Highlights
-
•
Engineered extracellular vesicles exert cytoprotective and anti-viral effects in a human lung epithelial line post-SARS-CoV-2 infection.
-
•
ASTEX modulate the transcriptome of infected cells primarily through suppression of mTOR signaling.
-
•
Attenuation of mTOR signaling inhibits SARS-CoV-2 replication and activating mTOR increases viral replication.
Keywords: Extracellular vesicles, SARS-CoV-2, Covid-19, Engineered cells, mTOR, Viral replication
Abbreviations: EVs, Extracellular vesicles; ASTEX, Activated specialized tissue effector extracellular vesicles; SARS-CoV-2, Severe acute respiratory syndrome coronavirus-2; Covid-19, Coronavirus disease 2019; mTOR, Mammalian target of rapamycin
Abstract
Effective treatment approaches for patients with COVID-19 remain limited and are neither curative nor widely applicable. Activated specialized tissue effector extracellular vesicles (ASTEX) derived from genetically-enhanced skin fibroblasts, exert disease-modifying bioactivity in vivo in models of heart and lung injury. Here we report that ASTEX antagonizes SARS-CoV-2 infection and its pathogenic sequelae. In human lung epithelial cells exposed to SARS-CoV-2, ASTEX is cytoprotective and antiviral. Transcriptomic analysis implicated the mammalian target of rapamycin (mTOR) pathway, as infected cells upregulated mTOR signaling and pre-exposure to ASTEX attenuated it. The implication of mTOR signaling was further confirmed using mTOR inhibition and activation, which increased and decreased viral load, respectively. Dissection of ASTEX cargo identifies miRs including miR-16 as potential inhibitors of mTOR signaling. The findings reveal a novel, dual mechanism of action for ASTEX as a therapeutic candidate for COVID-19, with synergistic antiviral and cytoprotective benefits.
Introduction
Nearly three years since the onset of the COVID-19 pandemic, therapeutic strategies remain limited [1,2] and unimpressive in terms of efficacy [3]. Current efforts at therapeutic development center around disruption of viral replication (i.e., anti-virals) and/or inhibition of inflammation (e.g., dexamethasone). Tissue reparative interventions are entirely lacking, and no agents have multiple synergistic properties. The urgency for novel therapeutic agents is accentuated by the possibility that SARS-CoV-2 may not be readily eradicated, given the emergence of variants as well as inadequate vaccination efforts in large segments of the global population.
Mesenchymal and stromal cell therapy has shown some promise in small clinical studies of COVID-19, based upon the cells’ anti-inflammatory and immunomodulatory properties, but several limitations restrict their applicability including therapeutic variability and paucity of long-term safety studies [4]. Cells work by secreting extracellular vesicles (EVs), nanosized lipid-bilayer particles which can be taken up locally and throughout the body. Due to their remarkable stability [5], EVs can be stored for extended periods, in conditions less rigorous than liquid nitrogen as is required for cells, and maintain their bioactivity without the need for potentially toxic cryopreservatives. Additionally, EVs can also circumvent delivery challenges associated with small biologics including cytokines, growth factors, and RNA [6]. Finally, EVs from therapeutic cell types which exert tissue-reparative effects including immunomodulation, anti-fibrosis would be clinically relevant in the context of Covid-19 pathology. Collectively, these advantages render EVs a promising approach to COVID-19 therapy.Therefore, we investigated the utility of EVs from therapeutically-engineered cells in limiting SARS-CoV-2 infection with the aim of developing a novel therapeutic against Covid-19.
Activated Specialized Tissue Effector EVs (ASTEX), are secreted by human neonatal skin fibroblasts reprogrammed to produce highly-effective EVs (by augmenting beta-catenin and the transcription factor gata4) [7]. In previous work, we demonstrated that ASTEX are tissue-reparative in myocardial injury [7], muscular dystrophy [7]. Most recently, we demonstrated that ASTEX attenuate lung inflammation and fibrosis in bleomycin-induced lung injury [8]. In a set of experiments designed to assess safety, we observed that exposure of a lung epithelial cell line to ASTEX before SARS-CoV-2 infection exerted both cytoprotective and antiviral effects. This dual mechanism of action (MoA) is therapeutically relevant to patients at various stages of COVID-19, where halting infection, as well as disease progression, is critical.
