Abstract
The use of the 16S rRNA gene for identification of nontuberculous mycobacteria (NTM) provides a faster and better ability to accurately identify them in addition to contributing significantly in the discovery of new species. Despite their associated problems, many rely on the use of public sequence databases for sequence comparisons. To best evaluate the taxonomic status of NTM species submitted to our reference laboratory, we have created a 16S rRNA sequence database by sequencing 121 American Type Culture Collection strains encompassing 92 species of mycobacteria, and have also included chosen unique mycobacterial sequences from public sequence repositories. In addition, the Ribosomal Differentiation of Medical Microorganisms (RIDOM) service has made freely available on the Internet mycobacterial identification by 16S rRNA analysis. We have evaluated 122 clinical NTM species using our database, comparing >1,400 bp of the 16S gene, and the RIDOM database, comparing ∼440 bp. The breakdown of analysis was as follows: 61 strains had a sequence with 100% similarity to the type strain of an established species, 19 strains showed a 1- to 5-bp divergence from an established species, 11 strains had sequences corresponding to uncharacterized strain sequences in public databases, and 31 strains represented unique sequences. Our experience with analysis of the 16S rRNA gene of patient strains has shown that clear-cut results are not the rule. As many clinical, research, and environmental laboratories currently employ 16S-based identification of bacteria, including mycobacteria, a freely available quality-controlled database such as that provided by RIDOM is essential to accurately identify species or detect true sequence variations leading to the discovery of new species.
The notion that sequence-based methodologies will take their place in routine clinical laboratories is an increasing reality. The initial cost of equipment, i.e., automated sequencers, can quickly be recovered with savings in personnel, time, and ultimately in health care costs. Laboratories are beginning to rely more on genotypic characterization, which is more specific and easier to standardize than conventional tests (14).
Leaders in the field of diagnostic mycobacteriology acknowledge the need for advanced methods for more accurate identification of mycobacteria to the species level (36, 39, 44). The number of members within the genus is highly underestimated, and of the 92 currently established species, as listed in J. P. Euzéby's: “List of bacterial names with standing in nomenclature” (http://www.bacterio.cict.fr/m/mycobacterium.html), many are considered potentially pathogenic. Biochemical characteristics have been well established for the most well-known Mycobacterium species, but with a rapid increase of newly described species, this becomes more and more complex, difficult, and perhaps obsolete. Also, many species that are biochemically inert or extremely slow growing contribute further to these problems.
The 16S rRNA gene is the most widely accepted gene used for bacterial identification. It has contributed greatly to the discovery of new species of the Mycobacterium genus, and it continues to serve as an important tool as an alternative to phenotypic identification methods. The direct sequencing of amplified DNA from the 16S rRNA gene of Mycobacterium species is well described (3, 21, 22, 32). Present sequencing technologies allow the acquisition of unambiguous and definite identification.
Currently, sequence-based identification is normally provided from a reference laboratory and may or may not be performed in parallel with phenotypic characterization or additional tests. The same applies to PCR-restriction fragment length polymorphism analysis (PRA) or high-performance liquid chromatography technology, also used as primary identification tools. As a reference laboratory for mycobacteriology, we commonly receive clinical isolates of nontuberculous mycobacteria (NTM) for species identification. To minimize turnaround time, 16S rRNA sequencing is performed on all isolates received and has become our primary tool for identification. Studies using 16S ribosomal DNA (rDNA) sequence-based identification methods for the identification of mycobacteria show that this technique is more rapid (hours versus weeks) and more accurate than conventional methods (18, 23, 28, 39). It also provides information on the taxonomic relatedness of new species, which may not be possible with other technologies.
As this technology is commonly employed for the identification and characterization of established and novel species, the number of 16S rRNA gene sequences submitted to public databases continues to grow rapidly. Many people have relied on these databases to identify species or establish new species. However, their use for identification is not without limitations. There are multiple problems with present sequence repositories, such as base errors, ambiguous base designation and incomplete sequences, which may not be evident to users and will often provide misleading results. To overcome these problems, Applied Biosystems (Foster City, Calif.) has developed the MicroSeq 16S rDNA Bacterial Identification System, a quality-controlled database containing over 1,200 bacterial type strains, including approximately 70 species of Mycobacterium. It also includes all materials required for amplification and sequencing of the full or partial 16S gene and a software package for analysis against the MicroSeq database. However, access to this database is on a cost per n uses basis and does not include all sequences of established species such as M. lentiflavum and M. haemophilum as well as many of the most recently described species. Consequently, users have often found discrepancies or inconclusive results (10, 28). This is not surprising, due to the overwhelming evidence that a much larger number of species exist than are currently validated. Of the 92 established mycobacterial species, one-third of them (n = 31) have been characterized in the last decade, with 19 being characterized since 1996 alone. Furthermore, the number of unique, uncharacterized mycobacterial 16S rRNA gene sequences submitted to public databases continues to increase at a much faster pace than publications are made available.
The Ribosomal Differentiation of Medical Microorganisms (RIDOM) service is currently in the process of making freely available a database much like that of MicroSeq. It comprises sequences determined in their laboratory, corresponding to bases 54 to 510 of the 16S rRNA gene of Escherichia coli. A database for Neisseriaceae and Moraxellaceae is available (16, 17), and more recently, a comprehensive mycobacteriology database has been made available (http://www.ridom.de). The RIDOM web-based service contains not only sequence data but also additional information such as clinical data, phenotypic and genotypic characteristics, and related references for each species, established as well as nonestablished.
In this work, we have determined the nearly complete 16S rRNA gene sequence data (>1,400 bp) for 121 American Type Culture Collection (ATCC) strains of mycobacterium species encompassing 92 species. The type strains among them were compared to those in public sequence repositories like GenBank, EMBL, and DDBJ, and also to the RIDOM database. New sequences have emerged from these reference strains, and these will be discussed. We have also determined the nearly complete 16S rRNA gene sequences of 122 clinical NTM species obtained in the last 2 years and have evaluated them using our database, comparing >1,400 bp of the 16S gene, and the RIDOM database, comparing ∼440 bp.
MATERIALS AND METHODS
Reference strains.
A total of 121 ATCC strains were sequenced for our 16S rDNA database. These included type strains from 80 species and 26 additional strains encompassing 18 of the species. They are listed in Tables 1 and 2. Twelve additional “species” obtained from the ATCC collection (three with two strains) for which the status is not validated were also tested (Table 3). All strains were grown in BACTEC 12B liquid medium and subcultured onto Lowenstein-Jensen slants or Middlebrook 7H10 agar plates. Organisms were incubated at 37°C, with the exception of M. ulcerans, M. marinum, M. burulii, and M. haemophilum, which were incubated at 30°C, and M. xenopi and M. botniense, which were incubated at 42°C. M. paratuberculosis was subcultured using 12B medium and Middlebrook 7H10 agar supplemented with Mycobactin J (Allied Monitor, Inc., Fayette, Mo.), whereas M. haemophilum was subcultured using 12B medium and Middlebrook 7H10 plates supplemented with hemin.
TABLE 1.
