Abstract
Companion diagnostic immunohistochemistry (IHC) tests are developed and performed without incorporating the tools and principles of laboratory metrology. Basic analytic assay parameters such as lower limit of detection (LOD) and dynamic range are unknown to both assay developers and end users. We solved this problem by developing completely new tools for IHC - calibrators with units of measure traceable to National Institute of Standards & Technology (NIST) Standard Reference Material (SRM) 1934. In this study, we demonstrate the clinical impact and opportunity for incorporating these changes into PD-L1 testing. Forty-one laboratories in North America and Europe were surveyed with newly-developed PD-L1 calibrators. The survey sampled a broad representation of commercial and laboratory-developed tests (LDTs). Using the PD-L1 calibrators, we quantified analytic test parameters that were previously only inferred indirectly after large clinical studies. The data show that the four FDA-cleared PD-L1 assays represent three different levels of analytic sensitivity. The new analytic sensitivity data explain why some patients’ tissue samples were positive by one assay and negative by another. The outcome depends on the assay’s lower limit of detection. Also, why previous attempts to harmonize certain PD-L1 assays were unsuccessful; the assays’ dynamic ranges were too disparate and did not overlap. PD-L1 assay calibration also clarifies the exact performance characteristics of LDTs relative to FDA-cleared commercial assays. Some LDTs’ analytic response curves are indistinguishable from their predicate FDA-cleared assay. IHC assay calibration represents an important transition for companion diagnostic testing. The new tools will improve patient treatment stratification, test harmonization, and foster accuracy as tests transition from clinical trials to broad clinical use.
INTRODUCTION
It is an axiom in laboratory science that accurate, reproducible testing requires assay calibrators having units of measure traceable to an accepted reference standard. These components – calibrators, traceable units of measure, and reference standards – are integral features of modern laboratory metrology. Hundreds of clinical laboratory reference standards, from amylase to zinc, support a corresponding number of clinical laboratory tests. These hundreds of higher order reference standards link to thousands of lower order standards - commercial calibrators – for regular use in clinical laboratories. Companion diagnostic IHC testing is an exception in not adopting these conventions. Despite their importance in cancer patient management, companion diagnostic IHC tests are still treated as “stains” rather than assays with metrologic standards.(1) With the recent description of NIST SRM 1934 as a universal IHC reference standard (2), we evaluated the impact of programmed death-ligand 1 (PD-L1) calibrators on laboratory testing.
PD-L1 IHC testing is a case study of the strengths and limitations of IHC companion diagnostic testing. A strength of PD-L1 testing is that all four Food and Drug Administration (FDA)-cleared IHC tests were shown to (variably) predict clinical responses to specific immune checkpoint inhibitors (ICI’s). For some patients, these ICI’s induce a striking augmentation of anti-tumor immunity that sometimes leads to dramatic clinical remissions. An important limitation, on the other hand, is that multiple predictive PD-L1 IHC assays were developed, each with varied, ill-defined performance characteristics, which are difficult to study and compare to each other. The four FDA-cleared companion/complementary (CDx) tests use different primary monoclonal antibodies, different automated instruments, different detection systems, different allowable pre-analytical conditions, different readout methods for assessing PD-L1 expression in tumor and/or inflammatory cells, and often different thresholds for positive vs. negative results. Adding as yet another layer of complexity, some laboratories develop PD-L1 laboratory-developed tests (LDTs) or use FDA-cleared tests for a non-corresponding ICI. Current IHC methods provide little insight into analytic sensitivity, as defined by the LOD and dynamic range. PD-L1 readouts provide no reference to the actual PD-L1 cellular protein concentration. PD-L1 calibrators offer the opportunity to define these variables and characterize the end result in terms of well-defined levels of analytic sensitivity.
Previously, a series of studies compared the performance of the various PD-L1 IHC tests to better understand how the tests relate to one another.(3–5) In these previously published studies comparing the various PD-L1 tests, analytic sensitivity was inferred indirectly by comparing staining results on a series of patient tumor samples or cell lines having unknown PD-L1 concentrations. In other words, analytic sensitivity was inferred in relative and descriptive terms. In this paper, we characterize the various approved PD-L1 tests, as well as some LDTs, in terms of the absolute PD-L1 protein concentration using an important new analytical tool.
Terminology.
The new reference materials described in this paper incorporate terminology that, although well-established in the field of laboratory medicine, is new to immunohistochemistry. For the sake of clarity, we define the terms.
“Analytic sensitivity” refers to the ability of an IHC test to detect a defined number of PD-L1 protein molecules. More sensitive tests will detect tumor cells expressing a lower number of PD-L1 molecules.
The “LOD” is the relevant measure of analytic sensitivity for qualitative assays like IHC.(6) The LOD is the lowest PD-L1 concentration that visibly stains, i.e., can be reliably distinguished from background. It also defines the lower boundary of the assay’s dynamic range.(7) Lower LODs indicate greater analytic sensitivity, in being able to detect fewer molecules of PD-L1. LOD is important because it is the threshold of cellular staining, directly affecting a pathologist’s readout.
The lower limit of quantification (LOQ) was not incorporated as a measure in this study but may be important for future quantitative IHC studies. It was not incorporated as a measure in this study because PD-L1 testing is qualitative in nature; each cell is judged as either positive or negative for PD-L1. Whereas LOD is the lower bound of the dynamic range, LOQ is the lower limit of the linear range. LOQ is often defined as the lowest analyte concentration that yields an assay precision with a coefficient of variation ≤ 20%.
The term “descriptive analytic sensitivity” refers to a recent practice of identifying cells/tissues with a relatively low level of analyte expression. Staining of these cells/tissues offers evidence that the assay likely has adequate sensitivity.(8) However, this is no substitute for an actual measured LOD and provides no insight into the assay dynamic range.
The “dynamic range” of a PD-L1 assay is the cellular PD-L1 concentration span from the LOD to the concentration that produces maximal staining.(7) Dynamic range is broader than the “linear range”. Whereas linear range incorporates the analyte concentration span that produces a corresponding proportional (linear) assay signal response, dynamic range also includes analyte concentrations at the high end showing non-linear increases in signal. Dynamic range also incorporates analyte concentrations at the low end where the precision is insufficiently poor to produce quantitative results but adequate for positive/negative test results. Dynamic range was selected as the relevant parameter in this study because PD-L1 testing is qualitative. Linear ranges apply to quantitative tests.
