Abstract
Background
The pathogenesis of ulcerative colitis (UC) is closely related to the gut microbiota. Moxibustion has been used to improve the inflammation and gastrointestinal dysfunctions in gastrointestinal disorders such as UC. In this study, we investigated whether moxibustion could improve the gut microbial dysbiosis induced by dextran sulphate sodium.
Methods
Twenty-five male rats were randomly assigned into five groups. The UC rat model was established by administering DSS solution. The rats in the moxibustion and normal rats with moxibustion groups were treated with moxibustion at Tianshu (bilateral, ST25) points, and the mesalazine group rats were treated with mesalazine once daily for 7 consecutive days. Disease activity index (DAI) and haematoxylin and eosin staining were used to evaluate the effect of moxibustion. Gut microbiota profiling was conducted by metagenomic high throughput sequencing technology. The gut microbiota composition, diversity and function were analyzed and compared using metagenomics methodologies.
Results
The DAI scores and histopathology scores in the moxibustion and mesalazine groups were significantly decreased compared with the UC group (P < 0.01). Moxibustion treatment increased abundance levels of Bacteroidetes, Actinobacteria, Ascomycota, Synergistetes and decreased abundance of Firmicutes, Proteobacteria. At the genus level, the abundance of Bacteroides, Bacteroides_bacterium_M7, Prevotella, Bacteroidales_bacterium_H2, were increased and Bacteroides_bacterium_H3, Parabacteroides, Porphyromonas, Alistipes, Parasutterella were decreased in the UC group in comparsion with those in the NG group. Moxibustion increased the abundance of Bacteroides and Bacteroides_bacterium_H3 and decreased Bacteroides_bacterium_M7, Prevotella, Bacteroidales_bacterium_H2. In UC group, the specie Bacteroides_massiliensis was negatively (P < 0.05) correlated with IL-23, Bacteroides_eggerthii_CAG109 and Bacteroides_eggerthii were negatively (P < 0.05) correlated with TGF-β. And the species Prevotella_sp_CAG1031 and Bacteroides_bacterium_H2 were significant positively (P < 0.05) correlated with IL-23. In addition, compare with the normal group, genes involved in certain metabolic pathways, such as energy production and conversion, amino acid transport and metabolism, carbohydrate transport and metabolism, were under-represented in the UC group, and these changes in the metabolic pathways could be reversed by moxibustion treatment and mesalazine treatment.
Conclusions
Our findings suggest that moxibustion treatment may protect the host from mucosal inflammation by modulating the intestinal microbiota community.
Keywords: Ulcerative colitis, Moxibustion, Gut microbiota, Metagenomic
Background
Ulcerative colitis (UC) is a chronic nonspecific intestinal inflammatory disease, which is clinical characterized by abdominal pain, diarrhea, rectal urgency and bleeding [1, 2]. The incidence of UC was steadily increasing with time around the world [3], and the patients generally have a reduced quality of life and are heavily burdened with medical costs [4]. However, the etiology and pathogenesis of the disease are not clear, and it is thought to be related to heredity, infection, immunity, environment and diet [5]. Evidence indicates that the intestinal microbiota is one of the key roles in the pathogenesis of UC [6–8].
Intestinal flora as the natural barrier of human body plays an important role in maintaining human health. Changes in internal and external environment can affect the structure of intestinal flora, if the intestinal environment is out of balance, such as bifidobacterium and lactobacillus significantly decreased, and bacteroides and escherichia coli increased in UC patients [9, 10], the intestinal bacteria and their metabolites continue to stimulate the host epithelial cells, affecting the intestinal mucosal immune system and inducing genetic predisposition in individuals with abnormal intestinal immune response, destructing of the structure and function of mucous membrane, eventually leading to the occurrence of UC.
Moxibustion, a common treatment in traditional Chinese medicine, is the burning of the herb moxa over acupuncture points. Modern experimental studies have shown that moxibustion has obvious advantage in adjusting immune function, which can correct abnormalities of cellular immune function, and improve, stabilize and coordinate the immune system [11, 12]. In the past 30 years, many scholars have conducted a lot of meaningful research on the mechanism of action of moxibustion in treating UC. It has been proved that moxibustion has a definite curative effect on UC [13–15], but its specific mechanism has not yet been fully understood. Our previous study found that Bifidobacterium, Lactobacillus were decreased and harmful bacteria E. coli, Fusobacterium were increased in the UC rats, and moxibustion can regulate the balance of those bacteria in UC rats [16]. To further understand the relationship between UC and intestinal flora, we used metagenomic high throughput sequencing technology to study the role of intestinal flora in the pathogenesis of UC and the role of moxibustion in the regulation of UC intestinal flora diversity.
Methods
Animals and experimental procedure
Twenty-five male Sprague–Dawley rats (weight, 180–220 g) were obtained from the Department of Laboratory Animal Science of Shanghai University of Traditional Chinese Medicine (No. SCXK (hu) 2012-0008). All animal experiments were carried out according to the Guidance Suggestions for the Care and Use of Laboratory Animals, formulated by the University Animal Care and Use Committee of Shanghai University of Traditional Chinese Medicine.
After one week of adaptive feeding, the rats were divided into five groups: Normal group (NG), UC model group (UC), moxibustion group (UC + MOX), mesalazine group (UC + MES), and normal rats with moxibustion group (NG + MOX), five rats were chosen in each group. UC model rats were established by 4% dextran sulphate sodium (DSS; Art.No.9011–18-1; MP Biomedical, CA, USA) solution for 7 days as described previously [17]. Moxibustion and mesalazine treatment will be started the next day after the completion of the model (Fig. 1). UC + MOX and NG + MOX: The Tianshu (bilateral, ST25) points were selected. The herb cakes were made of Chinese medicine powder and yellow wine into size of 0.6 cm in diameter and 0.45 cm in thickness. The moxa cone (diameter: 0.5 cm, high: 0.6 cm) was placed on the top of the herb cake, and the herb cake was placed on bilateral ST25 points, 2 moxa cones per time point for totally 7 days. UC + MES treated with mesalazine enteric coated tablet (Heilongjiang Tianhong Pharmaceutical Co., Ltd; Lot number: H20103359) dissolved in water by gavage at a daily dose of 300 mg/kg for 7 days. NG and UC groups only received the same fixation.
Fig. 1.
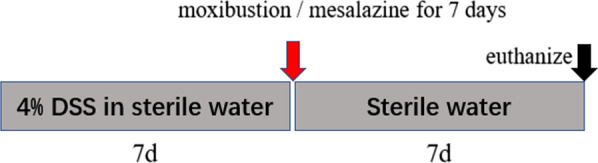
A schematic diagram of the experimental procedure
Disease activity index (DAI)
The severity of colonic inflammation in rats was evaluated by monitoring manifestations daily, such as body weight, stool consistency and rectal bleeding. The DAI score for each animal was performed according to the follow parameters: the rate of weight loss ((0, no change; 1, 1–5%; 2, 6–10%; 3, 11–15%; 4, > 15%), viscosity of stool (0, normal; 2, loose; 4, diarrhea) and status of hematochezia (0, normal; 2, positive occult blood test; 4, visible bleeding) [18]. The DAI score = (rate of weight loss + viscosity of stool + status of hematochezia)/3.