List of type strains of established mycobacterium species which were sequenced in our laboratory
| Species | Type strain | Code for result withn:
|
|||
|---|---|---|---|---|---|
| BLAST | RDP-II | RIDOM | |||
| M. abscessus | ATCC 19977 | 3 | 3 | 1a | |
| M. africanum | ATCC 25420 | 1b | 1b | 1b | |
| M. agri | ATCC 27406 | 4 | 4 | 1 | |
| M. aichiense | ATCC 27280 | 2 | 2 | 1 | |
| M. alvei | ATCC 51304 | 1 | 1 | 1 | |
| M. asiaticum | ATCC 25276 | 3 | 3 | 1 | |
| M. aurum | ATCC 23366 | 2 | 3 | 1 | |
| M. austroafricanum | ATCC 33464 | 1 | 1 | 1 | |
| M. avium subsp. avium | ATCC 25291 | 1c | 1c | 1c | |
| M. avium subsp. paratuberculosis | ATCC 19698 | 1c | 1c | 1c | |
| M. avium subsp. silvaticum | ATCC 49884 | 1c | 1c | 1c | |
| M. botniense | ATCC 700701 | 1 | 1 | 1 | |
| M. bovis | ATCC 19210 | 1b | 1b | 1b | |
| M. branderi | ATCC 51789 | 2 | 2 | 1 | |
| M. brumae | ATCC 51384 | 4 | 4 | 1 | |
| M. celatum | ATCC 51131 | NAd | NAd | NAd | |
| M. chelonae | ATCC 35752 | 3 | 2 | 1 | |
| M. chitae | ATCC 19627 | 2 | 2 | 1 | |
| M. chlorophenolicum | ATCC 49826 | 2 | 2 | 1 | |
| M. chubuense | ATCC 27278 | 3 | 2 | 1 | |
| M. confluentis | ATCC 49920 | 1 | 1 | 1 | |
| M. cookii | ATCC 49103 | 3 | 3 | 1 | |
| M. diernhoferi | ATCC 19340 | 3 | 3 | 1 | |
| M. duvalii | ATCC 43910 | 1 | 1 | 1 | |
| M. fallax | ATCC 35219 | 3 | 2 | 1 | |
| M. farcinogenes | ATCC 35753 | 2 | 2 | 1e | |
| M. flavescens | ATCC 14474 | 2 | 3 | 1f | |
| M. fortuitum | ATCC 6841 | 2 | 2 | 1g | |
| M. fortuitum subsp. acetamidolyticum | ATCC 35931 | 2g | 2g | 1g | |
| M. gadium | ATCC 27726 | 2 | 2 | 1 | |
| M. gastri | ATCC 15754 | 2h | 3h | 1h | |
| M. gilvum | ATCC 43909 | 2 | 1 | 1 | |
| M. goodii | ATCC 700504 | 3 | 3 | 1 | |
| M. gordonae | ATCC 14470 | 2 | 2 | 1 | |
| M. haemophilum | ATCC 29548 | 2 | 1 | 1 | |
| M. hassiacum | ATCC 700660 | 1 | 1 | 1 | |
| M. heidelbergense | ATCC 51253 | 2 | 2 | 1 | |
| M. hiberniae | ATCC 49874 | 2 | 2 | 1 | |
| M. interjectum | ATCC 51457 | 1 | 1 | 1 | |
| M. intermedium | ATCC 51848 | 2 | 2 | 1 |
| M. intracellulare | ATCC 13950 | 2 | 2 | 1 |
| M. kansasii | ATCC 12478 | 3h | 2h | 1 |
| M. komossense | ATCC 33013 | 2 | 2 | 1 |
| M. kubicae | ATCC 700732 | 3 | 3 | 1 |
| M. lentiflavum | ATCC 51985 | 1i | 1 | 1 |
| M. madagascariense | ATCC 49865 | 2 | 2 | 1 |
| M. mageritense | ATCC 700351 | 3 | 1 | 1 |
| M. malmoense | ATCC 29571 | 1 | 2 | 1 |
| M. marinum | ATCC 927 | 3 | 3 | 1j |
| M. microti | ATCC 19422 | 1b | 1b | 1b |
| M. moriokaense | ATCC 43059 | 4 | 4 | 1 |
| M. mucogenicum | ATCC 49650 | 3 | 3 | 1 |
| M. neoaurum | ATCC 25795 | 3 | 3 | 1 |
| M. nonchromogenicum | ATCC 19530 | 2 | 2 | 1 |
| M. obuense | ATCC 27023 | 2 | 2 | 1 |
| M. parafortuitum | ATCC 19686 | 1 | 1 | 1 |
| M. peregrinum | ATCC 14467 | 3 | 3 | 1k |
| M. phlei | ATCC 11758 | 3 | 2 | 1 |
| M. porcinum | ATCC 33776 | 4 | 4 | 1l |
| M. poriferae | ATCC 35087 | 4 | 4 | 1 |
| M. pulveris | ATCC 35154 | 4 | 4 | 1 |
| M. rhodesiae | ATCC 27024 | 4 | 4 | 1 |
| M. scrofulaceum | ATCC 19981 | 3 | 3 | 1 |
| M. senegalense | ATCC 35796 | 3 | 3 | 1e |
| M. septicum | ATCC 700731 | 3 | 3 | 1k |
| M. shimoidei | ATCC 27962 | 2 | 2 | 1 |
| M. simiae | ATCC 25275 | 2 | 3 | 1 |
| M. smegmatis | ATCC 19420 | 2 | 2 | 1 |
| M. sphagni | ATCC 33027 | 3 | 3 | 1 |
| M. szulgai | ATCC 35799 | 3 | 2 | 1 |
| M. terrae | ATCC 15755 | 2 | 2 | 1 |
| M. thermoresistibile | ATCC 19527 | 2 | 2 | 1 |
| M. tokaiense | ATCC 27282 | 4 | 4 | 1m |
| M. triplex | ATCC 700071 | 1 | 1 | 1 |
| M. triviale | ATCC 23292 | 2 | 2 | 1 |
| M. tuberculosis | ATCC 27294 | 1b | 1b | 1b |
| M. ulcerans | ATCC 19423 | 2 | 2 | 1j |
| M. vaccae | ATCC 15483 | 3 | 3 | 1 |
| M. wolinskyi | ATCC 700010 | 3 | 2 | 1 |
| M. xenopi | ATCC 19250 | 2 | 2 | 1 |
Both M. chelonae and M. abscessus gave 100% similarity.
Members of the M. tuberculosis complex (M. africanum, M. bovis, M. microti, and M. tuberculosis) are identical to each other and gave identical results.
M. avium, M. paratuberculosis, and M. silvaticum are identical to each other and gave identical results.
M. celatum was not tested against the databases (NA, not applicable). Please refer to text for explanation.
M. farcinogenes, M. fortuitum ATCC 49403 (third biovar) and M. senegalense gave 100% similarity.
RIDOM indicated 100% similarity with M. acapulcensis, which we had determined to be identical to three ATCC strains of M. flavescens, including the type strain. See Table 2.
Both M. fortuitum and M. fortuitum subsp. acetamidolyticum are identical to each other and gave identical results.
Although M. gastri and M. kansasii have identical sequences, which is reflected in RIDOM, BLAST results show M. gastri with a higher score, while RDP-II shows M. kansasii with a higher score.
Indicated as Mycobacterium sp. only. User must refer to the reference for species name.
Both M. marinum and M. ulcerans gave 100% similarity.
Both M. peregrinum and M. septicum gave 100% similarity.
Both M. porcinum and M. fortuitum ATCC 49404 (third biovar) gave 100% similarity.
Both M. tokaiense and M. murale gave 100% similarity.
Also shown is the evaluation of these strains against public databases using BLAST and RDP-II (dated 30 March 2001) and a quality-controlled database (RIDOM [dated 8 March 2001]). Corresponding codes for BLAST, RDP-II, and RIDOM results are as follows: 1, correctly identified by top score with 100% sequence similarity; 2, correctly identified by top score, but with ≤99% sequence similarity; 3, species present in database, but did not hold top score; 4, species not present in database. In general, over 1,400 bp of the gene was aligned in BLAST and RDP-II, whereas RIDOM aligns against E. coli bp 54 to 510.
TABLE 2.
Evaluation of ATCC strains sequenced in our laboratory that are not type strains but correspond to established speciesa
| Species | Reference strain no. | Analysis in comparison with the corresponding type strain |
|---|---|---|
| M. asiaticum | ATCC 25274 | 1 variation at bp 1020 |
| M. avium | ATCC 35717 | Identical to type strain |
| M. bovis | ATCC 35720 | Identical to type strain |
| ATCC 35726 | Identical to type strain | |
| M. branderi | ATCC 51788 | Identical to type strain |
| M. chelonae | ATCC 19237 | 3 variations at bp 999, 1008, 1039 (refer to text) |
| M. chitae | NCTC 10495 | Identical to type strain |
| M. diernhoferi | ATCC 19344 | Identical to type strain |
| M. flavescens | ATCC 23008 | Identical to type strain |
| ATCC 23033 | 18 variations from type strain | |
| M. fortuitum | ATCC 49403 | Identical to M. farcinogenes |
| ATCC 49404 | 1 variation from M. porcinum | |
| M. kansasii | ATCC 35775 | Identical to type strain |
| M. microti | ATCC 11152 | Identical to type strain |
| ATCC 35782 | Identical to type strain | |
| M. nonchromogenicum | ATCC 19531 | Identical to type strain |
| ATCC 35783 | Identical to type strain | |
| M. peregrinum | ATCC 23001 | Identical to type strain |
| ATCC 23015 | Identical to type strain | |
| M. porcinum | ATCC 33775 | Identical to type strain |
| M. scrofulaceum | ATCC 35786 | Identical to type strain |
| ATCC 35788 | 40 variations from type strain (corresponds to strain MCRO 33 [AF152559]) | |
| M. ulcerans | ATCC 35839 | Identical to type strain |
| ATCC 35840 | Identical to type strain | |
| M. vaccae | ATCC 25951 | Identical to type strain |
| M. xenopi | ATCC 25841 | Identical to type strain |
Sequences were compared to the type strains determined in our laboratory.
TABLE 3.
