Abstract
Accurate and sensitive detection of SARS-CoV-2 is an effective strategy for preventing the COVID-19 pandemic in the current absence of specific drug therapy. This study presents a novel enhanced substrate for label-free detection of respiratory viruses using surface-enhanced Raman Scattering. Sodium borohydride reduces silver ions to clustered silver nanoparticles to eliminate the disorganized peak signal of the traditional citrate reducing agent. Meanwhile, the study obtained the fingerprints and concentration-dependent curves of many respiratory viruses, including SARS-CoV-2, human adenovirus type 7, and H1N1 virus, with good linear relationships. The three viruses were also identified in serum and saliva within two minutes, combined with linear discriminant diagnostic analysis. Therefore, establishing this enhanced substrate is greatly valuable for the global response to the COVID-19 pandemic.
Keywords: Surface-enhanced Raman scattering, SARS-CoV-2, Enhanced substrate, Virus detection, Human adenovirus, Influenza virus
1. Introduction
Coronavirus is the largest known RNA virus with 80–120 nm diameter and 26–32 kb genome length [1]. The International Committee on Taxonomy of Viruses (ICTV) divided coronaviruses into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus [2]. Seven species, including two Alphacoronavirus (HCoV-229E and HCoV-NL63) and five Betacoronavirus (HCoV-OC43, SARS-CoV, HCoV-HKU1, MERS-CoV, and SARS-CoV-2) cause disease in humans. The HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1 species only cause mild infection [3], while SARS-COV [4], MERS-CoV [5], and SARS-CoV-2 [6] have caused several coronavirus outbreaks over the last two decades. The COVID-19 pandemic continues to ravage the world, with over 200 million human infections and four million deaths [7]. SARS-CoV-2 encodes 29 proteins [8], including four key structural proteins: nucleocapsid (N), spike (S), envelope (E), and membrane protein (M) [1], [9]. Although the SARS-CoV-2 structure is known, no effective treatment has been developed so far. Early detection, diagnosis, and isolation of infected persons remain the most important control tool against the COVID-19 pandemic.
Presently, the SARS-CoV-2 detection methods include nucleic acid detection and immunological method. The nucleic acid detection method includes polymerase chain reaction (PCR) and its derivative technology, such as nested real time-PCR, quantitative real-time PCR (qRT-PCR), droplet digital PCR (ddPCR), and loop-mediated isothermal amplification (LAMP). The qRT-PCR approach is the gold standard for detection and identification, a routine confirmatory test recommended by the World Health Organization (WHO) [10], [11]. The qRT-PCR sensitivity is 500–1000 copies/mL [12], while nested RT-PCR and ddPCR sensitivities are higher than qRT-PCR [13], [14]. However, the test requirements for nested RT-PCR and ddPCR techniques are high at the experimental site, time-consuming and complex.
Meanwhile, other factors, including the specimen quality, sampling time, experimental conditions, and the technical expertise of inspectors, affect nested RT-PCR and ddPCR test results. The frequent occurrences of false negatives jeopardize the authority of the nucleic acid tests. Therefore, immunological methods are usually applied to supplement SARS-CoV-2 testing since the errors in nucleic acid testing cannot be wholly avoided [15].
The immunological methods include enzyme-linked immunoadsorption (ELISA) [16], chemiluminescence [17], and colloidal gold immunochromatography [18]. ELISA is highly sensitive and is the most commonly used immunological method. The ELISA detection accuracy for the SARS-CoV-2 S and N protein antibodies increases with infection time, reaching over 90% [16], [19]. The disadvantage of ELISA is the false positives caused by cross-reaction. For example, some seasonal coronaviruses cross-react with the novel SARS-CoV-2 [20]. Thus, the shortfalls of nucleic acids and immunological systems emphasize the need for other detection methods.
For new and emergent infectious diseases with unknown pathogens, it is necessary to isolate and identify possible pathogens according to "Koch postulates" [21], then design specific probes suitable for nucleic acid or immunological detection, such as antibodies or DNA oligomers. The process has very high biosafety requirements, is time-consuming, and difficult to perform in the general laboratory. However, the ongoing COVID-19 pandemic demands a simple, reliable, low-cost virus detection method, a fundamental requisite for controlling the disease. The requisite also applies to other infectious diseases to reduce fatality among infected people.