Sample collection and pathological observation
At the end of the treatment, all of the rats were anesthetized by intraperitoneal injection of 2% pentobarbital sodium, and then sacrificed by cervical dislocation. The fecal samples were collected and immediately stored in freezing tubes at -80 °C until DNA extraction. Colon tissues fixed in 4% paraformaldehyde fixative solution were dehydrated, embedded in paraffin, cut into 4 μm sections for haematoxylin and eosin (HE) staining and pathological observation. The histological score of the HE-stained colon specimens was performed according to the follow parameters: damage/necrosis, inflammatory cell infiltration, submucosal edema, and hemorrhage of mucosa. Colonic gross damage scores were recorded according to the severity of changes: 0, no change; 1, mild; 2, moderate; 3, severe [19]. The histological score was calculated as the sum of scores assigned for damage/necrosis, inflammatory cell infiltration, submucosal edema, and hemorrhage of mucosa.
Bacterial genomic DNA extraction and sequencing
The Bacterial genomic DNA was extracted with QIAamp Fast DNA Stool Mini Kit (QIAGEN, Germany) from faecal samples as previously described [20]. DNA quality testing was assessed by 1% agarose gel electrophoresis. The metagenomic DNA library was prepared using PCR products according to the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs). The PCR products were evaluated using an Agilent 2100 Bioanalyser (Agilent Technologies, Santa Clara, CA, USA) with an Agilent DNA 1000 kit according to the manufacturer’s recommendations. Amplicon sequencing was performed using Illumina HiSeq2500 (Illumina, San Diego, CA, USA).
Quality control, alignment and assembly
Sequence contamination, such as the adaptor sequence, low quality bases and some impurity sequences, are often introduced during the sequencing. This data corruption must be removed to acquire the quality sequences for later analysis. The 3’ end of the read was trimmed and bases with average quality less than Q20 (the percentage of bases with a Phred value > 20) were removed. In addition, reads shorter than 60% of the original length were moved.
Then the clean reads were aligned with the NCBI database using SOAPaligner (version 2.21) by parameter of ‘–m 4 –r 2 –m 100 – × 1000’ for the detection of known bacteria, viruses, fungi and archaea [21]. The aligned reads were classified into six different levels including Kingdom, Phylum, Class, Order, Family, Species to count classification and abundance. The taxonomy profile was constructed at the above six different levels. In addition, trimmed high-quality metagenome reads were assembled using SOAPdenovo software (version 1.05) by different sizes of Kmer (51, 55, 59, 63) [22].
Gene prediction
Using MetaGeneMark (version 2.10) to predict the assembly results of the open reading frame (ORFs) and then screened the gene sequences with length greater than 100 bp and translated them into corresponding amino acid sequences. All predicted genes (identify is greater than 95%, coverage is greater than 90%) were clustered using CD-HIT [23]. The longest sequence of genes in each category was selected to construct a non-redundant gene sets for further analyses [24].
Functional annotation analysis
To identify the biological function and associated metabolic pathways for the differentially expressed genes, we conducted the Kyoto Encyclopedia of Genes and Genomes (KEGG) database using BLSAT (version 2.2.28+) to obtain the KEGG orthologue (KO) annotation information from KEGG database [25], and the abundance of Non-supervised Orthologous Groups (NOG) function in each sample was calculated by comparing the gene sequence with the evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) database [26].
Results
Effect of moxibustion on DSS induced UC rats
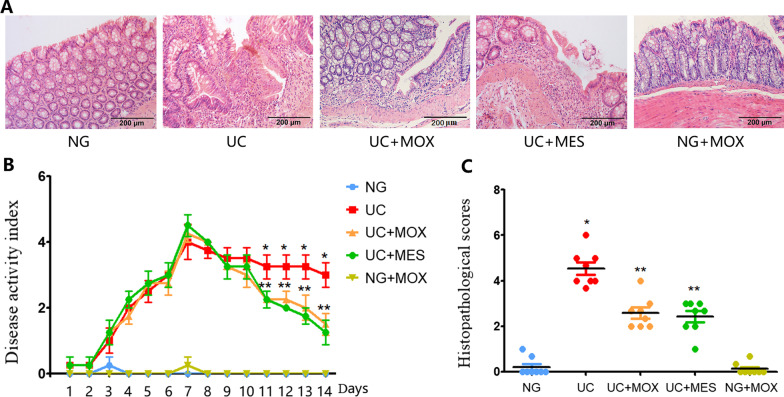
As shown in Fig. 2A, the colonic mucosal epithelium of the NG and NG + MOX group was intact. While the mucosal epithelium of UC rats was absent, ulcer, edema could be seen with a large amount of inflammatory cells infiltrations. Compared with the UC group, histopathological observation was improved in the UC + MOX and UC + MES group, the mucosal epithelium was restored, with healed ulcer and decreased inflammatory cells infiltration. Compared with the NG group, DAI score was significantly increased in the UC group (P < 0.01, Fig. 2B). There was no statistically significant difference in DAI scores between the NG group and the NG + MOX group. However, rats in the UC + MOX and UC + MES groups had lower DAI scores than the UC group (P < 0.01). The histopathology score in the UC group was significantly increased than that in the NG group (P < 0.01). Conversely, the histopathology scores in the UC + MOX and UC + MES groups were significantly decreased compared to the rats in the UC group (P < 0.01) (Fig. 2C).
Fig. 2.
The effect of moxibustion on DSS induced UC rats. A Histopathological observation (HE, × 200); B Disease activity index; C: Histopathological scores. *P < 0.01 vs. NG group; **P < 0.01 vs. UC group. NG: normal group; UC: UC model group; UC + MOX: moxibustion group; UC + MES: mesalazine group; NG + MOX: normal rats with moxibustion group
Taxonomic characterization of faecal microbiota
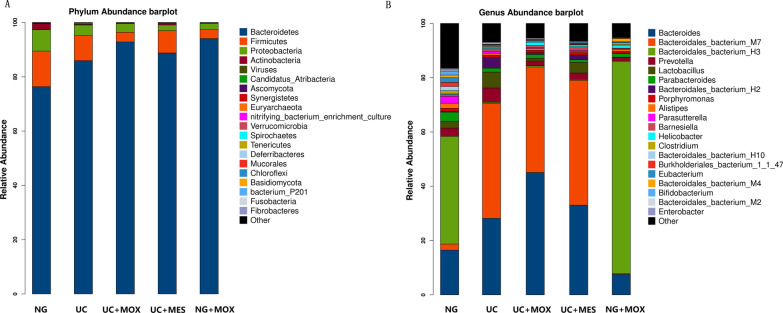
We got 192,389,261,700 base pair (bp) raw bases, with an average of 25,651,902 raw reads for each sample. After quality control, we obtained an average of 23,177,046 clean reads for each. Based on the clean reads, the composition of faecal microbiota communities was revealed at the phylum and genus level. Overall, Bacteroidetes, Firmicutes, Proteobacteria, Actinobacteria, Ascomycota, Synergistetes, Euryarchaeota, Verrucomicrobia, Spirochaetes, Mucorales, Basidiomycota and Fibrobacteres were detected in all samples. Among these phyla, Bacteroidetes, Firmicutes, Proteobacteria and Actinobacteria were the four most dominant phyla, accounting for more than 98% of microorganisms in all groups. In the UC group, Bacteroidetes increased and Firmicutes, Proteobacteria, Ascomycota, Actinobacteria, and Synergistetes decreased compared with NG rats. Compared with the UC group, the UC + MOX group had an increased abundance of Bacteroidetes, Actinobacteria, Ascomycota, Synergistetes and a decreased abundance of Firmicutes and Proteobacteria. In addition, Bacteroidetes, Actinobacteria, Ascomycota, Synergistetes were increased and Firmicutes, Proteobacteria and Deferribacteres were decreased in the UC + MES group compared with the UC group. Compared with the NG group, the NG + MOX group had a decreased abundance of Firmicutes, Proteobacteria and Actinobacteria and a increased abundance of Bacteroidetes (Fig. 3A).
