Abstract
The molecular fingerprints of 1,349 isolates of Mycobacterium bovis received between 1979 and August 2000 at Agence Française de Sécurité Sanitaire des Aliments (Afssa) have been obtained by spoligotyping. The majority of the isolates (1,266) were obtained from cattle living in France. An apparently high level of heterogeneity was observed between isolates. One hundred sixty-one spoligotypes were observed in total, of which 153 were from French isolates. The two predominant spoligotypes, designated BCG-like and GB54, accounted for 26 and 12% of the isolates, respectively. In addition, 84% of the spoligotypes were found fewer than 10 times. Analysis of the results by clustering and parsimony-based algorithms revealed that the majority of the spoligotypes were closely related. The predominant spoligotype was identical to that of the vaccine strain Mycobacterium bovis BCG, which was isolated in France at the end of the 19th century. Some spoligotypes were closely associated with restricted geographical areas. Interestingly, some spoligotypes, which were frequently observed in France, were also observed in neighboring countries. Conversely, few spoligotypes were common to France and England, and those that were shared were observed at very different frequencies. This last point illustrates the potential role for an international data bank, which could help trace the spread of M. bovis across national borders.
Bovine tuberculosis (TB) was endemic in France until the 1960s, with herd prevalence rates of 25% in 1955 (9). From this time onwards, a national program for TB control based on tuberculin skin testing with control of animal movements and total slaughter of infected herds was implemented. This control strategy resulted in a dramatic decrease in bovine tuberculosis leading to a herd prevalence rate of 0.09% in 1998 (2), suggesting that cattle are the most important reservoir, or even the sole reservoir, for Mycobacterium bovis in France. Due to the success of this control strategy, France was declared “officially free of bovine TB” by the European Commission (3).
The very low level of TB in cattle has resulted in the introduction of new control strategies. Consequently, there has been a progressive reduction in the use of skin testing, with an increasing emphasis on systematic sampling of suspect lesions identified at slaughterhouses for M. bovis isolate identification and molecular typing. New laboratory tools were therefore required in order to improve the traceability of the infections and identification of the origin of the outbreak (i.e., persistence in a herd, introduction of new animals from infected herds, or contamination from a neighboring infected herd).
Many new molecular techniques have been developed over recent years to aid the differentiation of isolates belonging to the Mycobacterium tuberculosis complex, which includes M. bovis. Among these techniques, the most widely used are spoligotyping (26), restriction fragment length polymorphism (RFLP) with different probes (IS6110, direct repeat [DR], and polymorphic guanine-cytosine-rich sequence [PGRS]), and variable number of tandem repeat (VNTR) typing (18, 19), which is presently considered a promising technique (28). RFLP with the IS6110 probe has proved to be very useful for the discrimination of M. tuberculosis isolates, which generally harbor a high copy number of this insertion sequence (IS) (21, 25, 39). However, for M. tuberculosis isolates with a low copy number of IS6110 (7, 22) and for the majority of M. bovis isolates from cattle which present only one copy of this IS (5, 6, 13, 26, 28, 37), spoligotyping has been shown to be more discriminating than IS61110 RFLP. DR-RFLP, which detects polymorphism within the same region of the chromosome as spoligotyping, is equally discriminating. PGRS-RFLP is considered to be the most discriminatory of these RFLP techniques for M. bovis (5, 12, 44, 45). But for all the RFLP techniques, DNA extraction is required. In addition, the technique itself is time-consuming and technically demanding, especially for PGRS-RFLP, and problems of reproducibility have been reported (15).
In contrast, spoligotyping is a rapid, simple, and reproducible technique which can be performed on cell lysates and even on clinical specimens (21, 34, 36). It is based on the polymorphism of a region called DR (23). The DR region is unique to bacteria belonging to the M. tuberculosis complex (4, 20, 23, 26, 42) and is constantly present in this group of mycobacteria (26, 30).
In the current protocols, spoligotyping involves the simultaneous detection of the presence or absence of 43 unique short DNA sequences (35 to 41 bp) called spacers. Spoligotyping is considered a very useful technique, at least at the level of a first screening (26, 35), and the results are produced in an intrinsically qualitative form, as the response for each spacer is either present or absent (15).
For these reasons, the first molecular typing technique applied at Agence Française de Sécurité Sanitaire des Aliments (Afssa) (formerly Centre National d'Eudes Vétérinaires et Alimentaires [CNEVA]), for M. bovis typing was spoligotyping. Spoligotyping of the majority of M. bovis isolates in the Afssa collection was performed in order to evaluate the biodiversity and distribution, in space and in time, of M. bovis populations isolated or identified in France. This paper presents our results for 1,349 isolates obtained from 1979 to August 2000.
MATERIALS AND METHODS
Mycobacterial isolates and strains. (i) M. bovis isolates.
Of the 1,349 M. bovis isolates obtained in this study from 1979 to August 2000, 1,266 were from France and 83 were from foreign countries or the French West Indies. Table 1 shows the number of isolates from France for which a spoligotype was obtained compared with the total number of isolates from France that were identified as M. bovis at Afssa by bacteriological methods. No isolates were collected during the years 1980 to 1982. In addition, for some years, like 1984 or 1986, the proportion of isolates available for spoligotyping was low.
TABLE 1.
Yearly evolution of numbers of isolates and spoligotypes
| Year | No. of isolates received | No. of M. bovis isolates | No. of typed M. bovis isolates (% of M. bovis isolates) | No. of spoligotypes |
|---|---|---|---|---|
| 1979a | 159 | 49 | 30 (61) | 14 |
| 1980b | 179 | 53 | 0 | |
| 1981b | 169 | 48 | 0 | |
| 1982b | 204 | 65 | 0 | |
| 1983c | 283 | 39 | 27 (69) | 12 |
| 1984c | 526 | 54 | 26 (48) | 13 |
| 1985c | 528 | 48 | 24 (50) | 11 |
| 1986a | 467 | 36 | 6 (17) | 6 |
| 1987 | 200 | 28 | 23 (82) | 9 |
| 1988 | 234 | 51 | 45 (88) | 17 |
| 1989 | 336 | 66 | 37 (56) | 10 |
| 1990 | 251 | 43 | 39 (91) | 16 |
| 1991 | 285 | 64 | 61 (95) | 18 |
| 1992 | 299 | 73 | 66 (90) | 28 |
| 1993 | 292 | 53 | 50 (94) | 25 |
| 1994 | 517 | 135 | 134 (99) | 39 |
| 1995 | 419 | 133 | 126 (95) | 40 |
| 1996 | 496 | 98 | 98 (100) | 33 |
| 1997 | 509 | 143 | 131 (92) | 40 |
| 1998 | 507 | 167 | 162 (97) | 46 |
| 1999 | 533 | 196 | 189 (96) | 52 |
| 2000d | 133 | 75 | 75 (100) | 22 |
| Total | 7,526 | 1,717 | 1,349 (79) |
Years for which many isolates could not be revivified.