Evaluation of nonestablished species from the ATCC collection: clarification of past-mentioned “species” of mycobacteriac
| Strain group | Species sequenced | Strain(s) | Sequencing result in comparison with our database | RIDOM result (%) |
|---|---|---|---|---|
| Identical to type strain of an established species | ||||
| “M. acapulcensis” | ATCC 14473T | Identical to M. flavescens ATCC 14474T and ATCC 23008 | M. acapulcensis ATCC 14473 (100) | |
| “M. brunense” | ATCC 23434 | Identical to M. avium, M. paratuberculosis, M. silvaticum | M. avium, M. paratuberculosis, M. silvaticum (100a) | |
| “M. burulii” | ATCC 25893, ATCC 25894 | Both identical to M. tuberculosis complex | M. caprae, M. tuberculosis, M. bovis, M. africanum, M. microti (100a) | |
| “M. lactis” | ATCC 27356 | Identical to M. hiberniae | M. hiberniae (100) | |
| “M. valentiae” | ATCC 29356 | Identical to M. duvalii | M. duvalii (100) | |
| With unique sequences | ||||
| “M. album” | ATCC 29676, ATCC 29677 | Unique; closest to M. wolinskyi, differing at E. coli bp 1006 and 1135 | M. wolinskyi (100a) | |
| “M. engbaekii” | ATCC 27353 | Unique; closest to M. hiberniae, differing at E. coli bp 305, 594, and 601 | M. engbaekii (100) | |
| “M. lacticola” | ATCC 9626T | Unique; closest to M. neoaurum | M. lacticola (100) | |
| “M. paraffinicum” | ATCC 12670 | Unique; closest to M. scrofulaceum | M. paraffinicum (100) | |
| “M. petroleophilum” | ATCC 21497T | Unique; closest to M. gilvum, differing by 11 bases | M. petroleophilum (99.88b) | |
| “M. seriolae” | ATCC 49159, ATCC 49160 | Unique; closest to M. ulcerans, differing at bases 95, 488, 967, 1005, and 1215 | M. marinum, M. ulcerans, 2 mismatchesa | |
| “M. shinshuense” | ATCC 33728T | Unique; closest to M. ulcerans, differing at bases 492 and 1288 | M. shinsuense (100) |
Strain or “species” not currently present in the RIDOM database.
The RIDOM sequence has a G at E. coli base 490, whereas our reference strain and one clinical strain have an R (G or A).
The species that have a unique sequence are included in the phylogenetic tree in Fig. 1 and are indicated by quotation marks.
Patient strains.
A total of 122 strains representing all NTM strains submitted for identification from January 1999 to February 2001 were tested using 16S rRNA-based identification. They are listed in Table 4. Specimens were normally submitted on solid medium, from which a DNA lysate could directly be prepared.
TABLE 4.
Evaluation of clinical NTM strains submitted to our laboratory in the span of 25 months
| Group | Species or identification | n | Comment |
|---|---|---|---|
| Corresponding with 100% similarity with a type strain sequence | M. abscessus | 1 | |
| M. avium or M. paratuberculosis | 4 | Phenotypically M. avium | |
| M. branderi | 5 | ||
| M. chelonae | 1 | ||
| M. fortuitum | 6 | ||
| M. gilvum | 1 | ||
| M. goodii | 6 | ||
| M. gordonae | 1 | ||
| M. kansasii or M. gastri | 6 | ||
| M. heckeshornense | 1 | ||
| M. lentiflavum | 7 | ||
| M. malmoense | 2 | ||
| M. marinum | 3 | ||
| M. obuense | 1 | ||
| M. peregrinum | 2 | ||
| M. petroleophilum | 1 | ||
| M. scrofulaceum | 1 | ||
| M. simiae | 2 | ||
| M. thermoresistibile | 4 | ||
| M. triplex | 1 | ||
| M. triviale | 1 | ||
| M. vaccae | 2 | ||
| M. xenopi | 2 | ||
| Subtotal | 61 | ||
| Divergent, but 16S rDNA sequence closely resembling that of an established species | M. interjectum-like | 1, 1 | 5- and 3-base divergence from type strain |
| M. gordonae-like | 1, 1, 1 | 3-, 3- and 5-base divergence from type strain; one of them shows salmon pigmentation | |
| M. gordonae group II (20) | 2 | 2-base divergence from type strain | |
| M. gordonae group III (20) | 1 | 1-base divergence from type strain | |
| M. xenopi-like | 1 | 1-base divergent; 1 Y at 213 | |
| M. xenopi-like | 1 | 27 ambiguities, 4 base variationsa | |
| M. kansasii subspecies (33) | 2 | 8-base divergence from type strain | |
| M. paraffinicum-like | 1, 1 | 2- and 3-base divergence from “M. paraffinicum” ATCC 12670 | |
| M. chelonae chemovar niacinogenes | 2 | 3-base divergence from type strain of M. chelonae | |
| M. novocastrense | 2 | 2-base divergence from type strain; phenotypically identical to M. flavescens | |
| M. neoaurum-like | 1 | 1-base divergence from type strain. | |
| Subtotal | 19 | ||
| Sequence corresponding to uncharacterized strain in GenBank | MCRO 6 [X93032] | 4 | 6-base divergence from M. nonchromogenicum |
| MCRO 8 [X93034] | 2 | 19-base divergence from M. lentiflavum; MACb probe positive | |
| MCRO 17 [X93028] | 5 | 6-base divergence from M. elephantis; 16 from M. pulveris | |
| Subtotal | 11 | ||
| Unique sequence, closest to | M. lentiflavum | 1, 2 | Differs by 15 and 17 bases; phenotypically similar to M. lentiflavum |
| M. austroafricanum | 1 | Differs by 35 bases | |
| M. fortuitum | 10 | Differs by 22 bases | |
| M. terrae | 1, 1, 2, 1 | Differs by 11, 12, 27, and 28 bases | |
| M. heckeshornense | 2 | Differs by 37 bases | |
| M. monacense | 1 | Differs by 14 bases | |
| M. chelonae | 1 | Differs by 21 bases; grew only on iron uptake medium | |
| M. bohemicum | 2 | Differs by 9 bases | |
| M. malmoense | 1 | Differs by 9 bases | |
| M. goodii | 1 | Differs by 10 bases | |
| M. triviale | 1 | Differs by 43 bases | |
| M. wolinskyi | 2 | Differs by 20 bases | |
| M. agri | 1 | Differs by 10 bases | |
| Subtotal | 31 |
The ambiguities in this isolate were represented by double peaks which corresponded to each base of M. heckeshornense and M. xenopi where the two species differ. Please refer to the text.
MAC, M. avium complex.
Preparation of DNA.
The equivalent of one large colony of organism was suspended in 1 ml of sterile TE buffer (Tris-HCl, 10 mM; EDTA, 1 mM) and boiled at 100°C for 10 min for mycobacterial inactivation. The organism was then mechanically lysed using a Mini Bead-Beater (Biospec Products, Bartlesville, Okla.) at maximum speed (set at 50) for 2 min (set at 12). The lysate was then centrifuged at 12,000 × g for 2 min to precipitate cellular debris, and the supernatant was transferred to a new sterile tube. DNA was quantitated using the PicoGreen dsDNA Quantitation Kit (Molecular Probes, Inc., Eugene, Oreg.) with a TD-700 Laboratory Fluorometer (Turner Designs, Sunnyvale, Calif.). The lysates were then stored at −20°C until required for PCR.
DNA amplification.
PCR amplification of the nearly complete 16S rRNA gene was performed for all isolates. Each reaction mixture contained approximately 10 ng of DNA; 2.5 mM MgCl2; 1× PCR buffer (Amersham Pharmacia Biotech, Baie d'Urfé, Quebec, Canada); a 200 μM concentration (each) of dCTP, dGTP, dATP, and dTTP; 1,000 pmol of each forward and reverse primer; and 1.25 U of Taq DNA polymerase (Amersham Pharmacia Biotech) in a final volume of 50 μl. Primers used were 8FPL (5′ AGT TTG ATC CTG GCT CAG 3′) and primer 1492 (5′ GGT TAC CTT GTT ACG ACT T 3′) (31), corresponding to E. coli 16S rRNA positions 8 to 27 and 1509 to 1491, respectively (5). The PCR was performed using the Perkin-Elmer GeneAmp PCR system 2400 with a cycle of 94°C for 5 min; 30 cycles of 94, 55, and 72°C for 1 min each; and a final extension at 72°C for 10 min, and was held at 4°C. Fifteen microliters of the PCR product was subjected to electrophoresis to ensure that successful amplification had occurred and that the correct band was obtained. The remaining PCR product was purified using MicroCon Centrifugal Filter Devices (Millipore Corporation, Nepean, Ontario, Canada), quantified using UV absorbance at 260 nm, and diluted to a concentration of 50 ng/μl for sequencing setup.