Materials and methods
EV preparation
ASTEX: Engineered fibroblasts that produce ASTEX are dubbed (ASTECs). ASTECs were prepared as described previously [7,8]. Briefly, primary neonatal skin fibroblasts were cultured in 10% FBS, 50 μg/mL gentamicin, and 2 mmol/L L-Glutamine in Iscove's Modified Dulbecco's medium (complete medium). Cells are transduced with two lentiviruses for activation of beta-catenin and gata4. Transduced cells are then subjected to antibiotic selection medium (complete medium with [5 µg/ml] puromycin and blasticidin [5 µg/ml]) for one week. After the selection period, only cells that have been transduced with both viruses remain and are expanded in complete media. ASTECs were conditioned in 100 ml of Iscove's Modified Dulbecco medium for fifteen days. At the end of the fifteen-day conditioning period, media were collected and sterile filtered using a 0.45 µm vacuum filter. Secreted EVs (ASTEX) were concentrated using a 100 kDa centrifugal filter (Millipore Sigma, cat# UFC901024).
TEV1: EVs from immortalized CDC-EVs (TEV1) were prepared as described previously [7,18]. Briefly Cardiosphere-derived cells were transduced with lentivirus particles containing shPeg1 (Santa Cruz, Cat # sc-61,315-V) and SV40 T+t (ABM, Cat # LV614) at an MOI of 20. 24 hrs post-transduction, viruses were removed, and fresh complete media was added for cell recovery for a further 24 hrs. Cells were then grown in selection media (puromycin 5 μg/ml). To generate EVs, transduced CDCs were cultured until confluence at passage 18 or older. The cells were washed with IMDM and were serum-free cultured at low oxygen (2% O2) overnight. Conditioned media were then collected, sterile filtered using a 0.45-μm filter, and the EVs were concentrated by ultrafiltration with a 100 KDa molecular weight cutoff filter (Millipore Sigma, cat# UFC901024).
Nanosight tracking analysis
Isolated EVs were diluted 1:100 in phosphate buffered saline, then quantified using Malvern Nanosight NS300 Instrument (Malvern Instruments). Acquisition parameters were kept constant throughout the study and include: camera levels of 15, detection level of 5. For each sample, a total of four (30 s) videos were recorded and analyzed.
Cryo-Scanning electron microscopy
Environmental Scanning electron microscopy (ESEM) of ASTEX was performed commercially using Covalent Metrology (Sunnyvale, CA). Briefly, images were acquired by drop casting 40 μL of concentrated EVs (100 kDa ultrafiltration) on a silicon coupon and imaging with the Apreo SEM.
Western blot
EV lysates and conditioned media were collected for ELISA and western blot, (respectively). Lysates were prepared from cell pellets by lysis using RIPA buffer (Millipore Sigma) supplemented with HALT protease inhibitor (Thermofisher Scientific) on ice for 30 min. Protein lysates were isolated by centrifugation at 14,000 rpm for 15 min at 4 °C. Protein concentration was measured using a DC™ Protein Assay kit (Bio-Rad). Membrane transfer was done using Turbo® Transfer System (BIO-RAD) following gel electrophoresis. Signal was detected using SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific). The following antibodies were used to probe for the targets provided in Table 2 below.
Table 2.
Gene expression assays.
| Assay Names | Species | Manufacturer | Assay Numbers |
|---|---|---|---|
| Ace | Human | Taqman gene expression assays (cat#4,331,182); Life Technologies | Hs00174179_m1 |
| ace2 | Human | Hs01085333_m1 | |
| adam10 | Human | Hs00153853_m1 | |
| adam17 | Human | Hs01041915_m1 | |
| pik3ca | Human | Hs00907954_m1 | |
| agt | Human | Hs01586213_m1 | |
| dectin1 | Human | Hs01902549_s1 | |
| n1* | SARS-CoV-2 | Integrated DNA Technologies | 2019-nCoV RUO Kit |
| n2* | SARS-CoV-2 |
*Viral copies of n1 and n2 genes were determined using standards included in the nCoV RUO Kit.
Cell culture
Calu-3 cells were commercially obtained (ATCC) and cultured in 10% fetal bovine serum in DMEM - Dulbecco's Modified Eagle Medium and 10 μg/ml gentamicin. Cells were passaged for up to five (1:2) passages.