The Surface-enhanced Raman scattering (SERS) is a simple, time-efficient, and cost-effective detection technology that is non-invasive, sensitive, and specific for detecting biological molecules such as nucleic acids and amino acids [22], [23]. Hence, the SERS technique is objective and more reliable than the other subjective detection methods [24]. Moreover, the SERS technique applied in clinical cases is simple, fast, reliable, and low-cost. SERS is currently used to detect various viruses [25], [26], including SARS-CoV-2, human adenovirus, and influenza virus. Researchers often bind specific antibodies to the nanoparticles with Raman activity to detect the target virus with improved detection sensitivity and specificity. For example, Wang et al. [27] designed a Fe3O4 @Ag nanoparticle that combines the influenza virus and adenovirus-specific antibodies to provide an extremely sensitive method for detecting both viruses. Besides binding antibodies, Zhang et al. [28] coated the nanoparticles with nucleic acid sequences of several viruses. They performed SERS detection on the complementary sequences of the tested samples to identify the viruses. Yang et al. [29] combined the ACE2 receptor on the gold nanoparticles, which specifically bind to the S protein of SARS-CoV-2, and detected the virus using SERS.
Besides using specific antibodies, nucleic acid sequences, or receptors as probes, SERS detection can employ other viral structures, including the sulfhydryl group that can specifically bind to gold. Eom et al. [30] used SERS to detect the oseltamivir-resistant mutant influenza strain by applying gold nanoparticles to bind to the viral mercapto group structure in infectious nasal secretions and saliva. However, designing such an enhanced substrate is a complex operation requiring unique packaging and specific probe-based preparations, which are difficult, non-universal, and expensive. Therefore, the logistical and technical details for using the SERS virus detection technology with stability, accuracy, sensitivity, universality, and low cost, require further exploration without serum and saliva background fluorescence interferences.
In this work, a new enhanced substrate was designed using sodium borohydride with no Raman activity to replace the traditional citrate root and reduce silver nanoparticles. The excessive sodium borohydride prevented silver oxide formation on the surface of silver nanoparticles and increased the SERS detection sensitivity. Moreover, the silver nanoparticles maximally bind to the amino group of the virus surface protein molecule, enhancing the virus particle Raman signal. The mixture of metal cation nanoparticles and the virus forms suitable virus hot spots. Moreover, the characteristic fingerprints of various viruses, including human adenovirus, SARS-CoV-2, and influenza virus, were indiscriminately investigated without labeling. The levels of viruses in serum and saliva were assessed by adding the stabilizing agent acetonitrile as an internal marker. When combined with the linear discriminant analysis (LDA), the SERS method rapidly diagnoses viruses with a minimum detection limit of 10 copies/test (PFU/ test) within 2 min. This detection method has broad application prospects for rapid, accurate, and low-cost detection of viral infections, applicable for controlling the worldwide COVID-19 pandemic.
2. Materials and methods
2.1. Virus acquisition and preparation
Formaldehyde inactivated novel coronavirus was purchased from Henan Provincial Key Laboratory of Immunology and Targeted Drugs, Laboratory Medicine of Xinxiang Medical University, and stored at 4 ℃ in aliquots of 107 copies/ mL. Human adenovirus type 7 (HAdV 7) was amplified by the A549 cell culture, and the recovered cell culture was purified by anion exchange chromatography (Biomiga, V1160, Biomiga Inc, CA, USA). The Reed-Muench method determined the titer of purified adenovirus, and β -propanolactone inactivated the purified adenovirus, stored at 4 ℃. The H1N1 influenza virus was cultured from Vero cells, and the Reed-Muench method determined its titer, while β -propanolactone inactivated the cell culture stored at 4 ℃.
2.2. Preparing the enhanced substrate and detection in PBS buffer
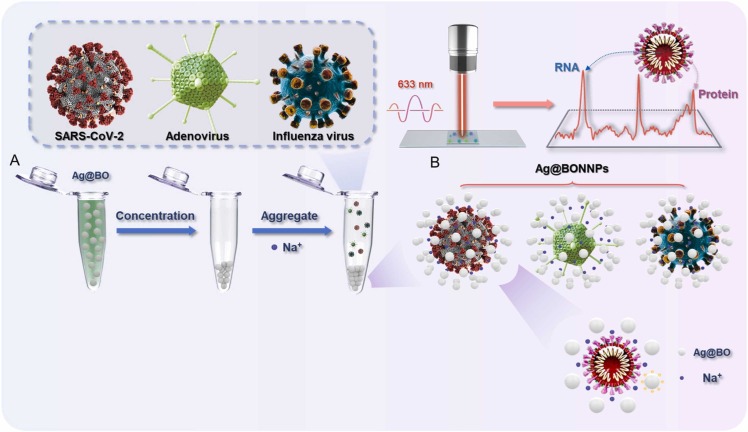
Fig. 1 shows the flow chart for preparing the enhanced substrate and SERS detection. A 5 mL silver nitrate solution (6.6 mg/ mL) was added to 495 mL sodium borohydride solution (0.133 mg/ mL) and stirred vigorously for 18 min, then centrifuged at 5500 rpm, 25 ℃ for 20 min. The supernatant was removed, and 20 μL of the silver sol was added to 2 μL acetonitrile, which was mixed with 10 μL of the virus sample and 24 μL sodium borohydride (0.05 M, pH=10). The SERS detection was performed using the WITec Alpha 300 R (Ulm, Germany) instrument after evenly stirring the solution. The laser wavelength was 633 nm and the scanning time was 60 s, the energy was 30 mW, and each test was accumulated once.