Fig. 3.
The effect of moxibustion on overall community structure at phylum and genus level. A at the phylum level, Bacteroidetes and Firmicutes were the most abundant taxa followed by Proteobacteria in five groups; B at the genus level, Bacteroides, Bacteroides_bacterium_M7, Bacteroides_bacterium_H3 were the most abundant taxa followed by Prevotella in five groups. NG: normal group; UC: UC model group; UC + MOX: moxibustion group; UC + MES: mesalazine group; NG + MOX: normal rats with moxibustion group
At the genus level, Bacteroides, Bacteroides_bacterium_M7, Bacteroides_bacterium_H3 followed by the Prevotella, Lactobacillus, Parabacteroides, Bacteroidales_bacterium_H2, Porphyromonas, Alistipes, Parasutterella, Barnesiella, Helicobacter, Clostridium, Bifidobacterium and Enterobacter were detected in all groups. Compared with the NG group, the relative abundance of Bacteroides, Bacteroides_bacterium_M7, Prevotella, Bacteroidales_bacterium_H2 and Barnesiella were increased and Bacteroides_bacterium_H3, Parabacteroides, Porphyromonas, Alistipes and Parasutterella reduced in the UC group. Interestingly, treatment of UC model rats with moxibustion led to a higher abundance of Bacteroides and Bacteroides_bacterium_H3 and a lower of Bacteroides_bacterium_M7, Prevotella, Bacteroidales_bacterium_H2 and Parasutterella. In addition, rats in the UC + MES group had a higher abundance of Bacteroides, Bacteroides_bacterium_M7, and Barnesiella and a lower abundance of Bacteroides_bacterium_H3, Prevotella, Parasutterella and Alistipes compared with the UC group (Fig. 3B).
A total of 5984 species have been identified, of which 2069 are core species distributed in all groups (Fig. 4). The average number of species detected in UC group was the highest (4926), while the lowest was found in NG group (2896). A total of 3611 species were detected in the UC + MOX rats, 3496 species in the UC + MES rats and 3830 species in the NG + MOX rats. Additionally, the UC group had the most unique species (800), even after treatment with moxibustion.
Fig. 4.
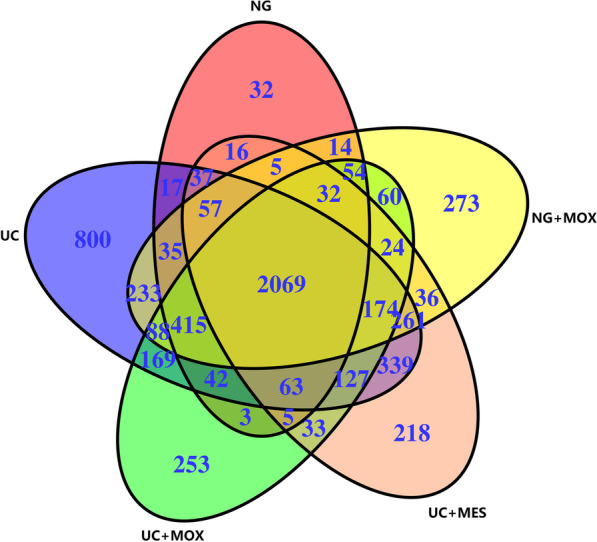
Summary of OTUs identified at the species level in all groups. There were 2069 core species were present in all groups. The UC group had the most unique species (800). NG: normal group; UC: UC model group; UC + MOX: moxibustion group; UC + MES: mesalazine group; NG + MOX: normal rats with moxibustion group
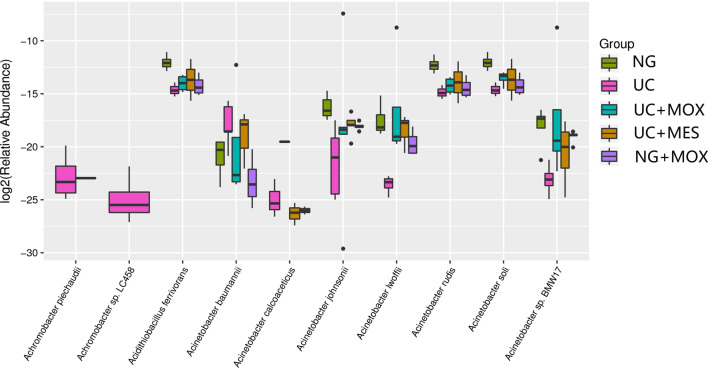
To gain further insight into the diversity of microbial community between groups, species-level differences was analysed (Fig. 5). Achromobacter piechaudii, Achromobacter sp.LC458, Acidithiobacillus ferrivorans, Acinetobacter baumannii, Acinetobacter calcoaceticus, Acinetobacter johnsonii, Acinetobacter lwoffii, Acinetobacter rudis, Acinetobacter soli and Acinetobacter sp.BMW17 were the most affected species. Additionally, Achromobacter piechaudii and Achromobacter sp.LC458 were the absolute dominant species in the UC group, suggesting that it may be related to the state of UC disease. Compared with the NG group, Achromobacter piechaudii, Achromobacter sp.LC458 and Acinetobacter baumannii were increased and Acidithiobacillus ferrivorans, Acinetobacter johnsonii, Acinetobacter lwoffii, Acinetobacter rudis, Acinetobacter soli and Acinetobacter sp. BMW17 were decreased in the UC model group. In addition, we found significantly lower abundance of Acinetobacter baumannii in the moxibustion group than in the UC group. We also noted that Acidithiobacillus ferrivorans, Acinetobacter johnsonii, Acinetobacter lwoffii, Acinetobacter rudis, Acinetobacter soli and Acinetobacter sp. BMW17 were increased in moxibustion, mesalazine and normal rats with moxibustion groups than UC group.
Fig. 5.
