerobic gram-negative bacteria. In: Isenberg H D, editor. Clinical microbiology procedures handbook. Washington, D.C.: American Society for Microbiology; 1992. pp. 1.19.1–1.19.110. [Google Scholar]
- 37.Speert D P, Campbell M E. Hospital epidemiology of Pseudomonas aeruginosa from patients with cystic fibrosis. J Hosp Infect. 1987;9:11–21. doi: 10.1016/0195-6701(87)90089-2. [DOI] [PubMed] [Google Scholar]
- 38.Spencer R C. The emergence of epidemic, multiple-antibiotic-resistant Stenotrophomonas (Xanthomonas) maltophilia and Burkholderia (Pseudomonas) cepacia. J Hosp Infect. 1995;30(Suppl.):453–464. doi: 10.1016/0195-6701(95)90049-7. [DOI] [PubMed] [Google Scholar]
- 39.Sun L, Jiang R-Z, Steinbach S, Holmes A, Campanelli C, Forstner J, Sajjan U, Tan Y, Riley M, Goldstein R. The emergence of a highly transmissible lineage of cbl+ Pseudomonas cepacia causing epidemics in North America and Britain. Nat Med. 1995;1:661–666. doi: 10.1038/nm0795-661. [DOI] [PubMed] [Google Scholar]
- 40.Van Couwenberghe C J, Cohen S H, Tang Y J, Gumerlock P H, Silva J., Jr Genomic fingerprinting of epidemic and endemic strains of Stenotrophomonas maltophilia (formerly Xanthomonas maltophilia) by arbitrarily primed PCR. J Clin Microbiol. 1995;33:1289–1291. doi: 10.1128/jcm.33.5.1289-1291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanholder R, Vanhaecke E, Ringoir S. Pseudomonas septicemia due to deficient disinfectant mixing during reuse. Int J Artific Org. 1992;15:19–24. [PubMed] [Google Scholar]
- 42.Vu-Thien H, Darbord J C, Moissenet D, Dulot C, Dufourcq J B, Marsol P, Garbarg-Chenon A. Investigation of an outbreak of wound infections due to Alcaligenes xylosoxidans transmitted by chlorhexidine in a burn unit. Eur J Clin Microbiol Infect Dis. 1998;17:724–726. doi: 10.1007/s100960050168. [DOI] [PubMed] [Google Scholar]
- 43.Vu-Thien H, Moissenet D, Valcin M, Dulot C, Tournier G, Garbarg-Chenon A. Molecular epidemiology of Burkholderia cepacia, Stenotrophomonas maltophilia, and Alcaligenes xylosoxidans in a cystic fibrosis center. Eur J Clin Microbiol Infect Dis. 1996;15:876–879. doi: 10.1007/BF01691221. [DOI] [PubMed] [Google Scholar]
- 44.Yao J D C, Conly J M, Krajden M. Molecular typing of Stenotrophomonas (Xanthomonas) maltophilia by DNA macrorestriction analysis and random amplified polymorphic DNA analysis. J Clin Microbiol. 1995;33:2195–2198. doi: 10.1128/jcm.33.8.2195-2198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]