Abstract
Salmonella enterica serotypes Derby, Mbandaka, Montevideo, Livingstone, and Senftenberg were among the 10 most prevalent serotypes isolated from farm animals in England and Wales in 1999. These serotypes are of potential zoonotic relevance; however, there is currently no “gold standard” fingerprinting method for them. A collection of isolates representing the former serotypes and serotype Gold Coast were analyzed using plasmid profiling, pulsed-field gel electrophoresis (PFGE), and ribotyping. The success of the molecular methods in identifying DNA polymorphisms was different for each serotype. Plasmid profiling was particularly useful for serotype Derby isolates, and it also provided a good level of discrimination for serotype Senftenberg. For most serotypes, we observed a number of nontypeable plasmid-free strains, which represents a limitation of this technique. Fingerprinting of genomic DNA by ribotyping and PFGE produced a significant variation in results, depending on the serotype of the strain. Both PstI/SphI ribotyping and XbaI-PFGE provided a similar degree of strain differentiation for serotype Derby and serotype Senftenberg, only marginally lower than that achieved by plasmid profiling. Ribotyping was less sensitive than PFGE when applied to serotype Mbandaka or serotype Montevideo. Serotype Gold Coast isolates were found to be nontypeable by XbaI-PFGE, and a significant proportion of them were found to be plasmid free. A similar situation applies to a number of serotype Livingstone isolates which were nontypeable by plasmid profiling and/or PFGE. In summary, the serotype of the isolates has a considerable influence in deciding the best typing strategy; a single method cannot be relied upon for discriminating between strains, and a combination of typing methods allows further discrimination.
Detailed strain identification is essential for the successful epidemiological investigation of Salmonella enterica outbreaks. Investigations have relied traditionally on serological methods and antibiograms. Phage typing has been also used for strain differentiation, but it is only available for a limited number of serotypes. In contrast, modern typing methods are based on characterization of the genotype of the organism. The basic premise of these typing systems is that epidemiologically related isolates are derived from the clonal expansion of a single precursor and share characteristics that differ from those of epidemiologically unrelated isolates. The usefulness of a particular characteristic (phenotypic or genotypic) for typing is related to its stability within a strain and its diversity within the species, reflecting the evolutionary genetic diversity arising from random, nonlethal mutations over time. Such mutations can be detected if they are seen to occur within a restriction site that determines a DNA fingerprint (15).
There is currently no “gold standard” typing system for Salmonella fingerprinting, particularly in the case of less commonly studied serotypes. Recent figures from the Salmonella surveillance program carried out at the Veterinary Laboratories Agency (MAFF) (7) indicated that Salmonella serotypes Derby, Mbandaka, Montevideo, Livingstone, and Senftenberg were among the 10 most prevalent serotypes isolated from farm animals in England and Wales in 1999. These serotypes have been found to be linked to human infection in previous reports. Serotype Derby has been found to be responsible for a number of human infections linked to meat (1, 5); serotype Mbandaka has been reported from cases of human infection in several countries (8, 19), and egg-based products were found to be the most frequently contaminated food groups. Serotype Montevideo has been a recognized causative organism of spontaneous abortion in sheep and foxhounds (3), as well as a cause of human infection linked with poor hygiene associated with food preparation (4, 25). Serotype Livingstone has been isolated from both food products and humans (16, 17, 22). Finally, serotype Gold Coast has also been identified in association with human infection (24), and it was included in our study.
Plasmid profile analysis has been used as a rapid method and has shown some success in the discrimination of Salmonella strains for several of these serotypes (6, 9, 20, 24). Also, previous studies conducted for serotype Enteritidis (11, 12) and serotype Montevideo (18) demonstrated the potential of ribotyping strain differentiation. Finally, pulsed-field gel electrophoresis (PFGE) has been widely used for Salmonella DNA fingerprinting (2, 13, 14, 26). This study focuses on the assessment of molecular methods (plasmid profiling, PFGE, and ribotyping) for intraserotype strain differentiation. The methods were applied to some of the most prevalent Salmonella serotypes isolated from animals in England and Wales. These serotypes are of potential zoonotic relevance based on previous reports; however, they have not been subjected to such intensive epidemiological study as the more commonly encountered serotypes (serotype Enteritidis or serotype Typhimurium) and therefore have no widely accepted, standardized protocol for discrimination below serotype level.
MATERIALS AND METHODS
Salmonella isolates.
Epidemiologically unrelated isolates from six different serotypes (serotype Derby, n = 12; serotype Mbandaka, n = 15; serotype Montevideo, n = 14; serotype Gold Coast, n = 15; serotype Senftenberg, n = 14; serotype Livingstone, n = 14) were selected to represent a diversity within each serotype based on antibiotic resistance, geographical site, and of date of isolation (1997, 1998, or 1999). All the Salmonella cultures were serotyped according to standard protocols (21). Isolates were screened for susceptibility to a panel of 16 antibiotics on Iso-Sensitest agar (catalog no. CM471; Oxoid, Basingstoke, Hampshire, United Kingdom) by a disk diffusion method (7). The following disks (Oxoid) were used: amikacin (10 μg), amoxicillin-clavulanic acid (30 μg), ampicillin (10 μg), apramycin (15 μg), chloramphenicol (10 μg), cefoperazone (30 μg), cefuroxime (30 μg), colistin (25 μg), furazolidone (15 μg), gentamicin (20 μg), nalidixic acid (30 μg), neomycin (10 μg), streptomycin (25 μg), sulfamethoxazole-trimethoprim (25 μg), tetracycline (10 μg) and triple sulfonamide (500 μg). Organisms with a zone diameter of less than 13 mm were classified as resistant.