Years for which no isolate could be found.
Years for which a high proportion of isolates was missing.
In the year 2000, isolates were collected from January to August.
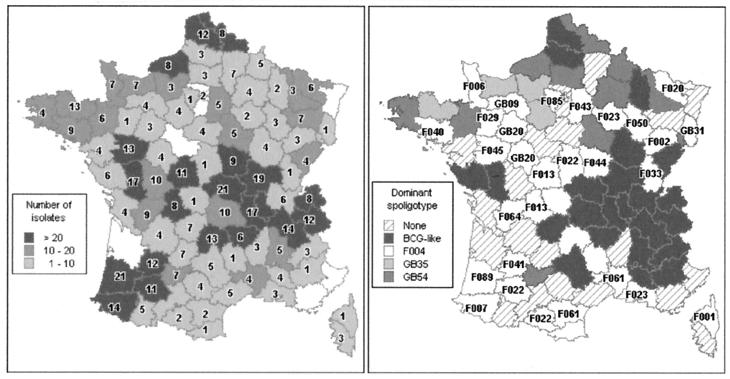
The geographical distribution of the French isolates for which a spoligotype was obtained is represented in Fig. 1. The distribution of all isolates by animal species showed that the majority of animals (1,230) were cattle (91.2%). In addition, 38 goats, 29 deer (all farm deer), 12 pets (3 dogs, 9 cats, 1 ferret, 1 rabbit), 3 sheep, 1 pig, and 14 wild animals represented the other species. With the exception of 2 wild boars, all wild animals were zoo animals (6 monkeys, 3 snow panthers, 1 puma, 1 tiger) or animals kept for experimental purposes (1 bison). The host species was unavailable for 22 isolates.
FIG. 1.
Geographical distribution of the French isolates and spoligotypes. (Left) Number of isolates and number of spoligotypes per department. For each department, the shade of grey indicates the number of isolates for which a spoligotype was identified; the number on the map indicates the number of spoligotypes found in that region. (Right) Dominant spoligotype per department. “None” corresponds to departments where two or more spoligotypes were codominant (i.e., were represented by the same number of isolates).
(ii) Reference strains.
The reference strains used in this study were M. tuberculosis H37Rv and M. bovis BCG P3 or ATCC 19210.
Spoligotyping.
The protocol developed by Kamberbeek et al. (26), as described by Aranaz et al. (6), was used for the spoligotyping of M. bovis isolates. Biodyne C membranes (Pall Gelman, Easthill, N.Y.) were prepared for reverse line blotting by the addition of 43 aminolink oligonucleotides by following the procedures recommended by the manufacturer of the oligonucleotides (Isogen Bioscience, Maarssen, The Netherlands). The isolates were tested as cell lysates. For the reference strains, we usually used purified DNA extracted according the technique described by Wilson et al. (43). In some cases, we used cell lysates obtained by heat treatment. The reference strains were systematically included in each hybridization assay.
Data processing.
The data were collected in Microsoft Access. The maps were constructed using the MapInfo geographical information system (Claritas ADDE, Boulogne, France).
Classification methods.
As discussed by Brittain et al. (D. Brittain, R. A. Skuce, and S. D. Neill, Abstr. 5th Int. Conf. Mycobacterium bovis, abstr., 2000), the “direction of evolution” of the DR region is very probably unidirectional, i.e., it occurs only by deletion of spacer regions, with the lack of contiguous spacers resulting from a succession of events or from a unique event. The objective of classification methods is to define groups and for molecular epidemiology studies, such groups should be stable over time. This requirement is better satisfied if the definition of a group is grounded on evolutionary arguments (16). Therefore, we have combined a classical cluster analysis method (which aim is only to identify groups of isolates on the basis of the degree of similarity of the DR area and does not allow any phylogenetic interpretation from the data), with a phylogenetic analysis using a parsimony-based method, which takes into account the unidirectional way of evolution of the DR region. The phylogram obtained using these methods can then be used to evaluate the significance (in evolutionary terms) of the groups defined using cluster analysis. Cluster analysis was conducted using the program NEIGHBOR from the PHYLIP package (17) with the option UPGMA, the distance matrix being computed using Dice coefficients. Phylogenetic reconstruction was conducted using the PAUP* software package (D. L. Swofford, Phylogenetic Analysis Using Parsimony [*and other methods], version 4; Sinauer Associates, Sunderland, Mass.), the lack of a spacer being considered a character. All changes were weighted equally and all character changes were considered irreversible (Irrev.Up), with the presence of a spacer being considered plesiomorphic. The heuristic method was selected with random sequences additions (100 replicates) and TBR branch swapping (options: DELTRAN optimization, MULPARS, and COLLAPSE 0-length branches). The trees were rooted with a BCG-like spoligotype (which presents the maximum number of spacers, i.e., 35 spacers) as an outgroup as a consequence of the irreversibility of the transformation of the character's state from presence to absence.
RESULTS
Global results.
We adopted the following nomenclature to name the different profiles obtained. In general, the letter F (for France) was used followed by a number, which was different for each different spoligotype obtained. Two exceptions were introduced: first, we used GB (for Great Britain, United Kingdom) plus a number when the profiles had been previously described in Great Britain; second, the term BCG-like was adopted for field isolates with a profile identical to the spoligotype of the vaccine M. bovis BCG strains.