Fig. 1.
(A): schematic presentation of the hot spots generated by aggregating sodium borohydride-induced silver nanoparticles. Ag@BO: Silver nanoparticles were obtained by reducing sodium borohydride. (B): Schematic diagram of the virus in the hot spots. Ag@BONNPs: The enhanced substrate formed by adding Na+ to the silver nanoparticles produced by sodium borohydride reduction.
2.3. Saliva and serum detection methods
The desired titer of virus particles was added to fetal bovine serum (or artificial saliva), used as the solvent, and the concentration was diluted from 1 × 105 PFU/ mL (copies/ mL) to 2 × 104 PFU/ mL (copies/ mL). The serum or saliva containing the virus was incubated at 56 ℃ for 30 min. Next, 20 μL of the silver sol was added to 2 μL acetonitrile, which was mixed with 10 μL of the virus sample and 24 μL of sodium borohydride. The other test conditions are the same as those of antibiotics in PBS buffer.
2.4. Machine learning
We obtained 1000 SERS spectra groups of each virus at the same concentration and 1200 Raman shifts (750–1750 cm−1) per each group as the LDA (Linear Discriminant Analysis) variables. All spectra were processed with LabSpec 6 involving baseline correction and normalization. The LDA was performed using the “FactomineR” package in the R environment. Moreover, the Raman spectra were projected onto the score map in proportion to the load using the “error ellipse” function to draw the error ellipse with 95% confidence. The LDA determined the main differences of virus Raman spectra, and the data obtained by the linear discriminant analysis were classified.
3. Results and discussion
Freshly prepared SARS-CoV-2 can facilitate SERS detection to obtain better fingerprints, but the process requires a high-level biosafety handled carefully, poses a threat to the health of researchers and biologically pollutes the equipment and environment. Therefore, we need a simple to prepare, safe, and universal model virus. The double-stranded DNA virus without an envelope, human adenovirus, produces three important structural proteins: hexon, penton, and spike. Like the SARS-CoV-2 S protein, the spike is prominent on the surface of the capsid and binds to the host-cell receptor [31], [32]. Additionally, the virus has a wide range of host specificity and tissue tropism and causes respiratory tract, eye, gastrointestinal tract, and other tissue infections, although often subclinical in healthy adults [33]. Thus, human adenovirus is a suitable model organism for virology research.