Sequencing of the 16S rRNA gene.
An ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) was used for the sequencing of the PCR product. The sequencing reaction required 4 ml of Premix, 3.2 pmol of sequencing primer, and 150 ng of the PCR product template in a total volume of 10 μl. Primers chosen allowed the sequencing of the nearly complete gene with overlap covering most regions, with a minimal set of primers required. These were ER11G (reverse: E. coli bp 359 to 341), ER10 (forward: E. coli bp 103 to 119) (45), 806R (reverse: E. coli bp 805 to 786) (31), pFr (reverse: E. coli bp 1073 to 1053), pFf (forward: E. coli bp 1053 to 1073) (12), and 1492. Additional primers were occasionally used to cover regions of ambiguity. The sequencing product was purified using the recommended Centricep columns (Princeton Separations, Adelphia, N.J.), followed by preparation for running onto the ABI PRISM 310 Genetic Analyzer, in accordance with the instructions of the manufacturer (Applied Biosystems).
The sequencing output from the ABI PRISM 310 Genetic Analyzer was analyzed using the accompanying DNA Sequence Analyzer computer software (Applied Biosystems). The Lasergene program (version 4.01; DNASTAR, Inc., Madison, Wis.) was used for sequence assembly, sequence alignment, and phylogenetic analysis. Multiple sequence alignments were determined using the CLUSTAL method algorithm. Analysis of clinical NTM strains was performed by comparing their sequences against the sequences of reference strains determined in our laboratory as well as select sequences obtained from GenBank.
Comparisons of type strains sequences with Internet-based services that include 16S rRNA gene sequence comparisons were performed using their default parameters of analysis. Sequence Match version 2.7 was performed in the Ribosomal Database Project (RDP-II) (Michigan State University, East Lansing) (25). The standard nucleotide-nucleotide analysis was used in the Basic Local Alignment Search Tool (BLAST). Similarity searches were also performed using RIDOM, a new user-interface hypertext based on FASTA and CLUSTAL W (16).
RESULTS
The 16S rRNA gene of Mycobacterium species.
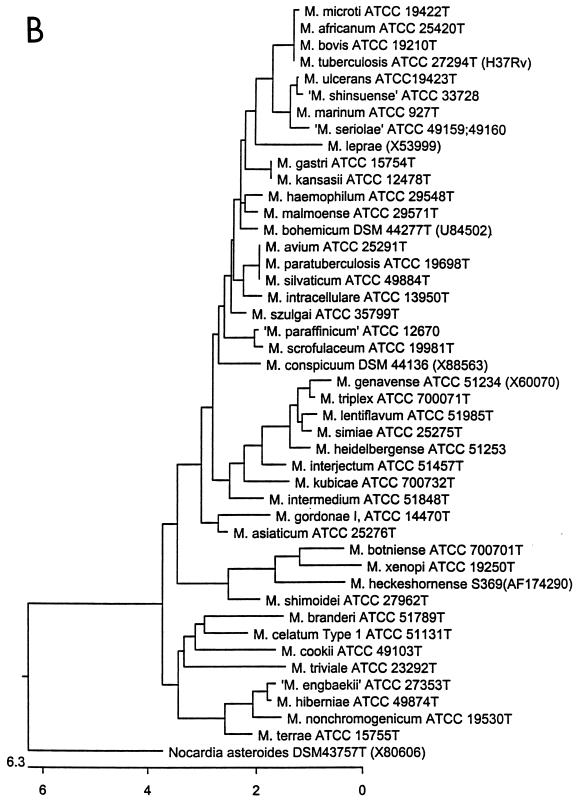
A phylogenetic tree was created that included the type strain of the 80 species determined in our laboratory, a representative sequence of each of the 12 nonestablished species, and 11 sequences from GenBank representing species not tested in our laboratory (Fig. 1). Initial analyses of these sequences showed that intraspecies variability in the 16S rRNA gene begins at E. coli bp position 69 with M. brumae and ends at bp 1462 in several species, with the exception of the M. leprae sequence from GenBank, which has a variation at bp 1466. The most variable regions begin at approximately bases 180 and 420 followed by smaller variable regions beginning at approximately bases 1010 and 1270. For an accurate determination of species similarities, all 5′ and 3′ ends were cut to identical positions along the gene, at E. coli bp 54 to 1470. Data for analysis in Fig. 1 therefore includes >1,400 bases of sequence data and all possible variations seen within all type strains of the genus.
FIG. 1.
Phylogenetic tree of mycobacteria, including one representative strain for each species. Multiple sequence alignments were determined using the CLUSTAL method algorithm in the Megalign component of the Lasergene program (version 4.01). (A) Phylogenetic tree inferring relationships among rapid growers of the Mycobacterium genus. (B) Phylogenetic tree inferring relationships among slow growers of the Mycobacterium genus. The tree was rooted using Nocardia asteroides as the outgroup sequence. Sequences were determined in our laboratory unless indicated by a GenBank accession number.
The lowest interspecies percent similarity among Mycobacterium species is 91.1%, between M. xenopi and M. chelonae. The majority of species, however, are >93% similar. Within rapid-grower species alone, the lowest interspecies similarity is 92.1%, between M. chelonae and M. hassiacum. Within slow growers, the lowest interspecies similarity is 91.9%, between M. xenopi and M. heidelbergense.
Of validated species, those which have identical 16S rRNA sequences include M. kansasii and M. gastri; M. avium and its subspecies M. paratuberculosis and M. silvaticum; and M. fortuitum and M. acetamidolyticum (a subspecies of M. fortuitum). M. tuberculosis, M. bovis, and M. africanum have identical 16S rRNA gene sequences, whereas all M. microti strains tested, i.e., ATCC 19422T, ATCC 35782, and ATCC 11152, have a one-base variation at position 1241 from the rest of the M. tuberculosis complex. If using the 5′ portion of the gene only (up to bp 510) for sequence analysis, such as with RIDOM or MicroSeq, the species that cannot be differentiated in addition the those mentioned above include M. abscessus and M. chelonae; M. marinum and M. ulcerans; M. senegalense, M. fortuitum ATCC 49403, and M. farcinogenes; M. porcinum and M. fortuitum ATCC 49404; M. tokaiense and M. murale; and M. septicum and M. peregrinum. They can, however, be differentiated in other regions of the gene.
Evaluation of BLAST, RDP-II, and RIDOM Internet-based programs.
To demonstrate the quality and accuracy of results provided from available databases, we submitted all type strain sequences determined in our laboratory for analysis using the NCBI database, the RDP-II, and most recently, the RIDOM database. A total of 79 type strains were “queried” (Table 1). The sequence of M. celatum ATCC 51131T determined in our laboratory was not included in the analysis because the sequence obtained indicated the presence of two gene sequences with numerous variations from each other, seemingly corresponding to both M. celatum type 1 and type 3 sequences.
For simplicity, results obtained by the three methods of analysis in Table 1 were assigned a code: 1, the correct species was given as the top score with 100% similarity; 2, the correct species was given the top score, but was not a perfect match; 3, the correct species was not given the top score; 4, the correct species was not in the database. All of our type strain sequences (n = 79) analyzed by RIDOM were correct, with 100% similarity. Where sequences are identical among a group of species, all species are listed as a perfect match. Contrarily, only 23% of species (n = 18) had a perfect match with sequences from GenBank and EMBL databases as determined by BLAST and 25% of species (n = 20) had a perfect match as determined by RDP-II. Of these, three belonged to the M. tuberculosis complex, whereas three belonged to M. avium and its subspecies. However, a user must be aware that the 16S rRNA gene is identical for all species within these groups, as only one species (that is of the best quality) will normally be given a top score, unlike searches performed using RIDOM, in which all identical matches will be listed equally as a top score.
The correct species was given the top score, although it was not a perfect match, to 38% (n = 30) of sequences entered in BLAST and 41% (n = 32) of sequences entered in RDP-II. Results ranged from a 98 to 99% similarity, with hamming distances as low as 20 in BLAST. Similarity scores were from 0.935 to 0.998 using RDP-II.
Results obtained in category 3 clearly showed that public sequence databases often offer an erroneous species as the best match for many queries. An overwhelming 29% (n = 23) and 24% (n = 19) of the type strains were not given top scores against GenBank sequences, using BLAST and RDP-II, respectively, despite the fact that these were present in the database. Top results were a mixture of alternate species or uncharacterized strains, with the accurate species often being low on the list of top matches due to their bad quality.