SARS-CoV-2
SARS-Related Coronavirus 2, Isolate USA-WA1/2020, was obtained from BEI Resources of the National Institute of Allergy and Infectious Diseases (NIAID). SARS-CoV-2 was passaged once in Vero-E6 cells and viral stocks were aliquoted and stored at −80 °C.
In vitro viral inhibition assays
SARS-CoV-2 viral assays were performed in UCLA BSL3 high containment facility. Calu-3 (ATCC HTB-55) cells were obtained from ATCC and cultured at 37 °C with 5% CO2 in Eagle's Minimum Essential Medium (EMEM) growth media with 10% fetal bovine serum and 100 units/ml penicillin. SARS-CoV-2 Isolate USA-WA1/2020 was obtained from BEI Resources of the National Institute of Allergy and Infectious Diseases (NIAID). Calu-3 cells were plated in 96-well plates (4 × 10^4 cells/well) and pretreated with ASTEX, TEV1, rapamycin (5 μM in DMSO; Millipore Sigma, cat# 553,210), MHY1485 (1 μM in DMSO; Millipore Sigma, cat# 500,554), remdesivir (10 μM in DMSO; Selleck Chemicals, cat# GS-5734), or saline (in triplicate, at indicated concentrations) for 1 hour before addition of SARS-CoV-2 (MOI 0.1). After 48 h post-infection (hpi) the cells and supernatant were isolated for RNA and protein extraction, and immunofluorescence staining.
RNA isolation and qRT-PCR
Cellular RNA was isolated using RNeasy Mini kit (Qiagen, Cat # 217,004). Reverse transcription was completed using a High-Capacity RNA-to-cDNA kit (Applied Biosystems, cat # 4,387,406), and qRT-PCR was performed using Taqman Fast Advanced Master Mix (Applied Biosystems, cat # 4,444,557) in QuantStudio™ 7 Flex Real-Time PCR System. RNA from EVs (for sequencing) was done using Norgen Urine Exosome Purification Kit (Norgen BioTek: cat #5770). Primers and their gene targets are described in Table 1 below:
Table 1.
Western blot antibodies.
| Antibody Names | Primary/Secondary | Company | Catalog Numbers |
|---|---|---|---|
| Mouse anti Human, CD63, Clone: H5C6 | Primary | BD Biosciences | 556,019 |
| Mouse Anti-Human CD81: Clone JS-81 | Primary | BD Biosciences | 555,675 |
| Rabbit TSG101 Polyclonal Antibody | Primary | Invitrogen | PA5–82,236 |
| Rabbit Anti-Human HSP90 (D69) | Primary | Abcam | ab61907 |
| Alix | Primary | Invitrogen | MA1–83,997 |
| Anti-Mouse IgG, HRP-linked | Secondary | Cell Signaling Technology | 7076S |
| Anti-Rabbit IgG, HRP-Linked Antibody | Secondary | Cell Signaling Technology | 7074S |
ELISA
For ELISA, conditioned media was clarified of cell debris by centrifugation at 3000 xg for 15 min followed by protein quantification as described above. IL-6 was quantified using Human IL-6 ELISA (sample type: cell culture supernatants/plasma/serum; Raybiotech, cat# ELH-IL6–2). The ectodomain of ACE2 was quantified using Human ACE2 PicoKine™ ELISA Kit (Boster Bio, cat# EK0997). All assays were performed following the manufacturer's instructions.
Total RNA sequencing
Library construction was performed using the SMARTer® Stranded Total RNA-Seq Kit (Takara Bio USA, Inc., Mountain View, CA). Briefly, total RNA samples were assessed for concentration using a Qubit fluorometer (ThermoFisher Scientific, Waltham, MA) and for quality using the 2100 Bioanalyzer (Agilent Biotechnologies, Santa Clara, CA). Total RNA was converted to cDNA and adapters for Illumina sequencing (with specific barcodes) are were added through PCR using only a limited number of cycles. The PCR products were purified, and then ribosomal cDNA was depleted. The cDNA fragments were further amplified with primers universal to all libraries. Lastly, the PCR products were purified once more to yield the final library. The concentration of the amplified library was measured with a Qubit fluorometer and an aliquot of the library was resolved on the Bioanalyzer. Sample libraries were multiplexed and sequenced on a NextSeq 500 platform (Illumina, San Diego, CA) using 75 bp single-end sequencing. On average, 50 million reads were generated from each sample.