Species-level differences of gut microbiota between groups. Achromobacter piechaudii and Achromobacter sp.LC458 were the absolute dominant species in the UC group. NG: normal group; UC: UC model group; UC + MOX: moxibustion group; UC + MES: mesalazine group; NG + MOX: normal rats with moxibustion group
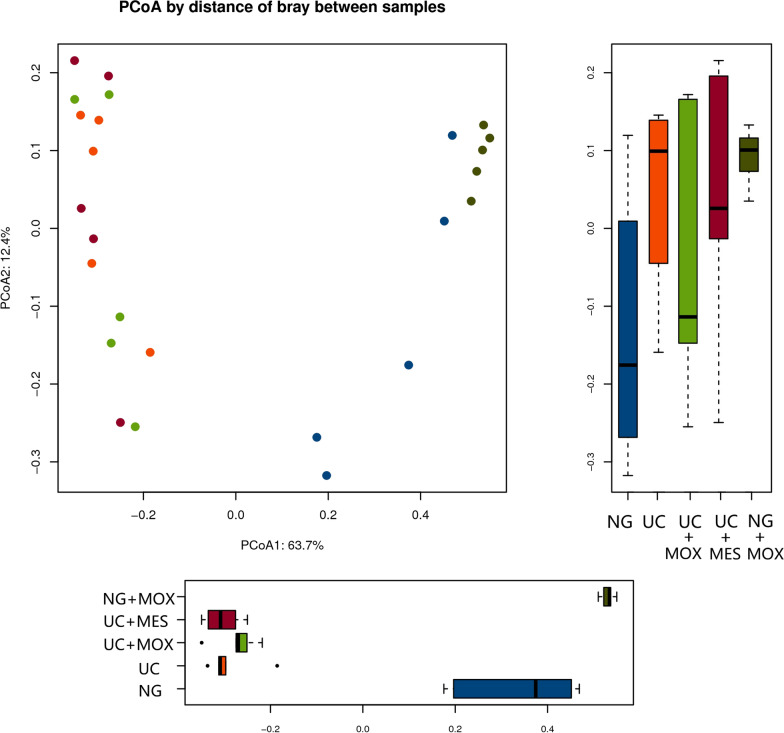
A principal coordinates analysis (PCoA) was performed to compare the composition of bacterial community within 25 samples, showed a clear separation of the community composition between normal rats and UC rats (Fig. 6). PCoA by distance of bray between samples explained 63.7% of variation in the first axes. The ordinations demonstrated that the disease state was the primary factor affecting the overall differences of bacterial communities between groups. The UC + MOX group was more similar to the UC + MES group on the first axis, whereas the NG + MOX group was more similar to the NG group. It was suggested that the diversity of gut microbiota was broken in UC rats. Furtherly, the flora community composition of UC rats was changed as well.
Fig. 6.
The effect of moxibustion on Beta diversity of gut microbiota. UC samples clustered separately from NG samples. The UC + MOX group was more similar to the UC + MES group on the first axis, whereas the NG + MOX group was more similar to the NG group. NG: normal group; UC: UC model group; UC + MOX: moxibustion group; UC + MES: mesalazine group; NG + MOX: normal rats with moxibustion group
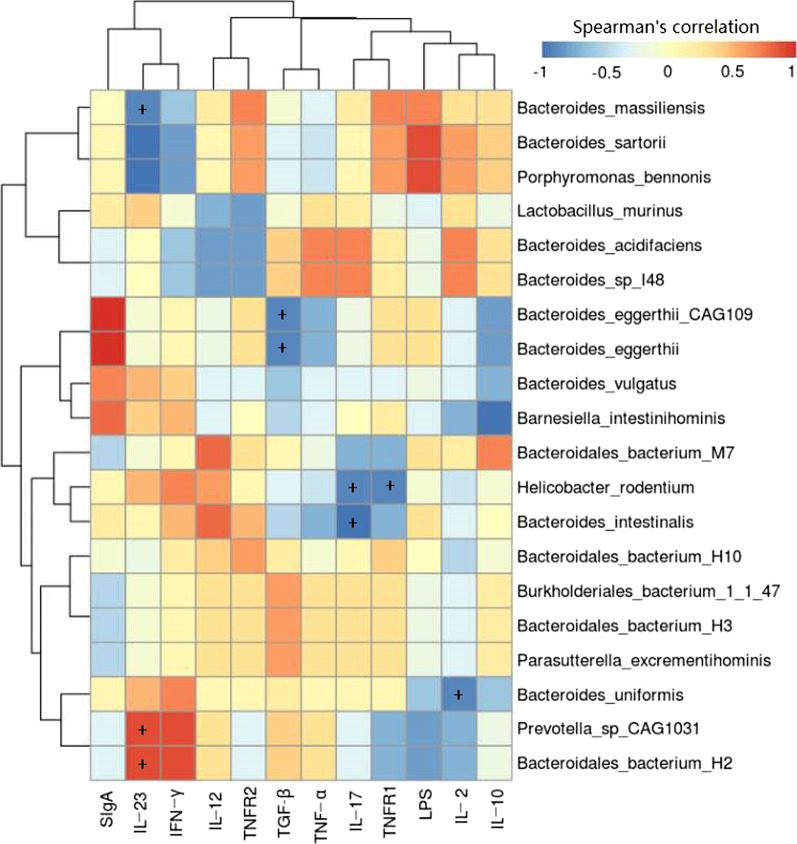
Correlation between the gut microbiota in species level and cytokines, LPS, SIgA in the UC model rats
Spearman correlation analysis between the gut microbiota and cytokines (IL-2, IL-10, IL-12, IL-17, IL-23, TGF-β, IFN-γ, TNF-α, TNFR1 and TNFR2), LPS, SIgA in serum in the UC model rats listed in Fig. 7. Our previous studies showed that compared with the UC rats, the levels of IL-6, IL-12, IL-17, IL-23, IFN-γ, LPS, TNF-α, TNFR1, TNFR2 in the moxibustion group were decreased and IL-2, TGF-β were increased [20]. From the figure, we found that Bacteroides_massiliensis was negatively (P < 0.05) correlated with IL-23, Bacteroides_eggerthii_CAG109 and Bacteroides_eggerthii were negatively (P < 0.05) correlated with TGF-β. A significant negative (P < 0.05) correlation also can be found between Helicobacter_rodentium and Bacteroides_intestinalis and IL-17. Helicobacter_rodentium was also negatively (P < 0.05) correlated with TNFR1, Bacteroides_uniformis was negatively (P < 0.05) correlated with IL-2. Additionally, IL-23 exhibited significant positive (P < 0.05) correlation with the species Prevotella_sp_CAG1031 and Bacteroides_bacterium_H2.
Fig. 7.
