Abstract
Novel PARP inhibitors with selective mode-of-action have been approved for clinical use. Herein, oxadiazole based ligands that are predicted to target PARP-1 have been synthesized and screened for the loss of cell viability in mammary carcinoma cells, wherein seven compounds were observed to possess significant IC50 values in the range of 1.4 to 25 µM. Furthermore, compound 5u, inhibited the viability of MCF-7 cells with an IC50 value of 1.4µM, when compared to Olaparib (IC50 = 3.2 µM). Compound 5s also decreased cell viability in MCF-7 and MDA-MB-231 cells with IC50 values of 15.3 and 19.2 µM, respectively. Treatment of MCF-7 cells with compounds 5u and 5s produced PARP cleavage, H2AX phosphorylation and CASPASE-3 activation comparable to that observed with Olaparib. Compounds 5u and 5s also decreased foci-formation and 3D Matrigel growth of MCF-7 cells equivalent to or greater than that observed with Olaparib. Finally, in silico analysis demonstrated binding of compound 5s towardsthe catalytic site of PARP-1, indicating that these novel oxadiazoles synthesized herein may serve as exemplars for the development of new therapeutics in cancer.
Keywords: oxadiazole, poly(ADP-ribose) polymerase, human breast cancer, nicotinamide
1. Introduction
Loss of function mutation of breast cancer genes 1/2 (BRCA1 and BRCA2) significantly increases the risk of development and progression of breast and other cancers [1]. Pre-clinical investigations determined that dysfunction of BRCA1/2 results in sensitization of cancer cells to the catalytic inhibition of Poly ADP-ribose polymerase (PARP), producing cell-cycle arrest followed by apoptosis [2]. This vulnerability (synthetic lethality) initially allowed pharmacological inhibition of PARP to be exploited in BRCA1/2 deficient cancer, which is now being extended to BRCA proficient cancer [3,4,5].
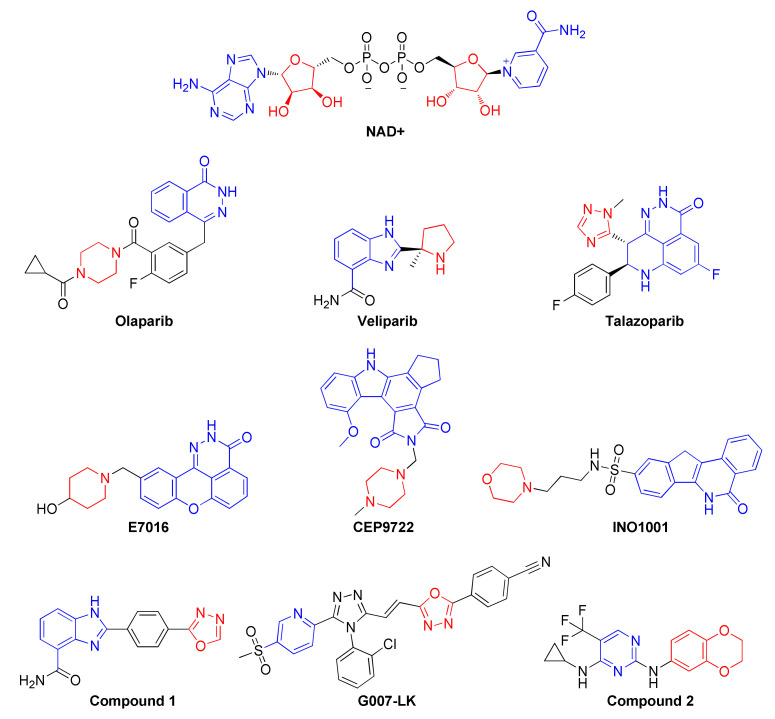
PARP1 is one member of a large family of structurally related proteins. PARP1 is a 113 kDa protein, which possesses three functional domains, namely the catalytic domain (CD), DNA-binding domain (DBD), and auto-modification domain (AMD) [6]. PARP-1 catalytically converts nicotinamide adenine dinucleotide (NAD+) into niacinamide and ADP-ribose, resulting in the transfer of ADP-ribose to downstream proteins modulating cellular functions. such as chromatin structure, gene transcription, DNA repair, and apoptosis [7]. Substrate mimetic compounds such as nicotinamide and 3-aminobenzamide were reported as weak PARP1 inhibitors with poor specificity, hence prompting chemists to design potent and specific PARP-1 inhibitors for clinical use [8,9,10]. To date, a number of PARP inhibitors (PARPis) with differing PARP trapping capacity have been clinically approved or received FDA Orphan Drug designation (Veliparib), including Olaparib, Talazoparib, and Rucaparib (Figure 1) [11].
Figure 1.
Design of a new generation of oxadiazoles, presumed to bind to the adenosine binding pocket of PARP1. The blue and red colored structures were used as effective PARP1 binding motifs.
However, both pre-clinical studies and clinical data demonstrate either intrinsic resistance to PARP inhibition or the development of acquired resistance to PARPi, resulting in limitation of the clinical response to the currently approved PARPis [12,13,14,15].
Replacement of the unsaturated heterocycles of many PARP inhibitors, such as niraparib, veliparib, E7016, CEP9722, and INO1001, with the saturated cyclic amine group generated a new generation of compounds that are reported to have low nano-molar inhibition of PARP activity and also display robust cellular activity [16,17,18]. In particular, compound 1 (Figure 1) bearing an oxadiazole moiety was observed to inhibit PARP1 activity with an IC50 as low as 1 nM resulting in a cellular EC50 of 3.7 nM; this was combined with an excellent pharmacokinetic profile [19]. Additionally, an oxadiazole containing a molecule called G007-LK (Figure 1) was reported as a potent inhibitor of tankyrases (TNKS) 1 and 2. G007-LK was co-crystallized with the PARP catalytic domain of TNKS2 and bound to the extended adenosine binding pocket of PARP, indicative of unique oxadiazole binding to PARP catalytic domains [20]. Recently, compound 2 (Figure 1) was also reported to inhibit the autophagy activating kinase 1 (ULK1) with an IC50 of 14 nM, and ULK1/2 inhibition in combination with Olaparib [21]. In continuation of the effort in the synthesis of novel oxadiazoles [22,23,24,25,26,27,28], herein synthesis of oxadiazoles with substitutions of pyridine, biphenyl, pyrimidine, and thiophene rings that target PARP1 and the efficacy of these compounds in BRCA wild-type estrogen receptor positive (ER+) and triple-negative breast cancer (TNBC) cells is reported.
2. Results and Discussion
2.1. Chemical Synthesis of Newer Oxadiazoles
3-bromobenzohydrazide 3a synthesis was undertaken by refluxing ethyl 3-bromobenzoate 2a and hydrazine, which was further reacted with different benzoic acids via intermolecular cyclization reaction using POCl3 as a cyclizing agent, and various oxadiazoles 4a–c were obtained. The resulting oxadiazoles 4a–c containing bromine were subjected to Suzuki coupling reactions with diverse boronic acids to obtain title compounds 5a–x (Scheme 1, Table 1). The synthesized oxadiazoles were characterized by 1H NMR, 13C NMR, LC-MS, HPLC, and melting points (Table 2 & refer to the Supplementary Materials).
Scheme 1.
Synthesis of novel substituted oxadiazoles from 5a–x. Ar/Het–Ar groups and structures of 4a–c are mentioned in Table 1.
Table 1.
List of Ar/Het.Ar groups and structures of 4a–c.
| Ar-B(OH)2/ Het.Ar-B(OH)2 | Structures of (4a–c) |
|---|---|
| (6-chloro-5-methylpyridin-3-yl)boronic acid |

|
| (4-(trifluoromethyl)phenyl)boronic acid | |
| (3-(cyclopentylcarbamoyl)phenyl)boronic acid | |
| (3-methoxyphenyl)boronic acid | |
| naphthalen-1-ylboronic acid | |
| pyridin-4-ylboronic acid | |
| pyrimidin-5-ylboronic acid | |
| (4,5-dimethylpyridin-3-yl)boronic acid | |
| pyridin-3-ylboronic acid | |
| (6-fluoro-5-methylpyridin-3-yl)boronic acid | |
| (2-fluoro-3-methylpyridin-4-yl)boronic acid | |
| (3-formylphenyl)boronic acid | |
| (5-acetylthiophen-2-yl)boronic acid |
Table 2.
List of newly synthesized oxadiazoles and their effect on cell viability of MCF–7 cells.
| Product 5a–x |
IC50 (μM) |
Product 5a–x |
IC50 (μM) |
|---|---|---|---|