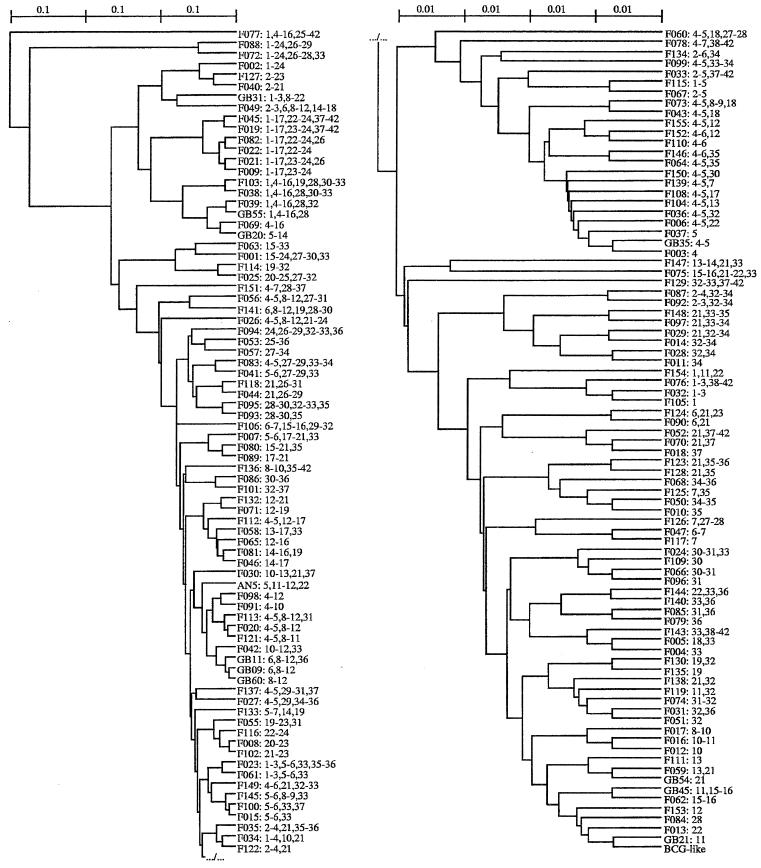
We obtained 161 spoligotypes from the 1,349 isolates typed in this study. The 1,266 isolates from France corresponded to 153 spoligotypes. The profiles corresponding to each spoligotype are given in Fig. 2. All profiles were typical of M. bovis, as described by many authors (24, 26, 30), with the absence of spacers 39 to 43 (which allows the distinguishing of M. bovis from M. tuberculosis) and the lack of spacers 3, 9, and 16.
FIG. 2.
Spoligotype phenogram obtained from 1,349 M. bovis isolates and corresponding patterns. For each spoligotype, the spacer deletions are indicated. Because all spoligotypes share the BCG-like spoligotype deletions (spacers 3, 9, 16, 39 to 43), these deletions are not included in each spoligotype description. Therefore, for example, the AN5 description (5, 11–12, 22) indicates the deletions of spacers 3, 5, 9, 11 to 12, 16, 22, and 39 to 43. The phenogram was generated using the UPGMA clustering method, with the between-spoligotypes distances being Dice coefficients. The scale indicates dissimilarity proportions. Due to the length of the phylogram, it has been fractionated into two parts, the upper right part representing the continuation of the lower left part.
The two most predominant spoligotypes, BCG-like and GB54, accounted for 38% of the French isolates (26 and 12% of the isolates, respectively). In most cases, we were unable to obtain precise data concerning the origins of the isolates. It is therefore not possible to exclude that some isolates were obtained from the same herd. In order to avoid possible bias due to an overestimation of some spoligotypes, the spoligotype frequencies were also calculated by counting only one isolate per herd (where the information was available and where the spoligotypes were identical). In this way, we obtained similar frequencies for BCG-like and GB54 (27 and 10%, respectively). For French isolates, 84% of the spoligotypes were present 10 times or fewer and 40% of spoligotypes were only found in one isolate.
Spatial distribution.
Figure 1, left, shows the number of spoligotypes found in each of the metropolitan French departments (French administrative subdivisions), and Fig. 1, right, shows the most frequent spoligotype per department. The most prevalent spoligotype, BCG-like, was widely distributed in France, with an apparent predilection for the southeastern areas, where it was the predominant spoligotype in most departments (Fig. 1, right). The next most prevalent spoligotype, GB54, was also present in many regions, as was GB35 (the next most frequently observed spoligotype). Initially, GB54 appeared to have been restricted to the north and northwest regions of France, and then it appeared to have extended into other regions of the country over time (data not shown). The majority of the spoligotyping patterns that have been observed in both Great Britain and in France were mainly restricted to northern French departments (Fig. 1, right). Conversely, the fourth most frequent spoligotype, F004, was isolated mostly in southern France. Some isolates were obtained from cattle imported into France or from samples sent from the French West Indies or from foreign countries. As shown in Table 2, the majority of the spoligotypes of these isolates were also observed in metropolitan French isolates (20 out of 28).
TABLE 2.
Description of isolates not originating in metropolitan France
| Spoligotype | No. of isolates from:
|
Total no. | |||||
|---|---|---|---|---|---|---|---|
| FWIa | Belgium | Spain | Italy | Mauritius | Tunisia | ||
| BCG-like | 1 | 3 | 20 | 2 | 26 | ||
| GB35 | 3 | 1 | 7 | 11 | |||
| GB54 | 2 | 7 | 1 | 10 | |||
| F088 | 3 | 3 | |||||
| F009 | 1 | 1 | |||||
| F040 | 1 | 1 | |||||
| F077 | 1 | 1 | |||||
| F127 | 1 | 1 | |||||
| GB09 | 1 | 1 | |||||
| Other | 4 | 9 | 14 | 1 | 28 | ||
| Total | 4 | 7 | 26 | 43 | 1 | 2 | 83 |
FWI, French West Indies.
Evolution with time.
The number of spoligotypes apparently increased with time, but this increase appeared to be related to the total number of isolates that were tested. There is a strong correlation between the number of isolates per year and the number of spoligotypes per year (Table 1). This was observed for most of the departments (Fig. 1, left), as departments from which many isolates were obtained were also those where spoligotype diversity was highest. Analysis of the change in frequency of the four most prevalent spoligotypes with time (data not shown) revealed that there was a slight decrease in the frequency of the spoligotype BCG-like and a corresponding increase in the frequency of GB54 over time.
Distribution by animal species.
All spoligotypes were isolated from cattle (Table 3). However, striking differences in species distribution were observed for spoligotypes GB54 and F040. Spoligotype GB54 was isolated from a wide range of animal species. Conversely, almost all isolates harboring the spoligotypes F040 were isolated from goats (14 out of 16).
TABLE 3.
Distribution of French and foreign isolates by animal species and by spoligotype
| Spoligtype | No. of isolates from:
|
Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Cattle | Goat | Deer | Wild animals | Pets | Sheep | Pig | Unknown | ||
| BCG-like | 329 | 13 | 1 | 6 | 1 | 6 | 356 | ||
| GB54 | 121 | 1 | 24 | 5 | 2 | 2 | 1 | 3 | 159 |
| GB35 | 81 | 1 | 1 | 3 | 86 | ||||
| F004 | 49 | 1 | 1 | 1 | 52 | ||||
| F040 | 2 | 14 | 16 | ||||||
| Other | 648 | 8 | 4 | 7 | 3 | 0 | 0 | 10 | 680 |
| Total | 1,230 | 38 | 29 | 14 | 12 | 3 | 1 | 22 | 1,349 |
Cluster analysis.
The distribution of the spoligotypes into groups by using a clustering method (UPGMA) is presented in Fig. 2. This phenogram shows that, apart from one minor group, the majority of the spoligotypes can be included in one major BCG-like group of similarity, which contains the BCG-like spoligotype. This highlights the high degree of homogeneity of the DR region among the isolates in our study.