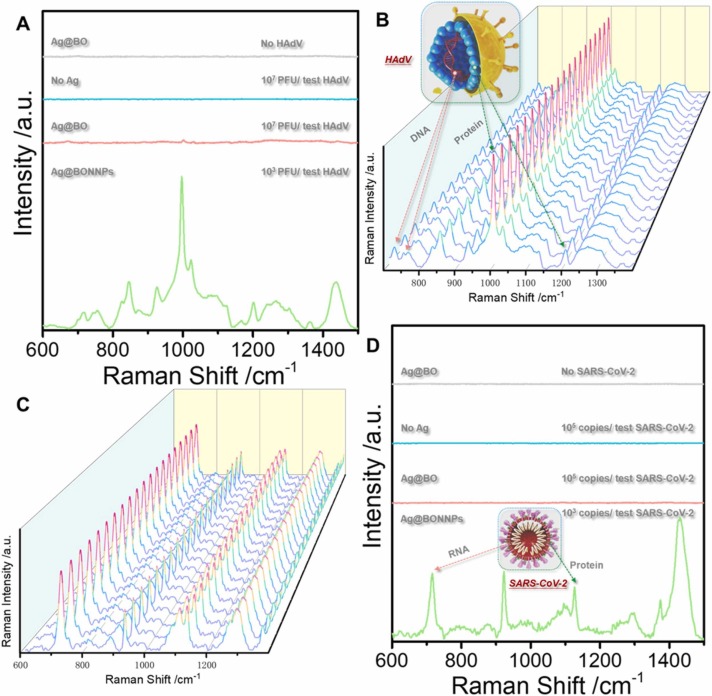
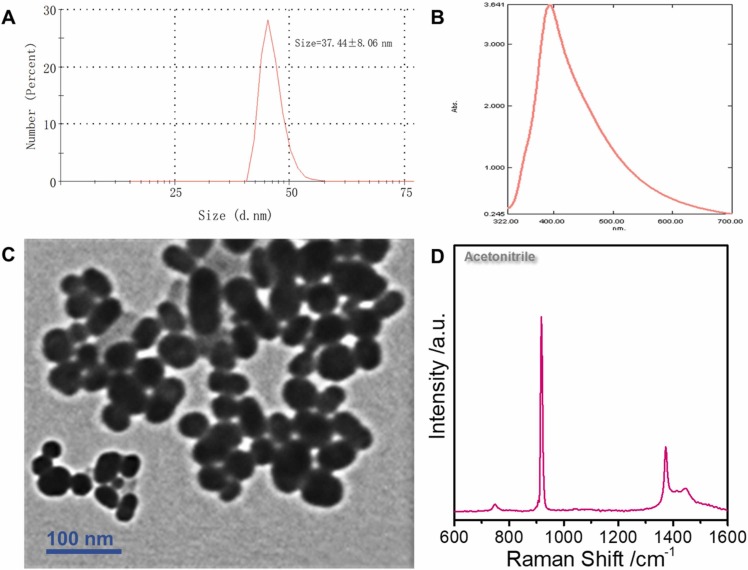
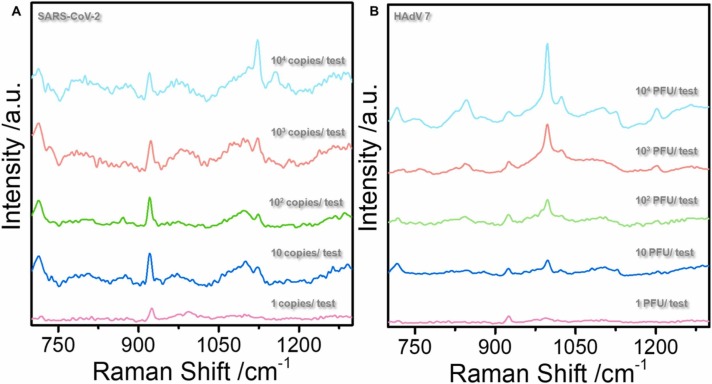
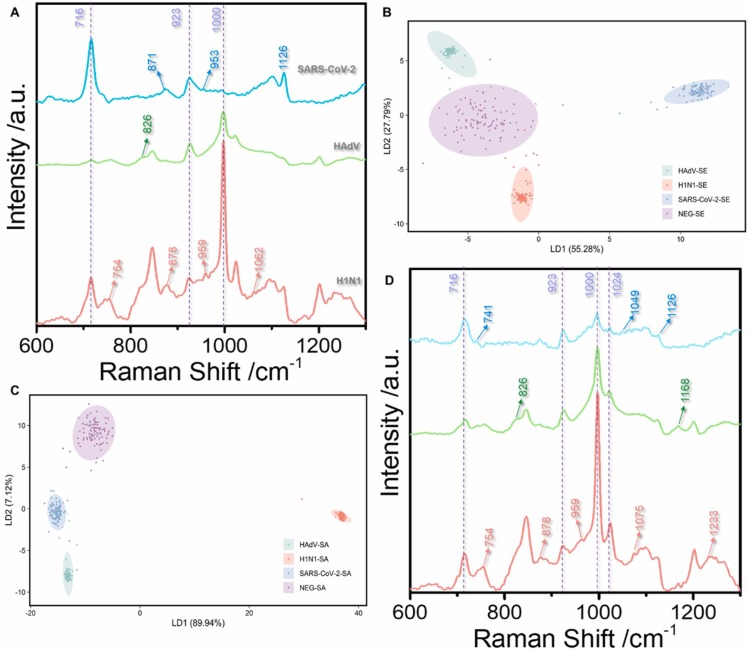
Previously, the main bottleneck for adopting the SERS technology for direct virus detection without labels was that the virus could not enter the "hot spot", had poor detection accuracy, and low sensitivity. Fig. 2A (blue line) shows the Raman spectrum of HAdV 7 at 107 PFU/ test, where the virus signal is undetectable. No viral signal was observed after adding silver nanoparticles (red line). Sodium borohydride was added to the reinforced substrate twice as a reducing agent and aggregator, respectively. First, sodium borohydride was used to reduce silver ions into silver nanoparticles. This approach replaced the traditional citric acid root reductant and avoided the citric acid root signal impurities. Besides, adding excess sodium borohydride can avoid the silver oxide formation on the surface of silver nanoparticles; thus, the virus can better enter the hot spots. The second addition of sodium borohydride mainly uses sodium ions as cationic aggregators. Moreover, the positive charge on the sodium ions can enhance the electromagnetic field of the silver nanoparticle enforcement system, inducing the nanoparticles to gather into hot spots; thus, enhancing the virus Raman signal. Fig. 2A (green line) shows the SERS map of HAdV 7 with the sodium borohydride aggregation agent. The signal peaks indicate protein (1000 cm−1) and nucleic acid structures (716 cm−1), and the supporting information shows specific peak positions. Similarly, the fingerprint of SARS-CoV-2 is shown (Fig. 2D). The DLS results show that the diameter of the silver nanoparticles is approximately 37.44 ± 8.06 nm ( Fig. 3A). However, the UV absorption shows that the maximum absorbance is approximately 396 nm (Fig. 3B). The TEM of the enhanced substrate is shown in Fig. 3C. Note that the thin film on the the surface of each silver nanoparticle may limit virus interaction with the silver nanoparticle surface. The pH of the whole probe is weakly alkaline (8.3) because sodium borohydride, an important reducing agent and aggregator in the preparation method, is alkaline and cannot exist under acidic conditions. Moreover, the weakly alkaline environment will neither affect virus particles nor interfere with virus detection. High reproducibility was observed in the random SERS spectra of 20 HAdV 7 and SARS-CoV-2 groups sampled at different times (Fig. 2B and C). Additionally, the characteristic structure of the virus was clearly observable even at the lowest detection concentration (10 PFU/ test (copies/ test)) ( Fig. 4A and B).