Plasmid analysis.
Plasmid DNA was isolated by the alkaline lysis method as described before (10). Samples were analyzed by electrophoresis in 1× Tris-borate-EDTA buffer at 150 V for 4 h on 0.8 and 1.5% agarose gels. The plasmid-containing strain Escherichia coli 39R861 and a supercoiled DNA ladder (Gibco BRL, Paisley, United Kingdom) were used to estimate plasmid sizes.
Restriction fragment length polymorphism analysis.
Genomic DNA was extracted from approximately 200 mg (wet weight) of bacteria, and then restriction enzyme digests (PstI-SphI) of Salmonella DNAs were prepared and fractionated by electrophoresis as described previously (12). Fractionated DNA was transferred to positively charged nylon membranes (Roche Molecular Biochemicals, Lewes, United Kingdom) using 0.4 mM NaOH in a vacuum blotting apparatus (Pharmacia Biotech, St. Albans, Hertsforshire, United Kingdom) connected to a variable pump set at 40 × 105 Pa for 1 h. Membranes were rinsed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and air- dried before DNA was fixed to the membranes by cross-linking under UV light. Membranes were prehybridized for 4 h at 42°C in 20 ml of DIG Easy Hyb (Roche Molecular Biochemicals). Plasmid pKK3535 carrying the rrnB rRNA operon from E. coli was extracted using a QIAfilter plasmid Midi purification kit (Qiagen, Crawley, United Kingdom) and labeled with digoxigenin-11-dUTP by a random primed DNA labeling technique using the DIG-High prime kit (Roche Molecular Biochemicals). Probes were denatured by boiling and added to fresh hybridization fluid at 20 ng/ml; hybridizations were performed overnight at 42°C in a Hybaid oven. The presence of the labeled probe was detected using the alkaline phosphatase-conjugated antibody DNA detection kit (Roche Molecular Biochemicals) and the chemiluminescent substrate disodium 3-(4-methoxyspiro[1,2-dioxetane-3,2′-{5′-chloro}tricyclo[3.3.1.13,7]decan]-4-yl) phenyl phosphate (CSPD) as recommended by the supplier. The images produced on X-ray film were computer analyzed using Gel Compar II software (version 1.01; Applied Maths, Kortrijk, Belgium). Molecular weights of the probed fragments were calculated by comparison with the external markers, and images from different gels were normalized accordingly. For the purposes of this study different PstI/SphI ribotypes (PS types) were allocated to strains when a genetic difference could be detected.
PFGE.
A single colony of each Salmonella isolate was incubated overnight at 37°C in 3-ml amounts of Luria-Bertani broth with moderate shaking. One-milliliter aliquots of the cultures were transferred into microcentrifuge tubes and washed twice with 1 ml of saline solution (0.85% [wt/vol] NaCl); finally cells were resuspended in 0.8 ml of saline solution and equilibrated at 40°C. This suspension was mixed in equal parts with molten 2% agarose (CleanCut; Bio-Rad, Hempstead, United Kingdom) and pipetted into disposable molds. Three of these agarose plugs were incubated overnight at 56°C in 2 ml of ES lysis buffer (0.5 M EDTA, 1% N-laurylsarcosine [Sigma, Poole, United Kingdom]) with proteinase K (Sigma) at a final concentration of 250 μg/ml. The next morning the lysis buffer was replaced with fresh ES buffer-proteinase K solution, and this was followed by a second overnight incubation at 56°C. Thereafter, DNA-containing-plugs were thoroughly washed in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8]) and stored at 4°C. Chromosomal DNA was digested with 30 U of XbaI (Promega, Southampton, United Kingdom), and PFGE was performed with a CHEF DRIII system (Bio-Rad) in 0.5× Tris-borate-EDTA extended-range buffer (Bio-Rad) (130 mM Tris, 45 mM boric acid, 2.5 mM EDTA) with recirculation at 14°C. DNA macrorestriction fragments were resolved on 1% agarose gels (PFGE-certified agarose [Bio-Rad]), and a lambda ladder pulsed-field gel marker (New England BioLabs, Hitchin, United Kingdom) was used as the size standard. Pulse times were ramped from 5 to 60 s during a 48-h run at 5.1 V/cm. The preparation and digestion of DNA from a proportion of the strains were repeated, and samples were electrophoresed under the same conditions to assess the reproducibility of the method. Macrorestriction patterns were compared with the use of Gel Compar II software. The molecular weights of the restriction fragments were calculated by comparison with the external markers, and images were normalized accordingly. Different profiles were assigned to XbaI-PFGE types (X types) in accordance with differences in the restriction patterns. A difference of at least one restriction fragment in the patterns was considered to be the criterion for discriminating between different clones or strains.
RESULTS
Plasmid profiling.