Eight established species do not currently have representative sequences in GenBank. These include M. agri, M. brumae, M. moriokaense, M. porcinum, M. poriferae, M. pulveris, M. rhodesiae, and M. tokaiense. Of these, five were easily recognized as unique, as hamming distances spanned from 20 to 38 bp in BLAST, corresponding to a score of 0.961 to 0.916 in RDP-II. The other three species had very few variations to their closest match, which would make it difficult for a user to determine whether they are in fact unique species. M. porcinum, a known animal pathogen, indicated a 5-bp difference from M. farcinogenes (AF055333). However, the same strain of M. farcinogenes sequenced in our laboratory indicated five variations with the GenBank sequence. Comparing sequences determined in our laboratory, only M. porcinum differs from M. farcinogenes by eight bases. M. rhodesiae differed from Mycobacterium sp. strain RJG11-135 (U30661) by 2 bp. Their closest known relative is M. aichiense. Results obtained for M. tokaiense indicate Mycobacterium sp. strain MA-112/96 as having the top score in both BLAST and RDP-II programs. If one chooses to obtain the corresponding reference, they will determine that this strain belongs to the newly described M. murale. In actuality, this sequence differs from our M. tokaiense sequence by only one base. All of the eight species not present in public databases were present in the RIDOM database.
There are approximately 21 sequences in GenBank named according to a Mycobacterium species that has not to date been validly published in a journal as a species. With their accession numbers and year of submission in parentheses, these are as follows: “M. anthracenicum” (Y15709, 1998), “M. bonickei” (AY01574, AY01580, AY01581, AY01582, 2001), “M. chromogen” (M29554, 1993), “M. doricum” (AF264700, 2000), “M. fluoroanthenovirans” (AJ276274, 2000), “M. fuerth” (AF316618, 2001), “M. genomospecies” (AY012573, AY012575, AY012576, AY012577, 2001), “M. holsaticum” (AJ310467, 2001), “M. houstonense” (AY012579, 2001) “M. immunogen” (AJ011771, 1998), “M. isoniacini”(X80768, 1997), “M. monacense” (AF107039, 1998), “M. ratisbonense” (AF055331, 1998; AJ271863, 2000), “M. sydneyiensis” (AF101243, 2000), and “M. tilburgii” (Z50172, 2001). The RIDOM database also contains some of these sequences, either determined in their laboratory or obtained from GenBank, with the accession number indicated.
Of those listed above, “M. bonickei,” “M. fuerth,” “M. genomospecies” (select strains), “M. ratisbonense,” and “M. sydneyiensis” have sequences that are identical to sequences in our database, whereas the others all appear to have unique sequences distinct from any established species to date. “M. fuerth” is identical to M. chelonae ATCC 19237 (named “chemovar niacinogenes”) determined in our laboratory. “M. sydneyiensis” has an sequence identical to that of the newly described M. heckeshornense, whereas “M. ratisbonense” sequences are identical to that of M. mucogenicum ATCC 49650. “M. houstonense” was assigned to the strain ATCC 49403 (currently M. fortuitum, third biovariant). Four submitted sequences named “M. bonickei” appear to be identical or nearly identical to M. fortuitum ATCC 49404, and one of them is stated as being strain ATCC 49939. The four sequences named “M. genomospecies” represented three different sequences. Two were identical to each other, and they were strains ATCC 49404 and ATCC 49935. Another was identical to the sequence of M. septicum, whereas the last sequence was ATCC 49938 (designated type strain) and was quite unique, being closest to M. neoaurum and M. diernhoferi. All sequences for “M. houstonense,” “M. bonickei,” and “M. genomospecies,” however, contained large gaps, and therefore the complete sequence could not be compared.
There is also a large number (∼80) of GenBank submissions designated as Mycobacterium species, with a great majority of them having a unique sequence. Their accuracy, however, is impossible to determine. Also, many of them are smaller sequence fragments. As best as possible, these were screened against the sequences determined in our database. Those that corresponded with an established species were eliminated, and those that appeared to be unique were further examined to ensure that their quality was acceptable, i.e., that there were no variations in areas conserved in 92 established species. At the moment, we included approximately 50 sequences from uncharacterized Mycobacterium sequences from public databases along with the reference strains from our laboratory to evaluate patient strains.
Evaluation of nonestablished species and other non-type strains.
Select ATCC strains identified as established species were determined to have divergent sequences from their type strain (Table 2). M. scrofulaceum ATCC 35788 was more closely associated with species related to M. simiae, including a short helix 18, characteristic of rapid growers and M. simiae (23). This strain also had an sequence identical to that of MCRO 33 (GenBank accession no. AF152559) (39) and M. simiae ATCC 15080 from RIDOM. M. flavescens ATCC 23033 had a unique sequence related to its type strain but was closer to M. novocastrense (U96747). M. asiaticum ATCC 25274 had one base variation from its type strain at position 1020. M. chelonae ATCC 19237, named M. chelonae chemovar niacinogenes in the ATCC collection, had three base differences from its type strain. Its sequence also corresponded to the submitted sequences in GenBank for “M. fuerth” (AF316618) and Fuerth 1999 (AF152558).
M. fortuitum ATCC 49403, of the sorbitol-positive subgroup of the M. fortuitum third biovariant complex, has 100% similarity to M. farcinogenes and M. senegalense when comparing the first 500 bases of the 16S gene, just as M. fortuitum ATCC 49404, of the sorbitol-negative subgroup, has 100% similarity to M. porcinum. Analyzing the complete gene reveals that ATCC 49403 is identical to M. farcinogenes and has four variations from M. senegalense at positions 1008, 1010, 1017, and 1019, whereas ATCC 49404 differs from M. porcinum by one base at position 1135.
We have determined the 16S rRNA gene of 12 species considered to date not established. (Table 3). Of the 12 nonvalidated species determined in our laboratory, only “M. paraffinicum” was present in the public databases, while 9 were present in RIDOM. Some had a sequence identical to that of an established species. “M. acapulcensis” had an identical sequence to that of strains of M. flavescens (ATCC 14474T, ATCC 23008, and ATCC 23033). “M. brunense” was identical to M. avium-M. paratuberculosis-M. silvaticum. “M. burulii” had an identical sequence to that of members of the M. tuberculosis complex. “M. lactis” was identical to M. hiberniae, and “M. valentiae” was identical to M. duvalii. The other seven nonestablished species had unique sequences: “M. album,” “M. engbaekii,” “M. lacticola,” “M. paraffinicum,” “M. petroleophilum,” “M. seriolae,” and “M. shinsuense.”
Evaluation of 122 clinical NTM strains.
The breakdown of analysis of 122 clinical strains submitted to our laboratory for identification was as follows: 61 strains had a sequence with 100% similarity to the type strain of an established species, 19 strains had some divergence but closely resembled a known species, 11 strains had sequences corresponding to uncharacterized strain sequences in public databases, and 31 strains represented unique sequences (Table 4).
Of the strains closely resembling an established species, one strain which phenotypically resembled M. xenopi had many double peaks in its sequence. A comparative alignment with sequences from M. xenopi and M. heckeshornense showed that this strain appeared to have a copy of both the M. xenopi and M. heckeshornense sequence. Two clinical isolates that were characterized with two distinct sequences were closely related, phenotypically and genotypically, to the M. interjectum type strain. Two clinical strains had an identical sequence to that of M. chelonae chemovar niacinogenes determined in our laboratory. Their biochemical profile combined results representative of both M. chelonae and M. abscessus.
A total of eleven strains corresponded to three sequences of uncharacterized strains from GenBank, namely MCRO 6 (n = 4), MCRO 8 (n = 2), and MCRO 17 (n=5).
A total of 31 strains held 18 unique sequences distinct from established and nonestablished species, as well as from strains uncharacterized from the public databases.
DISCUSSION
Our laboratory has determined the 16S rRNA gene sequences (>1,400 bp) of 121 ATCC strains of mycobacteria, encompassing 80 established species and 12 nonestablished species. Of the 92 established species currently existing, we did not determine the sequence for 12 of them: M. heckeshornense, M. bohemicum, M. conspicuum, M. elephantis, M. murale, M. novocastrense, M. hodleri, M. tuberculosis subsp. caprae, M. genavense, M. tusciae, M. lepraemurium, and M. leprae. The sequences determined in our laboratory, together with select sequences from GenBank that we did not have available (including the 12 established species, unpublished species, and strains not identified to the species level) comprised our database of mycobacterial sequences for the purpose of identification of clinical strains. While we are aware of the possible presence of sequence errors from GenBank data, we have evaluated them in comparison with close relatives and have determined them to be of acceptable quality, i.e., presenting no mismatches in conserved areas and no omissions or insertions out of place. We have chosen to combine these sets of sequences, providing the advantages of both the quality-controlled MicroSeq Kit and RIDOM as well as a large number of unique, but not-yet-characterized sequences commonly found in GenBank. The GenBank entries of these uncharacterized strains may or may not provide useful information regarding a particular specimen, i.e., clinical isolate or hint of pending publications of new species. Continuous expansion of our comprehensive database occurs with periodic searches for new GenBank entries and new specimens submitted. This has allowed us to confidently identify, in the least, species corresponding to a type strain sequence with a 100% similarity, in the same fashion as a MicroSeq kit (PE-Biosystems) and RIDOM, and made more evident which organisms did not have a sequence that perfectly matched available sequences. This does not occur when using public databases alone, as we have shown in Table 1. In addition, combining our sequences with select ones from GenBank does occasionally allow us to recognize a similarity with noncharacterized strains deposited in public databases with some, however limited, published information.