Fig. 2.
(A): The HAdV SERS spectra in different systems. The SERS Ag@BO spectrum without virus (gray line); The Raman spectrum of 107 PFU /test HAdV in PBS (blue line); The SERS spectrum of 107 PFU /test HAdV in silver nanoparticles (red line); The SERS spectrum of 103 PFU /test HAdV (green line) under the Ag@BONNPs method. (B) and (C) show the SERS spectra from 20 random groups of HadV (103 PFU /test) and SARS-CoV-2 (103 copies/ test) samples under the Ag@BONNPs method. (D): The SARS-CoV-2 SERS spectra in different systems. The SERS spectrum of Ag@BO without virus (gray line); The Raman spectrum of 105 copies /test SARS-CoV-2 sample in PBS (blue line); The SERS spectrum of 105 copies /test SARS-CoV-2 in silver nanoparticles (red line); The SERS spectrum of 103 copies /test SARS-CoV-2 (green line) under the Ag@BONNPs method. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
(A): The DLS of the novel enhanced substrate. (B) Ultraviolet absorption of the novel enhanced substrate. (C) The transmission electron microscope (TEM) of the SARS-CoV-2 enhanced substrate. (D) The acetonitrile SERS spectra.
Fig. 4.
(A) and (B) show the SARS-CoV-2 and HAdV 7 SERS spectra at different concentrations.
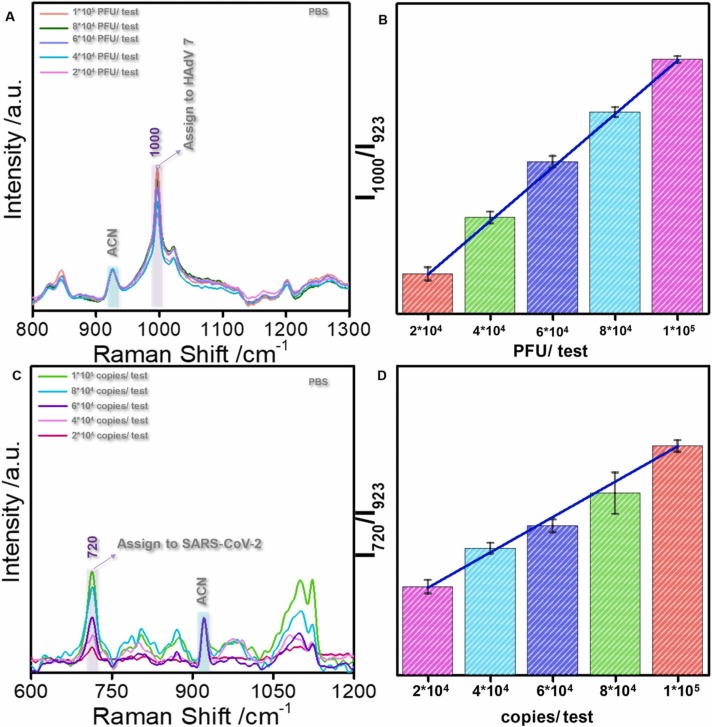
The relationship between variations in virus titer (copies) and Raman peak intensity was investigated to demonstrate the stability and reproducibility of this method. The SERS fingerprints of HAdV 7 with different titers in PBS showed that the peak intensity increased with increasing virus titer ( Fig. 5A). Subsequently, the 1000 cm−1 HAdV 7 peak position exhibited a linear relationship with the virus concentration. The error bar threshold was far below the required threshold for distinguishing different concentrations for quantitative virus identification (Fig. 5B). Similarly, the 716 cm−1 SARS-CoV-2 peak exhibited a linear relationship with the virus concentration (Fig. 5C). The error bar threshold was also below the required threshold for distinguishing different concentrations for quantitative virus identification (Fig. 5D). A simultaneous quantitative HadV7 analysis in saliva showed a linear relationship between the SERS peak intensity and virus concentration (Fig. S1). Therefore, the method presented in this study can quantify virus particles in saliva, an important prospect in clinical practice.