Data analysis for RNA sequencing
Raw reads obtained from RNA-Seq were aligned to the transcriptome using STAR (version 2.5.0) [19]/ RSEM (version 1.2.25) [20] with default parameters, using human GRCh38 (or mouse CRCm38) transcriptome reference downloaded from http://www.gencodegenes.org. Expression counts for each gene (TPM: transcripts per million) in all samples were normalized by the sequencing depth.
Immunocytochemistry
Cells were fixed with 4% PFA for 30–60 min at −20 °C. Cells were washed 3 times with PBS and permeabilized using blocking buffer (0.3% Triton X-100, 2% BSA, 5% Goat Serum, 5% Donkey Serum in 1 X PBS) for 1 h at room temperature. Subsequently, cells were incubated with anti-SARS-CoV-2 Spike antibody (Sino Biological, cat# 40,592-R0004; 1:200) or NFkB phosphor-p65 (pser536, Novus Biologicals; cat# NB100–82,088) antibody at 4 °C overnight. Cells were then washed 3 times with 1x PBS and incubated with fluorescence conjugated secondary antibodies (ThermoFisher Scientific): donkey anti-rabbit IgG Alexa Fluor 488 (cat# A21202) and Goat anti-mouse IgG Secondary Antibody, Alexa Fluor 555 (cat# A28180; 1:1000) for 1 h at room temperature. Nuclei were stained with DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride) (ThermoFisher Scientific; cat# D1306) at a dilution of 1:5000 in 1xPBS for 10 min. Cells were analyzed by fluorescence microscopy. Cells were counted using the DAPI signal. Five images per well were quantified for each condition using Cytation 5 imaging reader (BioTek).
mTOR phospho-antibody microarray
Postranscriptional effects of ASTEX on SARS-CoV-2-infected Calu-3 cells was performed using the mTOR phospho-antibody microarray (Full Moon Biosystems; cat# PMT138). Calu-3 cell lysates were isolated and quantified as described previously for western blot. The array was performed according to manufacturer instructions. Slides were read and analyzed in a blinded manner by Full Moon Biosystems.
Statistical analysis
GraphPad Prism 6.0 (GraphPad Software) was used to analyze the data. Comparison of three or more groups was performed using a one-way ANOVA followed by Tukey's post hoc multiple comparison test. Comparisons of two groups were analyzed using two-tailed unpaired t-tests with a confidence interval of 95%.
Results
ASTEX is cytoprotective and attenuates IL6 expression and NFkB activation in SARS-CoV-2 infection
Canonical wnt signaling is a central driver of the therapeutic potency of cells and their secreted EVs. Using this insight, we created Activated specialized tissue effector cells (ASTECs) by transducing neonatal fibroblasts with beta-catenin (ctnnb1) and the cardiac-specific transcription factor, gata4. ASTECs are then used as factories for therapeutic EVs’ production (ASTEX; Fig. S1). We have also immortalized cardiosphere-derived cells by transduction of SV40 small and large T-antigens along with knockdown of the Wnt regulator mest. Immortalized CDCs produce therapeutic EV-1 (TEV1). We decided to use TEV1 as an appropriate comparator to ASTEX as they are both EVs from genetically-engineered (and immortalized) cell types. Both EV sources have a comparable size distribution and EV output (Fig. SB-D). Finally, ASTEX and TEV1 both expressed conserved EV markers including CD63, CD81, Alix, HSP90, and TSG101; EV preparations were also negative for the cell-contamination marker calnexin (Fig. S1E). The implication of canonical wnt signaling in epithelial stem cell expansion and lung tissue regeneration rationalized our investigation of the therapeutic potential of these EVs in the context of SARS-CoV-2 infection.