The only divergent group included isolates from countries other than France. The F077 spoligotype was found in an isolate from Italy. The F088 plus F072 subgroup was found in Spanish isolates (F088) and in isolates from a French area very close to Spain (F072). The F040 subgroup (F040, F127, and F002) was found in Spanish cattle isolates (F040 and F127) and in a majority of French goats for F040, as already mentioned. Moreover, the F040 and F127 spoligotypes are very similar to those obtained in Spain from a majority of goats and from some cattle (4). Finally, the F009 subgroup (F009, F021, F022, F082, F019, and F045) was also found in Spanish isolates (F009) and in the French Pyrenees (F009, F022, and F045).
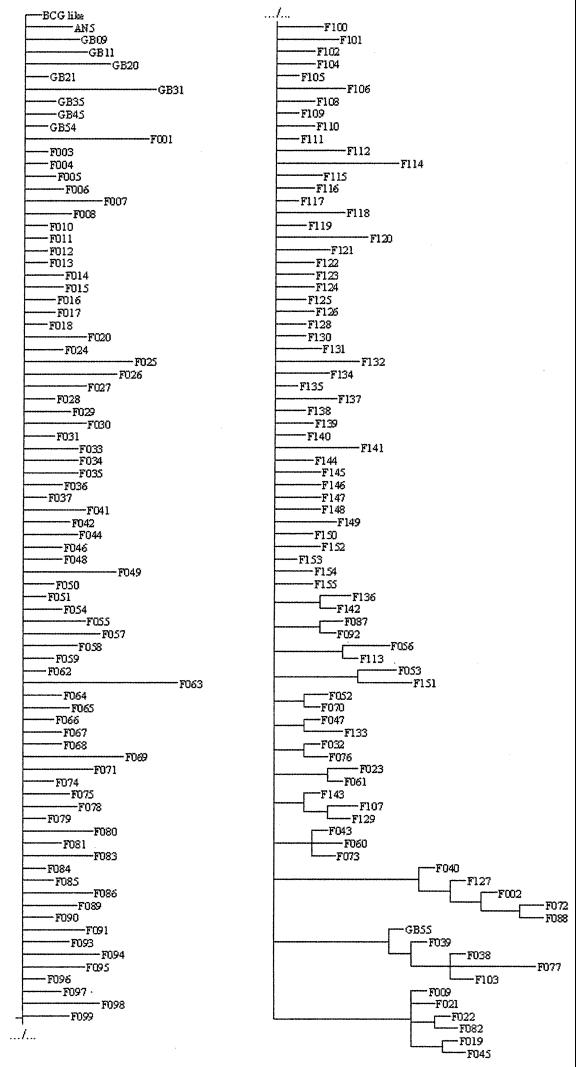
Phylogenetic analysis.
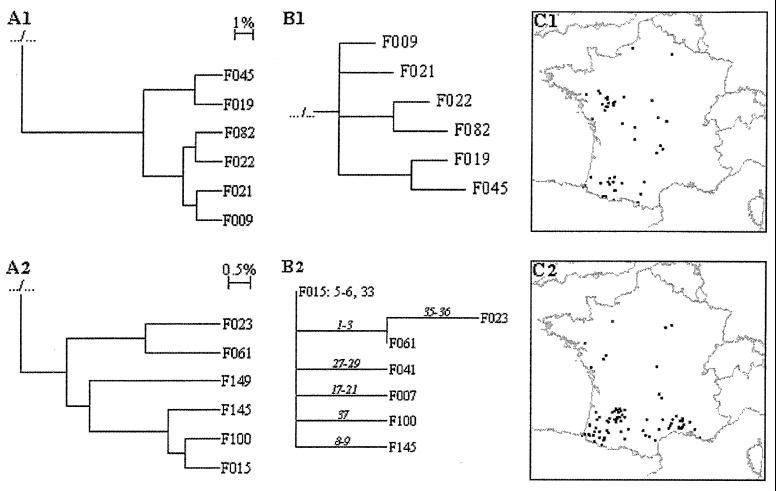
PAUP* did find four most parsimonious trees 312 steps long (CI = 0.11, RI = 0.70). The strict consensus of these trees (Fig. 3) was not resolved for nearly two-thirds of types, but in the remaining types, some subgroups were noticeable. For example, the F009 subgroup was found by this method (Fig. 4B1). Moreover, a detailed analysis of the four most parsimonious trees (data not shown), in conjunction with epidemiological data (i.e., the geographical distribution of the spoligotypes), allowed identification of a further subgroup, the F015 subgroup, composed of the F015, F100, F145, F061, and F023 spoligotypes. By assuming that the primary evolutionary event was the loss of one spacer (or of a block of contiguous spacers), we could identify a plausible evolutionary path for the F015 subgroups (Fig. 4B2). In this pathway, new strains were created by successive deletions occurring at distinct spacer regions from the parental state (F015), which harbors two deletion sites distinct from those of the BCG-like strain. The geographical distribution of the spoligotypes belonging to the F015 group was restricted to the South of France. A careful examination of the spoligotyping profiles and of the geographical distribution of the corresponding isolates (Fig. 4C2) allowed us to include two other spoligotypes, F007 and F041, in this subgroup. This was done on the basis of phylogenetic evidence (both spoligotypes share two distinct deletions with F015 and harbor an extra deletion at distinct sites) and of epidemiological evidence (both spoligotypes share the same geographical distribution as the one of the phylogram-derived F015 subgroups). Finally, the F015 spoligotype deletions include the loss of spacer 33. F004, which differs from BCG-like by the loss of spacer 33, is the fourth most common spoligotype in France; as for the other members of the F015 group, they were geographically restricted to southern French regions (Fig. 1, right). This suggests that the whole F015 subgroup could have evolved from the F004 spoligotype. Three of the four trees support divergence of the F004 branch just before that of the F015 subgroup.
FIG. 3.
Spoligotype phylogram obtained from 1,349 M. bovis isolates. Due to the length of the phylogram, it has been fractionated into two parts, the upper right part representing the continuation of the lower left part. For each spoligotype, the spacers' deletions can be found in the phenogram (Fig. 2). The phylogram was generated using PAUP*.
FIG. 4.
F009 and F015 groups. Trees for the F009 group (A1) and the F015 group (A2) were obtained using cluster analysis as done for Fig. 2. (B1 and B2) Trees were obtained using PAUP*. (B1) The tree for the F009 group was done as the one for Fig. 3. (B2) F015 group. The proposed evolutionary tree was derived from the phylogram (Fig. 3) and from geographical distribution. The parental spoligotype is indicated in plain letters (excluding typical M. bovis deletions); successive deletions are indicated in italics. (C1 and C2) Geographical distributions of the isolates of the F009 group (C1) and the F015 group (C2) are shown. Each dot corresponds to one isolate randomly positioned inside the department from which the isolate originated.