Fig. 5.
(A): The SERS spectra for HAdV in different PBS concentrations (2 *104-1 *105 PFU /test). (B): The bar graph of the I1000/I923 value corresponding to the HAdV change in PBS. (C): The SERS spectra for SARS-CoV-2 in different PBS concentrations (2 *104-1 *105 PFU /test). (D): The bar graph of the I720/I923 value corresponding to the SARS-CoV-2 change in PBS.
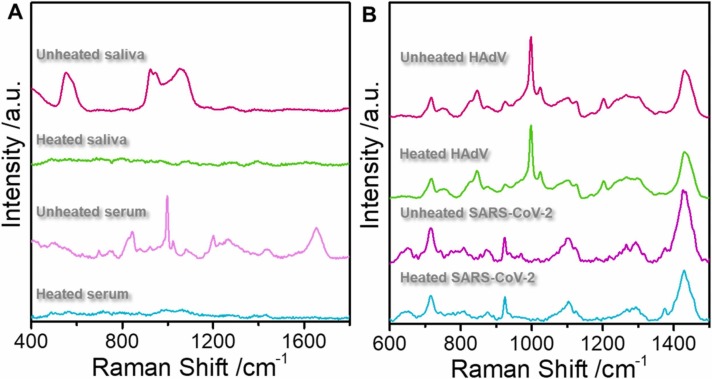
The current method was also applied to explore the SERS detection of viruses in saliva and serum. Fig. 6 shows the SERS spectra of heated and unheated saliva, serum, human adenovirus, and SARS-CoV-2. The multiple peaks in saliva and serum before heating affect the viral signal. However, these peaks are almost invisible after 30 min of heating at 56 ℃ because heating destroys proteins and other substances. However, heating did not change the virus SERS spectra because, at 56 ℃, the virus remained structurally stable. Fig. 7A and D show the spectra of HAdV 7, SARS-CoV-2, and H1N1 influenza viruses in saliva and serum, respectively. These spectra are consistent with the PBS buffer, suggesting that this new method can rapidly diagnose and detect viruses in biological samples. Moreover, the SERS spectra of the three viruses displayed different characteristic peaks, including 826 and 1168 cm−1 (HAdV 7), 741–871 cm−1, and 1049–1100 cm−1 (SARS-CoV-2), 960, 1221 cm−1, and 1103–1167 cm−1 (H1N1 influenza virus). Fig. 7B and 7C show the LDA of 1000 profile sets per virus molecule in serum and saliva, respectively, under the current experimental conditions. The three viruses are easily distinguished in the biological background based on the changes in the characteristic peak positions and the intensity of common peaks. The characteristic peak signal of the Raman spectrum was extracted from the noise using the LDA technique. Next, we applied LDA to the Raman spectra of serum and saliva with PBS buffer without virus as the control and implemented multiple Raman spectrum analyses on the three viruses. Eventually, the LDA process generated multiple points with different coordinate values. The multiple projection points of each cell had a clear, non-overlapping 95% confidence ellipse. The results suggest that the LDA approach detects the virus in serum and saliva and identifies the infection-causing virus.
Fig. 6.
(A): The saliva and serum SERS spectra obtained before and after heating at 56 ℃ for 30 min. The red line is unheated saliva, the green line is heated saliva, the purple line is unheated serum, and the blue-green line is heated serum. (B): The HadV and SARS-CoV-2 SERS spectra obtained before and after heating at 56 ℃ for 30 min. The red line is unheated HAdV, the green line is heated HAdV, the purple line is unheated SARS-CoV-2, and the blue-green line is heated SARS-CoV-2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
(A): The SERS spectra of three viruses (SARS-CoV-2, HAdV, and H1N1) in saliva. (B): The LDA technique classified the main characteristics of the SERS spectrum using serum-containing SARS-CoV-2, HAdV, and H1N1, respectively. The points in the score plot are repetitions of the same concentration (103 copies of the test or PFU/test). (C): The PCA method classified the main characteristics of the SERS spectra on saliva containing SARS-CoV-2, HAdV, and H1N1, respectively. The points in the score plot are repetitions of the same concentration (103 copies of the test or PFU/test). (D): The SERS spectra of three viruses (SARS-CoV-2, HAdV, and H1N1) in serum.