Heatmap of spearman’s correlation between the top 20 species and 12 inflammation-associated cytokines in UC group. N = 5; +P < 0.05. The different colors represent the range of (r) from − 1 to + 1. Orange indicates a perfect positive correlation, whereas blue indicates a perfect negative correlation
Functional annotation analysis
We used metagenomics methodologies to understand the functional capacity potential of gut microbiome in the UC progression. KEGG orthologs (KOs) is a taxonomic system of proteins (enzymes) that are highly similar in sequence. In total, 30,3740 KEGG genes were found and assigned to 38 KEGG pathways. The five functional modules were metabolism (62.81%), genetic information processing (16.45%), environment information processing (10.78%), cellular processes (4.33%), human diseases (3.81%) and organismal systems (1.82%). In five metabolic pathways, the most predominant functional categories were carbohydrate metabolism (44,838 genes, 14.76%), amino acid metabolism (34,340 genes, 11.31%), nucleotide metabolism (24,483 genes, 8.06%), energy metabolism (22,533 genes, 7.42%), translation (22,389 genes, 7.37%), membrane transport (20,333 genes, 6.69%) and replication and repair (15,827 genes, 5.21%) (Fig. 8A). Then we analyzed the differential KOs between NG and UC rats, and identified 33 KOs showing different abundance. Twenty out of these 33 KOs had the higher abundance in the NG rats, including those KOs associated with metabolism of cofactors and vitamins (K01633 and K03473), transport and catabolism (K04077), and carbohydrate metabolism (K15633). The other 13 KOs were enriched in UC rats, such as amino acid metabolism (K00215), translation (K02935), signaling molecules and interaction (K03699) (Fig. 8B). We also analyzed the differential KOs between UC and UC + MOX rats, and identified 12 KOs showing different abundance. Ten out of these 12 KOs had the higher abundance in the UC rats, including those KOs associated with nucleotide metabolism (K01933 and K01951) and membrane transport (K02471). The other 2 KOs were enriched in UC + MOX rats associated with Carbohydrate metabolism (K19265 and K00041) (Fig. 8C). Similarly, we found that there was 25 KOs showing different abundance between UC and UC + MES rats. Nineteen out of these 25 KOs had the higher abundance in the UC rats, including those KOs associated with metabolism of cofactors and vitamins (K01906), nucleotide metabolism (K01937, K01933, K01493, K01951 and K01429) and membrane transport (K02471, K10544, K02282 and K02006). The other 6 KOs were enriched in UC + MES rats associated with Carbohydrate metabolism (K19265 and K00041) and Amino acid metabolism (K00657) (Fig. 8D).
Fig. 8.
Classification of KEGG annotations of gut microbiota in all samples. A Summary of KEGG classification across all samples. B Differential KEGG orthologies (KOs) between NG and UC groups. C Differential KEGG orthologies (KOs) between UC and UC + MOX groups. D Differential KEGG orthologies (KOs) between UC and UC + MES groups. NG: normal group; UC: UC model group; UC + MOX: moxibustion group; UC + MES: mesalazine group; NG + MOX: normal rats with moxibustion group
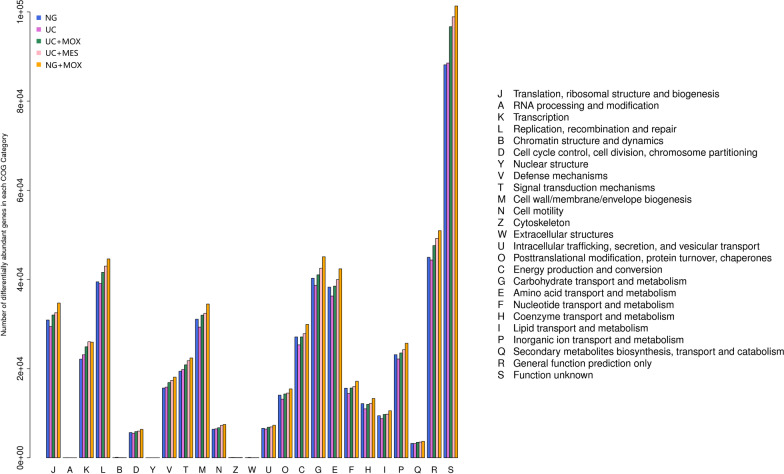
The EggNOG database is another common method for functional annotation using the Smith-Waterman alignment algorithm. We found that all genes were classified into 25 categories. Among these 25 functional classes, genes involved in certain metabolic pathways, such as energy production and conversion(25,339 and 27,102, respectively), amino acid transport and metabolism (36,275 and 38,286, respectively), nucleotide transport and metabolism (14,437 and 15,610, respectively), carbohydrate transport and metabolism (38,675 and 40,273, respectively), replication, recombination and repair (39,060 and 39,466, respectively), were under-represented in UC model rats compared with NG rats. It was suggested that the less vigorous microbial growth and metabolic stage in UC rats compared with that of normal rats. In addition, these changes in the metabolic pathways could be reversed by moxibustion treatment and mesalazine treatment (Fig. 9).
Fig. 9.
Classification of eggNOG annotations of gut microbiota in all samples. The capital letters on the X-axis represent different eggNOG categories. Y-axis shows the number of unigenes in each eggNOG category. NG: normal group; UC: UC model group; UC + MOX: moxibustion group; UC + MES: mesalazine group; NG + MOX: normal rats with moxibustion group
KEGG annotation exposes metabolic differences between UC and moxibustion treatment rats
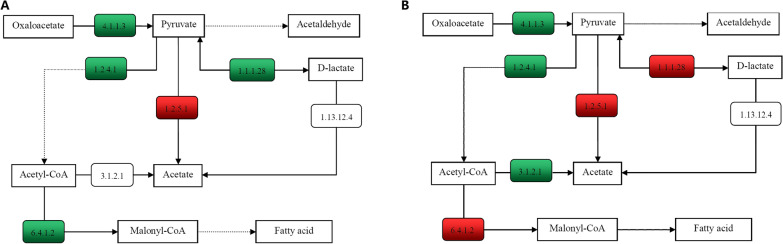
Marker genes involved in key processes of pyruvate metabolism that were significantly enriched in NG, UC or UC + MOX rats were annotated on KEGG pathway (Fig. 10). Oxaloacetate, which is an important intermediate in the tricarboxylic acid cycle, is used to produce pyruvate by oxaloacetate decarboxylase (EC 4.1.1.3). Compared with the NG group, pyruvate-producing genes (ECs 4.1.1.3 and 1.1.1.28) were reduced in the UC group. Compared with the UC group, pyruvate-producing genes (EC 1.1.1.28) was increased in the UC + MOX group. In the UC group, EC 1.2.4.1 involved in the reduction of pyruvate to acetyl-CoA which is a central metabolite of energy metabolism was also reduced compared with the NG group. EC 6.4.1.2 (Acetyl-CoA carboxylase), reduced in the UC group, was responsible for Acetyl-CoA to become malonyl-CoA which plays an important role in the biological synthesis of fatty acids and the biosynthesis of polyketones. Compared with the UC group, EC 6.4.1.2 (Acetyl-CoA carboxylase), increased in the UC + MOX group. In summary, the metabolism of fatty acids in the UC group was decreased compared with the NG group, and increased in UC + MOX group to some extent compared with the UC group.
Fig. 10.