DISCUSSION
Materials.
It is difficult to know if our sample, for which spoligotyping has been possible, taken globally, is representative of the population of M. bovis in France. However, apart from some years (like 1984 and 1986), it is representative of the M. bovis population isolated at Afssa. From 1988, the proportion of M. bovis isolates which could be typed increased, and the population used for this study is representative of the population of M. bovis strains isolated at Afssa.
The numbers of typed isolates varied greatly with the year of isolation. For some years the numbers are very low, especially for the earlier years covered by this study when we were unable to obtain spoligotyping data from stored material. The increase in absolute numbers of M. bovis isolates with time can appear paradoxical, as the incidence of TB in cattle dropped dramatically in France over the time period covered by this study due to the success of the national program for bovine TB control. In fact, as a consequence of this success, nonspecific skin test reactions increased, typical lesions at slaughter reduced, and the need for laboratory confirmation of all suspect lesions became increasingly necessary, resulting in the collection of a greater number of isolates.
The geographical distribution of our isolates appears very wide, although the prevalence of TB was higher in some regions. Most of the M. bovis isolates were obtained from cattle, reflecting the lack of a significant wildlife reservoir for M. bovis in France, at least for the period covered by this study. This assertion seems to be confirmed by the fact that once cattle TB was controlled in an infected area by classical test and slaughter measures focusing on cattle, TB disappeared from the area. In addition, when new outbreaks occurred some years later in such an area, they could be explained by contiguous infection or by the uncontrolled purchase of an infected cow.
Spoligotyping technique.
Our results confirm that spoligotyping is a very easy and rapid technique and does not require DNA extraction. For all our isolates, we used cell lysates. Our work has also allowed us to confirm the repeatability of the test, as we tested 183 isolates (13.5%) at least two times, with the same spoligotype obtained in all cases.
As we have not had the opportunity to test successive isolates taken at different times from the same animal, it is not possible from our work to form any conclusion concerning the stability of this technique. But, some elements are suggestive of such a stability, at least for the short-term and at a horizontal level; for example, in many herds where a high number of isolates were taken, the isolates did harbor the same spoligotype. In one case, the same spoligotype was isolated from two farms with clear epidemiological links at an interval of 7 years. Conversely, some data suggest that a switch could sometimes occur by deletion of a spacer in the same herd (the deletion of spacer 33 was suggested in two herds, where spoligotypes BCG-like and F004 where both present). However, we have still to confirm that this is not due to the coexistence of more than one isolate in one herd, which may not be a rare event (10) (G. Ferretti, personal communication).
Other authors have underlined the stability of the DR region (10, 20, 34, 38). Alito et al. (1) and Niemann et al. (29) have shown that its stability is higher than that of IS6110 (14), and a study of medieval remains is consistent with such a stability, at least at the level of the species profile of the DR region (40). With 161 spoligotypes obtained for 1,349 isolates, the discriminatory power of the test has to be considered very good for a technique which is essentially considered to be a screening technique. Roring et al. (35) have suggested that spoligotyping is of value for epidemiological studies of M. bovis. In our case, it has been very helpful for some epidemiological analysis, essentially in cases where nonfrequent spoligotypes where involved (data not shown). However, it is clear that other typing techniques are required for further discrimination, especially for those spoligotypes that have a high frequency in some epidemiological circumstances.
Classification methods.
Parsimony-based methods assume that each character (the presence or absence of each spacer) evolves independently (the elementary evolutionary event here being the loss of one spacer). This is of course not true if we consider that spacers may be lost directly by blocks. Therefore, before drawing conclusions, we evaluated the robustness of both phenogram- and phylogram-derived groups by using the geographical location of the spoligotypes (a given group being more or less restricted to a given area). In the case of the F015 subgroup, such an analysis led us to exclude one spoligotype of the phenogram-derived group and to add two extra spoligotypes to the phylogram-derived F015 subgroup (Fig. 4B2 and C2).
Spatial distribution.
For the most prevalent spoligotypes, like BCG-like, GB54, and even GB35, which have a rather wide distribution, there is obviously a need for additional techniques based on independent markers in order to allow epidemiological studies. In the case of frequent spoligotypes that have a limited geographical extension, like F004, the appearance of isolates with this spoligotype in areas from where they were absent can to a certain extent help to trace the origin of the infection. But the possibility of a switch from BCG-like, still to be demonstrated, could limit the value of this apparent “geographical advantage.” The precise identification of a close association between an area and a spoligotype could be particularly helpful for epidemiological purposes. Such a geographical clustering has already been described in England for some spoligotypes (16). Our data show for example a very close association between F041 and Department 47 (Lot-et-Garonne) and between F064 and Department 16 (Charente).
Comparison with spoligotypes obtained in other countries.
The absence of a bank of spoligotype data makes it difficult to compare our profiles with those obtained in other countries. However, the spoligotypes in France are rather different from those obtained in Great Britain, as only 10 spoligotypes out of 161 were also observed in Great Britain. This represents only 6.2% of the spoligotypes observed in France and approximately 16% of the spoligotypes known in Great Britain. The predominant spoligotype in Great Britain, corresponding to our GB09 spoligotype, is present in France at a very low level (0.7% of the isolates, with only 11 cases). Conversely, it is dominant in countries that have had traditional links with Great Britain, such as Australia, Canada, Iran (12), the Republic of Ireland (11), and countries from South America, especially Argentina (45).
The second most predominant spoligotype in France, GB54, was also observed in Great Britain, but at a low level. It is also present in some other countries, sometimes in a high proportion of isolates. In Spain, GB54 (called Sp7) was the most frequent of the spoligotypes in the study of Aranaz et al. (6), accounting for 59 out of 129 isolates from cattle (46%). The third most prevalent spoligotype, GB35, is also common in Italy (37).
Conversely, it is striking that the predominant spoligotype in France, BCG-like has never been detected in Great Britain. However, the BCG-like spoligotype is frequent in M. bovis isolates from some other Latin European countries, like Italy (Ferretti, personal communication) (37) and Spain (6, 24). Our results have confirmed the presence of this BCG-like spoligotype in M. bovis isolates from Belgium, Italy, and Spain and also from Tunisia, which has had historical and commercial links with France from more than one century (Table 2). The presence of rare spoligotypes (like F077) or special subgroups of spoligotypes (e.g., those found in Spanish isolates) also underlines the possible existence of some autochthonous strains in those countries, which could have been maintained due to some particular ecological environments.