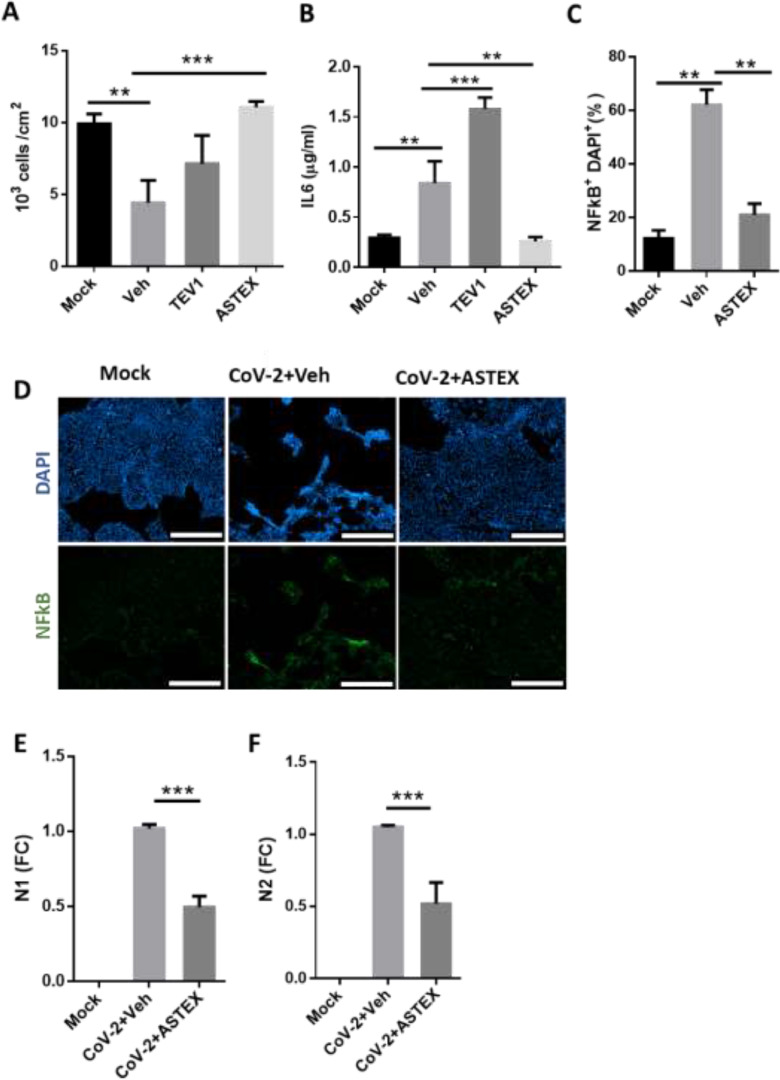
Calu-3 is a human lung epithelial line that is highly permissive to SARS-CoV-2 infection even at low multiplicities of infection (MOI). As such it is a valuable tool for in vitro screening of anti-SARS-CoV-2 compounds [9]. In this model, we tested the ability of therapeutic EVs to protect cells from SARS-CoV-2-induced inflammation and cytotoxicity. To this end, Calu-3 cells were pre-treated with ASTEX and, for equivalent comparison, EVs from immortalized cardiosphere-derived cells (therapeutic EV 1; TEV1), or vehicle before exposure to SARS-CoV-2 (MOI: 0.1) or saline (mock). Two days post-infection, only cells pre-treated with ASTEX had significantly improved survival compared to vehicle; a trend was observed with TEV1 (Figs. 1A and S2A). Additionally, both ASTEX and TEV1 suppressed ACE2 receptor shedding (Fig. S2B). Indeed, gene expression (fold change; FC) of the receptor sheddases ADAM10 and ADAM17 were reduced in ASTEX-treated cells whereas only the former was reduced in TEV1-treated cells. This suggests that ADAM10 is critical to infection-mediated ACE2 receptor shedding (Fig. S2C,D); receptor loss may not be directly cytoprotective. Interestingly ACE2 expression was reduced in both ASTEX and TEV1-treated groups, while ACE was reduced only in the ASTEX group (Fig. S2E,F). No significant differences were observed in the expression of angiotensinogen (Fig. S2G). In an in vivo model, preservation of ACE2/ACE signaling may attenuate downstream effects of RAS dysregulation seen with COVID-19. Cytoprotection was also coupled with attenuated inflammation as shown by reduced interleukin 6 production (IL6; Fig. 1B) and nuclear factor k beta (NFκB) expression (Fig. 1C,D) in the ASTEX treated cells following infection. Most surprisingly, ASTEX impaired viral infection as shown by reduction in copy number of the SARS-CoV-2 nucleoproteins N1 and N2 (Fig. 1E,F). In conclusion, ASTEX exerts cytoprotective, and antiviral activities in SARS-CoV-2 infected lung epithelial cells. These effects are mediated, at least in part by inhibition of IL6 expression and NFkB activation.