Knowing the dynamic range is important because of considerations as to whether the differences in analytical sensitivity among the various assays can potentially be compensated by adjusting the cutoffs in the readouts (e.g., Tumor Proportion Score (TPS) or Combined Positive Score (CPS)). For example, a 20% positive cells cutoff on a highly sensitive stain may be equally predictive of patient responses as a 5% positive cells cutoff on a lower sensitivity test. This is because analytic sensitivity can have a profound effect on the percent positive cells. IHC stains with greater sensitivity result in higher percentages of stained cells. The effects of IHC analytic sensitivity on test results can be so profound as to completely change the diagnostic test result, as previously demonstrated for estrogen receptor testing.(2) If the dynamic ranges of the tests show significant overlap, then adjusting the threshold of positivity may have the potential to stratify patients into treatment groups in an equivalent manner.
MATERIALS AND METHODS
Recently, we described the first system of IHC reference materials that provide for quantitative characterization of IHC protocol performance.(2, 9–11) The new system incorporates units of measure traceable to Standard Reference Material 1934 at the National Institute of Standards and Technology (NIST).(2) This new system of measurement permits quantitative characterization of IHC assays, with precise measurements of LOD and dynamic range, expressed as the number of molecules per cell equivalent detected by an IHC assay. In this study, we apply the Boston Cell Standards (BCS) calibrators to PD-L1 testing, to directly measure and compare the analytic performance of the four FDA-cleared CDx assays and various LDTs.
Two types of PD-L1 BCS calibrator slides, for primary antibodies binding to the intracellular and extracellular PD-L1 domains, were manufactured and distributed to 41 laboratories in the U.S., Canada (Canadian Biomarker Quality Assurance (CBQA)), the Netherlands, and Belgium. The two PD-L1 BCS calibrators are distinguished by the portion of the PD-L1 protein that is attached to the calibrator.
PD-L1 calibrator – intracellular domain.
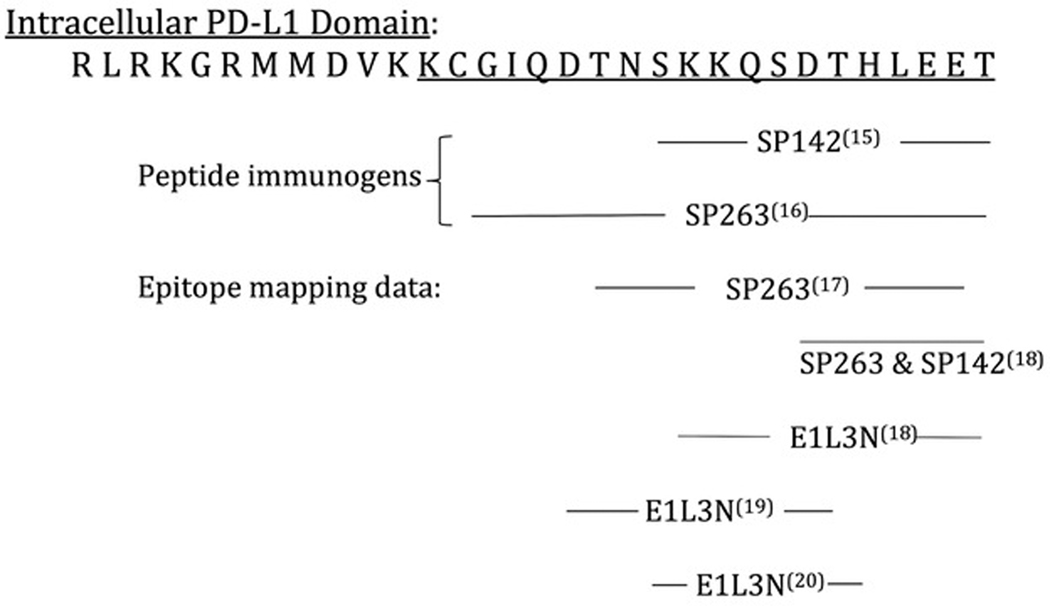
This BCS calibrator incorporates a peptide spanning most of the intracellular domain of PD-L1. We have previously described the use of peptides that incorporate an epitope as controls or calibrators in lieu of a native protein.(12–14) The PD-L1 intracellular domain peptide includes the epitopes of monoclonal antibodies (mAbs) SP142, SP263, E1L3N, ZR3, and 73–10. The peptide was purchased from CS Bio, Menlo Park, CA and is 93% pure based on mass spectroscopy analysis by the manufacturer. The remaining 7% is comprised of similar peptides that, due to less than 100% incorporation during synthesis, may randomly lack an individual amino acid. Most of these slightly truncated peptides still likely incorporate the relevant epitopes.
Figure 1 shows a (single letter amino acid) representation of the PD-L1 intracellular domain. With minor modifications, the peptide used for the BCS calibrator is underlined. In addition, Fig. 1 shows available epitope mapping information for 3 PD-L1 mAbs – SP142, SP263, and E1L3N, with cited references. For each antibody, a span of amino acids is identified that, based on epitope mapping data, include the epitope. For SP263 and E1L3N, epitope mapping data from multiple sources is shown, each with different but overlapping data. The actual linear epitope will be found within the region of overlap.
Fig. 1. PD-L1 BCS calibrator, intracellular domain.
The epitope location data for various primary antibodies is shown. The amino acids are represented using the single letter amino acid code. The peptide calibrator incorporates the underlined amino acid sequence. The epitopes for the SP142, SP263, and E1L3N mAbs are found within the indicated regions.
The intracellular domain BCS calibrator was manufactured by covalently attaching a peptide to cell sized (7 – 8 micron diameter) glass microbeads, as previously described.(11, 21, 22) This coupling reaction was performed at ten different peptide concentrations, resulting in PD-L1 concentrations at regularly spaced intervals. The peptide incorporates a fluorescein molecule to establish traceability of measurement to NIST SRM 1934, as previously described.(2) For the intracellular domain PD-L1 BCS calibrators, those concentrations are 34,000 – 2,200,000 molecules of PD-L1 peptide per microbead.
PD-L1 calibrator – extracellular domain.
PD-L1 tests incorporating monoclonal antibodies 22C3 and 28–8 were evaluated using extracellular domain (ECD) BCS calibrators. These two mAbs recognize epitopes in the extracellular domain (ECD) of PD-L1 that, in our experience, cannot be represented as linear epitopes. Recent data indicate that these two epitopes are at least partly glycosylation-dependent.(18) Therefore, a recombinant ECD protein is used as the calibrator. Fluorescein-conjugated, purified recombinant PD-L1 ECD with a C-terminal poly-histidine tail, produced in HEK293 (human embryonic kidney) cells, was purchased from GenScript, Piscataway, NJ. The PD-L1 ECD protein was covalently coupled to cell sized glass microbeads, as described above. The PD-L1 concentration per microbead was measured by interpolating the fluorescence intensity against a calibration curve traceable to NIST SRM 1934, as previously described.(2) The resulting molecular concentration of fluorescein was then divided by the fluorescein:protein ratio, as measured spectrophotometrically. The concentration of PD-L1 ECD protein per microbead was 2,200 – 600,000 molecules.