Table 1 shows the distribution of plasmid types for the isolates included in the study. The plasmid profile type comprised a numeral, indicating the number of plasmids observed, followed by a letter, corresponding to the order in which the type was encountered. Plasmid profiling of the 12 serotype Derby isolates produced 12 distinct types with one to six plasmids. Of the 15 serotype Mbandaka isolates subjected to plasmid profiling, 7 did not harbor plasmids and the remaining 8 isolates were differentiated into seven distinct types harboring one or two plasmids. Limited success was observed when the technique was applied to serotype Montevideo isolates: only 6 of the 14 isolates were shown to harbor plasmids, and six distinct profile types were observed among these, with one to three plasmids present. Of the 15 serotype Gold Coast isolates tested, 5 did not carry plasmids, and the remaining 10 isolates were differentiated into seven distinct types with one to three plasmids. Five of the 14 serotype Livingstone isolates studied did not harbor plasmids, and the remaining 9 isolates were differentiated into nine distinct profiles with one to five plasmids.
TABLE 1.
Results of DNA fingerprinting of Salmonella isolates from different serotypes
| Serotype | Reference no. | Ribotype (PS type) | PFGE (X type) | Plasmid profilea | Combined typeb | Antibiotic resistancec |
|---|---|---|---|---|---|---|
| Derby | 64/97 | D-PS1 | D-X6 | 2A | D1 | Tr |
| 1817/97 | D-PS6 | D-X6 | 6A | D2 | Sfr Sur Tr | |
| 14/97 | D-PS7 | D-X11 | 4A | D3 | Sf Su Tr | |
| 74/98 | D-PS9 | D-X3 | 2B | D4 | Tr | |
| 370/98 | D-PS5 | D-X5 | 1A | D5 | Sensitive | |
| 1160/98 | D-PS2 | D-X7 | 4B | D6 | Sf Su Tr | |
| 152/98 | D-PS3 | D-X8 | 4C | D7 | Sf Su | |
| 1144/99 | D-PS10 | D-X1 | 4D | D8 | A C S Su Tr | |
| 2984/99 | D-PS4 | D-X2 | 4E | D9 | A Sf Su Tr | |
| 106/99 | D-PS8 | D-X4 | 4F | D10 | Cx Sf Su Tr | |
| 3602/99 | D-PS2 | D-X9 | 3A | D11 | S Tr | |
| 3165/99 | D-PS1 | D-X10 | 3B | D12 | Sf Su Tr | |
| Mbandaka | 8467/97 | M-PS5 | M-X10 | No plasmids | M1 | Sensitive |
| 82/97 | M-PS3 | M-X6 | No plasmids | M2 | Sensitive | |
| 1865/97 | M-PS1 | M-X3 | No plasmids | M3 | Nar | |
| 5446/97 | M-PS2 | M-X13 | 2C | M4 | S Sf Su Tr | |
| 4595/97 | M-PS1 | M-X5 | 2D | M5 | Sf Su Tr | |
| 1473/98 | M-PS6 | M-X12 | 1B | M6 | Ap G S Sf Sur | |
| 175/98 | M-PS1 | M-X9 | No plasmids | M7 | S Sur | |
| 1666/98 | M-PS1 | M-X4 | No plasmids | M8 | Nar | |
| 578/98 | M-PS1 | M-X2 | 1C | M9 | Sensitive | |
| 228/98 | M-PS1 | M-X1 | 1D | M10 | Sf Sur | |
| 965/99 | M-PS1 | M-X6 | No plasmids | M11 | Sensitive | |
| 990/99 | M-PS1 | M-X7 | 2E | M12 | Sfr Sur Tr | |
| 3313/99 | M-PS4 | M-X11 | 1C | M13 | A S Su Tr | |
| 2536/99 | M-PS1 | M-X8 | 1E | M14 | Sf Sur | |
| 4561/99 | M-PS1 | M-X6 | No plasmids | M11 | Sensitive | |
| Montevideo | 2820/97 | Mo-PS3 | Mo-X1 | 1C | Mo1 | S Su Tr |
| 83/97 | Mo-PS3 | Mo-X2 | No plasmids | Mo2 | Sensitive | |
| 8101/97 | Mo-PS3 | Mo-X3 | No plasmids | Mo3 | Sensitive | |
| 3719/97 | Mo-PS3 | Mo-X5 | No plasmids | Mo4 | Cr | |
| 7832/97 | Mo-PS4 | Mo-X1 | No plasmids | Mo5 | Sensitive | |
| 2689/98 | Mo-PS1 | Mo-X1 | No plasmids | Mo6 | Cr | |
| 7029/98 | Mo-PS1 | Mo-X6 | No plasmids | Mo7 | Sensitive | |
| 4981/98 | Mo-PS1 | Mo-X7 | 3C | Mo8 | Nar | |
| 27/98 | Mo-PS1 | Mo-X7 | 2F | Mo9 | Nar | |