4. Conclusions
This work developed a new label-free virus detection method using sodium borohydride as the reducing agent and aggregator. Acetonitrile ensured the formation of a high-quality hot spot while maintaining the stability of the enhanced substrate. Additionally, the method obtained a high signal-to-noise ratio, good reproducibility of the virus SERS signal, and high sensitivity for direct detection of the virus particle. Using acetonitrile as the internal standard, the linear relationship between the virus titer and the SERS spectra confirmed that saliva detection is reliable without background fluorescence interference. This new approach has three key points (1) The approach obtained a signal for detecting the virion, and the relationship between the virus concentration in the saliva background and the SERS spectra; (2) The virus detection limit reaches 10 PFU/test (copies/test); (3) Sodium borohydride functions as a reducing agent and aggregator to avoid generating miscellaneous citrate root peak signals. Altogether, this new detection method is suitable for clinical tests and confirmatory diagnosis of patients in the incubation period, thus controlling worldwide outbreaks. The method has also promoted the application of SERS for virus detection, improved the disposal capacity for emergent and newly emerging viral infectious diseases. Besides, it facilitated the analysis of the interaction between virus and host cells, which is currently ongoing in our laboratory.
Supporting information
Virus sample preparation methods; enhanced substrate synthesis method; the SERS spectra obtained for HAdV in saliva at different concentrations; specific peak positions.
CRediT authorship contribution statement
Zhe Zhang: Conceptualization, Methodology, Resources, Investigation, Writing – original draft, Writing – reviewing and editing Shen Jiang: Methodology, Resources, Investigation, Writing – original draft, Formal analysis Xiaotong Wang: Validation, Investigation, Data curation, Writing – reviewing and editing Tuo Dong: Visualization, Resources, Data curation Yunpeng Wang: Resources, Data curation Dan Li: Resources Xin Gao: Resources, Zhangyi Qu: Resources, Investigation. Yang Li: Conceptualization, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the Introduce high-level talent incentive project, China (No. 0103–31021200052).
Biographies
Zhe Zhang is a student of Harbin Medical University, whose research direction is the detection of microorganisms by surface enhanced Raman spectroscopy;
Shen Jiang is a student of Harbin Medical University, whose research direction is synthesis of enhanced substrate and biological sample detection;
Xiaotong Wang is a teacher of Harbin Medical University, whose research direction is the detection of macromolecules by surface enhanced Raman spectroscopy;
Tuo Dong is a teacher of Harbin Medical University, whose research direction is the detection of infectious diseases and microbiology research;
Yunpeng Wang is a teacher of Harbin Medical University, whose research direction is the detection of pathogenic microorganisms and drug resistance research;
Dan Li is a student of Guizhou University, whose research direction is signal enhancement of surface -enhanced Raman spectroscopy;
Xin Gao is a teacher of Guizhou University, whose research direction is condensed matter physics;
Zhangyi Qu is a teacher of Harbin Medical University, whose research direction is the detection of infectious diseases and microbiology research;
Yang Li is a teacher of Harbin Medical University, and his research direction is surface-enhanced Raman spectroscopy biological analysis.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2022.131568.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Chan J.F., Kok K.H., Zhu Z., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su S., Wong G., Shi W., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marra M.A., Jones S.J., Astell C.R., et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 5.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/nejmoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Coronavirus (COVID-19) Dashboard, data last updated: 2021/7/16, 7:21pm CEST. 〈https://covid19.who.int/〉.