PD-L1 testing survey.
The PD-L1 intracellular and extracellular domain calibrators with the aforementioned range of concentrations were applied to microscope slides in a 5 × 3 array as previously described.(2) Each calibrator spot on the slide in the 5 × 3 array incorporates approximately 5000 peptide- or protein-coated (test) microbeads. The calibrators, applied to microscope slides, were sent by regular mail at ambient temperature. Each participating laboratory stained the slides using their own PD-L1 assay protocols and then returned them to a central site, either in The Netherlands, Canada, or Boston, MA. Tissue samples were also included in the Canadian part of the survey, but on separate slides sent along with the PD-L1 calibrator slides.
LOD measurement.
BCS calibrators returned to Boston or Canada were photographed using a Zeiss Axioskop microscope fitted with a Spot Imaging Solutions Insight Gigabit CCD camera (Diagnostic Instruments Inc., Sterling Heights, MI). For calibrators managed by the Dutch central site, calibrators were scanned (Philips UFS, The Netherlands) and the images were sent to Boston Cell Standards for analysis. The details of microbead stain intensity quantification are described elsewhere.(9, 11) Briefly, stain intensity is quantified in an algorithm running in MatLab. Mean stain intensity data after image segmentation are normalized by expressing each as a ratio to a smaller color standard microbead that is also present in every image. This normalization standardizes the measurements, compensating for any variability in microscopy from day-to-day. For each PD-L1 assay, the maximum stain intensity is set at 100% and the other stain intensity data are expressed as a percentage of that maximum. This way, all of the PD-L1 assays are graphed on the same 0 – 100% scale.
From the stain intensity data associated with each calibrator concentration, we calculated the LOD for each of the PD-L1 stains. The method for calculating LOD was previously described.(2) Briefly, the LOD is characterized as the PD-L1 concentration associated with a stain intensity that is 3SD above the mean of a sample that has an antigenically irrelevant analyte. This simplified calculation is appropriate because the standard deviations associated with blank calibrators were identical to the standard deviations of low-positive calibrators. Assay dynamic ranges were also calculated from the same stain intensity data from image analysis and represented as analytic response curves.
PD-L1 LDT (E1L3N) staining methods.
Two laboratories stained for PD-L1 using the E1L3N primary antibody. Both purchased the antibody from Cell Signaling Technology (Danvers MA). A first lab used the antibody at a 1:500 dilution with a high pH antigen retrieval solution for 30 minutes, Leica Biosystems Bond III and Leica polymer detection system. A second lab used the antibody at 1:100 with a citrate buffered antigen retrieval solution for 20 minutes, on a Dako Autostainer with a Roche HRP multimer detection system.
RESULTS
Lower limit of detection (LOD) assay comparisons.
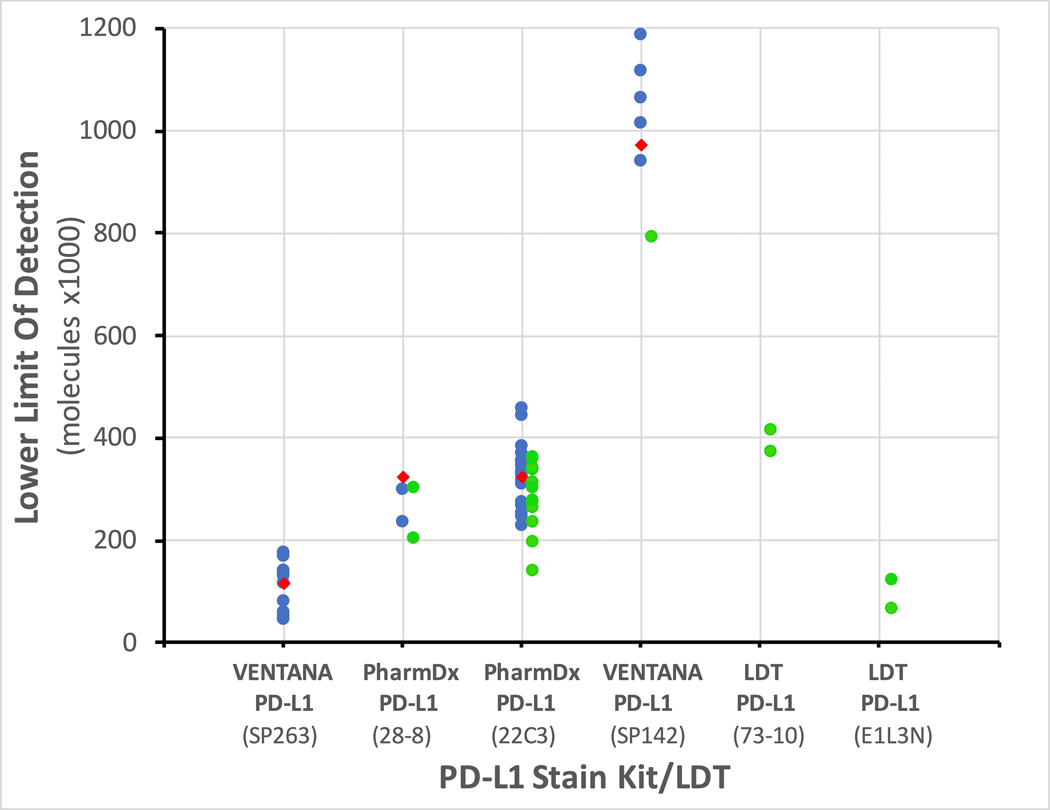
Figure 2 depicts the differences in LOD among the four FDA-cleared commercial PD-L1 tests and several LDTs. These LODs reflect the analytic sensitivity of the entire assay, not just the primary antibody. The data are from 59 PD-L1 assays in 41 different laboratories. Each dot is a separate PD-L1 LOD measurement. The LODs are color-coded: blue is an FDA-cleared kit, green is an LDT. Our LOD measurements show that the four FDA-cleared tests appear to have been developed at three different analytic sensitivity levels. The previous Blueprint studies (4, 5) identified two analytic sensitivity levels but our findings agree with a large meta-analysis.(3) Among the FDA-cleared PD-L1 kits, the VENTANA PD-L1 (SP263) assay was the most sensitive, with an LOD below 200,000 molecules per cell equivalent. It was closely followed by PD-L1 IHC 28–8 pharmDx and PD-L1 IHC 22C3 pharmDx, both showing LODs in the 200,000 – 400,000 molecules per cell equivalent range. The VENTANA PD-L1 (SP142) assay was substantially less sensitive, with LODs in the 800,000 – 1,000,000 molecules per cell equivalent range.