|
160.00 |

|
31.53 |

|
205.10 |

|
21.88 |

|
160.90 |

|
43.47 |

|
345.00 |

|
33.22 |

|
74.80 |

|
76.40 |

|
>100 |

|
30.85 |

|
27.13 |

|
15.33 |

|
182.40 |

|
265.20 |

|
41.23 |

|
1.45 |

|
18.45 |

|
85.30 |

|
84.00 |

|
23.14 |

|
25.04 |

|
>100 |
2.2. Efficacy of Oxadiazoles in Breast Cancer Cells
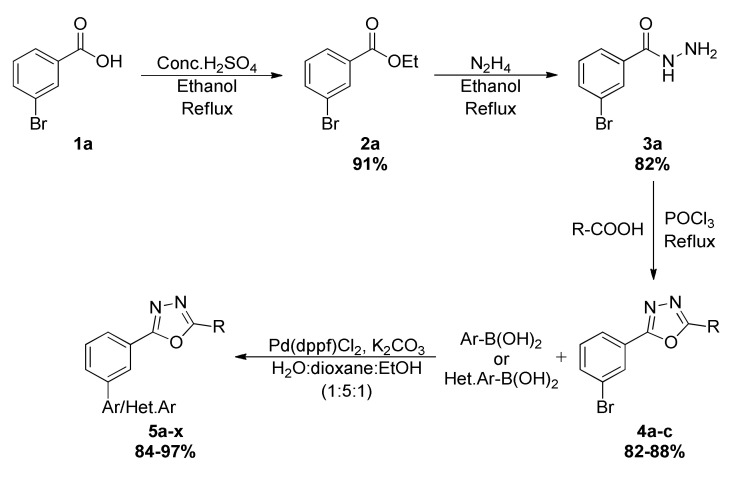
Initially, the newly synthesized oxadiazoles were examined in cell viability studies in human breast cancer (MCF-7) cells using Alamar Blue assays [29,30]. As internal control, Tamoxifen and Doxorubicin produced loss of viability of MCF-7 cells with IC50 values of 1.69 and 0.24 µM, respectively. The results of the assay revealed that compounds such as 5h, 5m, 5p, 5r, 5s, 5u, and 5w inhibited the viability of MCF-7 cells with varied IC50 values of 21.8, 27.1, 18.4, 25.0, 15.3, 1.4, and 23.1 µM, respectively. All of the compounds that bear a pyridine ring produced significant loss of viability of MCF-7 cells. The compound 5u exhibited IC50 values of 1.4 and 29.3 µM against MCF-7 and MDA-MB-231 cells, respectively, which was significant when compared to Olaparib (3.2 and 70.2 µM). The better activity of the compound 5u could be attributed to the presence of the naked benzaldehyde group. One more compound, 5s,was found to be effective againstboth MCF-7 and MDA-MB-231 cells where IC50 values of 15.3 and 19.2 µM, respectively (Figure 2). In addition to the lipophilic structures of these tested compounds, the presence of polar and active groups in the lead structures was found to decrease viability in human breast cancer cells.
Figure 2.
Lead compounds 5u, 5s, and Olaparib produce loss of viability of ER+ breast cancer (MCF-7) and triple-negative breast cancer (MDA-MB-231) cells. MCF-7 and MDA-MB-231 were treated with 5u, 5s and Olaparib (0, 0.01, 0.1, 1, 10, 100 µM) for 72 h, respectively. The viability of MCF-7 (A) and MDA-MB-231 cells (B) were analyzed using a microplate reader to measure the fluorescence of Alamar Blue reagent (excitation wavelength is between 530–560 nm and emission wavelength is 590 nm). The results are means of triplicate determinations.
2.3. Newer Oxadiazoles Inhibited the Catalytical Activity of PARP1 In Vitro
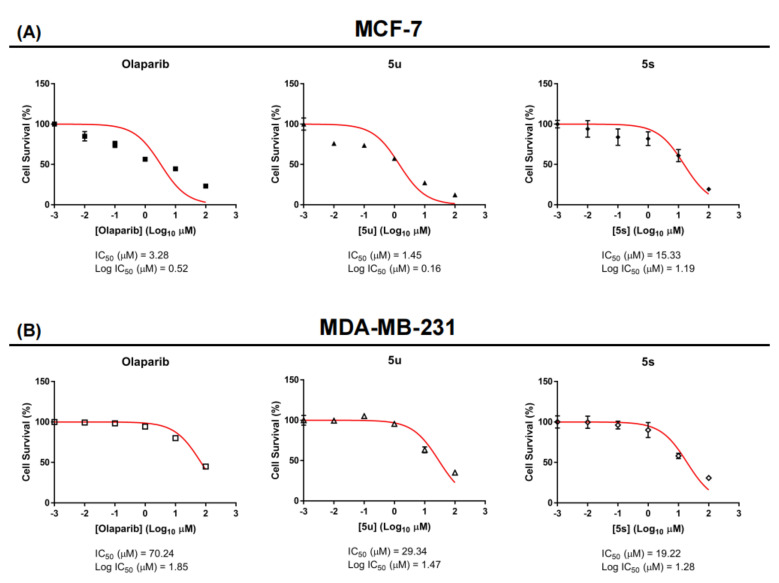
The most active oxadiazoles (5u and 5s) were tested for their capacity to inhibit PARP1 activity using the PARP/Apoptosis Universal Colorimetric Assay Kit, as described [31,32]. The screening of these compounds was performed at concentrations of 0.01, 0.1, 1, 10, and 100 µM in triplicate (n = 3) measurements, in which the compound 5u showed 4.82, 6.84, 45.39, 54.18, and 82.25% inhibition and 5s showed 2.38, 10.58, 21.33, 64.99, and 76.50% inhibition, respectively. The positive control, 3-Aminobenzamide, inhibited PARP1 activity by 0.50, 6.17, 23.32, 56.63, and 85.74% at the tested concentrations, respectively (Figure 3).
Figure 3.
Inhibition of PARP1 catalytic activity by oxadiazoles and 3-aminobenzamide. Graphical representation of the colorimetric readout of the inhibition of catalytic activity of PARP−1 by oxadiazoles and 3-aminobenzamide.
2.4. Compounds 5u and 5s Increased PARP1 Cleavage, Phospho-H2AX Levels and CPP32 (Caspase-3) Activation in Breast Cancer Cells
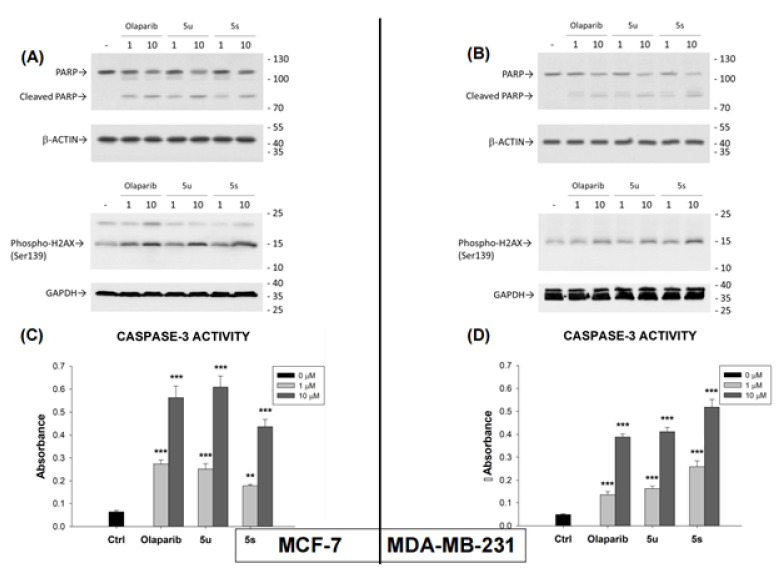
As 1,3,4-oxadiazoles have been previously reported to cleave endogenous PARP1, the effects of compounds 5u, 5s, and Olaparib on PARP1 cleavage in MCF-7 and MDA-MB-231 cells were examined [33]. For this purpose, MCF-7 (Figure 4A) and MDA-MB-231 (Figure 4B) cells were incubated with compounds 5u or 5s or Olaparib at equivalent concentrations. Equivalent levels of cleaved PARP1 were observed in both cell lines after treatment with compounds 5u or 5s or Olaparib.
Figure 4.
The effect of compounds 5u, 5s, and Olaparib on PARP1 cleavage (upper panel of A,B) phosphorylation of H2A-X (Ser139) (lower panel of A,B), and CASPASE-3 activity (C,D) in MCF-7 and MDA-MB-231 cells. Cancer cells were treated with or without compounds 5u, 5s,and Olaparib (1 or 10 µM). The protein levels after treatment were observed by western blot analysis. GAPDH was used as input control. The data are expressed as mean ± SEM. ** p ≤ 0.01; *** p ≤ 0.001.
Furthermore, phosphorylation of the histone variant H2AX on Ser139, forming pH2AX, is an early response of a cell to DNA double-strand breaks [34]. Stress-induced DNA damage results in rapid phosphorylation of Ser139 of H2AX by PI3K-like kinases such as DNA-PK, ATM, and ATR [35]. Olaparib treatment has also been reported to significantly increase the level of p-H2AX [36,37]. Treatment of MCF7 and MDA-MB-231 cells with compound 5u or 5s or Olaparib produced equivalent increases in the level of p-H2AX in both cell lines (Figure 4). CPP32 (CASPASE-3) is a prototypical caspase that is activated during apoptosis [38,39,40]. CPP32 activity was measured after treatment of both cell lines with compounds 5u or 5s or Olaparib (Figure 4C,D). Again, similar results were observed with all three compounds in both cell lines.
2.5. Compounds 5u and 5s Inhibit Oncogenicity and Decreases 3D Growth of Breast Cancer Cells
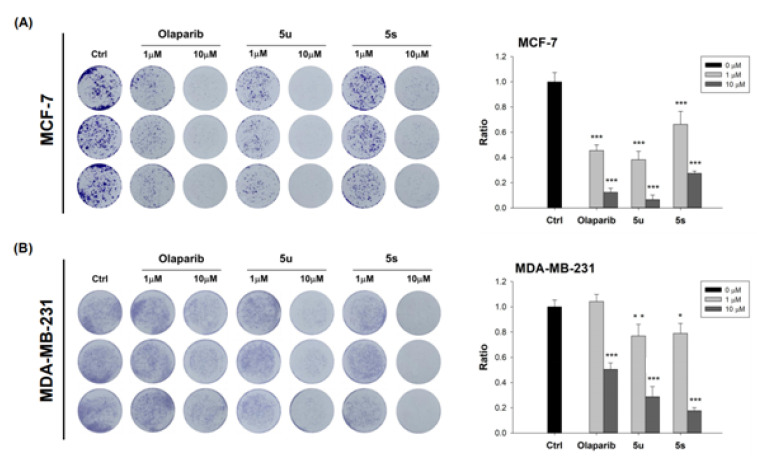
MCF-7 and MDA-MB-231 cells were first cultured in monolayers in 24-well plates for foci formation assays and treated with compounds 5u, 5s, or Olaparib. The foci formed by MCF-7 (Figure 5A) and MDA-MB-231 (Figure 5B) cells were decreased in a dose-dependent manner, approximately equivalently, by all three compounds. Hence, compounds 5u and 5s inhibit the oncogenicity of both ER+ and triple-negative breast cancer cells.
Figure 5.
Compounds 5u, 5s, and Olaparib inhibited the foci formation of MCF-7 (left panel of A) and MDA-MB-231 (left panel of B) cells. Visualization of foci formed was achieved by use of Crystal Violet. Inhibition of foci formation was also quantified (right panels of both A,B) The data are expressed as mean ± SEM. * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001.
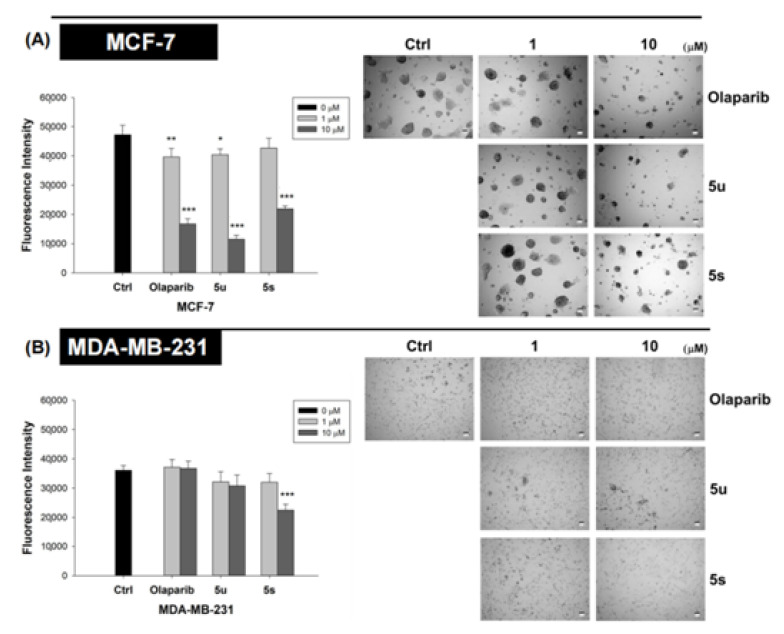
To examine the efficacy of these compounds in an ex vivo organoid model, we grew both MCF-7 and MDA-MB-231 cells in 3D Matrigel®. Again, compounds 5u and 5s exhibited roughly equivalent efficacy to Olaparib in inhibiting 3D growth of both cancer cell lines (Figure 6).
Figure 6.
Compounds 5u, 5s, and Olaparib inhibited the growth of MCF-7 (A) and MDA-MB-231 (B) breast cancer cells in 3-D Matrigel. Cells were grown in a medium containing 4% Matrigel. Cells were treated with vehicles or compounds 5s, 5u, or Olaprib (1 or 10 µM), after colonies had formed. Compounds were added into the culture medium every 3 days for a total of 2 cycles. Alamar Blue assay was used for quantification of live cells. The data are expressed as mean ± SEM. * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001.