The purpose of performing sequence similarity searches using the 16S rDNA sequence of type strains of mycobacterium species that we had determined in our laboratory was to evaluate the accuracy of results obtained by such programs as BLAST, RDP-II, and RIDOM. Using only type strains eliminates any possible errors of species identification due to initial strain misidentification. As indicated in Table 1, all type strains corresponded with 100% similarity with those of RIDOM. As anticipated, a great portion of results obtained from comparisons with GenBank and EMBL submissions, the basis for both NCBI and RDP-II, were inaccurate (Table 1), with abundant ambiguities, sequence gaps, and proven errors. Although BLAST and RDP-II acquire their sequences from the same databases, differences in search results are derived from the fact that BLAST searches against all available sequences, whereas RDP-II acquires only select GenBank sequences and incorporates them in their own database against which searches are made.
The high proportion of misleading results acquired from public databases is not surprising, as submissions are not peer reviewed. Similarity searches can significantly deter the user from the true identification of an organism, even if the organism sequence is present in the databases. The sequences of the majority of well-known mycobacterial species were submitted in the early 1990s, when methods used may not have had the capabilities of providing the quality sequences easily obtained today. Inconsistent sequence ends, noncharacterized or ambiguous entries, pseudogaps, and insertions will indicate a percent similarity of less than 100% for what should be identical sequences. Therefore, a result of 98 or 99% similarity can either result from a “perfect species match” with a poor quality sequence, or a divergence of up to 30 bp between two species. In BLAST, several GenBank sequences contain large sequence gaps in the submission. This can lead to misidentification, because even if sequences are correct, a highest score may be given to a sequence with 99% similarity against one with a 100% match if the fragment size of the query was longer. In some cases, species names are not indicated in the title, but are stated in the corresponding reference (M. peregrinum, M. branderi, M. lentiflavum, and M. tusciae), which may lead a user to refer to the next-closest sequence of a well-established species.
Eight type strains of established species from the ATCC currently do not have representative sequences in public databases. However, these are present in the RIDOM database. An additional 12 species (nonestablished) from the ATCC collection were also evaluated, resulting in some new 16S rDNA sequences, where their validation as distinct species would be deemed acceptable in conjunction with some additional characterization, and some were identical to previously established species. There are also several “species” names found in public databases that are not published in any journal as an established species to date. Many of them that were present in GenBank 1 or 2 years ago have been recently published. We anticipate and hope that the same will occur for other submitted sequences from new mycobacterium species.
A great proportion of mycobacterial sequences in GenBank do not have a designated species name. Some correspond closely to an established species (4), while a large proportion appear to be unique. With the 92 currently established species, along with approximately 50 unique sequence types belonging to uncharacterized strains and nonestablished species in GenBank, and 18 new sequences derived from clinical strains submitted to our laboratory, there may be more than 160 mycobacterial species that exist. There is no indication that additional unique sequences will not continue to be found. With the advent of so many newly described species associated with disease, these cannot be bypassed.
Even when working with a “perfect” 16S rRNA database, similarity results are not necessarily as clear-cut as expected. It is generally true that each species holds a unique and stable 16S rRNA gene sequence. However, there is no magic number as to how many differences in the 16S rRNA gene are required to differentiate between species. It has been suggested that strains are the same species if they have fewer than 5 to 15 base differences in the 16S rRNA gene or that they are related species if they differ by at least 1.5% (13). This is not the case with mycobacteria. A difference of 5 nucleotides within the complete 16 SrRNA gene of a mycobacterium along with clear phenotypic differences confidently indicates a genetically unique and distinct taxon (22). However, the same holds true in several cases with less sequence variation between some of the species.
Microheterogeneity within a species, well described for M. gordonae, appears to be restricted to one to two base differences from each other in the region of bp 100 to 300 in four documented sequences (20). In addition, we have identified from clinical strains two other M. gordonae sequences, differing by three and five bases from the type strain. Further variations are found between bases 463 and 478 that would be detected using RIDOM or MicroSeq. One peculiar isolate, NRCM 00-146, would suggest a sixth sequence type for M. gordonae, having only three variations from the type strain. However, colony pigment and morphology suggested otherwise, as the colonies appeared rough and salmon colored like some species related to the M. terrae complex. Contamination by M. gordonae was ruled out. Variations among the six sequences described were not detected beyond base 478. Other species which are known to have several 16S rRNA sequence types include M. bohemicum (43), M. celatum (7), M. interjectum (24), M. lentiflavum (40), and M. mucogenicum (38).
The establishment of new species in cases where the 16S rRNA genes are very similar may be influenced by the clinical relevance. For example, M. goodii, which was previously indistinguishable diagnostically from M. smegmatis and has only a four-base difference in the 16S rRNA gene, was established as a new species due to its different susceptibility pattern (6). The establishment of other mycobacterial species closely related to one another is possible with the inclusion of various algorithms that can include mycolic acids, PRA and susceptibility patterns, epidemiological factors, and various molecular markers.
The RIDOM program provides a general interpretation with query results, for example, the species is not present in their database if similarity with the best match is ≤97%, or “no close relative available” if similarity with the best match is ≤96%. However, a similarity of 98 to 99% suggests “your sequence likely derives from this species,” which is difficult to evaluate, since an ∼450 bp comparison with as much as a nine-base variation will result in 98%. In general, members of the Mycobacterium genus are closely related to each other and closely related species may differ only by a few bases or not at all.
Other factors which can contribute to intraspecies variability include the presence of two 16S rRNA gene alleles in the genome of many mycobacteria, generally but not exclusively in rapid growers. Slow growers are known to have one copy (1), although some exceptions have been documented, such as for M. terrae and M. celatum (27, 30). Similarly, M. chelonae and M. abscessus are an exception to the rule that rapid growers have two copies, as they have only one copy (11). When present in multiple copies, rRNA operons are generally identical or very similar (9). We have observed base positions in some strains which had double peaks, suggesting the presence of two gene copies: R (A and G) at bp 490 in M. petroleophilum, R at bp 93 in M. poriferae, and Y (C and T) at bp 474 in two clinical M. flavescens-like strains. Generally, no double peaks were observed in the great majority of the rapid growers sequenced in our laboratory.
Alternatively, we have observed significant variations between what we suspect were two copies present in the type strain of M. celatum and a clinical strain resembling M. xenopi. Upon closer evaluation of the double peaks in sequence alignments, we determined that M. celatum ATCC 51131 contained the sequences of both M. celatum type 1 and M. celatum type 3, exactly as previously determined for a clinical strain of M. celatum (30). The M. xenopi-like isolate contained 29 double peaks, corresponding exactly to 29 of the 39 base differences in ∼1,450 bp of the 16S rDNA between M. xenopi and M. heckeshornense. In the other 10 positions, the isolate corresponded to the sequence of one species or the other. This large difference in gene copy sequences within a strain has also been documented for a clinical isolate of M. terrae (27): the two 16S rRNA gene sequences which were obtained from separate clones diverged by 18 bp from each other in an ∼1,030-base fragment and corresponded to the sequences of MCRO 16 (X93027) and MCRO 24 (X93031) (39). Our observations regarding our type strain of M. celatum and our clinical M. xenopi-like isolate, and the observations of Ninet et al. with a clinical M. terrae-like isolate may suggest that if an organism contains two 16S sequence copies, this may not necessarily be detected. In each of the three cases, the sequences were published individually elsewhere. During the PCR process, one copy may have prevailed over the other, resulting in a lack of ambiguous bases.
Point mutations in the 16S rRNA gene of several mycobacterium species has also been associated with resistance to a number of antibiotics. Several single-base mutations within the gene have been associated with streptomycin resistance in M. tuberculosis (26), while a single 16S rRNA mutation confers resistance to amikacin and other 2-deoxystreptamine aminoglycosides in M. tuberculosis (35), M. abscessus, and M. chelonae (29).