Fig. 2. Lower limit of detection (LOD) of various PD-L1 assays (x axis).
Lower numbers (on the y axis) equate to greater sensitivity. Each dot represents a separate IHC laboratory test. Blue dots depict FDA-cleared assays in clinical laboratories, green dots for laboratory-developed tests (LDTs), and red diamonds for FDA-cleared assays as performed by a reference laboratory. Tissue staining in Fig. 2 was performed by these reference labs. For enhanced clarity, the LDT data are positioned slightly to the right of the vertical lines.
LOD influences patient test results.
A previous national study of estrogen receptor testing demonstrated that IHC tests with lower LODs result in a higher number of positive patient test results.(2) The same is true for PD-L1. The FDA-cleared commercial assays with lower LODs (Fig. 2) are the assays associated with more PD-L1 positive test results.(3) Figure 3 shows an example. Serial sections of the same tumor sample were stained using the four FDA-cleared kits. The images in Fig. 3 are from two reference labs whose LODs (x1000) are shown with red diamonds in Fig. 2. The images are arranged with the most sensitive assay, the VENTANA PD-L1 (SP263), in the upper left-hand corner (Fig. 3A) and the least sensitive, the VENTANA PD-L1 (SP142) assay, in the lower right (Fig. 3D). An LOD of 90 (x1000, Fig. 3A) resulted in the highest stain intensity and highest number of positive cells. LODs of 324 (x1000, Fig. 3B) and 322 (x1000, Fig. 3C) showed similar numbers of positive cells, but the latter is slightly obscured by cytoplasmic, non-specific, staining (Fig. 3C). The highest LOD of 974 (x1000, Fig. 3D) reveals only a single PD-L1+ cell. These findings were expected and, in fact, were implicit assumptions in previous studies using patient tissue staining to indirectly infer assay analytic sensitivity.(4, 5) This example illustrates the importance of LOD on the surgical pathologist readout.
Fig. 3. Photomicrographs of PD-L1 staining on 4 commercial assays.
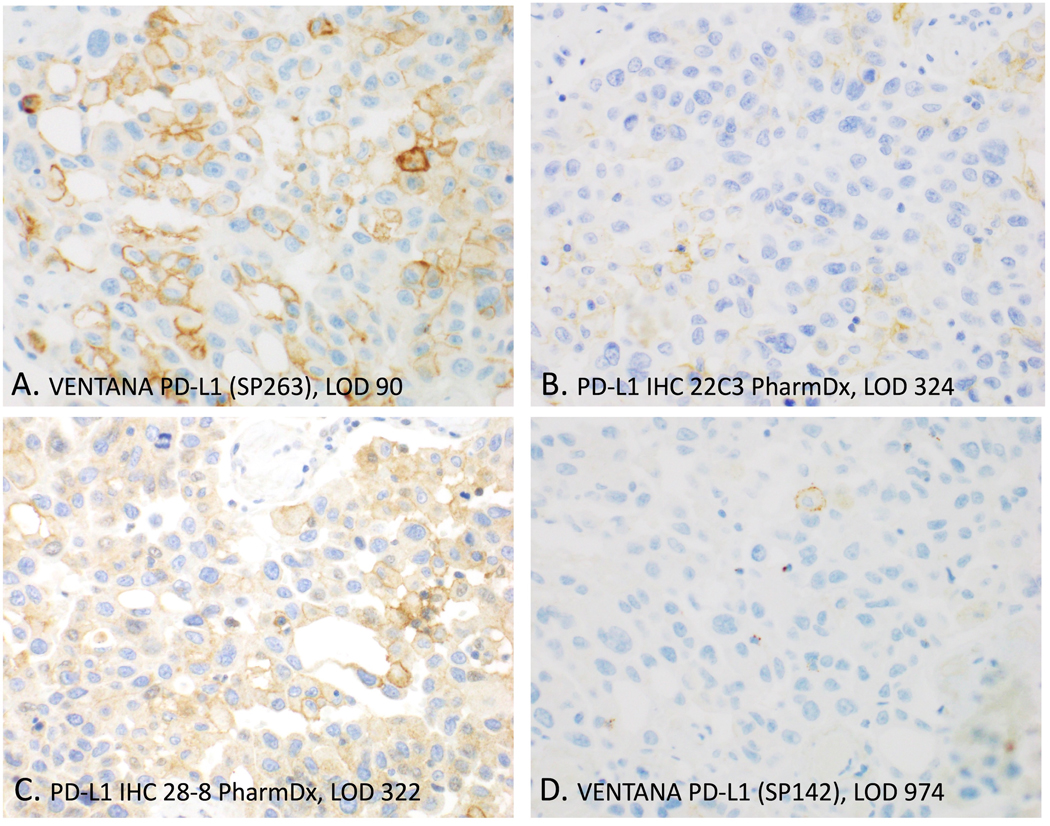
These are serial sections of the same tissue sample and stained by the reference labs (red diamonds of Fig. 1). Overall, high analytic sensitivity (as represented by low LODs) was associated with more PD-L1 positive cells and stronger staining. LODs are x1000 molecules PD-L1 per cell equivalent.
Dynamic ranges of FDA-cleared assays.