2.6. In Silico Analysis of the Interaction of Compound 5s with the PARP1 Catalytic Domain
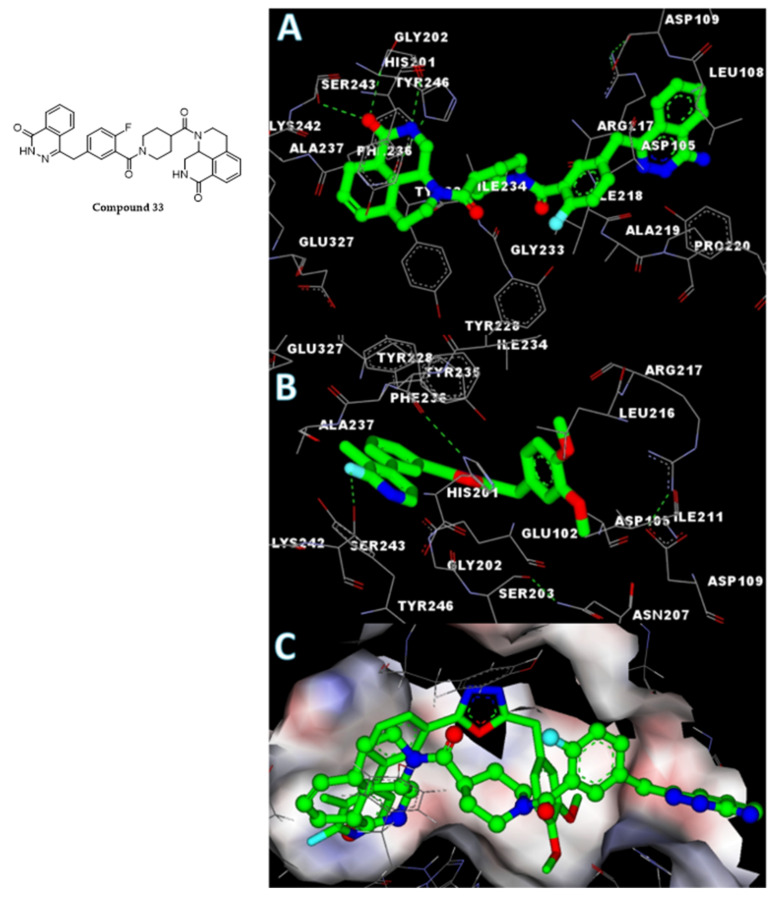
To understand the interaction of the novel oxadiazoles with the PARP1 catalytic domain, the co-crystal structure of compound 33 [(9aR)-1-[(1-{2-fluoro-5-[(4-oxo-3,4-dihydrophthalazin-1-yl)methyl]benzoyl}piperidin-4-yl)carbonyl]-1,2,3,8,9,9a-hexahydro-7H-benzo[de][1,7]naphthyridin-7-one] with the PARP1 catalytic domain (PDB ID: 4HHY) was analyzed [41]. Accelrys DS version 2.5 was used for the molecular docking utilizing the previously reported protocol [42,43]. Compound 33 and the active oxadiazole compound 5s were docked towards the catalytic domain of PARP1. The analysis of the results indicated that the core region of compound 33 called benzo[de][1,7]naphthyridin-7-one was occupying the active site of the catalytic domain of PARP1, which was similar when compared to the phenylic-pyridine group of active compound 5s, which showed high negative CDOCKER energy. The co-crystal ligand compound 33 appears to possess higher negative CDOCKER energy of 45.8 kcal/mole, when compared to the most active compound 5s, whose high negative CDOCKER energy was recorded as 28.1 kcal/mole. The detailed molecular interactions between the amino acid residues of the PARP1 catalytic domain and compound 33 or compound 5s are shown in Figure 7. The detailed molecular interactions of compound 5s, which exhibited higher binding affinities with the PARP1 catalytic domain by having stable H-bonds with Ser243, were also influenced by three intramolecular H-bonds between His201-Ala237, Asp105-Arg217, and Ser203-Asn207. p-p stacking of compound 5s with Tyr228, Tyr235, Phe236, His201, and Tyr246 by fluoro, a methyl-substituted pyridinyl-phenyl group contributed to the net and higher negative CDOCKER energy of the ligand and enzyme complex.
Figure 7.
Structure of compound 33 and in silico molecular interaction studies of co-crystal ligand ((A), compound 33) and compound 5s (B) with the PARP catalytic domain. The surface view of merged ligands is also presented for clarity (C). The protein bound ligands were presented in stick (compound 5s) and ball and stick (compound 33) mode for better visualization.
3. Materials and Methods
All chemicals and solvents were purchased from Sigma-Aldrich (Bangalore, India). The completion of reactions was monitored by pre-coated silica gel TLC plates and hexane/ethyl acetate used as an eluent. 1H and 13C NMR were recorded on an Agilent NMR spectrophotometer (400 MHz); TMS was used as an internal standard and CDCl3 and DMSO were used as solvents; chemical shifts are expressed as ppm. Mass spectra and HPLC were collected on Waters (Xevo G2-XS QTof) (Wilmslow, UK) and Acquity UPLC (waters column: Sunfire C18, 4.6 × 250 mm) (Milford, MA, USA), respectively.
3.1. General Procedure for the Synthesis of 2,5-Disubstituted-1,3,4-oxadiazole
A mixture of benzoic acid (1a) (1 mmol), and concentrated sulfuric acid (1 mmol) in 10 mL of ethanol was stirred and refluxed for 7 h. After completion of the reaction, the reaction mass was cooled and ethanol was removed by high vacuum pressure. Then, the reaction mixture was diluted with ethyl acetate and neutralized by bicarbonate solution. The organic layer was separated out and dried over anhydrous sodium sulphate, and concentrated under vacuum; ethyl benzoate was obtained as a white solid. To the ethyl benzoate (2a) (1 mmol), hydrazine hydrate (1 mmol) and 15 mL of ethanol were added and refluxed for 5 h. The completion of reaction was monitored by TLC and the traces of hydrazine hydrate were washed with water; acid hydrazide was obtained as a solid. To a mixture of acid hydrazide (3a) (1 mmol) and substituted acids (1 mmol), 6 mL of POCl3 was added and refluxed at 80 °C for 8 h. The completion of the reaction was monitored by TLC. The reaction mass was quenched with crushed ice and neutralized by potassium carbonate to pH 8. The solid obtained was filtered, washed with water, and subjected to column chromatography (hexane/ethyl acetate eluent)/recrystallized with suitable solvents to obtain pure oxadiazoles. 4a–c (1 mmol), substituted aryl/het.aryl boronic acids (Ar/Het.Ar-B(OH)2) (1.2 mmol) and potassium carbonate (3 mmol) were taken in a sealed tube containing H2O:dioxane:EtOH (1:5:1) solvent. The above mixture was stirred for 15 min at RT under an inert atmosphere (N2 atm). Then, a catalyst (Pd(dppf)Cl2) (0.1 mmol) was added to the above reaction mass and heated to 120 °C for 45 min. Aryl-substituted 1,3,4-oxadiazole (5a–x) was obtained and purified through the chromatographic technique using hexane/ethyl acetate as an eluent.
3.2. 2-(3-Bromophenyl)-5-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-1,3,4-oxadiazole (4a)
Off white solid; 82% yield.
3.3. 2-(3-Bromophenyl)-5-(4-methoxybenzyl)-1,3,4-oxadiazole (4b)
Off white solid; MP: 68–70 °C; 88% yield; 1H NMR (DMSO, 400 MHz): δ 8.01 (s, 1H), 7.89 (d, J = 7.6 Hz, 1H), 7.76 (d, J = 7.6 Hz, 1H), 7.56–7.42 (m, 1H), 7.26 (d, J = 8.4 Hz, 2H), 6.88 (d, J = 8.4 Hz, 2H), 4.24 (s, 3H), 3.69 (s, 3H); 13C NMR (DMSO, 100 MHz): δ 166.7, 163.3, 158.9, 135.0, 132.1, 130.5 (2C), 129.1, 126.5, 125.9, 125.86, 122.8, 114.6 (2C), 55.5, 30.4; MS: 344.02, m/z = 345.00 [M + H]+, 346.99 [M + 2H]+.
3.4. 2-(3-Bromophenyl)-5-(3,4-dimethoxybenzyl)-1,3,4-oxadiazole (4c)
Off white solid; MP: 74–76 °C; 84% yield; 1H NMR (DMSO, 400 MHz): δ 8.04 (s, 1H), 7.92 (d, J = 8 Hz, 1H), 7.79 (d, J = 8 Hz, 1H), 7.58–7.44 (m, 1H), 6.96 (s, 1H), 6.95–6.79 (m, 2H), 4.24 (s, 2H), 3.71 (s, 3H), 3.69 (s, 3H); 13C NMR (DMSO, 100 MHz): δ 166.6, 163.3, 149.3, 148.5, 135.0, 132.1, 129.1, 126.9, 126.0, 125.9, 122.8, 121.4, 113.2, 112.5, 56.0, 30.8; MS: 374.03, m/z = 375.01 [M + H]+, 377.01 [M + 2H]+.
3.5. 2-(3-(6-Chloro-5-methylpyridin-3-yl)phenyl)-5-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-1,3,4-oxadiazole (5a)
Off white solid; MP: 164–162 °C; 91% yield; 1H NMR (CDCl3, 400 MHz): δ 8.50 (s, 1H), 8.28 (s, 1H), 7.82 (s, 1H), 7.72–7.61 (m, 4H), 8.13 (d, J = 8 Hz, 1H), 6.99 (d, J = 8.8 Hz, 1H), 4.33 (s, 4H), 2.48 (s, 3H); 13CNMR (CDCl3, 100 MHz): δ 164.6,163.8, 161.2, 146.9, 145.2, 143.9, 137.9, 137.7, 134.6, 132.7, 130.0, 129.9, 126.5, 125.3, 124.9, 120.7, 118.1, 117.9, 116.9, 116.2, 64.6, 64.2, 19.8; MS: 405.09, m/z = 406.06 [M + H]+.