Functional variations in pyruvate metabolism. A between NG and UC groups, the numbers in the red box represent genes in the UC group that were more abundant than in the normal group, whereas the green box represent genes that were less abundant in the UC group. B between UC and UC + MOX groups, the numbers in the red box represent genes in the UC + MOX group that were more abundant than in the UC group, whereas the green box represent genes that were less abundant in the UC + MOX group. NG: normal group; UC: UC model group; UC + MOX: moxibustion group
Discussion
Our body harbours enormous quantities of microorganisms that reside in the gut, skin, nasal, oral cavities and vagina [27]. The microbiota colonized in the gut is one of the most important habitats and can regulates the intestinal mucosal barriers, promotes energy intake, absorption and storage, mediates development of the host immune system, and prevents propagation of pathogenic microorganisms [28–31]. Previous studies have shown that dysbiosis of the gut microbiota can cause a large spectrum of human diseases, such as inflammatory bowel disease, colorectal cancer, obesity, type 1diabetes [8, 32–34]. In this study, our data showed that there were some differences between normal and ulcerative colitis rats in the gut microbiota, which is consistent with the previous study. Previous studies have shown that moxibustion is a safe and effective treatment for UC [15, 35–37], but its specific mechanism has not yet been fully understood. Therefore, we used metagenomic sequencing to determine the effects of moxibustion treatment on the microbial compositions and functional variations in a UC rat model.
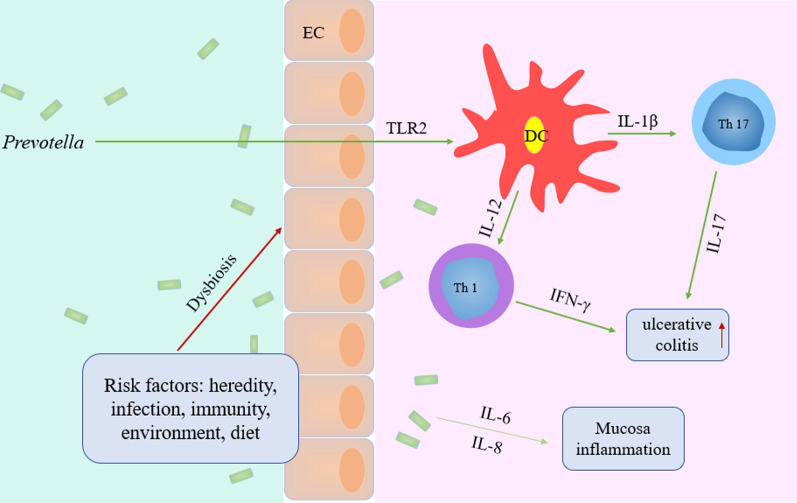
We found that Bacteroidetes and Firmicutes were the dominant taxa at phylum level, followed by Proteobacteria in all groups, it was consistent with our previous study using 16S rRNA amplicon sequencing [20]. Previous studies have found many bacteria associated with UC, and some of our findings are consistent with previous studies. Studies reported that Prevotella predominantly activate Toll‐like receptor 2, leading to production of T helper type 17 (Th17) polarizing cytokines and Th1 by dendritic cells, including IL‐23, IL‐1 and IFN-γ [38]. Furthermore, Prevotella has been identified to be linked with elevated IL-6, IL-8, IL-17 and CCL20 producing cells in the mucosa [38, 39] and enriched in IBD patients [40] (Fig. 11). Consistent with the studies, we found that the relative abundance of Prevotella was increased in UC rats compared with HC rats, indicating an association with ulcerative colitis. It was indicated that Prevotella may be an important pathological organism involved in ulcerative colitis by promoting chronic inflammation. In our study, the relative abundance of Parabacteroides was decreased in UC rats compared with HC rats. Studies have found that Parabacteroides can increase the level of gamma-aminobutyric acid (GABA) in the brain [41]. GABA is an important nerve signal substance in the mature central nervous system (CNS). Emerging evidence indicates that GABA has many beneficial effects such as ameliorating immune and inflammatory response. GABA inhibits pro-Inflammatory cytokine such as IL-12, IFN-γ, and TNF-α production, while significantly increasing tissue levels of IL-10 in DSS-induced colitis [42]. Therefore, the abnormality of the relative abundance of Parabacteroides may be the key to the pathogenesis of UC, this appears to be a novel finding that will require additional research.
Fig. 11.
Prevotella‐mediated inflammation in DSS colitis. Prevotella stimulate the release of IL‐1β and IL-12 by dendritic cells (DC) through TLR2, which in turn mediates IL‐17 and IFN-γ production by Th17 and Th1 cells. Dysbiosis‐associated increase in Th17 and Th1 immune responses may affect ulcerative colitis
Notably, we detected a significant higher abundance of Acinetobacter baumannii in UC rats. Acinetobacter baumannii, a gram-negative bacterium, is an opportunistic pathogen capable of causing a variety of infections (pulmonary, wound, skin, urinary tract, and bloodstream infections) in susceptible hosts [43, 44]. At the species level, the Achromobacter piechaudii and Achromobacter sp.LC458 were the absolute dominant species in the UC group and barely detectable in other groups. In previous studies, researchers have reported that Achromobacter piechaudii can act as an opportunistic pathogen cause certain human diseases [45, 46]. Accordingly, we can infer that those two bacterium may be related to the state of UC disease and warrant further investigation. Compared with normal rats, the abundance of some species such as Acidithiobacillus ferrivorans, Acinetobacter johnsonii were decreased in UC rats. In addition, the relative abundance levels of several opportunistic pathogenic species were increased in the UC rats. Thus, there was a significant difference in community composition between normal rats and UC rats, which was confirmed by PCoA.
Interestingly, we found that moxibustion and mesalazine treatment decreased the abundance levels of Prevotella. Prevotella‐mediated mucosal inflammation leads to accumulation of intestinal inflammatory mediators and bacterial products, which in turn may affect the occurrence and development of ulcerative colitis. Thus, we have reason to believe that moxibustion could adjust the inflammatory response of ulcerative colitis by the changes of the intestinal microbiota community. We also found that moxibustion treatment increased the abundance levels of Bacteroides and decreased Prevotella abundance. Bacteroides has been found to induce CD4 + T cells by enhancing anti-inflammatory IL-10 and suppressing pro-inflammatory IL-17 production through secreting polysaccharide A which is not only able to prevent but also cure experimental colitis in animals [47, 48]. Prevotella can stimulate inflammatory cytokines, such as IL-17 [38], Bacteroides can up-regulate anti-inflammatory IL-10 and down-regulate inflammatory cytokines IL-17. Our previous studies showed that compared with the UC rats, the levels of IL-6, IL-12, IL-17, IL-23, IFN-γ, LPS, TNF-α, TNFR1, TNFR2 in the moxibustion group were decreased and IL-2, TGF-β were increased [20]. It suggested that moxibustion treatment may protect the host from mucosal inflammation by modulating the intestinal microbiota community (such as Prevotella and Bacteroides) to down-regulating inflammatory cytokines and up-regulating anti-inflammatory cytokines. In addition, spearman correlation analysis between the intestinal flora and cytokines such as IL-6, IL-12 indicated that the intestinal flora is closely related to the secretion and expression of cytokines in ulcerative colitis, and they interacted with each other to regulate immune function in the body. Moxibustion can adjust the balance of intestinal microecology and restore the immune system, so as to ameliorate DSS-induced ulcerative colitis. Moxibustion can regulate the intestinal flora through multi-faceted and multi-level synergistic effects, but the specific mechanism is still unclear, and further research is needed. At present, the research on the effect of moxibustion on the intestinal flora is mainly focuses on regulating the intestinal microbiota by regulating brain-gut peptides and inflammatory response [49]. In this study, we found that moxibustion can promote the repair of mucous membrane in UC rats and improve the microbiome. However, the underlying mechanisms may require further study through animal and cell experiments. In the future, we can pay more attention to the warming effect of moxibustion, acupoint specificity, acupoint combination and different intervention measures to better study the specific mechanism of moxibustion in regulating intestinal flora.