| 1703/98 | Mo-PS2 | Mo-X9 | 1F | Mo10 | Sensitive | |
| 1018/99 | Mo-PS1 | Mo-X1 | No plasmids | Mo6 | Sensitive | |
| 571/99 | Mo-PS1 | Mo-X4 | No plasmids | Mo11 | Cr | |
| 4769/99 | Mo-PS1 | Mo-X8 | 2G | Mo12 | Nar | |
| 4254/99 | Mo-PS1 | Mo-X10 | 1G | Mo13 | Tr | |
| Gold Coast | 1581/97 | G-PS6 | No plasmids | G1 | S Tr | |
| 6235/97 | G-PS6 | 1E | G2 | Sf Su Tr | ||
| 2801/97 | G-PS6 | 2C | G3 | Ap S Sf Sur | ||
| 1209/97 | G-PS7 | 3D | G4 | A C S Sf Su Tr | ||
| 210/97 | G-PS8 | No plasmids | G5 | Sensitive | ||
| 4695/98 | G-PS2 | 2H | G6 | A Ne T S Sf Sur | ||
| 1409/98 | G-PS3 | 1E | G7 | S Sf Su Tr | ||
| 1050/98 | G-PS3 | 1E | G7 | Sf Sur | ||
| 782/98 | G-PS8 | No plasmids | G5 | Sensitive | ||
| 5898/98 | G-PS8 | No plasmids | G5 | Sensitive | ||
| 758/99 | G-PS1 | 2I | G8 | Tr | ||
| 8818/99 | G-PS3 | No plasmids | G9 | Sensitive | ||
| 4172/99 | G-PS4 | 1G | G10 | Ar | ||
| 1088/99 | G-PS5 | 1E | G11 | Sfr | ||
| 4560/99 | G-PS9 | 1A | G12 | Cr | ||
| Senftenberg | 3298/97 | S-PS4 | S-X2 | 3C | S1 | Cr |
| 2589/97 | S-PS4 | S-X3 | 3E | S2 | Sr | |
| 486/97 | S-PS6 | S-X3 | 1H | S3 | Nar | |
| 1110/97 | S-PS5 | S-X7 | 1I | S4 | Sr Sur | |
| 6122/97 | S-PS4 | S-X9 | 1C | S5 | Sensitive | |
| 71/98 | S-PS2 | S-X3 | No plasmids | S6 | Sensitive | |
| 2343/98 | S-PS6 | S-X3 | 2J | S7 | Nar | |
| 923/98 | S-PS6 | S-X3 | 1H | S8 | Nar | |
| 1227/98 | S-PS1 | S-X8 | 1J | S9 | Nar | |
| 30/99 | S-PS1 | S-X1 | 1I | S10 | Cr | |
| 3826/99 | S-PS6 | S-X4 | 1H | S11 | Nar | |
| 2546/99 | S-PS7 | S-X5 | 2J | S12 | Nar | |
| 2184/99 | S-PS8 | S-X6 | 2K | S13 | Nar | |
| 472/99 | S-PS3 | S-X2 | 1K | S14 | Cr | |
| Livingstone | 30/97 | L-PS3 | 3F | L1 | Sensitive | |
| 193/97 | L-PS3 | L-X3 | 3G | L2 | Sensitive | |
| 7122/97 | L-PS3 | L-X2 | No plasmids | L3 | Sensitive | |
| 2484/97 | L-PS3 | L-X5 | No plasmids | L4 | Sensitive | |
| 3102/97 | L-PS3 | L-X4 | 3H | L5 | Sensitive | |
| 768/98 | L-PS2 | L-X1 | No plasmids | L6 | Sensitive | |
| 3553/98 | L-PS1 | L-X5 | 3I | L7 | Sensitive | |
| 5775/98 | L-PS5 | 1I | L8 | Sensitive | ||
| 2319/98 | L-PS8 | 2G | L9 | Sensitive | ||
| 8882/99 | L-PS1 | L-X6 | 3J | L10 | Tr | |
| 6356/99 | L-PS1 | L-X7 | 5A | L11 | Tr | |
| 5807/99 | L-PS4 | 1L | L12 | Sensitive | ||
| 58/99 | L-PS6 | No plasmids | L13 | Cr | ||
| 7468/99 | L-PS7 | No plasmids | L14 | Sensitive |
The numeral in the plasmid profile represents the number of plasmids in the strain.
The combined type assigned to the strains represents a combination of results from the different typing methods.
Isolates were resistant to the antibiotic(s) listed unless otherwise noted. Antibiotics: A, ampicillin; Ak, amikacin; Ap, apramycin; Ax, amoxicillin-clavulanic acid; C, chloramphenicol; Ce, cefoperazone; Ct, colistin; Cx, cefuroxine; F, furazolidine; G, gentamicin; Na, nalidixic acid; Ne, neomycin; S, streptomycin; Sf, sulfamethoxazole-trimethoprim; Su, compound sulfonamides; T, tetracycline. The classification sensitive indicates sensitivity to all antibiotics tested.
Types 1A, 1C, 1E, 2C, 1G, 1I, 2G, and 3C were present in isolates belonging to two or more of the serotypes in the study (Table 2). All the remaining types were only found within specific serotypes. Table 2 shows the distribution of the different plasmids (molecular weights) for each plasmid profile encountered.
TABLE 2.