How do we confidently identify an organism that is shown to have one or more variations in comparison to a known strain? If microheterogeneity occurs, how can we tell whether the strain in question is the same species, a subspecies, or a new species? The word “microheterogeneity” may give an impression that minor variations within a species are random and with little significance. However, these seem to be consistent from one strain to another. Unless there are strong phenotypic differences, this is difficult to determine without further testing the true status of these strains. Even then, extensive analysis using multiple strains would be required to determine the significance of new sequences, some of which may be of clinical or epidemiological interest. Ambiguities must be examined on a case by case basis, and due to a lack of complete mycobacterial 16S rRNA gene sequence databases or alternative molecular information, some results will often remain inconclusive. It is imperative that sequence databases be of the best quality to overcome potential problems due to the relatively high similarity among mycobacterial species. The success of RIDOM in obtaining perfect matches with all established species suggests that even with instances of microheterogeneity in certain groups of organisms, the majority of isolates do correspond exactly (with 100% similarity) to a type strain sequence.
Other genes have been studied in mycobacteriology for the identification of NTM, mainly to overcome the high similarity of 16S rRNA gene sequences within the mycobacterium genus. These include hsp65, dnaJ, the 32-kDa gene, recA, rpoB, sodA, and the 16S-23S spacer regions (2, 19, 34, 37, 41, 42, 46). These genes are generally less conserved and often provide more insight on questionable associations between two strains based on 16S rRNA data, such as for M. gastri and M. kansasii. However, the impact or significance of the greater rate of intraspecies variability in these genes has not extensively been studied. In addition, since none of these genes have as comprehensive a database as for the 16S rRNA gene to date, problems with identification would arise with less well-established and novel species in addition to the higher rate of apparent intraspecies divergence common in these genes. At this time, conclusive sequence-based identification using alternative gene targets occurs generally if it corresponds exactly to a type strain. For this reason, the low mutation rate of the 16S rRNA gene continues to be an advantage. Studies of potential gene targets must encompass many species with well-characterized strains.
While this study mostly focuses on diagnostic mycobacteriology, it is important to remember the true goal, which is to determine clinical relevance and proper treatment when disease with NTM species occurs. Since there are 92 species currently considered valid, what is the significance of the countless other mycobacterial sequences obtained from various laboratories? We have certainly not unearthed all existing mycobacterium species. The advent of molecular methods for identification, particularly sequence-based methods, has exponentially increased the discovery and validation of new mycobacterium species in the last few years, many of which have a clinical story. We also cannot assume that prior to more recent technologies all species were correctly identified, as these were routinely identified by biochemical methods and new species likely have often been identified as the closest known species as opposed to a unique organism. There is no standard antibiotic regimen for NTM, and due to the various susceptibility profiles of these species, accurate identification is essential. Misidentification results in delay of appropriate antibiotic therapy, which may contribute to the death of some patients (8, 47). Unfortunately, there are so many species, and only the ones best known to cause disease, such as M. tuberculosis and M. avium in human immunodeficiency virus patients, have a suggested standardized methodology for susceptibility testing with a clinical correlation. To recognize new species and describe as best as possible their assumed clinical significance as well as treatment outcome if applicable is an important step towards a better understanding of NTM species. Even if strains are not fully characterized, published cases of diseases caused by NTM species that include the 16S rDNA data (15) would then ensure recognition of these same species if they were detected elsewhere and would also lead to the validation of a new species. Clinical information as well as antibiotic susceptibility characteristics help slowly fill the large gap in the knowledge of NTM diseases. Prospective or retrospective analyses of difficult-to-identify organisms using molecular identification will further contribute to growing databases. This approach has been taken both in the context of a global laboratory evaluation of mycobacterial specimens (39, 44) and on a case-specific basis, leading up to the designation of new species.
A quality-controlled database such as that provided by RIDOM or MicroSeq is critical to the evaluation of the impact of accurate mycobacterial species identification on clinical diagnosis and treatment. While it appears that molecular-based identification methods without conventional testing are adequate, perhaps key biochemical tests are deemed necessary in certain instances when trying to differentiate genotypically similar species. It is also important to have knowledge of which species are identical based on the first 500 bases of the 16S rDNA gene, which is the most efficient approach in 16S-based identification, and to be given options to further differentiate. While RIDOM suggests to undertake ITS1 sequencing when more than one match results from a query, another suggestion is to amplify the complete 16S rRNA gene, being no greater effort than amplifying a shorter fragment, and using alternate primers with the same template to sequence relevant regions. Alternatively, 16S rDNA and ITS1 amplification could be incorporated in one PCR as they are adjacent to each other. The broad nature of ribosomal primers that amplify all bacteria has an advantage in that while this work discusses mainly our experience with mycobacteria, the ideas generally apply to all members of the kingdom. We have detected five strains of Tsukamurella species (data not included) submitted to our laboratory as NTM species due to the weak acid-fast nature.
The applications of sequence-based methodologies are countless, from an ever-increasing database of genes used for identification as well as molecular antibiotic resistance detection. Furthermore, the technology is simple and easy to implement in most laboratories. Manufacturers continue to develop various models to accommodate low or high throughput and various molecular protocols that include typing and sequencing. While the MicroSeq and RIDOM databases provide excellent results for the majority of mycobacterial isolates, RIDOM is not only freely accessible but also significantly more comprehensive. The RIDOM services for which there has long been a need will become an essential tool for all laboratories with sequencing capacities, not only in mycobacteriology but also, as their database expands, in all of diagnostic bacteriology.
REFERENCES
- 1.Bercovier H, Kafri O, Sela S. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem Biophys Res Commun. 1986;136:1136–1141. doi: 10.1016/0006-291x(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 2.Blackwood K S, He C, Gunton J, Turenne C Y, Wolfe J, Kabani A M. Evaluation of recA sequences for identification of Mycobacterium species. J Clin Microbiol. 2000;38:2846–2852. doi: 10.1128/jcm.38.8.2846-2852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boddinghaus B, Rogall T, Flohr T, Blocker H, Bottger E C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottger E C, Kirschner P, Springer B, Zumft W. Mycobacteria degrading polycyclic aromatic hydrocarbons. Int J Syst Bacteriol. 1997;47:247. [Google Scholar]
- 5.Brosius J, Palmer M L, Kennedy P J, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown B A, Springer B, Steingrube V A, Wilson R W, Pfyffer G E, Garcia M J, Menendez M C, Rodriguez-Salvado B, Jost KC, Jr, Chiu S H, Onyi G O, Bottger E C, Wallace R J., Jr Mycobacterium wolinskyi sp. nov. and Mycobacterium goodii sp. nov., two new rapidly growing species related to Mycobacterium smegmatis and associated with human wound infections: a cooperative study from the International Working Group on Mycobacterial Taxonomy. Int J Syst Bacteriol. 1999;49:1493–1511. doi: 10.1099/00207713-49-4-1493. [DOI] [PubMed] [Google Scholar]
- 7.Bull T J, Shanson D C, Archard L C, Yates M D, Hamid M E, Minnikin D E. A new group (type 3) of Mycobacterium celatum isolated from AIDS patients in the London area. Int J Syst Bacteriol. 1995;45:861–862. doi: 10.1099/00207713-45-4-861. [DOI] [PubMed] [Google Scholar]
- 8.Bux-Gewehr I, Hagen H P, Rusch G S, Feurle G E. Fatal pulmonary infection with Mycobacterium celatum in an apparently immunocompetent patient. J Clin Microbiol. 1998;36:587–588. doi: 10.1128/jcm.36.2.587-588.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cilia V, Lafay B, Christen R. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol Biol Evol. 1996;13:451–461. doi: 10.1093/oxfordjournals.molbev.a025606. [DOI] [PubMed] [Google Scholar]