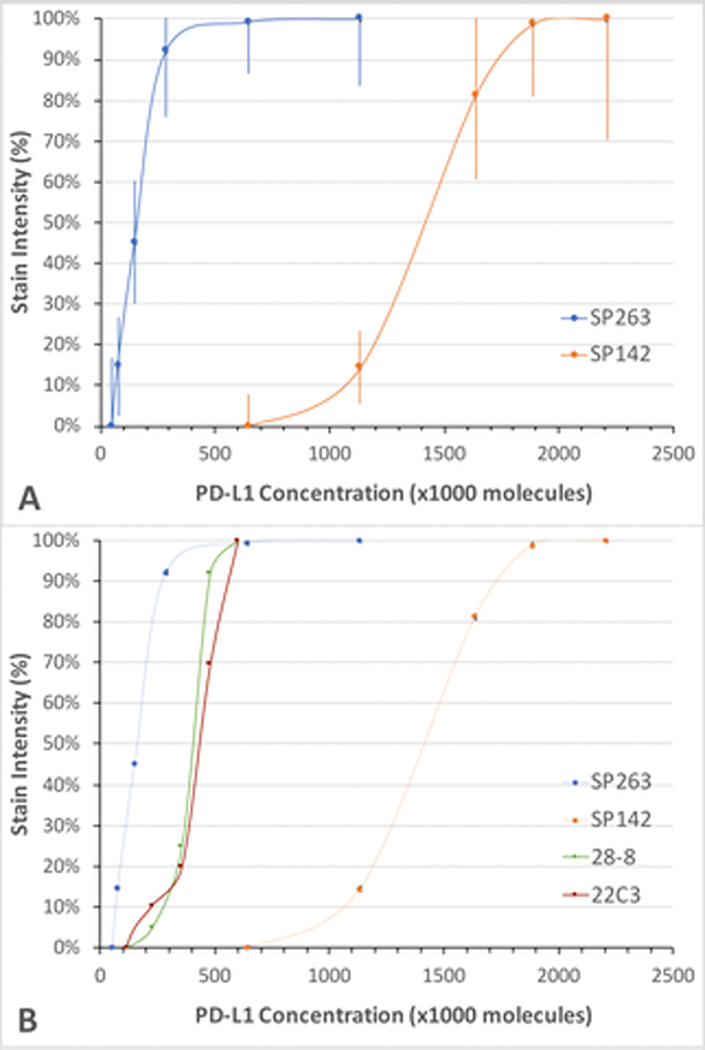
Whereas LOD identifies the analyte concentration threshold for staining, dynamic range describes stain intensity across a concentration range. Therefore, whereas LOD affects the percent positive cells, dynamic range provides greater insight into whether IHC assays can be harmonized. Dynamic range will also be important if stain intensity, rather than just the presence of stain, is important in tumor scoring. Figure 4A illustrates the analytic response curves for the two FDA-cleared PD-L1 immunohistochemical assays that recognize the PD-L1 intracellular domain – SP142 and SP263. The curves illustrate the aggregate data from 5 IHC laboratories running the VENTANA PD-L1 (SP142) assay and 17 IHC laboratories running the VENTANA PD-L1 (SP263) assays. The error bars represent the standard deviation of stain intensity among the pool of laboratories, i.e., lab-to-lab variability. The data show a very large difference between the two assays; there is no overlap in their assay dynamic range. The lowest PD-L1 concentration that begins to register any visibly detectable stain with the VENTANA PD-L1 (SP142) assay is at the staining plateau of the VENTANA PD-L1 (SP263) assay. The data demonstrate that there are PD-L1 tumor concentrations that can be strongly positive with the VENTANA PD-L1 (SP263) assay and negative with the VENTANA PD-L1 (SP142) assay. The data explain why it is not possible to harmonize the two assays by adjusting the readout cutpoints (Discussion).
Fig. 4. Consensus analytic response curves of FDA-cleared PD-L1 assays.
In each panel, the stain intensity (y axis) is expressed as a percentage of the maximum stain intensity for each assay and graphed as a function of PD-L1 concentration (x axis). A. Performance characteristics of the VENTANA SP263 and SP142 PD-L1 assays, correlating the generation of visible signal at various PD-L1 concentrations. Each dot is the mean ± SD of the pool of laboratories participating in the survey. B. Performance characteristics of the PharmDx 28–8 and 22C3 PD-L1 assays. For comparison, the analytic response curves shown in panel A are included.
Figure 4B shows the analytic response curves of the other two FDA-cleared PD-L1 assays, PD-L1 IHC 28–8 pharmDx and PD-L1 IHC 22C3 pharmDx. The data for the PD-L1 IHC 22C3 pharmDx assay are generated from 17 laboratories using the 22C3 FDA-cleared kit and 5 laboratories for the PD-L1 IHC 28–8 pharmDx assay. Figure 4B shows that these two assays had nearly identical analytic performance. Their analytic sensitivity is less than that for VENTANA PD-L1 (SP263) assay but much greater than VENTANA PD-L1 (SP142) assay. Like the VENTANA PD-L1 (SP263) assay, they also have little overlap with the analytic performance of VENTANA PD-L1 (SP142) assay.
Dynamic ranges of laboratory-developed tests (LDTs).
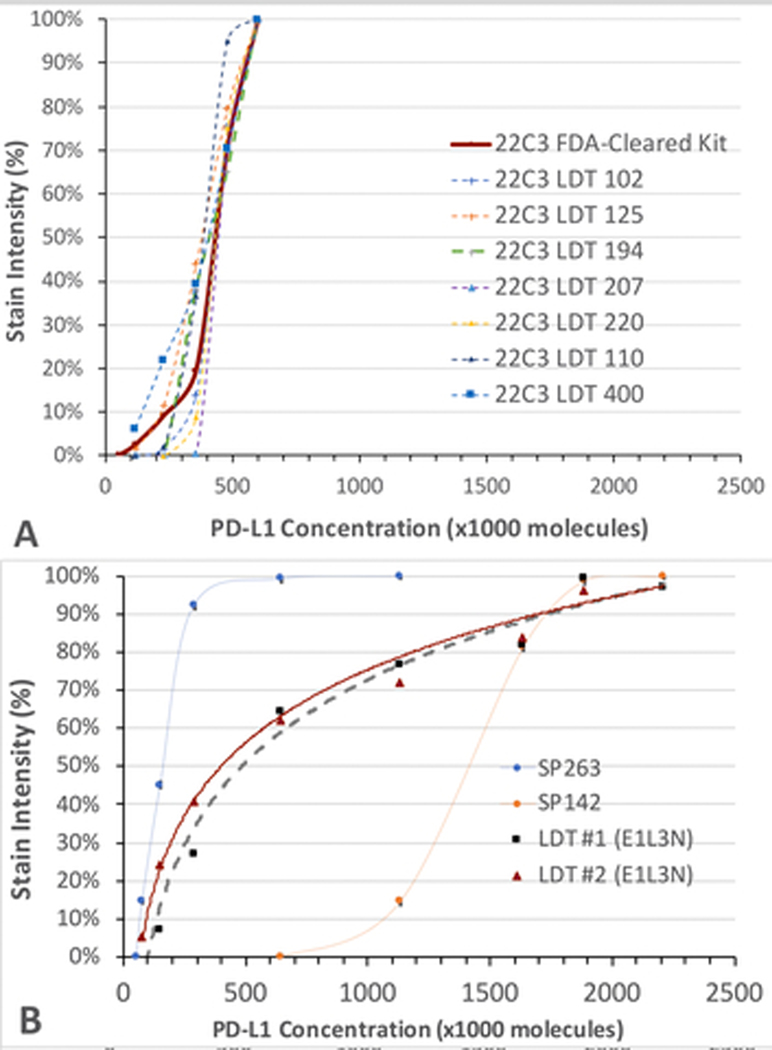
Principally for reasons of cost, some laboratories use PD-L1 LDTs. Since these tests were never calibrated against clinical outcomes, it is especially important to assess each LDT’s analytic performance against the analytic performance of the FDA-cleared PD-L1 assay that it is intended to replace. In Fig. 5A, 7 LDTs that use the 22C3 monoclonal antibody (mAb, dashed lines) are compared to the FDA-cleared kit (solid red line) using the same mAb. These LDTs were specifically developed for the same purpose as the FDA-approved CDx PD-L1 IHC 22C3 pharmDx. The laboratories performed clinical validation in order to provide evidence that they are fit-for-purpose; these laboratories demonstrated >90% positive percent agreement (PPA) and negative percent agreement (NPA) to the CDx assay.(23) Their analytic response curves, as shown in the dashed lines, are almost identical to the FDA-cleared kit (solid red line). This finding corroborates the contention that centralized development of LDTs for predictive biomarkers does work.