Results of plasmid profiles of 84 Salmonella isolates
| Plasmid type | Size(s) of plasmid(s) (kb) | Salmonella serotype(s) |
|---|---|---|
| 1A | 83 | Derby, Gold Coast |
| 1B | 7.4 | Mbandaka |
| 1C | 110 | Mbandaka, Montevideo, Senftenberg |
| 1D | 9.3 | Mbandaka |
| 1E | 6.9 | Mbandaka, Gold Coast |
| 1F | 3.7 | Montevideo |
| 1G | 46 | Montevideo, Gold Coast |
| 1H | 2.1 | Senftenberg |
| 1I | 4.1 | Senftenberg, Livingstone |
| 1J | 2.4 | Senftenberg |
| 1K | 2.5 | Senftenberg |
| 1L | 2.6 | Livingstone |
| 2A | 95.8, 3.3 | Derby |
| 2B | 20, 3.7 | Derby |
| 2C | 6.9, 3.7 | Mbandaka, Gold Coast |
| 2D | 83, 46 | Mbandaka |
| 2E | 150, 4.1 | Mbandaka |
| 2F | 46, 6.4 | Montevideo |
| 2G | 46, 4.1 | Montevideo, Livinstone |
| 2H | 5.6, 4.3 | Gold Coast |
| 2I | 6.9, 3.4 | Gold Coast |
| 2J | 3.3, 2.1 | Senftenberg |
| 2K | 4.1, 3.3 | Senftenberg |
| 3A | 95.8, 3.9, 3.4 | Derby |
| 3B | 95.8, 6.9, 3.4 | Derby |
| 3C | 46, 6.4, 4.1 | Montevideo, Senftenberg |
| 3D | 46, 5.6, 4.8 | Gold Coast |
| 3E | 46, 6.1, 2.4 | Senftenberg |
| 3F | 4.1, 3.3, 2.4 | Livingstone |
| 3G | 110, 83, 3.3 | Livingstone |
| 3H | 5.3, 5, 4.1 | Livingstone |
| 3I | 4.6, 3.3, 2.5 | Livingstone |
| 3J | 4.3, 3.3, 2.4 | Livingstone |
| 4A | 6.9, 4.8, 3.9, 3.3 | Derby |
| 4B | 95.8, 6.9, 3.9, 3.3 | Derby |
| 4C | 110, 8.3, 6.9, 3.3 | Derby |
| 4D | 95.8, 4.3, 3.9, 3.4 | Derby |
| 4E | 95.8, 6.9, 3.9, 3.4 | Derby |
| 4F | 110, 20, 3.7, 3.3 | Derby |
| 5A | 46, 4.3, 4.1, 3.3, 2.4 | Livingstone |
| 6A | 95.8, 8.3, 6.9, 3.7, 3.3, 2.9 | Derby |
PstI-SphI ribotyping.
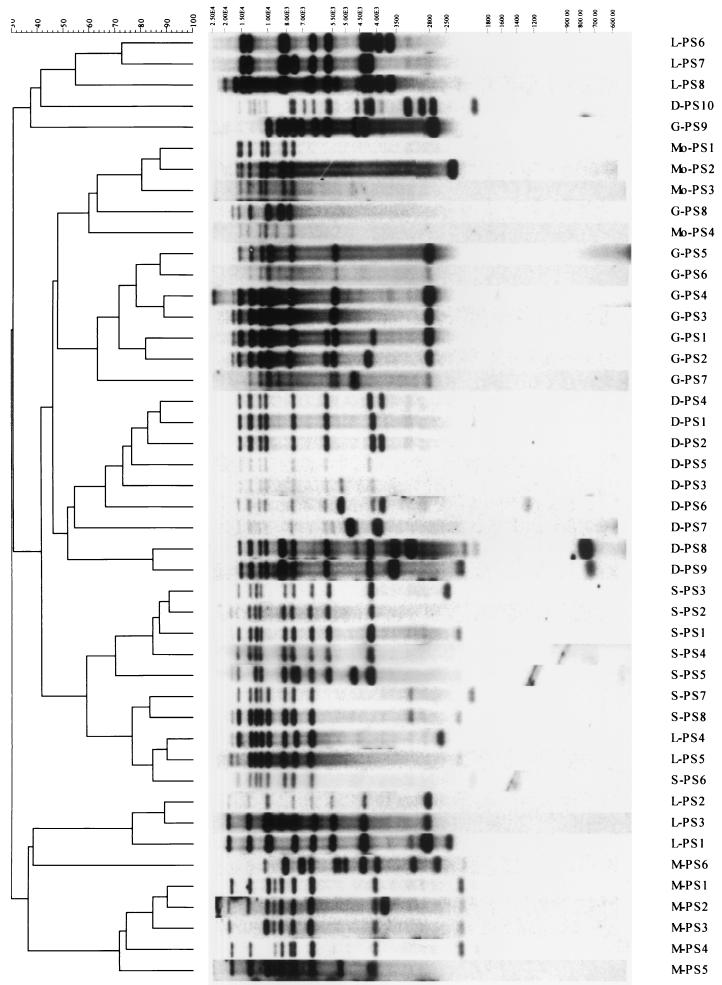
Table 1 shows a summary of ribotypes determined for the isolates included in the study. This technique differentiated serotype Derby isolates into 10 different ribotypes (D-PS types), serotype Mbandaka isolates into 6 M-PS types, serotype Montevideo isolates into 4 Mo-PS types, serotype Gold Coast isolates into 9 G-PS types, serotype Senftenberg isolates into 8 S-PS types, and serotype Livingstone isolates into 8 L-PS types. Figure 1 represents a dendrogram with all ribotypes for the isolates included in the study. None of the patterns produced appeared in more than one serotype tested.
FIG. 1.
Dendrogram generated by the Gel Compar II software showing the relationship of 45 representative fingerprints (PS types) for Salmonella isolates from England (serotype Derby, n = 12; serotype Mbandaka, n = 15; serotype Montevideo, n = 14; serotype Gold Coast, n = 15; serotype Senftenberg, n = 14; serotype Livingstone, n = 14). The analysis of the bands generated was performed using the Dice coefficient and unweighted pair group method with arithmetic averages.
XbaI-PFGE.