3.6. 2-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-5-(4′-(trifluoromethyl)-[1,1′-biphenyl]-3-yl)-1,3,4-oxadiazole (5b)
Brown solid; MP: 160–162 °C; 92% yield; 1H NMR (CDCl3, 400 MHz): δ 8.23–8.19 (m, 2H), 7.96–7.86 (m, 2H), 7.68–7.62 (m, 2H), 7.61–7.44 (m, 4H), 6.97 (d, J = 8 Hz, 1H), 4.31–4.28 (m, 4H); 13C NMR (CDCl3, 100 MHz): δ 164.5, 163.9, 149.1, 148.2, 146.9, 143.9, 138.9, 135.5, 134.6, 130.2, 129.9, 125.4, 124.9, 123.7, 120.7, 118.1, 116.9, 116.2, 64.6, 64.2; MS: 424.10, m/z = 425.09 [M + H]+.
3.7. N-Cyclopentyl-3′-(5-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-1,3,4-oxadiazol-2-yl)-[1,1′-biphenyl]-3-carboxamide (5c)
Off white solid; MP: 186–188 °C; 93% yield; 1H NMR (CDCl3, 400 MHz): δ 8.26 (s, 1H), 8.05–8.01 (m, 2H), 7.75–7.70 (m, 3H), 7.61–7.59 (m, 2H), 7.50–7.47 (m, 1H), 7.57–7.47 (m, 1H), 6.96 (d, J = 8 Hz, 1H), 6.44 (d, J = 4 Hz, 1H), 4.45–4.40 (m, 1H), 4.31–4.28 (m, 4H), 2.12–2.02 (m, 2H), 1.75–1.68 (m, 2H), 1.66–1.56 (m, 2H), 1.55–1.52 (m, 2H); 13C NMR (CDCl3, 100 MHz): δ 167.1, 164.4, 164.0, 146.8, 143.9, 141.3, 140.2, 135.7, 130.3, 129.9, 129.6, 129.1, 126.3, 125.9, 125.8, 125.4, 124.5, 120.7, 118.0, 116.9, 116.2, 64.6, 64.2, 51.9, 33.2, 23.9; MS: 467.18, m/z = 468.20 [M + H]+.
3.8. 2-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-5-(3′-methoxy-[1,1′-biphenyl]-3-yl)-1,3,4-oxadia-zole (5d)
Off white solid; MP: 118–120 °C; 93% yield; 1H NMR (CDCl3, 400 MHz): δ 8.32 (s, 1H), 8.08 (d, J = 8 Hz, 1H), 7.74 (d, J = 8 Hz, 1H), 7.66–7.56 (m, 1H), 7.41–7.73 (m, 1H), 7.25–7.22 (m, 1H), 7.17 (s, 1H), 7.00–6.93 (m, 1H), 4.33–4.32 (m, 4H), 3.88 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 164.4, 164.2, 160.1, 146.8, 143.9, 142.1, 141.4, 130.3, 129.9, 129.5, 125.7, 125.5, 124.5, 120.7, 119.7, 118.1, 117.1, 116.2, 113.3, 112.9, 64.6, 64.2, 55.4; MS: 386.12, m/z = 387.14 [M + H]+.
3.9. 2-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-5-(3-(naphthalen-1-yl)phenyl)-1,3,4-oxadiazole (5e)
Off white solid; MP: 156–158 °C; 97% yield; 1H NMR (CDCl3, 400 MHz): δ 8.23–8.19 (m, 1H), 7.95–7.86 (m, 3H), 7.67–7.45 (m, 9H), 6.97 (d, J = 8 Hz, 1H), 4.30–4.29 (m, 4H); 13C NMR (CDCl3, 100 MHz): δ 164.4, 164.2, 146.8, 143.9, 141.8, 138.8, 137.6, 133.8, 133.2, 131.4, 129.1, 128.3, 127.1, 126.4, 125.9, 125.8, 125.6, 125.1, 120.7, 118.0, 117.0, 116.2, 64.6, 64.2; MS: 406.13, m/z = 407.12 [M + H]+.
3.10. 2-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-5-(3-(pyridin-4-yl)phenyl)-1,3,4-oxadiazole (5f)
Brown solid; MP: 194–196 °C; 95% yield; 1H NMR (CDCl3, 400 MHz): δ 8.71–8.70 (m, 2H), 8.36 (s, 1H), 8.16 (d, J = 8 Hz, 1H), 7.78 (d, J = 8 Hz, 1H), 7.64–7.61 (m, 3H), 7.58–7.56 (m, 2H), 6.98 (d, J = 8 Hz, 1H), 4.32–4.31 (m, 4H); 13C NMR (CDCl3, 100 MHz): δ 164.5, 163.7, 150.4, 147.1, 146.9, 143.9, 139.2, 129.9, 127.2, 125.3, 124.9, 121.7, 120.7, 118.1, 116.9, 116.2, 64.6, 64.2; MS: 357.11, m/z = 358.09 [M + H]+.
3.11. 2-(4-Methoxybenzyl)-5-(3-(pyrimidin-5-yl)phenyl)-1,3,4-oxadiazole (5g)
Pink solid; MP: 90–92 °C; 86% yield; 1H NMR (CDCl3, 400 MHz): δ 9.24 (s, 1H), 8.99 (s, 2H), 8.21 (s, 1H), 8.09 (d, J = 6 Hz, 1H), 7.75–7.59 (m, 2H), 7.28 (d, J = 7.2 Hz, 2H), 6.88 (d, J = 6.8 Hz, 2H), 4.23 (s, 2H), 3.78 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 165.9, 164.4, 159.1, 158.0, 154.9 (2C), 135.3, 133.2, 130.2, 130.0, 129.9 (2C), 127.2, 125.6, 125.2 (2C), 114.4 (2C), 55.3, 31.1; MS: 344.36, m/z = 345.08 [M + H]+.
3.12. 2-(3,4-Dimethoxybenzyl)-5-(3-(pyrimidin-5-yl)phenyl)-1,3,4-oxadiazole (5h)
Brown solid; MP: 98–100 °C; 90% yield; 1H NMR (CDCl3, 400 MHz): δ 9.24 (s, 1H), 8.99 (s, 2H), 8.22 (s, 1H), 8.08 (d, J = 6.8 Hz, 1H), 7.77–7.59 (m, 2H), 6.95–6.78 (m, 3H), 4.23 (s, 2H), 3.85 (s, 6H); 13C NMR (CDCl3, 100 MHz): δ 161.2, 159.7, 153.2, 150.2, 144.5, 143.8, 130.5, 128.5, 125.5, 125.3, 124.0, 122.4, 121.2, 120.5, 116.3, 107.2, 106.8, 51.2, 26.8; MS: 374.39, m/z = 375.09 [M + H]+.
3.13. 2-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-5-(3-(pyrimidin-5-yl)phenyl)-1,3,4-oxadiazole (5i)
Off white solid; MP: 190–192 °C; 91% yield; 1H NMR (CDCl3, 400 MHz): δ 9.26 (s, 1H), 9.03 (s, 2H), 8.31 (s, 1H), 8.18 (d, J = 8 Hz, 1H), 7.74–7.62 (m, 4H), 6.98 (d, J = 8 Hz, 1H), 4.32–4.27 (m, 4H); 13C NMR (CDCl3, 100 MHz): δ 164.6, 163.5, 157.9, 154.9, 146.9, 143.9, 135.4, 133.4, 130.4, 129.9, 127.2, 125.3, 125.2, 120.7, 118.1, 116.8, 116.2, 64.2, 64.0; MS: 358.10, m/z = 359.11 [M + H]+.
3.14. 2-(3,4-Dimethoxybenzyl)-5-(3-(4,5-dimethylpyridin-3-yl)phenyl)-1,3,4-oxadiazole (5j)
Brown solid; MP: 90–92 °C; 90% yield; 1H NMR (CDCl3, 400 MHz): δ 8.56 (s, 1H), 8.19 (s, 1H), 7.97 (d, J = 7.6 Hz, 1H), 7.69 (d, J = 8.0 Hz, 1H), 7.64 (s, 1H), 7.62–7.48 (m, 1H), 6.96–6.74 (m, 3H), 4.22 (s, 2H), 3.86 (s, 3H), 3.85 (s, 3H), 2.54 (s, 3H), 2.35 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 165.6, 164.9, 156.7, 149.3, 148.5, 144.5, 139.0, 135.7, 133.0, 131.6, 130.0, 129.7, 126.1, 125.9, 125.2, 124.6, 121.0, 111.9, 111.5, 55.9, 31.5, 22.2, 19.2; MS: 401.46, m/z = 402.12 [M + H]+.
3.15. 2-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-5-(3-(4,5-dimethylpyridin-3-yl)phenyl)-1,3,4-oxadiazole (5k)
Off white solid; MP: 198–200 °C; 97% yield; 1H NMR (CDCl3, 400 MHz): δ 8.60 (s, 1H), 8.29 (S, 1H), 8.09 (d, J = 8 Hz, 1H), 7.73–7.60 (m, 5H), 6.99 (d, J = 8 Hz, 1H), 4.33–4.32 (m, 4H), 2.56 (s, 3H), 2.37 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 164.5, 163.9, 156.8, 146.8, 144.6, 143.9, 139.1 135.7, 133.1, 131.6, 129.9, 129.7, 125.9, 125.2, 124.7, 120.7, 118.1, 117.0, 116.2, 64.6, 64.2, 19.2; MS: 385.14, m/z = 386.15 [M + H]+.
3.16. 2-(4-Methoxybenzyl)-5-(3-(pyridin-3-yl)phenyl)-1,3,4-oxadiazole (5l)
Brown thick mass; 84% yield; 1H NMR (CDCl3, 400 MHz): δ 8.85 (s, 1H), 8.61 (s, 1H), 8.20 (s, 1H), 8.06–7.86 (m, 2H), 7.74–7.52 (m, 2H), 7.39 (s, 1H), 7.26 (s, 2H), 6.87 (s, 2H), 4.22, (s, 2H), 3.77 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 165.8, 164.8, 159.0, 148.9, 148.0, 138.7, 135.5, 134.6, 130.2, 129.9 (2C), 126.4, 125.7, 125.4 (2C), 124.8, 123.7, 114.4 (2C), 55.3, 31.1; MS: 343.38, m/z = 344.11 [M + H]+.
3.17. 2-(3,4-Dimethoxybenzyl)-5-(3-(pyridin-3-yl)phenyl)-1,3,4-oxadiazole (5m)
Yellow solid; MP: 78–80 °C; 88% yield; 1H NMR (CDCl3, 400 MHz): δ 8.86 (s, 1H), 8.63 (s, 1H), 8.21 (s, 1H), 8.02 (d, J = 7.2 Hz, 1H), 7.91 (d, J = 7.6 Hz, 1H), 7.71 (d, J = 7.2 Hz, 1H), 7.58 (t, J = 7.6 Hz, 1H), 7.40 (d, J = 4.8 Hz, 1H), 6.96–6.77 (m, 3H), 4.22 (s, 2H), 3.86 (s, 3H), 3.85 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 165.7, 164.8, 149.3, 149.0, 148.5, 148.1, 138.7, 135.5, 134.5, 130.2, 129.8, 126.4, 126.1, 125.4, 124.8, 123.7, 121.0, 111.9, 111.5, 55.9, 31.5; MS: 373.40, m/z = 406.12 [M + CH3OH + H]+.