Fig. 5. Individual analytic response curves of PD-L1 LDTs.
Panel A shows 7 LDTs using the 22C3. Panel B shows 2 LDTs using the E1L3N primary antibody. This is the same as the LDTs PD-L1 (E1L3N) in Fig. 2. In each panel, the stain intensity as a percentage of the maximum (y axis) is graphed as a function of PD-L1 concentration (x axis). The FDA-cleared consensus curves are also included as a comparison for the corresponding extra-cellular (panel A) or intracellular (panel B) domain calibrators.
Figure 5B shows the analytic response curves of 2 other LDTs that both use the E1L3N primary antibody. In one instance, the assay is used only for research purposes and was not tested for concordance with an FDA-cleared assay. Data about the validation of the other are not available. Whereas the LODs of these LDTs are similar to the VENTANA PD-L1 (SP263), the dynamic range is broader. Therefore, the two assays are likely to show similar numbers of positive cells (because of similar LODs) but the stain intensities may differ (because of different dynamic ranges).
DISCUSSION
In the broad field of laboratory medicine, traceable reference materials are the gold standard for verifying analytic assay performance. This principle applies not only to quantitative but also qualitative assays, which incorporate defined analyte concentration thresholds separating positive from negative test results. The introduction of calibrators to IHC represents a fundamental departure from traditional practice and is new to many surgical pathologists. Like many other reference materials, BCS calibrators are prepared from purified analyte in a synthetic matrix. The PD-L1 calibrators are in the form of either a peptide (intracellular domain) or recombinant protein (extracellular domain), attached to a cell surrogate – a cell-sized glass microbead. Detailed explanations and photographs were previously published.(2, 11, 21, 24) The microbeads adhere to the glass slide and are subjected to all of the steps in staining, from de-waxing/hydration and antigen retrieval at the beginning to counterstaining and coverslipping at the end. This is their first application to PD-L1 testing, an assay that has already been the subject of intensive study. The data illustrate what can be learned:
The commercial FDA-cleared assays exist at three analytic sensitivity levels.
The two Roche (Ventana) assays, with mAbs SP142 and SP263, were at the opposite extremes of analytic sensitivity, without overlap of their analytic response curves. The two Agilent assays, using mAbs 22C3 and 28–8, were nearly identical to each other and fall intermediate to the extremes of the other two assays. These findings are slightly different from earlier studies, which inferred that the commercial assay incorporating the SP263 antibody is equivalent to those incorporating the 22C3 and 28–8 antibodies.(4, 5, 25) The previous studies, using a split-sample study design, may not have identified subtle differences in analytic sensitivity among assays because: (a) the PD-L1 concentrations in tissue samples are unknown, and (b) there is imprecision associated with manual readouts by pathologists. Previous studies indirectly inferred PD-L1 assay analytic sensitivity and the interchangeability of assays by using a large number of tissue samples or cell lines with unknown concentrations of PD-L1.(3–5, 25, 26) Assuming that the PD-L1 concentrations in these samples were randomly distributed, they collectively represent the spectrum of PD-L1 concentrations. By comparing the percentage of positive cases or positive cells for each assay, the relative analytic sensitivities for various assays were inferred. To have detected the higher analytic sensitivity of the VENTANA PD-L1 (SP263) assay, the previous studies would needed to have tested a sufficient number of samples with PD-L1 concentrations that fall above the LOD of the VENTANA PD-L1 SP263 assay and below that of the pharmDx 22C3 or 28–8 assays. Since the patient samples included in the studies had unknown PD-L1 concentrations, there was no way to identify them in advance. Also, any imprecision in the pathologist readout adds further “noise” to the system, obscuring true differences in analytic sensitivity. By virtue of the use of greater sample numbers, a recent meta analysis (3) was able to identify the higher analytic sensitivity of the VENTANA PD-L1 (SP263) assay. With calibrators, it is relatively simple to directly measure and compare analytic sensitivity.
The dynamic range data explain the inability to harmonize assays.
The data in Fig. 4 may explain published findings from the IMpassion130 study, describing an inability to harmonize the VENTANA PD-L1 (SP142) assay with either the VENTANA PD-L1 (SP263) assay or the PD-L1 IHC 22C3 pharmDx assay by adjusting the readout thresholds.(27) This inability to harmonize would be expected because their analytic response curves are highly disparate, showing no overlap. Many PD-L1 low/moderate expressing tumors that are detected with the SP263 or 22C3 assays will be below the LOD for the SP142 assay. The SP142 assay will fail to detect them regardless of the readout criteria.
The LOD data explain differences in patient test results.
For example, the IMpassion130 study found that the SP142-positive tumors are a subset of the SP263-positive or 22C3-positive groups.(27, 28) The same is true for the Blueprint studies.(4, 5) This falls in line with the hierarchy of analytic sensitivity shown in Figs. 2 and 4. Only high cellular concentrations of PD-L1 will be positive for the SP142 assay. The Impassion130 data also underscore the point that a highly sensitive assay is not necessarily better as a predictive IHC biomarker assay. In that study, the benefit for the atezolizumab regimen was driven predominantly by the (less sensitive) SP142-positive subgroup.(28, 29)
LDTs can reproduce the analytic performance of an FDA-cleared assay.
Figure 5A illustrates the analytic performance of seven LDTs using the 22C3 primary antibody, all of which were validated (using patient samples) for equivalence to the PD-L1 IHC 22C3 pharmDx assay.(23) Figure 5A shows that analytic performance of all seven is indistinguishable from the FDA-cleared PD-L1 IHC 22C3 pharmDx assay. Used in this fashion, PD-L1 BCS calibrators may find utility as a quick and inexpensive check for assay analytical performance equivalence before confirmation with large numbers of patient samples.