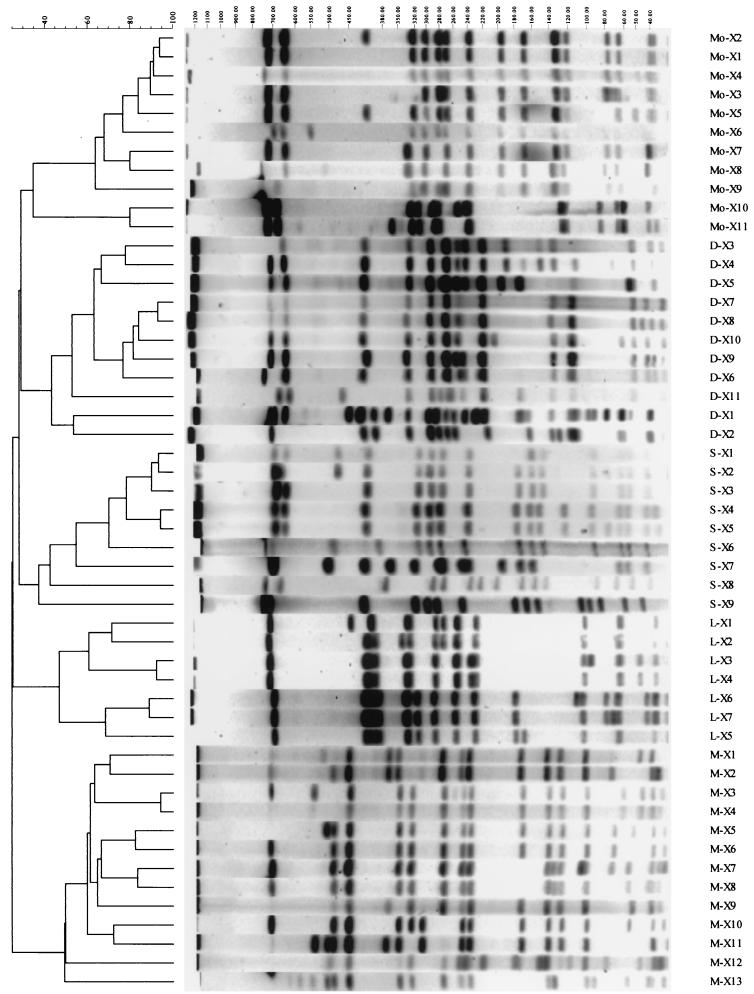
Table 1 shows a summary of PFGE types encountered for the isolates included in the study. This method successfully differentiated serotype Derby into 11 distinct profiles (X types), serotype Mbandaka into 13 X types, serotype Montevideo into 11 X types, and serotype Senftenberg into 11 X types. Serotype Gold Coast isolates were found to be nontypeable by this method, with no fragments produced for any isolate. Also, limited success was obtained when PFGE was applied to serotype Livingstone isolates, with 6 of 14 isolates producing no fragments, although the remaining 8 isolates where differentiated into 7 distinct profiles. Figure 2 represents a dendrogram with all PFGE types for the isolates included in the study.
FIG. 2.
Dendrogram generated by the Gel Compar II software showing the relationship of 51 representative fingerprints (X types) for Salmonella isolates from England (serotype Derby, n = 12; serotype Mbandaka, n = 15; serotype Montevideo, n = 14; serotype Gold Coast, n = 15; serotype Senftenberg, n = 14; serotype Livingstone, n = 14). The analysis of the bands generated was performed using the Dice coefficient and unweighted pair group method with arithmetic averages.
Combined types.
The use of each of the typing methods identified different groups of clones. Therefore, the results could be combined to obtain an overall fingerprint type (Table 1). With the combination of results described above, most of the unrelated isolates for all the serotypes included in this study were identified as a different clone (Table 1). Two serotype Mbandaka isolates had an identical genetic fingerprint (strain M11), two serotype Montevideo isolates had an identical genetic fingerprint (strain Mo6), two serotype Gold Coast isolates had an identical genetic fingerprint (strain G7), and finally three serotype Gold Coast isolates had an identical genetic fingerprint (strain G5).
DISCUSSION
A collection of Salmonella isolates (n = 84) representing commonly isolated serotypes in the United Kingdom was analyzed using three methods aimed at demonstrating polymorphisms in the plasmid and genomic DNA. The success of the three molecular methods in identifying polymorphisms was different for each serotype. Also, within serotypes, the types obtained by each of the methods did not coincide, and a combination of results allowed further discrimination.
Most of the serotypes from this study showed a degree of variation in plasmid number and molecular weight. However, the discriminatory power of the method was observed to vary between serotypes. Most plasmid profile types were shown to be serotype specific, with only some of them (1A, 1C, 1E, 1G, 1I, 2C, 2G, and 3C) being repeated in two or more serotypes. From all serotypes with the exception of serotype Derby, a number of strains did not carry plasmids and were nontypeable by plasmid profiling. This represents a serious limitation for the use of this typing method for some serotypes. This method was particularly useful for serotype Derby isolates, for which each isolate presented a different plasmid profile. It also provided a good level of discrimination for serotype Senftenberg. Previous studies have found plasmid profiling to be of use in intraserotype differentiation (6, 20, 24), although many of these were performed prior to the widespread implementation of the more advanced methods of PFGE and ribotyping. Few recently published articles involve the use of plasmid profiling as a stand-alone procedure. Although this and other studies show that plasmid profiling is not the most sensitive method, the technique does hold significant advantages, particularly the short time in which the procedure can be performed. The rapidity, combined with the relative simplicity of the procedure and basic apparatus required, makes it adequate as an initial procedure that may be used by laboratories which are less able to perform more complicated methods. The inherent mobility of the plasmid DNA suggests instability of the characteristic under scrutiny. This is a limitation which must be recognized in epidemiological research and has brought into question what can be regarded as a suitable plasmid profile for analysis. Some studies (6) regard the presence of a single, identical plasmid as sufficient proof that isolates are identical and therefore epidemiologically related. Other studies (15) suggest that numerous plasmids must be present and regard the presence of a single plasmid insufficient as representative of a clone. Results from this study show that plasmid profiling alone may not be sufficient to accurately identify clones.