Conclusions
In conclusion, based on the analysis of the Illumina Hiseq platform, moxibustion improved the richness and diversity of microbial community in rats with ulcerative colitis, and after moxibustion, the composition and diversity of the intestinal flora of the UC group were restored. Opportunistic pathogens decreased (such as Prevotella), while the abundance of Bacteroides was elevated in various degrees. Thus, moxibustion treatment could protect the host from mucosal and ameliorate intestinal inflammation of DSS-induced ulcerative colitis by modulating the intestinal microbiota community.
Acknowledgements
We would like to thank the Realbio Genomics Institute in this study for Sequencing data analysis. We thank all the authors for their wonderful collaboration.
Abbreviations
- UC
Ulcerative colitis
- DSS
Dextran sulphate sodium
- ORFs
Open reading frame
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NOG
Non-supervised Orthologous Groups
- PCoA
Principal coordinates analysis
Authors' contributions
WXM and WLY conceived and designed the study; QQ and LYN wrote the main manuscript text; LSY, ZLS and CZ analyzed the data; WHG and LHR revised the manuscript critically. All authors read and approved the final manuscript.
Funding
This work was supported by grants from Natural Science Foundation of Shanghai (Grant Number 20ZR1453100); The National Natural Science Foundation of China (Grant Number 81473758, 81674074, 81973955); Three-year Action Plan Project of Shanghai Traditional Chinese Medicine Development (Grant Number ZY (2018-2020)-CCCX-2004-01) and the National Basic Research Program of China (973 program) (Grant Number 2015CB554501) for financial support.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the NCBI repository, accession: PRJNA795867.
Declarations
Ethics approval and consent to participate
The protocol of animal experiments was approved by the University Animal Care and Use Committee of Shanghai University of Traditional Chinese Medicine and performed strictly according to the guideline on animal experimentation.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qin Qi, Ya-Nan Liu and Si-Yi Lv have contributed equally to this work
Contributor Information
Qin Qi, Email: 719928895@qq.com.
Ya-Nan Liu, Email: 5527lyn@163.com.
Si-Yi Lv, Email: lvsiyi641@163.com.
Huan-Gan Wu, Email: wuhuangan@126.com.
Lin-Shuang Zhang, Email: 953275321@qq.com.
Zhan Cao, Email: caozhan1987@163.com.
Hui-Rong Liu, Email: lhr_tcm@139.com.
Xiao-Mei Wang, Email: wxm123@vip.sina.com.
Lu-Yi Wu, Email: luyitcm@163.com.
References
- 1.Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389(10080):1756–1770. doi: 10.1016/S0140-6736(16)32126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feuerstein JD, Moss AC, Farraye FA. Ulcerative Colitis. Mayo Clin Proc. 2019;94(7):1357–1373. doi: 10.1016/j.mayocp.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 3.Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390(10114):2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 4.Niewiadomski O, Studd C, Hair C, Wilson J, McNeill J, Knight R, et al. Health care cost analysis in a population-based inception cohort of inflammatory bowel disease patients in the first year of diagnosis. J Crohns Colitis. 2015;9(11):988–996. doi: 10.1093/ecco-jcc/jjv117. [DOI] [PubMed] [Google Scholar]
- 5.Zatorski H, Sałaga M, Zielińska M, Piechota-Polańczyk A, Owczarek K, Kordek R, et al. Experimental colitis in mice is attenuated by topical administration of chlorogenic acid. Naunyn Schmiedebergs Arch Pharmacol. 2015;388(6):643–651. doi: 10.1007/s00210-015-1110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ananthakrishnan AN, Bernstein CN, Iliopoulos D, Macpherson A, Neurath MF, Ali RAR, et al. Environmental triggers in IBD: a review of progress and evidence. Nat Rev Gastroenterol Hepatol. 2018;15(1):39–49. doi: 10.1038/nrgastro.2017.136. [DOI] [PubMed] [Google Scholar]
- 7.Mentella MC, Scaldaferri F, Pizzoferrato M, Gasbarrini A, Miggiano GAD. Nutrition, IBD and gut microbiota: a review. Nutrients. 2020;12(4):944. doi: 10.3390/nu12040944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glassner KL, Abraham BP, Quigley EMM. The microbiome and inflammatory bowel disease. J Allergy Clin Immunol. 2020;145(1):16–27. doi: 10.1016/j.jaci.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Astó E, Méndez I, Audivert S, Farran-Codina A, Espadaler J. The efficacy of probiotics, prebiotic inulin-type fructans, and synbiotics in human ulcerative colitis: a systematic review and meta-analysis. Nutrients. 2019;11(2):293. doi: 10.3390/nu11020293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui L, Guan X, Ding W, Luo Y, Wang W, Bu W, et al. Scutellaria baicalensis Georgi polysaccharide ameliorates DSS-induced ulcerative colitis by improving intestinal barrier function and modulating gut microbiota. Int J Biol Macromol. 2021;166:1035–1045. doi: 10.1016/j.ijbiomac.2020.10.259. [DOI] [PubMed] [Google Scholar]
- 11.Song G, Fiocchi C, Achkar JP. Acupuncture in inflammatory bowel disease. Inflamm Bowel Dis. 2019;25(7):1129–1139. doi: 10.1093/ibd/izy371. [DOI] [PubMed] [Google Scholar]
- 12.Han Y, Ma TM, Lu ML, Ren L, Ma XD, Bai ZH. Role of moxibustion in inflammatory responses during treatment of rat ulcerative colitis. World J Gastroenterol. 2014;20(32):11297–11304. doi: 10.3748/wjg.v20.i32.11297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan X, Ma F, Yu Y, Du D, Wang Z, Chen C, et al. Effects of herb-partitioned moxibustion for ulcerative colitis: a protocol for systematic review and meta-analysis. Medicine (Baltimore) 2020;99(31):e21319. doi: 10.1097/MD.0000000000021319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee DH, Kim JI, Lee MS, Choi TY, Choi SM, Ernst E. Moxibustion for ulcerative colitis: a systematic review and meta-analysis. BMC Gastroenterol. 2010;10:36. doi: 10.1186/1471-230X-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei D, Xie L, Zhuang Z, Zhao N, Huang B, Tang Y, et al. Gut microbiota: a new strategy to study the mechanism of electroacupuncture and moxibustion in treating ulcerative colitis. Evid Based Complement Alternat Med. 2019;2019:9730176. doi: 10.1155/2019/9730176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Ma Z, Cui YH, Dong HS, Zhao JM, Dou CZ, et al. Effects of herb-partitioned moxibustion on the miRNA expression profiles in colon from rats with DSS-induced ulcerative colitis. Evid Based Complement Alternat Med. 2017;2017:1767301. doi: 10.1155/2017/1767301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang XM, Lu Y, Wu LY, Yu SG, Zhao BX, Hu HY, et al. Moxibustion inhibits interleukin-12 and tumor necrosis factor alpha and modulates intestinal flora in rat with ulcerative colitis. World J Gastroenterol. 2012;18(46):6819–6828. doi: 10.3748/wjg.v18.i46.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma TM, Xu N, Ma XD, Bai ZH, Tao X, Yan HC. Moxibustion regulates inflammatory mediators and colonic mucosal barrier in ulcerative colitis rats. World J Gastroenterol. 2016;22(8):2566–2575. doi: 10.3748/wjg.v22.i8.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y, Ding G, Zheng H, Lü T, Ma Z, Wu H, et al. Effect of herb-partitioned moxibustion on dopamine levels and dopamine receptor 1 expression in the colon and central nervous system in rats with Crohn's disease. J Tradit Chin Med. 2019;39:356–363. [PubMed] [Google Scholar]