Animal species.
The case of GB54 suggests that isolates with this spoligotype may have a wider host range than isolates with different spoligotypes. This is suggested by the fact that even if the prevalence of BCG-like isolates is higher in France, this spoligotype is less frequent in the cattle-plus-goats group than in groups of the other animal species, compared to the prevalence of GB54, with a very significant difference (χ2 = 54, P < 0.001). This difference was still highly significant (χ2 = 30.5, P < 0.001) when we removed the isolates belonging to known outbreaks in order to suppress the possible bias linked to an overestimation of one category of species.
In Italy, GB54 is present in wild boars, also suggesting a capacity of the strains presenting this spoligotype to adapt to different animal species (6).
The case of “F040-like” isolates (F040, F088, and F127) is particularly interesting to consider. These isolates present many similarities with the Spanish goat isolates identified as belonging to a new group, which would create a new subspecies, M. tuberculosis subsp. caprae (4). Isolates with the F040 spoligotype have also been isolated, essentially from French goats and from Spanish cows. Those presenting the F088 spoligotype have been isolated from Spanish samples. The F127 isolate has also a Spanish origin. These spoligotypes are almost identical (F040 and F127 belong to the same subgroup; Fig. 2 and 3) and are very similar to the Spanish goat spoligotype [N. Haddad, A. Ostyn, B. Durand, C. Karoui, J. Inwald, S. Hughes, M. F. Thorel, and G. Hewinson, Abstr. 30th IUATLD World Conf. Lung Health, abstr. 199-PD, Int. J. Tuberc. Lung Dis., 3(Suppl. 1):S203, 1999]. Studies are in progress in order to determine if our F040-like isolates have in common with the Spanish goat isolates other genetic characteristics which would allow to integrate them in the proposed new subspecies.
Clustering and phylogenetic results.
The most striking feature of the spoligotype profiles of M. bovis isolates in France is the high level of diversity of the spoligotypes, which is associated with a high degree of homogeneity of the main group designed by clustering. This is illustrated by two elements. First, the total number of spoligotypes is very high, relative to the total number of typed isolates, when compared to many other locations, especially islands like Great Britain (10) or Australia (12) and developing countries like Cameroon (31) or Tanzania (27). Second, the frequency of the two main spoligotypes (BCG-like and GB54) is low compared to studies in other countries. For example, the two main spoligotypes in Great Britain account for 70% of the isolates (10; R. G. Hewinson, unpublished data). The most frequent spoligotype was found in 52% of the isolates in the Republic of Ireland (11), in 39.3% (Ferretti, personal communication) to 66.6% (37) of the isolates in Italy, and in 46% of cattle isolates in Spain (6). In a study conducted with 273 isolates from Australia, Canada, and Iran, 88% of the isolates belonged to the same spoligotype (12).
On a practical point of view, this tends to show that spoligotyping is appropriate for M. bovis differentiation in the case of France. These results illustrate the fact that the discriminatory power of the different typing techniques available for M. bovis varies with the country (5). To explain this diversity of the spoligotypes in France, and in the meantime the homogeneity of the BCG-like group, several explanations can be proposed. First, it is possible that the high diversity of breeds and of ecological situations may have exerted local selective pressure on BCG-like strains, particularly on the DR region. Secondly, the high propensity of France to exchange cattle, due to its geographical situation and due to a tradition of trade, may have allowed the introduction of cattle from neighboring countries. These countries share with France many similar features, which could explain the existence of a relatively low degree of divergence between many isolates present in these countries and those described in France (Fig. 2). Finally, it is possible that the competition between M. bovis strains was reduced by the TB control program. The data comparing the biodiversity of M. tuberculosis strains in developed and developing countries (41) suggest that when the prevalence of TB is high, a dominant strain tends to exclude the others. Conversely, in countries where the prevalence of human TB is low, more types can be present. This observation seems to be confirmed in our case, in the context of cattle TB due to M. bovis.
The fact that the majority of our spoligotypes are included in the same major group, including BCG-like, favors the hypothesis that the spoligotype BCG-like may be a parental spoligotype. To confirm this hypothesis, it would be interesting to investigate the genetic proximity between French isolates with the BCG-like spoligotype and the M. bovis BCG strains, especially those most closely related to the original M. bovis isolate from which BCG was derived (8), using other genetic markers. As the BCG-like profile is the most conserved within the M. bovis subspecies in terms of the number of spacers, the hypothesis of an ancestral status of this profile is also corroborated by the accepted concept that the evolution of the DR region occurs by spacer deletions (15). It has also been suggested in the case of Cameroon that a BCG-like strain has constituted the parental strain for the M. bovis strains presently isolated in Cameroon, due to the similarity of the dominant spoligotype in this country with BCG-like (31).
Using a parsimony-based method, we could notice on the one hand that the spoligotypes lacking a limited and uncharacteristic number of spacers (especially one spacer) are included in a “constellation” of spoligotypes, for which no definite link is possible to establish with other spoligotypes with more deletions. On the other hand, there is also a high level of incertitude in the case of some of the more-deleted spoligotypes; in this case, the high number of possible events (from only one big deletion to a series of limited or unique deletions) makes it very difficult to determine their precise origin(s). The use of other independent genetic markers remains necessary to go deeper into the knowledge of phylogenetic relationships between M. bovis isolates.
In conclusion, our results are the first, to our knowledge, which concern more than 1,300 isolates of M. bovis collected during a period of 20 years (excluding 1980 to 1982). Our results tend to show that, at least in France, there has been a dominant spoligotype, BCG-like, for a long time, and these results tend to confirm the global stability of the DR region (10, 29, 34, 38), especially compared to those of other markers, like IS6110 (29). But, our results are also suggestive of a progressive evolution of spoligotypes with time. This is illustrated by the slight relative decrease with time of BCG-like isolates, by the very high number of spoligotypes in the BCG-like group, and by the possible switch from BCG-like to F004 in two herds. This would confirm that the evolution of the DR region occurs by successive deletions (15). This theory is highly compatible with the very plausible hypothesis that the more ancient isolates in France would be characterized by the BCG-like spoligotype from which the other French spoligotypes could be derived.
For a more practical point of view, it is interesting to observe that for French isolates the spoligotyping technique offers a rather high level of discrimination compared to some other countries (10, 12, 27, 31). As a screening technique, it appears to be very useful and can even be used for the spatial (geotyping) and temporal (chronotyping) traceability of outbreaks, to a certain extent, at least if rare spoligotypes are involved. It would be interesting to compare its performances to that of other available techniques, like RFLP with the PGRS or pUCD probes (32, 33) or like VNTR typing. The use of these additional techniques will be necessary for more precise epidemiological applications, especially for those isolates presenting a very common spoligotype, like BCG-like or GB54.