Previous studies (11, 12) recognized that ribotyping with restriction endonucleases PstI and SphI provided good discrimination among strains of serotype Enteritidis, and this enzyme combination was therefore applied to the isolates under scrutiny in our own study. Also, PFGE with restriction endonuclease XbaI has been widely recognized as a sensitive method for molecular fingerprinting in several Salmonella serotypes (12, 13, 14, 26). Fingerprinting of genomic DNA from the isolates subjected to study by ribotyping and PFGE produced a significant variation in results, depending on the serotype of the strain. Both PstI/SphI ribotyping and XbaI-PFGE provided a similar degree of strain differentiation for serotype Derby and serotype Senftenberg, only marginally lower than that achieved by plasmid profiling. Ribotyping was shown to be less sensitive than PFGE when applied to serotype Mbandaka or serotype Montevideo. Salmonella serovar Gold Coast isolates were found to be nontypeable by XbaI-PFGE, and a significant proportion of them were found to be plasmid free, a finding in common with a number of serotype Livingstone isolates that were nontypeable by plasmid profiling and/or PFGE. Data obtained by this study suggested that PstI/SphI ribotyping may be successfully used to demonstrate polymorphisms within these two serotypes and that a combination with plasmid profiling may give an appropriate level of discrimination.
In summary, pulsed-field gel electrophoresis gave better strain differentiation for serotypes Derby, Mbandaka, Montevideo, and Senftenberg than the other methods alone. Cluster analysis of the profiles obtained showed a clear serotype differentiation in separate clusters (Fig. 2). Serotype Gold Coast and some isolates of serotype Livingstone were found to be nontypeable by this technique. In this study, no relationship between plasmid profile, PFGE type, and ribotype was identified, and a combination of results from the different methods provided a high degree of strain differentiation for all the serotypes included in the study.
Data produced by this study showed that for a majority of cases PFGE and ribotyping methods had enhanced discriminatory ability compared to plasmid profiling. While PFGE and ribotyping methods are more powerful, they are also considerably more time-consuming than plasmid profiling and require more advanced techniques and equipment. Many epidemiological typing studies have used PFGE as a basis of identification of clones in Salmonella. This method has been proven to be highly discriminatory when applied to some serotypes, although the criteria for analysis still appears to differ between studies. A recent study (23) suggested criteria for interpretation of PFGE data that were aimed to a very specific situation (discrete sets of isolates obtained during nosocomial infections spanning relatively short periods [1 to 3 months]) and that were never regarded to be the basis for universal interpretation of PFGE patterns. However, many workers have applied the former as universal criteria for restriction pattern interpretation. Standardization of protocols and methods for analysis would aid reproducibility between laboratories and aid the flow of epidemiological information. This “flow” can be improved by the implementation of image acquisition and analysis software (such as the Gel Compar II software utilized in this study). Software of this type would allow a degree of standardization between institutes with the subsequent implementation of a commonly recognized format for data.
The findings of this study, together with previously published studies, suggest that the serotype of the isolates may have a considerable influence in deciding the best typing strategy and that a single method cannot be relied upon for discriminating between strains. The most reliable and effective approach to fingerprinting of Salmonella for epidemiological investigations is a combination of methods. Such genetic information, used in conjunction with antibiotic resistance profiles, would help to detect the emergence of potential new strains by genetic variation and spread of antimicrobial resistance among existing strains. It is also important to remember that for the validation of any fingerprinting method it is essential to include good standard epidemiological information in the studies.
ACKNOWLEDGMENTS
This work was supported by the Ministry of Agriculture, Fisheries, and Food (United Kingdom).
We gratefully acknowledge M. Altwegg, who provided plasmid pKK3535.