- 20.Qi Q, Liu YN, Jin XM, Zhang LS, Wang C, Bao CH, et al. Moxibustion treatment modulates the gut microbiota and immune function in a dextran sulphate sodium-induced colitis rat model. World J Gastroenterol. 2018;24(28):3130–3144. doi: 10.3748/wjg.v24.i28.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M, Kiyohara C, et al. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol. 2009;56(1):80–87. doi: 10.1111/j.1574-695X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- 22.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 23.Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513(7516):59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 24.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40(Database issue):D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soldavini J, Kaunitz JD. Pathobiology and potential therapeutic value of intestinal short-chain fatty acids in gut inflammation and obesity. Dig Dis Sci. 2013;58(10):2756–2766. doi: 10.1007/s10620-013-2744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16(10):605–616. doi: 10.1038/s41575-019-0173-3. [DOI] [PubMed] [Google Scholar]
- 29.Okumura R, Takeda K. Maintenance of intestinal homeostasis by mucosal barriers. Inflamm Regen. 2018;38:5. doi: 10.1186/s41232-018-0063-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolodziejczyk AA, Zheng D, Elinav E. Diet-microbiota interactions and personalized nutrition. Nat Rev Microbiol. 2019;17(12):742–753. doi: 10.1038/s41579-019-0256-8. [DOI] [PubMed] [Google Scholar]
- 31.Koh A, Bäckhed F. From association to causality: the role of the gut microbiota and its functional products on host metabolism. Mol Cell. 2020;78(4):584–596. doi: 10.1016/j.molcel.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Peterson CT, Sharma V, Elmén L, Peterson SN. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin Exp Immunol. 2015;179(3):363–377. doi: 10.1111/cei.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosca AE, Iesanu MI, Zahiu CDM, Voiculescu SE, Paslaru AC, Zagrean AM. Capsaicin and gut microbiota in health and disease. Molecules. 2020;25(23):5681. doi: 10.3390/molecules25235681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al Bander Z, Nitert MD, Mousa A, Naderpoor N. The gut microbiota and inflammation: an overview. Int J Environ Res Public Health. 2020;17(20):7618. doi: 10.3390/ijerph17207618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Zhang Q, Deng Y, Deng G, Huang F, Zhou Y, et al. Efficacy and safety of heat-sensitive moxibustion in the treatment of ulcerative colitis: A protocol for a systematic review and meta-analysis. Medicine (Baltimore) 2021;100(4):e24078. doi: 10.1097/MD.0000000000024078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang N, Zhang Q, Xie L, Li C, Zhuang Z, Lin S, et al. Electroacupuncture and moxibustion regulate hippocampus glia and mitochondria activation in DSS-induced colitis mice. Evid Based Complement Alternat Med. 2020;2020:2530253. doi: 10.1155/2020/2530253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin YY, Zhao JM, Ji YJ, Ma Z, Zheng HD, Huang Y, et al. Typical ulcerative colitis treated by herbs-partitioned moxibustion: a case report. World J Clin Cases. 2020;8(8):1515–1524. doi: 10.12998/wjcc.v8.i8.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151(4):363–374. doi: 10.1111/imm.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B, Du P, Du Y, Zhao D, Cai Y, Yang Q, et al. Luteolin alleviates inflammation and modulates gut microbiota in ulcerative colitis rats. Life Sci. 2021;269:119008. doi: 10.1016/j.lfs.2020.119008. [DOI] [PubMed] [Google Scholar]
- 40.Tao JH, Duan JA, Jiang S, Feng NN, Qiu WQ, Ling Y. Polysaccharides from Chrysanthemum morifolium Ramat ameliorate colitis rats by modulating the intestinal microbiota community. Oncotarget. 2017;8(46):80790–80803. doi: 10.18632/oncotarget.20477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell. 2018;173(7):1728–1741.e13. doi: 10.1016/j.cell.2018.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma X, Sun Q, Sun X, Chen D, Wei C, Yu X, et al. Activation of GABAA receptors in colon epithelium exacerbates acute colitis. Front Immunol. 2018;9:987. doi: 10.3389/fimmu.2018.00987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antunes LC, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71(3):292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 44.Cho GS, Li B, Rostalsky A, Fiedler G, Rösch N, Igbinosa E, et al. Diversity and antibiotic susceptibility of acinetobacter strains from milk powder produced in Germany. Front Microbiol. 2018;9:536. doi: 10.3389/fmicb.2018.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krause ML, Sohail MR, Patel R, Wittich CM. Achromobacter piechaudii bloodstream infection in an immunocompetent host. Am J Case Rep. 2012;13:265–267. doi: 10.12659/AJCR.883527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filipic B, Malesevic M, Vasiljevic Z, Lukic J, Novovic K, Kojic M, et al. Uncovering differences in virulence markers associated with achromobacter species of CF and Non-CF origin. Front Cell Infect Microbiol. 2017;7:224. doi: 10.3389/fcimb.2017.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang YC, Ching YH, Chiu CC, Liu JY, Hung SW, Huang WC, et al. TLR2 and interleukin-10 are involved in Bacteroides fragilis-mediated prevention of DSS-induced colitis in gnotobiotic mice. PLoS ONE. 2017;12(7):e0180025. doi: 10.1371/journal.pone.0180025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sittipo P, Lobionda S, Choi K, Sari IN, Kwon HY, Lee YK. Toll-like receptor 2-mediated suppression of colorectal cancer pathogenesis by Polysaccharide A From Bacteroides fragilis. Front Microbiol. 2018;9:1588. doi: 10.3389/fmicb.2018.01588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang WY, Liang FX, Song AQ, Huang Q, Chen R. Current situation and thinking on the intestinal microflora regulation with acupuncture and moxibustion. Zhen Ci Yan Jiu. 2019;44(1):71–4. Chinese. 10.13702/j.1000-0607.180118 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the NCBI repository, accession: PRJNA795867.