At the European and even international level, our study shows that it is very useful to compare the spoligotypes observed in different countries. The constitution of an international data bank and the development of collaborative studies would allow the elaboration of an evolving map of bovine TB in the world. This would help to better understand the dynamics of this disease by integrating trade data, regional specificity, like the involvement of wild reservoirs, and some intrinsic factors, like the capacity of the DR region to evolve with time.
ACKNOWLEDGMENT
We thank very much Guillaume Lecointre, Institut de Systématique, IFR CNRS 1541, Museum National d'Histoire Naturelle, for kindly welcoming us on his MAC and for allowing us to use his personal version of PAUP*.
REFERENCES
- 1.Alito A, Morcillo N, Scipioni S, Dolmann A, Romano M I, Cataldi A, van Soolingen D. The IS6110 restriction fragment polymorphism in particular multidrug-resistant Mycobacterium tuberculosis strains may evolve too fast for reliable use in outbreak investigation. J Clin Microbiol. 1999;37:788–791. doi: 10.1128/jcm.37.3.788-791.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. Direction Générale de l'Alimentation (ed.). Paris, France: Ministère de l'Agriculture et de la Pêche; 2000. Enquête statistique annuelle 1998. [Google Scholar]
- 3.Anonymous. Commission Decision C(2000)-4069 of 11 July 2000 amending the decision 99/467/CE establishing the officially tuberculosis-free status of bovine herds of certain member states or regions of member states. Eur Commun Off J. 2000;L176:51. [Google Scholar]
- 4.Aranaz A, Liebana E, Gomez-Manpaso E, Galan J C, Cousins D, Ortega A, Blasquez J, Baquero F, Mateos A, Suarez G, Dominguez L. Mycobacterium tuberculosis subsp. caprae subsp. nov.: a taxonomic study of a new member of the Mycobacterium tuberculosis complex isolated from goats in Spain. Int J Syst Bacteriol. 1999;49:1263–1273. doi: 10.1099/00207713-49-3-1263. [DOI] [PubMed] [Google Scholar]
- 5.Aranaz A, Liebana E, Mateos A, Dominguez L, Cousins D. Restriction Fragment Length Polymorphism and spacer oligonucleotide typing: a comparative analysis of fingerprinting strategies for Mycobacterium bovis. Vet Microbiol. 1998;61:311–324. doi: 10.1016/s0378-1135(98)00192-8. [DOI] [PubMed] [Google Scholar]
- 6.Aranaz A, Liebana E, Mateos A, Dominguez L, Vidal D, Domingo M, Gonzolez O, Rodriguez-Ferri E F, Bunschoten A E, van Embden J D A, Cousins D. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: a tool for studying epidemiology of tuberculosis. J Clin Microbiol. 1996;34:2734–2740. doi: 10.1128/jcm.34.11.2734-2740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer J, Andersen A B, Kremer K, Miorner H. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J Clin Microbiol. 1999;37:2602–2606. doi: 10.1128/jcm.37.8.2602-2606.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behr M A, Small P M. A historical and molecular phylogeny of BCG strains. Vaccine. 1999;17:915–922. doi: 10.1016/s0264-410x(98)00277-1. [DOI] [PubMed] [Google Scholar]
- 9.Benet J J. Pour en finir avec la tuberculose. Bull Groupements Techniques Vet. 2001;2001:417–424. [Google Scholar]
- 10.Clifton-Hadley R S, Inwald J, Archer J, Hughes S, Palmer N, Sayers A R, Sweeney K, van Embden J D A, Hewinson R G. Recent advances in DNA fingerprinting using spoligotyping—epidemiological applications in bovine tuberculosis. Brit Cattle Vet Assoc. 1998;6:79–82. [Google Scholar]
- 11.Costello E, O'Grady D, Flynn O, O'Brien R, Rogers M, Quigley F, Egan J, Griffin J. Study of restriction fragment length polymorphism analysis and spoligotyping for epidemiological investigation of Mycobacterium bovis infection. J Clin Microbiol. 1999;37:3217–3222. doi: 10.1128/jcm.37.10.3217-3222.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cousins D, Williams S, Liebana E, Aranaz A, Bunschoten A, van Embden J, Ellis T. Evaluation of four DNA typing techniques in epidemiological investigations of bovine tuberculosis. J Clin Microbiol. 1998;36:168–178. doi: 10.1128/jcm.36.1.168-178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cousins D V, Williams S N, Dawson D J. Tuberculosis due to Mycobacterium bovis in the Australian population: DNA typing of isolates, 1970–1994. Int J Tuberc Lung Dis. 1999;3:722–731. [PubMed] [Google Scholar]
- 14.de Boer A S, Borgdorff M W, de Haas P E, Nagelkerke N J, van Embden J D, van Soolingen D. Analysis of the rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient isolates. J Infect Dis. 1999;180:1238–1244. doi: 10.1086/314979. [DOI] [PubMed] [Google Scholar]
- 15.Durr P A, Clifton-Hadley R S, Hewinson R G. Molecular epidemiology of bovine tuberculosis. I. Mycobacterium bovis genotyping. Rev Sci Tech Off Int Epizoot. 2000;19:675–688. doi: 10.20506/rst.19.3.1241. [DOI] [PubMed] [Google Scholar]
- 16.Durr P A, Clifton-Hadley R S, Hewinson R G. Molecular epidemiology of bovine tuberculosis. II. Applications of genotyping. Rev Sci Tech Off Int Epizoot. 2000;19:689–701. doi: 10.20506/rst.19.3.1240. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. Phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 18.Filliol I, Ferdinand S, Negroni L, Sola C, Rastogi N. Molecular typing of Mycobacterium tuberculosis based on variable tandem DNA repeats used alone and in association with spoligotyping. J Clin Microbiol. 2000;38:2520–2524. doi: 10.1128/jcm.38.7.2520-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frothingham R, Meeker-O'Connell W A. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 20.Goguet de la Salmonière Y O, Li H O, Torrea G, Bunschoten A, van Embden J D A, Gicquel B. Evaluation of spoligotyping in a study of transmission of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:2210–2214. doi: 10.1128/jcm.35.9.2210-2214.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goyal M, Lawn S, Afful B, Acheampong J W, Griffin G, Shaw R. Spoligotyping in molecular epidemiology of tuberculosis in Ghana. J Infect. 1999;38:171–175. doi: 10.1016/s0163-4453(99)90246-3. [DOI] [PubMed] [Google Scholar]