REFERENCES
- 1.Barrel R A. Isolations of salmonellas from humans and foods in the Manchester area: 1981–1985. Epidemiol Infect. 1987;3:277–284. doi: 10.1017/s0950268800062038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender J B, Hedberg C W, Boxrud D J, Besser J M, Wicklund J H, Smith K E, Osterholm M T. Use of molecular subtyping in surveillance for Salmonella enterica serotype Typhimurium. N Engl J Med. 2001;344:189–195. doi: 10.1056/NEJM200101183440305. [DOI] [PubMed] [Google Scholar]
- 3.Caldow G L, Graham M M. Abortion in foxhounds and a ewe flock associated with Salmonella Montevideo infection. Vet Rec. 1998;142:138–139. doi: 10.1136/vr.142.6.138. [DOI] [PubMed] [Google Scholar]
- 4.Cartwright K A, Evans B G. Salmon as a food-poisoning vehicle-two successive Salmonella outbreaks. Epidemiol Infect. 1988;101:249–257. doi: 10.1017/s0950268800054169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chau P Y, Shortridge K F, Huang C T. Salmonella in pig carcasses for human consumption in Hong Kong: a study on the mode of contamination. J Hyg (London) 1977;78:253–260. doi: 10.1017/s002217240005614x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crichton P B, Old D C, Taylor A, Rankin S C. Characterisation of strains of Salmonella serotype Livingstone by multiple typing. J Med Microbiol. 1996;44:325–331. doi: 10.1099/00222615-44-5-325. [DOI] [PubMed] [Google Scholar]
- 7.Evans S, Kidd S. Salmonella in livestock production. London, United Kingdom: Veterinary Laboratories Agency, Ministry of Agriculture, Fisheries and Food; 1999. p. 129. [Google Scholar]
- 8.Fantasia M, Pontello M, Filetici E, Aureli P. Salmonella Mbandaka isolated in Italy, 1979–1986. Microbiologica. 1989;12:49–54. [PubMed] [Google Scholar]
- 9.Holmberg S D, Osterholm M T, Senger K A, Cohen M L. Drug resistant Salmonella from animals fed antimicrobials. N Engl J Med. 1984;311:617–622. doi: 10.1056/NEJM198409063111001. [DOI] [PubMed] [Google Scholar]
- 10.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landeras E, Gonzalez-Hevia M A, Mendoza M C. Molecular epidemiology of Salmonella serotype Enteritidis. Relationship between food, water and pathogenic strains. Int J Food Microbiol. 1998;43:81–90. doi: 10.1016/s0168-1605(98)00092-0. [DOI] [PubMed] [Google Scholar]
- 12.Liebana E, Garcia-Migura L, Breslin M F, Davies R H, Woodward M J. Diversity of Salmonella enterica serotype Enteritidis from English poultry farms assessed by multiple genetic fingerprinting. J Clin Microbiol. 2001;39:154–161. doi: 10.1128/JCM.39.1.154-161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukinmaa S, Schildt R, Rinttila T, Siitonen A. Salmonella Enteritidis phage types 1 and 4: pheno- and genotypic epidemiology of recent outbreaks in Finland. J Clin Microbiol. 1999;37:2176–2182. doi: 10.1128/jcm.37.7.2176-2182.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyytikainen O, Koort J, Ward L, Schildt R, Ruutu P, Japisson E, Timonen M, Siitonen A. Molecular epidemiology of an outbreak caused by Salmonella enterica serovar Newport in Finland and the United Kingdom. Epidemiol Infect. 2000;124:185–192. doi: 10.1017/s0950268899003696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maslow J N, Mulligan M E, Arbeit R D. Molecular epidemiology: Application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–164. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 16.Old D C, Porter-Boveri M, Munro D S. Human infection in Tayside, Scotland due to Salmonella serotype Livingstone. J Med Microbiol. 1994;40:134–140. doi: 10.1099/00222615-40-2-139. [DOI] [PubMed] [Google Scholar]
- 17.Old D C, McLaren I M, Wray C. A possible association between Salmonella Livingstone strains from man and poultry in Scotland. Vet Rec. 1995;137:544. doi: 10.1136/vr.137.21.544. [DOI] [PubMed] [Google Scholar]
- 18.Old D C, Chisholm S A, Crichton P B, Taylor A. Grouping of Salmonella enterica serotype Montevideo strains by ribotyping and IS200 profiling. Epidemiol Infect. 2000;124:375–382. doi: 10.1017/s0950268899004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlandella B M, Conti F, Marranzano M, Foti M, Daidone A, Leopizzi N. Isolation of S. Mbandaka from a child seen at the University of Messina pediatric clinic. G Batteriol Virol Immunol. 1993;85:12–19. [PubMed] [Google Scholar]
- 20.Schmidt T J, Barret J S, Schrader J S, Scherach C S, McGee H B, Feldman R A, Brenner D J. Salmonellosis associated with marijuana. A multi-state outbreak traced by plasmid fingerprinting N. Engl J Med. 1982;306:1249–1253. doi: 10.1056/NEJM198205273062101. [DOI] [PubMed] [Google Scholar]
- 21.Shipp C R, Rowe B. A mechanical microtechnique for Salmonella serotyping. J Clin Pathol. 1980;33:595–597. doi: 10.1136/jcp.33.6.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steffen W. Salmonella Livingstone outbreak at a children's health resort. Z Gesamte Hyg. 1984;30:222–223. [PubMed] [Google Scholar]
- 23.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Threllfall E J, Hall M L M, Rowe B. Salmonella Gold-coast from outbreaks of food-poisoning in the British Isles can be differentiated by plasmid profiles. J Hyg Camb. 1986;97:115–122. doi: 10.1017/s0022172400064408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Threlfall E J, Hampton M D, Ward L R, Richardson I R, Lanser S, Greener T. Pulsed field gel electrophoresis identifies an outbreak of Salmonella enterica serotype Montevideo infection associated with a supermarket hot food outlet. Commun Dis Public Health. 1999;2:207–209. [PubMed] [Google Scholar]
- 26.Valdezate S, Echeita A, Diez R, Usera M A. Evaluation of phenotypic and genotypic markers for characterisation of the emerging gastroenteritis pathogen Salmonella Hadar. Eur J Clin Microbiol Infect Dis. 2000;19:275–281. doi: 10.1007/s100960050475. [DOI] [PubMed] [Google Scholar]