3.18. 2-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-5-(3-(pyridin-3-yl)phenyl)-1,3,4-oxadiazole (5n)
Off white solid; MP: 138–140 °C; 95% yield; 1H NMR (CDCl3, 400 MHz): δ 8.91 (s, 1H), 8.65 (s, 1H), 8.32 (s, 1H), 7.96 (d, J = 8 Hz, 1H), 7.74–7.66 (m, 4H), 7.42 (s, 1H), 6.99 (d, J = 8 Hz, 1H), 4.33 (s, 4H); 13C NMR (CDCl3, 100 MHz): δ 164.5, 163.9, 149.1, 148.2, 146.9, 143.9, 138.9, 135.5, 134.6, 130.2, 129.9, 126.4, 125.4, 124.9, 123.7, 120.7, 118.1, 116.9, 116.2, 64.6, 64.2; MS: 357.11, m/z = 358.09 [M + H]+.
3.19. 2-(3-(6-Fluoro-5-methylpyridin-3-yl)phenyl)-5-(4-methoxybenzyl)-1,3,4-oxadiazole (5o)
Off white solid; MP: 86–88 °C; 91% yield; 1H NMR (CDCl3, 400 MHz): δ 8.25 (s, 1H), 8.17 (s, 1H), 8.00 (d, J = 7.6 Hz, 1H), 7.82 (d, J = 8.8 Hz, 1H), 7.67 (d, J = 7.6 Hz, 1H), 7.64–7.50 (m, 1H), 7.29 (d, J = 8.4 Hz, 2H), 6.89 (d, J = 8.4 Hz, 2H), 4.24 (s, 2H), 3.79 (s, 3H), 2.37 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 165.8, 164.7, 163.3, 159.1, 143.0, 142.8, 140.3, 140.25, 137.9, 133.6, 130.0, 129.9 (2C), 129.8, 126.2, 125.7, 125.3, 124.8, 119.9, 119.6, 114.4 (2C), 55.3, 31.1, 14.5; MS: 375.39, m/z = 376.11 [M + H]+.
3.20. 2-(3,4-Dimethoxybenzyl)-5-(3-(6-fluoro-5-methylpyridin-3-yl)phenyl)-1,3,4-oxadiazole (5p)
Brown solid; MP: 91–93 °C; 95% yield; 1H NMR (CDCl3, 400 MHz): δ 8.14–8.00 (m, 2H), 7.96 (s, 1H), 7.66–7.52 (m, 1H), 7.46 (d, J = 7.6 Hz, 1H), 7.07 (d, J = 5.2 Hz, 1H), 6.96–6.77 (m, 3H), 4.22 (s, 2H), 3.86 (s, 3H), 3.85 (s, 3H), 2.20 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 165.8, 164.8, 163.3, 149.3, 148.6, 142.9, 142.8, 140.4, 140.3, 137.9, 133.6, 130.9, 130.1, 129.8, 126.2, 126.0, 125.3, 121.0, 120.0, 119.7, 111.9, 111.5, 55.9, 31.5, 14.5; MS: 405.42, m/z = 406.12 [M + H]+.
3.21. 2-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-5-(3-(6-fluoro-5-methylpyridin-3-yl)phenyl)-1,3,4-oxadiazole (5q)
Off white solid; MP: 144–146 °C; 94% yield; 1H NMR (CDCl3, 400 MHz): δ 8.27 (d, J = 8 Hz, 1H), 8.11 (d, J = 8 Hz, 1H), 7.86 (d, J = 8 Hz, 1H), 7.70–7.59 (m, 2H), 6.99 (d, J = 8 Hz, 1H), 4.33–4.32 (m, 4H); 13C NMR (CDCl3, 100 MHz): δ 164.5, 163.8, 163.3, 146.9, 143.9, 143.1, 142.9, 140.4, 137.9, 133.7, 130.0, 129.9, 126.2, 125.3, 124.9, 119.7, 118.1, 116.9, 116.2, 115.9, 64.6, 64.2, 14.6; MS: 389.11, m/z = 390.09 [M + H]+.
3.22. 2-(3-(2-Fluoro-3-methylpyridin-4-yl)phenyl)-5-(4-methoxybenzyl)-1,3,4-oxadiazole (5r)
White solid; MP: 94–96 °C; 87% yield; 1H NMR (CDCl3, 400 MHz): δ 8.14–8.01 (m, 2H), 7.96 (s, 1H), 7.67–7.53 (m, 1H), 7.47 (d, J = 8 Hz, 1H), 7.28 (d, J = 8.8 Hz, 2H), 7.09 (d, J =5.2 Hz, 1H), 6.88 (d, J = 8.4 Hz, 2H), 4.23 (s, 2H), 3.79 (s, 3H), 2.21 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 166.0, 164.6, 163.9, 159.1, 152.9, 144.2, 144.1, 139.0, 131.7, 129.9 (2C), 129.3, 126.7, 125.5, 124.3, 122.3, 122.28, 117.6, 117.3, 114.4 (2C), 55.3, 31.1, 11.9; MS: 375.39, m/z = 376.09 [M + H]+.
3.23. 2-(3,4-Dimethoxybenzyl)-5-(3-(2-fluoro-3-methylpyridin-4-yl)phenyl)-1,3,4-oxadiazole (5s)
Brown solid; MP: 84–86 °C; 94% yield; 1H NMR (CDCl3, 400 MHz): δ 8.24 (s, 1H), 8.17 (s, 1H), 7.99 (d, J = 7.6 Hz, 1H), 7.82 (d, J = 8.8 Hz, 1H), 7.67 (d, J = 8.4 Hz, 1H), 7.64–7.50 (m, 1H), 6.97–6.78 (m, 3H), 4.23 (s, 2H), 3.86 (s, 3H), 3.85 (s, 3H), 2.36 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 165.8, 164.6, 163.9, 152.8, 149.3, 148.6, 144.3, 144.1, 139.0, 131.7, 130.8, 129.3, 126.7, 126.0, 122.3, 122.26, 121.0, 117.6, 117.3, 111.9, 111.5, 55.9, 31.5, 11.9; MS: 405.42, m/z = 406.13 [M + H]+.
3.24. 2-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-5-(3-(2-fluoro-3-methylpyridin-4-yl)phenyl)-1,3,4-oxadiazole (5t)
Off white solid; MP: 128–130 °C; 91% yield; 1H NMR (CDCl3, 400 MHz): δ 8.17 (d, J = 8 Hz, 1H), 8.10–8.06 (m, 1H), 7.65–7.60 (m, 3H), 7.49–7.47 (m, 1H), 7.11 (d, J = 4 Hz, 1H), 6.99–6.87 (m, 1H), 4.33–4.30 (m, 4H), 2.23 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 164.6, 163.9, 163.6, 161.6, 152.9, 146.9, 144.4, 143.9, 139.1, 131.6, 129.4, 129.0, 126.8, 124.6, 122.3, 120.7, 120.5, 118.1, 117.6, 117.3, 116.9, 116.2, 114.0, 64.6, 64.2, 14.1; MS: 389.11, m/z = 390.13 [M + H]+.
3.25. 3′-(5-(4-Methoxybenzyl)-1,3,4-oxadiazol-2-yl)-[1,1′-biphenyl]-3-carbaldehyde (5u)
Yellow thick mass; 91% yield; 1H NMR (CDCl3, 400 MHz): δ 10.09 (s, 1H), 8.26 (s, 1H), 8.12 (s, 1H), 8.01 (d, J = 7.6 Hz, 1H), 7.95–7.82 (m, 2H), 7.76 (d, J = 7.6 Hz, 1H), 7.70–7.51 (m, 2H), 7.29 (d, J = 8.4 Hz, 2H), 6.89 (d, J = 8.4 Hz, 2H), 4.24 (s, 2H), 3.78 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 192.1, 165.9, 164.9, 159.0, 140.8, 140.6, 137.0, 133.0, 130.3, 129.9 (2C), 129.72, 129.7, 129.3, 128.0, 126.3, 125.7, 125.4, 124.6, 114.4 (2C), 55.3, 31.1; MS: 370.40, m/z = 371.19 [M + H]+.
3.26. 3′-(5-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-1,3,4-oxadiazol-2-yl)-[1,1′-biphenyl]-3-carbal-dehyde (5v)
Light yellow solid; MP: 158–160 °C; 94% yield; 1H NMR (CDCl3, 400 MHz): δ 10.11 (s, 1H), 8.35 (s, 1H), 8.16–8.11 (m, 1H), 7.93–7.90 (m, 1H), 7.80–7.78 (m, 1H), 7.67–7.60 (m, 5H), 7.42–7.38 (m, 1H), 7.00–6.98 (m, 1H), 4.34–4.30 (m, 4H); 13C NMR (CDCl3, 100 MHz): δ 192.1, 164.5, 163.9, 146.9, 143.9, 140.9, 140.7, 137.0, 133.1, 130.3, 129.8, 129.7, 129.3, 128.2, 126.3, 125.4, 120.5, 118.1, 116.1, 64.6, 64.2; MS: 384.11, m/z = 385.12 [M + H]+.
3.27. 1-(5-(3-(5-(4-Methoxybenzyl)-1,3,4-oxadiazol-2-yl)phenyl)thiophen-2-yl)ethanone (5w)
Off white solid; 80–82 °C; 90% yield; 1H NMR (CDCl3, 400 MHz): δ 8.27 (s, 1H), 7.97 (d, J = 6.4 Hz, 1H), 7.76 (d, J = 6.4 Hz, 1H), 7.67 (s, 1H), 7.57–7.35 (m, 2H), 7.29 (d, J = 6.8 Hz, 2H), 6.89 (d, J = 6.8 Hz, 2H), 4.23 (s, 2H), 3.79 (s, 3H), 2.57 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 190.4, 165.9, 164.5, 159.1, 150.8, 143.9, 134.3, 133.3, 129.9, 129.8 (2C), 129.2, 127.0, 125.7, 124.9, 124.8, 124.3, 114.4 (2C), 55.3, 31.1, 26.6; MS: 390.45, m/z = 391.05 [M + H]+.
3.28. 1-(5-(3-(5-(3,4-Dimethoxybenzyl)-1,3,4-oxadiazol-2-yl)phenyl)thiophen-2-yl)ethanone (5x)
Brown solid; 102–104 °C; 95% yield; 1H NMR (CDCl3, 400 MHz): δ 8.26 (s, 1H), 7.98 (s, 1H), 7.81–7.61 (m, 2H), 7.57–7.34 (m, 2H), 6.87 (d, J = 12.4 Hz, 3H), 4.23 (s, 2H), 3.86 (s, 6H), 2.56 (s, 3H); 13C NMR (CDCl3, 100 MHz): δ 190.5, 165.8, 164.5, 150.7, 149.3, 148.6, 143.9, 134.3, 133.3, 130.8, 129.9, 129.2, 127.0, 126.1, 124.8, 124.3, 121.0, 111.9, 111.5, 55.9, 31.5, 26.6; MS: 420.48, m/z = 421.06 [M + H]+.
3.29. MCF-7 and MDA-MB-231 Cell Viability Assay