- 8.Bukkitgar S.D., Shetti N.P., Aminabhavi T.M. Electrochemical investigations for COVID-19 detection-A comparison with other viral detection methods. Chem. Eng. J. 2021;420 doi: 10.1016/j.cej.2020.127575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naqvi A.A.T., Fatima K., Mohammad T., et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys. Acta Mol. Basis Dis. 2020;1866(10) doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z., Tong Y., Wu J., et al. Chinese expert consensus on the nucleic acid detection of SARS-CoV-2. Ann. Transl. Med. 2020;8(24):1631. doi: 10.21037/atm-20-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu L., Wu S., Hao X., et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 2020;66(7):975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J., Cai K., Zhang R., et al. Novel one-step single-tube nested quantitative real-time PCR assay for highly sensitive detection of SARS-CoV-2. Anal. Chem. 2020;92(13):9399–9404. doi: 10.1021/acs.analchem.0c01884. [DOI] [PubMed] [Google Scholar]
- 14.Suo T., Liu X., Feng J., et al. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020;9(1):1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou G., Zhao Q. Perspectives on therapeutic neutralizing antibodies against the Novel Coronavirus SARS-CoV-2. Int J. Biol. Sci. 2020;16(10):1718–1723. doi: 10.7150/ijbs.45123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Clinical performance of different SARS-CoV-2 IgG antibody tests. J. Med Virol. 2020;92(10):2243–2247. doi: 10.1002/jmv.26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai X.F., Chen J., Li Hu J., et al. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of coronavirus disease 2019. J. Infect. Dis. 2020;222(2):189–193. doi: 10.1093/infdis/jiaa243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montesinos I., Gruson D., Kabamba B., et al. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W., Liu L., Kou G., et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58(6):e00461–20. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng K.W., Faulkner N., Cornish G.H., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370(6522):1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pascale V., Mark A., Sansonetti P.J. Pathogens, microbiome and the host: emergence of the ecological Koch’s postulates. FEMS Microbiol Rev. 2018;42(3):273–292. doi: 10.1093/femsre/fuy003. [DOI] [PubMed] [Google Scholar]
- 22.Zong C., Xu M., Xu L.J., et al. Surface-enhanced raman spectroscopy for bioanalysis: reliability and challenges. Chem. Rev. 2018;118(10):4946–4980. doi: 10.1021/acs.chemrev.7b00668. [DOI] [PubMed] [Google Scholar]
- 23.Langer J., Jimenez de Aberasturi D., Aizpurua J., et al. Present and future of surface-enhanced Raman scattering. ACS nano. 2020;14(1):28–117. doi: 10.1021/acsnano.9b04224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ralbovsky N.M., Lednev I.K. Towards development of a novel universal medical diagnostic method: Raman spectroscopy and machine learning. Chem. Soc. Rev. 2020;49(20):7428–7453. doi: 10.1039/D0CS01019G. [DOI] [PubMed] [Google Scholar]
- 25.Shrivastav Anand M., Cvelbar Uroš., Abdulhalim Ibrahim. A comprehensive review on plasmonic-based biosensors used in viral diagnostics. Commun. Biol. 2021;4(1):70. doi: 10.1038/s42003-020-01615-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shvalya Vasyl, Filipič Gregor, Zavašnik Janez, et al. Surface-enhanced Raman spectroscopy for chemical and biological sensing using nanoplasmonics: the relevance of interparticle spacing, and surface morphology. Appl. Phys. Rev. 2020;7(3) doi: 10.1063/5.0015246. [DOI] [Google Scholar]
- 27.Wang C., Wang C., Wang X., et al. Magnetic SERS strip for sensitive and simultaneous detection of respiratory viruses. ACS Appl. Mater. Interfaces. 2019;11(21):19495–19505. doi: 10.1021/acsami.9b03920. [DOI] [PubMed] [Google Scholar]
- 28.Zhang D., Huang L., Liu B., Ge Q., Dong J., Zhao X. Rapid and ultrasensitive quantification of multiplex respiratory tract infection pathogen via lateral flow microarray based on SERS nanotags. Theranostics. 2019;9(17):4849–4859. doi: 10.7150/thno.35824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y., Peng Y., Lin C., et al. Human ACE2-functionalized gold “virus-trap” nanostructures for accurate capture of SARS-CoV-2 and single-virus SERS detection. Nanomicro Lett. 2021;13(1):109. doi: 10.1007/s40820-021-00620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eom G., Hwang A., Kim H., et al. Diagnosis of tamiflu-resistant influenza virus in human nasal fluid and saliva using surface-enhanced Raman scattering. ACS Sens. 2019;4(9):2282–2287. doi: 10.1021/acssensors.9b00697. [DOI] [PubMed] [Google Scholar]
- 31.Rafie K., Lenman A., Fuchs J., Rajan A., Arnberg N., Carlson L.A. The structure of enteric human adenovirus 41-A leading cause of diarrhea in children. Sci. Adv. 2021;7(2):eabe0974. doi: 10.1126/sciadv.abe0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao H., Song Y., Chen Y., et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183(3) doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez-Illana M., Martínez M., Condezo G.N., et al. Cryo-EM structure of enteric adenovirus HAdV-F41 highlights structural variations among human adenoviruses. Sci. Adv. 2021;7(9):eabd9421. doi: 10.1126/sciadv.abd9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material