PD-L1 inter-laboratory variances as measured with calibrators mirror proficiency testing fail rates.
The data scatter in Fig. 2 and the error bars in Fig. 4A both reveal standard deviations of approximately 10 – 20% around each mean. These data mirror the approximately 20% fail rates for laboratory PD-L1 testing.(30) Most proficiency testing failures are due to insufficient analytic sensitivity.(31) By providing a direct measure of analytic sensitivity, calibrators may be helpful to IHC laboratories in ensuring accurate testing.
Study limitations.
Calibrators provide a direct measure of analytic variables relating to the assay itself, up to and including the point of producing a stained slide. Analytic variables are a major source of IHC errors.(32) Calibrators do not evaluate the accuracy of pathologist read-outs or pre-analytic variables, which need to also be addressed if a laboratory is to report accurate test results.
Another limitation relates to the commutability of reference materials. The introduction of the first traceable PD-L1 IHC reference materials introduces this topic for the first time. Commutability refers to the ability of a reference material to accurately mirror the analyte as it exists in a patient sample.(33) The evaluation of commutability in reference materials is covered in CLSI document EP30-A.(34) In the context of PD-L1 IHC, it refers to whether the staining of calibrators, with purified peptide or protein PD-L1 analytes, is identical to the staining of native PD-L1 in the patient’s biopsy. For example, the 22C3 and 28–8 mAbs are partly dependent on glycosylation.(18) Our PD-L1 extracellular domain protein was produced in a human embryonic cell line, which glycosylates transfected proteins. If the tumor or infiltrating immune cells demonstrate different patterns of glycosylation, rendering the bioengineered PD-L1 slightly different than PD-L1 in tissue sections, then it might affect antibody affinity and result in a different analytic response curve. We did not attempt to evaluate commutability; the tools to do so (as per CLSI guidelines) in IHC do not presently exist. Nonetheless, we believe it likely that the calibrators are commutable because:
The extracellular PD-L1 domain calibrator data for the two PD-L1 assays that target the extracellular domain (PD-L1 IHC 22C3 pharmDx and the PD-L1 IHC 28–8 pharmDx) approximately mirror the data as reported using patient tissue samples.(4, 5, 25)
The intracellular PD-L1 calibrator peptide is nearly identical to the peptide that was used as an immunogen for generating the mAbs. The intracellular calibrator data also mirror published findings using tissue sections.(4, 5, 25)
Future benefits.
The most important benefit of incorporating reference materials into IHC CDx testing is something that could not be evaluated in this study. We expect that analytic reference materials will dramatically improve the identification of reproducible cellular expression thresholds distinguishing responder from non-responder patient groups. This is especially impactful for predictive biomarkers because without characterization and direct monitoring of analytical sensitivity, obtaining accurate and reproducible test results is challenging.(32) In fact, without analytic test characterization, the optimal threshold distinguishing patient responders from non-responders to a candidate drug may not even be within the measuring range of the test. This is exemplified by the completely different dynamic ranges of the VENTANA PD-L1 (SP142) and VENTANA PD-L1 (SP263) assays (Fig. 4A).
Integration into clinical laboratory use.
BCS calibrators offer IHC laboratory directors a new tool to improve analytic test performance. For PD-L1 and other CDx testing for protein expression by IHC, the ability to measure and monitor analytic sensitivity will improve clinical test accuracy (diagnostic sensitivity and specificity). To achieve that, calibrators will be helpful:
During initial assay validation, to verify adequate analytic sensitivity.
When starting a new reagent lot, to verify that the new reagent is equally potent as the previous.
After major instrument repairs or replacement of sub-systems.
To verify correlation of multiple instruments, all performing the same stain at a single site.
During a problem investigation.
To determine the optimal dilution of a concentrated antibody.
Periodically, such as monthly, to verify continued test accuracy.
For assay developers, CDx IHC testing, reference materials will also facilitate methodology transfer from clinical trials to clinical IHC laboratories for predictive and prognostic IHC biomarkers.
ACKNOWLEDGMENTS
The authors also are grateful to the many laboratories in the U.S., Canada, the Netherlands, and Belgium for their participation.
FUNDING STATEMENT
Research reported in this paper was supported in part by the National Cancer Institute of the National Institutes of Health under award number R44CA213476 (to SAB).
Footnotes
COMPETING INTERESTS
NAH declares no competing financial interests. EET serves as an advisory board member for Merck, Pfizer, BMS, Seagen, Roche, AstraZeneca, and Agilent. S.R.S., K.V., and S.A.B. are principals at Boston Cell Standards, where some of the work was conducted, and are shareholders in the company. Boston Cell Standards holds a patent and other patent applications on the technology used in the study.