MCF-7 and MDA-MB-231 (2000) cells were cultured in MEM or Leibovitz’s L-15 medium enriched with 2% FBS and maintained in a humidified atmosphere of 5% CO2 at 37 °C [44]. DMSO dissolved compounds were kept as a stock solution, and diluted with a cell culture medium to the desired concentration. Cancer cells (4 × 103) were grown overnight in 96-well plates, cultured, and treated with oxadiazoles or Olaparib at 0, 0.01, 0.1, 10, 100, and 1000 µM concentrations for 72 h. The inhibitory effect of the compounds was assessed using Alamar Blue reagent.
3.30. Assay of PARP Activity
PARP activity was measured using the PARP/Apoptosis Universal Colorimetric Assay Kit (4677-096-K, R&D Systems Ltd., Abingdon, UK) [45]. The enzyme inhibition assay uses the histone proteins immobilized 96-well plate, which measures the incorporation of ADP-ribose (biotin-tagged), followed by detection using streptavidin-conjugated horse-radish peroxidase, and finally the color development after the addition of substrate. Oxadiazoles were incubated with the recombinant human PARP enzyme prior to detection.
3.31. Western Blot Analysis
Western blot analysis was performed to determine the levels of PARP1 cleavage and p-H2AX in MCF-7 and MDA-MB-231 cells using previously reported methods. Cells at 50–60% confluence were treated with compounds and incubated for 60 h in 2% FBS. MCF-7 and MDA-MB-231 cells were also treated with Olaparib, 5u, or 5s and the levels of cleaved PARP1 and p-H2A-X were determined using the respective antibodies.
3.32. Caspase-3 Activity Assay
Caspase-3 activity was measured by Caspase-Glo® 3 assay kit (Promega Madison, WI, USA) according to the manufacturer’s instructions. MCF-7 and MDA-MB-231 cells were seeded in 96-well plates and incubated overnight. Serum-free media or media with compounds were added to cells after 12 h. After a 24h incubation period, Caspase-Glo® 3 reagent and media (1:1) was added into each well and incubated for 30 min in the dark. Luminescence activity (absorbance) was measured using a TECAN reader.
3.33. Foci Formation Assay
MCF-7 and MDA-MB-231 cells were plated in six-well plates in 2 mL media with or without compounds and cultured. When foci had formed, MCF-7 and MDA-MB-231 cells were washed with PBS and fixed with ice-cold methanol for 10 min at 4°C. Methanol was removed and crystal violet solution was used to stain foci at room temperature for 10 min. The resultant solution was removed; immobilized MCF-7 and MDA-MB-231 cells were rinsed with distilled water and dried at room temperature.
3.34. Molecular Docking Analysis
The molecular docking analysis was performed using the CDOCKER program of Accelrys DS version 2.5 [46]. The co-crystal structures of compound 33 and the PARP1 catalytic domain (PDB ID: 4HHY) were retrieved from RCSB and the binding site of the co-crystal ligand was defined. The protein preparation tool was applied and water was removed from the crystal. The CFF force field was applied to the protein. The lowest energy ring conformation was kept for each compound. We docked compound 33 and compound 5s (Figure 7) towards the catalytical domain of PARP1 using a docking protocol of the CDOCKER program. A cut-off based on the docking score of reference PARP-1 inhibitors was used, and ligands with the highest score were subjected to visualization using Accelrys graphic interface.
3.35. Statistical Analysis
Data were expressed as mean ± SD and statistical analysis was performed using unpaired Student’s t-test. A p-value of less than 0.05 was considered statistically significant. The graphical presentations were prepared using GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA, USA). p < 0.05 was considered statistically significant, * p < 0.05; ** p < 0.005.
4. Conclusions
In conclusion, newer and diverse structure added-oxadiazoles were designed and synthesized, leading to the identification of structures that produce loss of cell viability in breast cancer cells. Furthermore, the tested oxadiazoles showed dose-dependent inhibition of the catalytic activity of PARP-1 in a cell-free system. Additionally, functional studies of the tested compounds revealed that they may be an initial structure in the drug-development process, wherein inhibition of PARP-1 activity is desirable.
Supplementary Materials
The following are available online, 1H NMR, 13C-NMR, mass spectra of synthesized molecules and their IC50 values determined against human breast cancer cells.
Author Contributions
Conceptualization, D.V., S.R., P.E.L., J.-R.Y. and B.B.; methodology, D.V., S.S.G., D.D. and B.B.; data curation, S.R. and V.P.; validation, V.P.; formal analysis, S.R., V.P., J.-R.Y., B.B. and P.E.L.; software, resources, investigation, visualization, supervision, project administration, funding acquisition, writing, P.E.L. and B.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Council of Scientific and Industrial Research, DBT-NER, and the Vision Group on Science and Technology (CESEM), Government of Karnataka. This work was also supported by the Shenzhen Key Laboratory of Innovative Oncotherapeutics (ZDSYS20200820165400003) (Shenzhen Science and Technology Innovation Commission), China; the Shenzhen Development and Reform Commission Subject Construction Project ([2017]1434), China; the Overseas Research Cooperation Project (HW2020008) (Tsinghua Shenzhen International Graduate School), China; the Tsinghua University Stable Funding Key Project (WDZC20200821150704001); the Shenzhen Bay Laboratory (21310031), China; and the TBSI Faculty Start-up Funds, China. D.V. and S.S.G. thank the DST PhD Fellowship from KSTePS, Karnataka, and the University of Mysore, for providing the SC/ST fellowship, respectively.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
The reported PARP inhibitors are available from Dr. Basappa Lab.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Economopoulou P., Dimitriadis G., Psyrri A. Beyond BRCA: New hereditary breast cancer susceptibility genes. Cancer Treat. Rev. 2015;41:1–8. doi: 10.1016/j.ctrv.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Weil M.K., Chen A.P. PARP inhibitor treatment in ovarian and breast cancer. Curr. Probl. Cancer. 2011;35:7–50. doi: 10.1016/j.currproblcancer.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 4.Oh S.Y., Rahman S., Sparano J.A. Perspectives on PARP inhibitors as pharmacotherapeutic strategies for breast cancer. Expert Opin. Pharmacother. 2021;22:981–1003. doi: 10.1080/14656566.2021.1876662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsumoto K., Nishimura M., Onoe T., Sakai H., Urakawa Y., Onda T., Yaegashi N. PARP inhibitors for BRCA wild type ovarian cancer; gene alterations, homologous recombination deficiency and combination therapy. Jpn. J. Clin. Oncol. 2019;49:703–707. doi: 10.1093/jjco/hyz090. [DOI] [PubMed] [Google Scholar]