- 22.Goyal M, Saunders N A, van Embden J D A, Young D B, Shaw R J. Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment length polymorphism. J Clin Microbiol. 1997;35:647–651. doi: 10.1128/jcm.35.3.647-651.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groenen P, Bunschoten A, van Soolingen D, van Embden J D A. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 24.Guiterrez M, Samper S, Gavigan J A, Garcia Marin J F, Martin C. Differentiation by molecular typing of Mycobacterium bovis strains causing tuberculosis in cattle and goats. J Clin Microbiol. 1995;33:2953–2956. doi: 10.1128/jcm.33.11.2953-2956.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horgen L, Sola C, Devallois A, Goh K S, Rastogi N. Follow up of Mycobacterium tuberculosis transmission in the French West Indies by IS6110-DNA fingerprinting and DR-based spoligotyping. FEMS Immunol Med Microbiol. 1998;21:203–212. doi: 10.1111/j.1574-695X.1998.tb01167.x. [DOI] [PubMed] [Google Scholar]
- 26.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazwala R, Sinclair K, Challans J, Kambarage D M, Sharp J M, van Embden J D A, Daborn C J, Nyange J. Zoonotic importance of Mycobacterium tuberculosis complex organisms in Tanzania: a molecular biology approach. Rabat, Morocco: Actes Editions; 1997. [Google Scholar]
- 28.Kremer K, van Soolingen D, Frothingham R, de Haas W H, Hermans P W M, Martin C, Palittapongarnpim P, Plikaytis B B, Riley L W, Yakrus M A, Musser J M, van Embden J D A. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niemann S, Richter E, Rüsch-Gerdes S. Stability of Mycobacterium tuberculosis IS6110 restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial isolates from patients with drug-resistant tuberculosis. J Clin Microbiol. 1999;37:409–412. doi: 10.1128/jcm.37.2.409-412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niemann S, Richter E, Rüsch-Gerdes S. Differentiation among members of the Mycobacterium tuberculosis complex by molecular and biochemical features: evidence for two pyrazinamide-susceptible subtypes of M. bovis. J Clin Microbiol. 2000;38:152–157. doi: 10.1128/jcm.38.1.152-157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Njanpop-Lafourcade B M, Inwald J, Ostyn A, Durand B, Hughes S, Thorel M F, Hewinson G, Haddad N. Molecular typing of Mycobacterium bovis isolates from the Cameroon. J Clin Microbiol. 2001;39:222–227. doi: 10.1128/JCM.39.1.222-227.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Brien R, Danilowicz B S, Bailey L, Flynn O, Costello E, O'Grady D, Rogers M. Characterization of the Mycobacterium bovis restriction fragment length polymorphism DNA probe pUCD and performance comparison with standard methods. J Clin Microbiol. 2000;38:3362–3369. doi: 10.1128/jcm.38.9.3362-3369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Brien R, Flynn O, Costello E, O'Grady D, Rogers M. Identification of a novel DNA probe for strain typing Mycobacterium bovis by restriction fragment length polymorphism analysis. J Clin Microbiol. 2000;38:1723–1730. doi: 10.1128/jcm.38.5.1723-1730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian L, van Embden J D A, van der Zanden A G M, Weeltevreden E F, Duanmu H, Douglas J T. Retrospective analysis of the Beijing family of Mycobacterium tuberculosis in preserved lung tissues. J Clin Microbiol. 1999;37:471–474. doi: 10.1128/jcm.37.2.471-474.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roring S, Brittain D, Bunschoten A E, Hughes M S, Skuce R A, van Embden J D, Neill S D. Spacer oligotyping of Mycobacterium bovis isolates compared to typing by restriction fragment length polymorphism using PGRS, DR and IS6110 probes. J Clin Microbiol. 1998;36:111–120. doi: 10.1016/s0378-1135(98)00178-3. [DOI] [PubMed] [Google Scholar]
- 36.Roring S, Hughes M S, Skuce R A, Neill S D. Simultaneous detection and strain differentiation of Mycobacterium bovis directly from bovine tissue specimens by spoligotyping. Vet Microbiol. 2000;74:227–236. doi: 10.1016/s0378-1135(00)00189-9. [DOI] [PubMed] [Google Scholar]
- 37.Serraino A, Marchetti G, Sanguinetti V, Rossi M C, Zanoni R G, Catozzi L, Bandera A, Dini W, Mignone W, Franzetti F, Gori A. Monitoring of transmission of tuberculosis between wild boars and cattle: genotypical analysis of strains by molecular epidemiology techniques. J Clin Microbiol. 1999;37:2766–2771. doi: 10.1128/jcm.37.9.2766-2771.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soini H, Pan X, Amin A, Graviss E A, Siddiqui A, Musser J M. Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping. J Clin Microbiol. 2000;38:669–676. doi: 10.1128/jcm.38.2.669-676.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sola C, Horgen L, Devallois A, Rastogi N. Combined numerical analysis based on the molecular description of Mycobacterium tuberculosis by four repetitive sequence-based DNA typing systems. Res Microbiol. 1998;149:349–360. doi: 10.1016/s0923-2508(98)80440-3. [DOI] [PubMed] [Google Scholar]
- 40.Taylor G, Goyal M, Legge A J, Shaw R J, Young D. Genotypic analysis of Mycobacterium tuberculosis from medieval human remains. Microbiology. 1999;145:899–904. doi: 10.1099/13500872-145-4-899. [DOI] [PubMed] [Google Scholar]
- 41.van Soolingen D, Qian L, de Haas P E W, Douglas J T, Traore H, Portaels F, Qing H Z, Enkhsaikan D, Nymadawa P, van Embden J D A. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol. 1995;33:3224–3228. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viana-Niero C, Guiterrez C, Sola C, Filliol I, Boulhhbal F, Vincent V, Rastogi N. Genetic diversity of Mycobacterium africanum clinical isolates based on IS6110-restriction fragment polymorphism analysis, spoligotyping, and variable number tandem repeats. J Clin Microbiol. 2001;39:57–65. doi: 10.1128/JCM.39.1.57-65.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. 3rd ed. Vol. 1. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1989. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z, Ijaz K, Bates J H, Eisenach K D, Cave M D. Spoligotyping and polymorphic GC-rich repetitive sequence fingerprinting of Mycobacterium tuberculosis strains having few copies of IS6110. J Clin Microbiol. 2000;38:3572–3576. doi: 10.1128/jcm.38.10.3572-3576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zumarraga M, Martin C, Samper S, Alito A, Latini O, Bigi F, Roxo E, Cicuta M E, Errico F, Ramos M C, Cataldi A, van Soolingen D, Romano M I. Usefulness of spoligotyping in molecular epidemiology of Mycobacterium bovis-related infections in South America. J Clin Microbiol. 1999;37:296–303. doi: 10.1128/jcm.37.2.296-303.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]