REFERENCES
- 1.Taylor C. Growing pains: from a stain to an assay. Appl Immunohistochem Mol Morphol 27, 325–326 (2019) [DOI] [PubMed] [Google Scholar]
- 2.Torlakovic E, Sompuram S, Vani K, Wang L, Schaedle A, DeRose P, et al. Development and validation of measurement traceability for in situ immunoassays. Clin Chem 67, 763–771 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torlakovic E, Lim H, Adam J, Barnes P, Bigras G, Chan A, et al. “Interchangeability” of PD-L1 immunohistochemistry assays: a meta-analysis of diagnostic accuracy. Mod Pathol 33, 4–17 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsao M, Kerr K, Kockx M, Beasley M-B, Borczuk A, Botling J, et al. PD-L1 immunohistochemistry comparability study in real-life clinical samples: Results of Blueprint Phase 2 project. J Thor Oncol 13, 1302–1311 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch F, McElhinny A, Stanforth D, Ranger-Moore J, Jansson M, Kulangara K, et al. PD-L1 Immunohistochemistry Assays for Lung Cancer: Results from Phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J Thor Oncol 12, 208–222 (2017) [DOI] [PubMed] [Google Scholar]
- 6.Tholen D, Linnet K, Kondratovich M, Armbruster D, Garrett P, Jones R, et al. EP17-A Protocols for determination of limits of detection and limits of quantitation; Approved guideline. Vol 24(34). Clinical Laboratory Standards Institute: Wayne, PA, 2004. [Google Scholar]
- 7.International Union of Pure and Applied Chemistry (IUPAC). Compendium of chemical terminology, 2nd Ed. (the “Gold Book”). Blackwell Scientific Publiications: Oxford, 1997. [Google Scholar]
- 8.Cheung C, D’Arrigo C, Dietel M, Francis G, Fulton R, Gilks C, et al. Evolution of quality assurance for clinical immunohistochemistry in the era of precision medicine. Part 4: Tissue tools for quality assurance in immunohistochemistry. Appl Immunohistochem Mol Morphol 25, 227–230 (2017) [DOI] [PubMed] [Google Scholar]
- 9.Sompuram S, Vani K, Schaedle A, Balasubramanian A, Bogen S. Quantitative assessment of immunohistochemistry laboratory performance by measuring analytic response curves and limits of detection. Arch Pathol Lab Med 142, 851–862 (2018) [DOI] [PubMed] [Google Scholar]
- 10.Sompuram S, Vani K, Schaedle A, Balasubrmanian A, Bogen S. Selecting an optimal positive IHC control for verifying antigen retrieval. J Histochem Cytochem 67, 275–289 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sompuram S, Vani K, Tracey B, Kamstock D, Bogen S. Standardizing immunohistochemistry: A new reference control for detecting staining problems. J Histochem Cytochem 63, 681–690 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sompuram S, Kodela V, Ramanathan H, Wescott C, Radcliffe G, Bogen S. Synthetic peptides identified from phage-displayed combinatorial libraries as immunodiagnostic assay surrogate quality control targets. Clin Chem 48, 410–420 (2002) [PubMed] [Google Scholar]
- 13.Sompuram S, Vani K, Hafer L, Bogen S. Antibodies Immunoreactive With Formalin-Fixed Tissue Antigens Recognize Linear Protein Epitopes. Am J Clin Pathol 125, 82–90 (2006) [PubMed] [Google Scholar]
- 14.Sompuram S, Vani K, Bogen S. A Molecular Model of Antigen Retrieval Using a Peptide Array. Am J Clin Pathol 125, 91–98 (2006) [PubMed] [Google Scholar]
- 15.Kowanetz M, Koeppen H, Boyd Z, Liao Z, Zhu Y, Vennapusa B, et al. (2016). Anti-PD-L1 antibodies and diagnostic uses thereof. (U.S. Patent Application 20160009805 A1) United States Patent and Trademark Office [Google Scholar]
- 16.Couto F, Liao Z, Zhu Y. (2015). PD-L1 antibodies and uses thereof. (U.S. Patent Application 20150346208 A1) United States Patent and Trademark Office [Google Scholar]
- 17.Kintsler S, Cassataro M, Drosch M, Holenya P, Knuechel R, Braunschweig T. Expression of programmed death ligand (PD-L1) in different tumors. Comparison of several current available antibody clones and antibody profiling. Ann Diag Pathol 41, 24–37 (2019) [DOI] [PubMed] [Google Scholar]
- 18.Lawson N, Dix C, Scorer P, Stubbs C, Wong E, Hutchinson L, et al. Mapping the binding sites of antibodies utilized in programmed cell death ligand-1 predictive immunohistochemical assays for use with immuno-oncology therapies. Mod Pathol 33, 518–530 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sompuram S. PD-L1 epitope mapping (unpublished data). (2019) [Google Scholar]
- 20.Schats K, Van Vre E, Schrijvers D, De Meester I, Kockx M. (Poster) Epitope mapping of PD-L1 primary antibodies (28–8, SP142, SP263, E1L3N). J Clin Oncol 35, 3028–3028 (2017) [Google Scholar]
- 21.Vani K, Sompuram S, Schaedle A, Balasubramanian A, Bogen S. Analytic response curves of clinical breast cancer IHC tests. J Histochem Cytochem 65, 273–283 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vani K, Sompuram S, Naber S, Goldsmith J, Fulton R, Bogen S. Levey-Jennings analysis uncovers unsuspected causes of immunohistochemistry stain variability. Appl Immunohistochem Mol Morphol 24, 688–694 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torlakovic E, Albadine R, Bigras G, Boag A, Bojarski A, Cabanero M, et al. Canadian multicenter project on standardization of programmed death-ligand 1 immunohistochemistry 22C3 laboratory-developed tests for Pembrolizumab therapy in NSCLC. J Thor Oncol 15, 1328–1337 (2020) [DOI] [PubMed] [Google Scholar]
- 24.Vani K, Sompuram S, Schaedle A, Balasubramanian A, Pilichowska M, Naber S, et al. The importance of epitope density in selecting a positive IHC control. J Histochem Cytochem 65, 463–477 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratcliffe M, Sharpe A, Midha A, Barker C, Scott M, Score P, et al. Agreement between Programmed Cell Death Ligand-1 Diagnostic Assays across Multiple Protein Expression Cutoffs in Non–Small Cell Lung Cancer. Clin Cancer Res 23, 3585–3591 (2017) [DOI] [PubMed] [Google Scholar]
- 26.McLaughlin J, Gang H, Schalper K, Carvajal-Hausdorf D, Pelekanou V, Rehman J, et al. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small cell lung cancer. JAMA Oncology 2, 46–54 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rugo H, Loi S, Adams S, Schmid P, Schneeweiss A, Iwata CBH, et al. Exploratory analytical harmonization of PD-L1 immunohistochemistry assays in advanced triple-negative breast cancer: A retrospective substudy of IMpassion130. San Antonio Breast Cancer Symposium 2019 Meeting Abstracts. Cancer Res 80, Abstract PD1–07 (2020) [Google Scholar]
- 28.Schmid P, Rugo H, Adams S, Schneeweiss A, Barrios C, Iwata H, et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 21, 44–59 (2020) [DOI] [PubMed] [Google Scholar]
- 29.Saleh N. PD-L1: A biomarker with baggage. OncologyLive 21, (Jan 30, 2020) [Google Scholar]
- 30.NordiQC. Assessment Run C8 2020 PD-L1 Keytruda. In: Aalborg, Denmark, 2020. [Google Scholar]
- 31.Vyberg M, Nielsen S. Proficiency testing in immunohistochemistry - experiences from Nordic Immunohistochemical Quality Control (NordiQC). Virchows Arch 468, 19–29 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogen S. A root cause analysis into the high error rate for clinical immunohistochemistry. Appl Immunohistochem Mol Morphol 27, 329–338 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braga F, Panteghini M. Commutability of reference and control materials: an essential factor for assuring the quality of measurements in laboratory medicine. Clin Chem Lab Med 57, 967–973 (2019) [DOI] [PubMed] [Google Scholar]
- 34.CLSI. Characterization and qualification of commutable reference materials for laboratory medicine (EP30-A); Approved guideline. Clinical and Laboratory Standards Institute: Wayne, PA, 2010. [Google Scholar]