- 6.Krishnakumar R., Kraus W.L. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol. Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morales J., Li L., Fattah F.J., Dong Y., Bey E.A., Patel M., Gao J., Boothman D.A. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit. Rev. Eukaryot. Gene Expr. 2014;24:15–28. doi: 10.1615/CritRevEukaryotGeneExpr.2013006875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandya K.G., Patel M.R., Lau-Cam C.A. Comparative study of the binding characteristics to and inhibitory potencies towards PARP and in vivo antidiabetogenic potencies of taurine, 3-aminobenzamide and nicotinamide. J. Biomed. Sci. 2010;17((Suppl. S1)):S16. doi: 10.1186/1423-0127-17-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fongmoon D., Shetty A.K., Basappa, Yamada S., Sugiura M., Kongtawelert P., Sugahara K. Chondroitinase-mediated degradation of rare 3-O-sulfated glucuronic acid in functional oversulfated chondroitin sulfate K and E. J. Biol. Chem. 2007;21:36895–36904. doi: 10.1074/jbc.M707082200. [DOI] [PubMed] [Google Scholar]
- 10.Nirvanappa A.C., Mohan C.D., Rangappa S., Ananda H., Sukhorukov A.Y., Shanmugam M.K., Sundaram M.S., Nayaka S.C., Girish K.S., Chinnathambi A., et al. Novel Synthetic Oxazines Target NF-κB in Colon Cancer In Vitro and Inflammatory Bowel Disease In Vivo. PLoS ONE. 2016;11:e0163209. doi: 10.1371/journal.pone.0163209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCann K.E. Advances in the use of PARP inhibitors for BRCA1/2-associated breast cancer: Talazoparib. Future Oncol. 2019;15:1707–1715. doi: 10.2217/fon-2018-0751. [DOI] [PubMed] [Google Scholar]
- 12.Henneman L., van Miltenburg M.H., Michalak E.M., Braumuller T.M., Jaspers J.E., Drenth A.P., de Korte-Grimmerink R., Gogola E., Szuhai K., Schlicker A., et al. Selective resistance to the PARP inhibitor olaparib in a mouse model for BRCA1-deficient metaplastic breast cancer. Proc. Natl. Acad. Sci. USA. 2015;112:8409–8414. doi: 10.1073/pnas.1500223112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russo A.L., Kwon H.C., Burgan W.E., Carter D., Beam K., Weizheng X., Zhang J., Slusher B.S., Chakravarti A., Tofilon P.J., et al. In vitro and in vivo radiosensitization of glioblastoma cells by the poly (ADP-ribose) polymerase inhibitor E7016. Clin. Cancer Res. 2009;15:607–612. doi: 10.1158/1078-0432.CCR-08-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Awada A., Campone M., Varga A., Aftimos P., Frenel J.S., Bahleda R., Gombos A., Bourbouloux E., Soria J.C. An open-label, dose-escalation study to evaluate the safety and pharmacokinetics of CEP-9722 (a PARP-1 and PARP-2 inhibitor) in combination with gemcitabine and cisplatin in patients with advanced solid tumors. Anticancer Drugs. 2016;27:342–348. doi: 10.1097/CAD.0000000000000336. [DOI] [PubMed] [Google Scholar]
- 15.Paldino E., Cardinale A., D’Angelo V., Sauve I., Giampà C., Fusco F.R. Selective Sparing of Striatal Interneurons after Poly (ADP-Ribose) Polymerase 1 Inhibition in the R6/2 Mouse Model of Huntington’s Disease. Front. Neuroanat. 2017;11:61. doi: 10.3389/fnana.2017.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tong Y., Bouska J.J., Ellis P.A., Johnson E.F., Leverson J., Liu X., Marcotte P.A., Olson A.M., Osterling D.J., Przytulinska M., et al. Synthesis and evaluation of a new generation of orally efficacious benzimidazole-based poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors as anticancer agents. J. Med. Chem. 2009;52:6803–6813. doi: 10.1021/jm900697r. [DOI] [PubMed] [Google Scholar]
- 17.Priya B.S., Swamy S.N., Tejesvi M.V., Basappa, Sarala G., Gaonkar S.L., Naveen S., Prasad J.S., Rangappa K.S. Synthesis, characterization, antimicrobial and single crystal X-ray crystallographic studies of some new sulfonyl, 4-chloro phenoxy benzene and dibenzoazepine substituted benzamides. Eur. J. Med. Chem. 2006;41:1262–1270. doi: 10.1016/j.ejmech.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Lee J.H., Mohan C.D., Basappa S., Rangappa S., Chinnathambi A., Alahmadi T.A., Alharbi S.A., Kumar A.P., Sethi G., Ahn K.S., et al. The IκB Kinase Inhibitor ACHP Targets the STAT3 Signaling Pathway in Human Non-Small Cell Lung Carcinoma Cells. Biomolecules. 2019;9:875. doi: 10.3390/biom9120875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voronkov A., Holsworth D.D., Waaler J., Wilson S.R., Ekblad B., Perdreau-Dahl H., Dinh H., Drewes G., Hopf C., Morth J.P., et al. Structural basis and SAR for G007-LK, a lead stage 1,2,4-triazole based specific tankyrase 1/2 inhibitor. J. Med. Chem. 2013;56:3012–3023. doi: 10.1021/jm4000566. [DOI] [PubMed] [Google Scholar]
- 20.Ren H., Bakas N.A., Vamos M., Chaikuad A., Limpert A.S., Wimer C.D., Brun S.N., Lambert L.J., Tautz L., Celeridad M., et al. Design, Synthesis, and Characterization of an Orally Active Dual-Specific ULK1/2 Autophagy Inhibitor that Synergizes with the PARP Inhibitor Olaparib for the Treatment of Triple-Negative Breast Cancer. J. Med. Chem. 2020;63:14609–14625. doi: 10.1021/acs.jmedchem.0c00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malojirao V.H., Girimanchanaika S.S., Shanmugam M.K., Sherapura A., Dukanya, Metri P.K., Vigneshwaran V., Chinnathambi A., Alharbi S.A., Rangappa S., et al. Novel 1,3,4-oxadiazole Targets STAT3 Signaling to Induce Antitumor Effect in Lung Cancer. Biomedicines. 2020;8:368. doi: 10.3390/biomedicines8090368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dukanya, Shanmugam M.K., Rangappa S., Metri P.K., Mohan S., Basappa, Rangappa K.S. Exploring the newer oxadiazoles as real inhibitors of human SIRT2 in hepatocellular cancer cells. Bioorg. Med. Chem. Lett. 2020;30:127330. doi: 10.1016/j.bmcl.2020.127330. [DOI] [PubMed] [Google Scholar]
- 23.Mohan C.D., Anilkumar N.C., Rangappa S., Shanmugam M.K., Mishra S., Chinnathambi A., Alharbi S.A., Bhattacharjee A., Sethi G., Kumar A.P., et al. Novel 1,3,4-Oxadiazole Induces Anticancer Activity by Targeting NF-κB in Hepatocellular Carcinoma Cells. Front. Oncol. 2018;8:42. doi: 10.3389/fonc.2018.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey V., Wang B., Mohan C.D., Raquib A.R., Rangappa S., Srinivasa V., Fuchs J.E., Girish K.S., Zhu T., Bender A., et al. Discovery of a small-molecule inhibitor of specific serine residue BAD phosphorylation. Proc. Natl. Acad. Sci. USA. 2018;115:E10505–E10514. doi: 10.1073/pnas.1804897115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang M., Wang B., Chong Q.Y., Pandey V., Guo Z., Chen R.M., Wang L., Wang Y., Ma L., Kumar A.P., et al. A novel small-molecule inhibitor of trefoil factor 3 (TFF3) potentiates MEK1/2 inhibition in lung adenocarcinoma. Oncogenesis. 2019;8:65. doi: 10.1038/s41389-019-0173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anusha S., Mohan C.D., Ananda H., Baburajeev C.P., Rangappa S., Mathai J., Fuchs J.E., Li F., Shanmugam M.K., Bender A., et al. Adamantyl-tethered-biphenylic compounds induce apoptosis in cancer cells by targeting Bcl homologs. Bioorg. Med. Chem. Lett. 2016;26:1056–1060. doi: 10.1016/j.bmcl.2015.12.026. [DOI] [PubMed] [Google Scholar]