- 10.Cloud J, Rosenberry H, Neal H, Turenne C, Jama M, Hillyard D, Carroll K. Identification of mycobacteria using 16S rDNA sequencing, abstr. C-237. Abstr. 101st Gen. Meet. Am. Soc. Microbiol. Washington, D.C.: American Society for Microbiology; 2001. [Google Scholar]
- 11.Domenech P, Menendez M C, Garcia M J. Restriction fragment length polymorphisms of 16S rRNA genes in the differentiation of fast-growing mycobacterial species. FEMS Microbiol Lett. 1994;116:19–24. doi: 10.1111/j.1574-6968.1994.tb06669.x. [DOI] [PubMed] [Google Scholar]
- 12.Edwards U, Rogall T, Blocker H, Emde M, Bottger E C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox G E, Wisotzkey J D, Jurtshuk P. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 14.Fredericks D N, Relman D A. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin Microbiol Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodwin A, Lumb R, Patkin M, Bastian I. Isolation of a novel mycobacterium from an adolescent with cervical lymphadenitis. Eur J Clin Microbiol Infect Dis. 1998;17:516–518. doi: 10.1007/BF01691137. [DOI] [PubMed] [Google Scholar]
- 16.Harmsen D, Rothgänger J, Singer C, Albert J, Frosch M. Intuitive hypertext-based molecular identification of micro-organisms. Lancet. 1999;353:291. doi: 10.1016/S0140-6736(98)05748-1. [DOI] [PubMed] [Google Scholar]
- 17.Harmsen D, Singer C, Rothganger J, Tonjum T, de Hoog G S, Shah H, Albert J, Frosch M. Diagnostics of Neisseriaceae and Moraxellaceae by ribosomal DNA sequencing: ribosomal differentiation of medical microorganisms. J Clin Microbiol. 2001;39:936–942. doi: 10.1128/JCM.39.3.936-942.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holberg-Petersen M, Steinbakk M, Figenschau K J, Jantzen E, Eng J, Melby K K. Identification of clinical isolates of Mycobacterium spp. by sequence analysis of the 16S ribosomal RNA gene. Experience from a clinical laboratory. APMIS. 1999;107:231–239. [PubMed] [Google Scholar]
- 19.Kim B J, Lee S H, Lyu M A, Kim S J, Bai G H, Chae G T, Kim E C, Cha C Y, Kook Y H. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB) J Clin Microbiol. 1999;37:1714–1720. doi: 10.1128/jcm.37.6.1714-1720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirschner P, Bottger E C. Microheterogeneity within rRNA of Mycobacterium gordonae. J Clin Microbiol. 1992;30:1049–1050. doi: 10.1128/jcm.30.4.1049-1050.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirschner P, Bottger E C. Species identification of mycobacteria using rDNA sequencing. In: Parish T, Stoker N G, editors. Methods in molecular biology. 101. Mycobacteria protocols. Totowa, N.J: Humana Press Inc.; 2000. pp. 349–361. [DOI] [PubMed] [Google Scholar]
- 22.Kirschner P, Meier A, Bottger E C. Genotypic identification and detection of mycobacteria—facing novel and uncultured pathogens. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 173–190. [Google Scholar]
- 23.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F C, Bottger E C. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lumb R, Goodwin A, Ratcliff R, Stapledon R, Holland A, Bastian I. Phenotypic and molecular characterization of three clinical isolates of Mycobacterium interjectum. J Clin Microbiol. 1997;35:2782–2785. doi: 10.1128/jcm.35.11.2782-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maidak B L, Cole J R, Lilburn T G, Parker C T, Jr, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier A, Kirschner P, Bange F C, Vogel U, Bottger E C. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: mapping of mutations conferring resistance. Antimicrob Agents Chemother. 1994;38:228–233. doi: 10.1128/aac.38.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ninet B, Monod M, Emler S, Pawlowski J, Metral C, Rohner P, Auckenthaler R, Hirschel B. Two different 16S rRNA genes in a mycobacterial strain. J Clin Microbiol. 1996;34:2531–2536. doi: 10.1128/jcm.34.10.2531-2536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel J B, Leonard D G B, Pan X, Musser J M, Berman R E, Nachamkin I. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J Clin Microbiol. 2000;38:246–251. doi: 10.1128/jcm.38.1.246-251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prammananan T, Sander P, Brown B A, Frischkorn K, Onyi G O, Zhang Y, Bottger E C, Wallace R J., Jr A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J Infect Dis. 1998;177:1573–1581. doi: 10.1086/515328. [DOI] [PubMed] [Google Scholar]
- 30.Reischl U, Feldmann K, Naumann L, Gaugler B J, Ninet B, Hirschel B, Emler S. 16S rRNA sequence diversity in Mycobacterium celatum strains caused by presence of two different copies of 16S rRNA gene. J Clin Microbiol. 1998;36:1761–1764. doi: 10.1128/jcm.36.6.1761-1764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 32.Rogall T, Flohr T, Bottger E C. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol. 1990;136:1915–1920. doi: 10.1099/00221287-136-9-1915. [DOI] [PubMed] [Google Scholar]
- 33.Ross B C, Jackson K, Yang M, Sievers A, Dwyer B. Identification of a genetically distinct subspecies of Mycobacterium kansasii. J Clin Microbiol. 1992;30:2930–2933. doi: 10.1128/jcm.30.11.2930-2933.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth A, Fischer M, Hamid M E, Michalke S, Ludwig W, Mauch H. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S–23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol. 1998;36:139–147. doi: 10.1128/jcm.36.1.139-147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sander P, Prammananan T, Bottger E C. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol Microbiol. 1996;22:841–848. doi: 10.1046/j.1365-2958.1996.01532.x. [DOI] [PubMed] [Google Scholar]
- 36.Shinnick T M, Good R C. Mycobacterial taxonomy. Eur J Clin Microbiol Infect Dis. 1994;13:884–901. doi: 10.1007/BF02111489. [DOI] [PubMed] [Google Scholar]
- 37.Soini H, Viljanen M K. Diversity of the 32-kilodalton protein gene may form a basis for species determination of potentially pathogenic mycobacterial species. J Clin Microbiol. 1997;35:769–773. doi: 10.1128/jcm.35.3.769-773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Springer B, Bottger E C, Kirschner P, Wallace R J., Jr Phylogeny of the Mycobacterium chelonae-like organism based on partial sequencing of the 16S rRNA gene and proposal of Mycobacterium mucogenicum sp. nov. Int J Syst Bacteriol. 1995;45:262–267. doi: 10.1099/00207713-45-2-262. [DOI] [PubMed] [Google Scholar]
- 39.Springer B, Stockman L, Teschner K, Roberts G D, Bottger E C. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol. 1996;34:296–303. doi: 10.1128/jcm.34.2.296-303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Springer B, Wu W K, Bodmer T, Haase G, Pfyffer G E, Kroppenstedt R M, Schroder K H, Emler S, Kilburn J O, Kirschner P, Telenti A, Coyle M B, Bottger E C. Isolation and characterization of a unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J Clin Microbiol. 1996;34:1100–1107. doi: 10.1128/jcm.34.5.1100-1107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takewaki S, Okuzumi K, Manabe I, Tanimura M, Miyamura K, Nakahara K, Yazaki Y, Ohkubo A, Nagai R. Nucleotide sequence comparison of the mycobacterial dnaJ gene and PCR-restriction fragment length polymorphism analysis for identification of mycobacterial species. Int J Syst Bacteriol. 1994;44:159–166. doi: 10.1099/00207713-44-1-159. [DOI] [PubMed] [Google Scholar]
- 42.Telenti A, Marchesi F, Balz M, Bally F, Bottger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torkko P, Suomalainen S, Iivanainen E, Suutari M, Paulin L, Rudback E, Tortoli E, Vincent V, Mattila R, Katila M L. Characterization of Mycobacterium bohemicum isolated from human, veterinary, and environmental sources. J Clin Microbiol. 2001;39:207–211. doi: 10.1128/JCM.39.1.207-211.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wayne L G, Good R C, Bottger E C, Butler R, Dorsch M, Ezaki T, Gross W, Jonas V, Kilburn J, Kirschner P, Krichevsky M I, Ridell M, Shinnick T M, Springer B, Stackebrandt E, Tarnok I, Tarnok Z, Tasaka H, Vincent V, Warren N G, Knott C A, Johnson R. Semantide- and chemotaxonomy-based analyses of some problematic phenotypic clusters of slowly growing mycobacteria, a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int J Syst Bacteriol. 1996;46:280–297. doi: 10.1099/00207713-46-1-280. [DOI] [PubMed] [Google Scholar]
- 45.Widjojoatmodjo M N, Fluit A C, Verhoef J. Molecular identification of bacteria by fluorescence-based PCR-single-strand conformation polymorphism analysis of the 16S rRNA gene. J Clin Microbiol. 1995;33:2601–2606. doi: 10.1128/jcm.33.10.2601-2606.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zolg J W, Philippi S S. The superoxide dismutase gene, a target for detection and identification of mycobacteria by PCR. J Clin Microbiol. 1994;32:2801–2812. doi: 10.1128/jcm.32.11.2801-2812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zurawski C A, Cage G D, Rimland D, Blumberg H M. Pneumonia and bacteremia due to Mycobacterium celatum masquerading as Mycobacterium xenopi in patients with AIDS: an underdiagnosed problem? Clin Infect Dis. 1997;24:140–143. doi: 10.1093/clinids/24.2.140. [DOI] [PubMed] [Google Scholar]