- 27.Rangappa K.S., Basappa. New cholinesterase inhibitors: Synthesis and structure–activity relationship studies of 1,2-benzisoxazole series and novel imidazolyl-d2-isoxazolines. J. Phys. Org. Chem. 2005;18:773–778. doi: 10.1002/poc.936. [DOI] [Google Scholar]
- 28.Basappa, Kavitha C.V., Rangappa K.S. Simple and an efficient method for the synthesis of 1-[2-dimethylamino-1-(4-methoxy-phenyl)-ethyl]-cyclohexanol hydrochloride: (+/−) venlafaxine racemic mixtures. Bioorg. Med. Chem. Lett. 2004;14:3279–3281. doi: 10.1016/j.bmcl.2004.03.098. [DOI] [PubMed] [Google Scholar]
- 29.Satoh M., Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 30.Gilardini Montani M.S., Prodosmo A., Stagni V., Merli D., Monteonofrio L., Gatti V., Gentileschi M.P., Barilà D., Soddu S. ATM-depletion in breast cancer cells confers sensitivity to PARP inhibition. J. Exp. Clin. Cancer Res. 2013;32:95. doi: 10.1186/1756-9966-32-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deepti Sharma L.D.F., Sivaraman Padavattan C.R., Geifman-Shochat S., Liu C.F., Davey C.A. PARP1 exhibits enhanced association and catalytic efficiency with γH2A.X-nucleosome. Nat. Commun. 2019;10:5751. doi: 10.1038/s41467-019-13641-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohan C.D., Srinivasa V., Rangappa S., Mervin L., Mohan S., Paricharak S., Baday S., Li F., Shanmugam M.K., Chinnathambi A., et al. Trisubstituted-Imidazoles Induce Apoptosis in Human Breast Cancer Cells by Targeting the Oncogenic PI3K/Akt/mTOR Signaling Pathway. PLoS ONE. 2016;11:e0153155. doi: 10.1371/journal.pone.0153155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mah L.J., El-Osta A., Karagiannis T.C. γH2AX: A sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 34.Datta R., Banach D., Kojima H., Talanian R.V., Alnemri E.S., Wong W.W., Kufe D.W. Activation of the CPP32 protease in apoptosis induced by 1-beta-D-arabinofuranosylcytosine and other DNA-damaging agents. Blood. 1996;88:1936–1943. doi: 10.1182/blood.V88.6.1936.bloodjournal8861936. [DOI] [PubMed] [Google Scholar]
- 35.Ji J., Zhang Y., Redon C.E., Reinhold W.C., Chen A.P., Fogli L.K., Holbeck S.L., Parchment R.E., Hollingshead M., Tomaszewski J.E., et al. Phosphorylated fraction of H2AX as a measurement for DNA damage in cancer cells and potential applications of a novel assay. PLoS ONE. 2017;12:e0171582. doi: 10.1371/journal.pone.0171582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang M., Qi L., Li L., Li Y. The caspase-3/GSDME signal pathway as a switch between apoptosis and pyroptosis in cancer. Cell Death Discov. 2020;6:112. doi: 10.1038/s41420-020-00349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vo P.H.T., Nguyen T.D.T., Tran H.T., Nguyen Y.N., Doan M.T., Nguyen P.H., Lien G.T.K., To D.C., Tran M.H. Cytotoxic components from the leaves of Erythrophleumfordii induce human acute leukemia cell apoptosis through caspase 3 activation and PARP cleavage. Bioorg. Med. Chem. Lett. 2021;31:127673. doi: 10.1016/j.bmcl.2020.127673. [DOI] [PubMed] [Google Scholar]
- 38.Woo M., Hakem R., Soengas M.S., Duncan G.S., Shahinian A., Kägi D., Hakem A., McCurrach M., Khoo W., Kaufman S.A., et al. Essential contribution of caspase 3/CPP32 to apoptosis and its associated nuclear changes. Genes Dev. 1998;12:806–819. doi: 10.1101/gad.12.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye N., Chen C.H., Chen T., Song Z., He J.X., Huan X.J., Song S.S., Liu Q., Chen Y., Ding J., et al. Design, synthesis, and biological evaluation of a series of benzo[de][1,7]naphthyridin-7(8H)-ones bearing a functionalized longer chain appendage as novel PARP1 inhibitors. J. Med. Chem. 2013;56:2885–2903. doi: 10.1021/jm301825t. [DOI] [PubMed] [Google Scholar]
- 40.Chikkegowda P., Pookunoth B.C., Bovilla V.R., Veeresh P.M., Leihang Z., Thippeswamy T., Padukudru M.A., Hathur B., Kanchugarakoppal R.S., Madhunapantula S.V. Design, Synthesis, Characterization, and Crystal Structure Studies of Nrf2 Modulators for Inhibiting Cancer Cell Growth In Vitro and In Vivo. ACS Omega. 2021;6:10054–10071. doi: 10.1021/acsomega.0c06345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashwini N., Garg M., Mohan C.D., Fuchs J.E., Rangappa S., Anusha S., Swaroop T.R., Rakesh K.S., Kanojia D., Madan V., et al. Synthesis of 1,2-benzisoxazole tethered 1,2,3-triazoles that exhibit anticancer activity in acute myeloid leukemia cell lines by inhibiting histone deacetylases, and inducing p21 and tubulin acetylation. Bioorg. Med. Chem. 2015;23:6157–6165. doi: 10.1016/j.bmc.2015.07.069. [DOI] [PubMed] [Google Scholar]
- 42.Bao Y., Oguz G., Lee W.C., Lee P.L., Ghosh K., Li J., Wang P., Lobie P.E., Ehmsen S., Ditzel H.J., et al. EZH2-mediated PP2A inactivation confers resistance to HER2-targeted breast cancer therapy. Nat. Commun. 2020;11:5878. doi: 10.1038/s41467-020-19704-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhardwaj V., Tan Y.Q., Wu M.M., Ma L., Zhu T., Lobie P.E., Pandey V. Long non-coding RNAs in recurrent ovarian cancer: Theranostic perspectives. Cancer Lett. 2021;502:97–107. doi: 10.1016/j.canlet.2020.12.042. [DOI] [PubMed] [Google Scholar]
- 44.Basappa B., Chumadathil Pookunoth B., Shinduvalli Kempasiddegowda M., Knchugarakoppal Subbegowda R., Lobie P.E., Pandey V. Novel Biphenyl Amines Inhibit Oestrogen Receptor (ER)-α in ER-Positive Mammary Carcinoma Cells. Molecules. 2021;26:783. doi: 10.3390/molecules26040783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poh H.M., Chiou Y.S., Chong Q.Y., Chen R.M., Rangappa K.S., Ma L., Zhu T., Kumar A.P., Pandey V., Basappa, et al. Inhibition of TFF3 Enhances Sensitivity-and Overcomes Acquired Resistance-to Doxorubicin in Estrogen Receptor-Positive Mammary Carcinoma. Cancers. 2019;11:1528. doi: 10.3390/cancers11101528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Basappa, Sugahara K., Thimmaiah K.N., Bid H.K., Houghton P.J., Rangappa K.S. Anti-tumor activity of a novel HS-mimetic-vascular endothelial growth factor binding small molecule. PLoS ONE. 2012;7:e39444. doi: 10.1371/journal.pone.0039444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.