Fig. 1.
ASTEX exert cytoprotective and anti-viral effects in vitro. (A) SARS-CoV-2-infected Calu-3 cells pre-treated with ASTEX show enhanced survival, reduced IL-6 secretion (B), and reduced NFkB activation (C, D) compared to vehicle (Veh)- or EVs from immortaliced CDC EVs (TEV-1) treated groups (Mock: uninfected cells). ASTEX also exerted anti-viral effects as shown by reduced copy numbers of SARS-CoV-2 nucleoproteins N1 (E) and N2 (F). (A-F; n=three biological replicates per group). Comparisons between two groups were done using a Student's T test and comparisons between three or more were done using One Way ANOVA with Tukey's test for multiple comparisons; *p < 0.05, **p < 0.01, ***p < 0.001 using a 95% CI. Scale bar for D: 500 μm.
mTOR is activated during SARS-CoV-2 infection and repressed by ASTEX
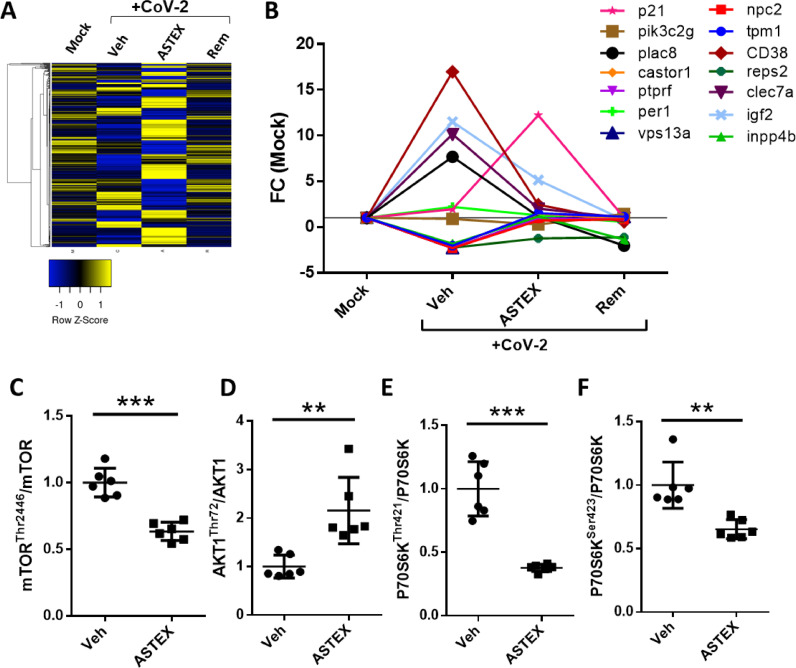
To investigate the mode of action (MoA) whereby ASTEX inhibits SARS-CoV-2 infection, we performed a transcriptomic analysis of cells exposed to ASTEX, vehicle, or positive control (remdesivir). SARS-CoV-2 infection-induced dramatic changes in the transcriptome of host cells (Fig. 2A). Cells exposed to ASTEX, however, counteracted those changes: they retained expression of cytoskeletal proteins (Fig. S3A), and normal levels of integrin transcripts compared to vehicle (Fig. S3B). Integrin expression is an important factor in viral entry as integrin receptors have been shown to cloak the KGD binding site necessary for viral uptake. Surprisingly, ASTEX also activated a robust inflammasome response but with reduced levels of the pro-apoptotic mediator caspase 1 (Fig. S3C). By contrast, ASTEX suppressed interferon-signaling genes (Fig. S3D) and pro-inflammatory cytokine/chemokine expression and restored transforming growth factor (tgfb) gene expression levels compared to vehicle (Fig. S3E). The largest observed effect on a pathway, altered by SARS-CoV-2 infection and modulated by ASTEX, centers on the mammalian target of rapamycin (mTOR) (Fig. 2B). Coronaviruses and influenza viruses promote replication through cap-independent protein synthesis via enhanced mTOR signaling. Furthermore, mounting evidence implicates mTOR activation in SARS-CoV-2 infection [12,13] and its inhibition reduces viral replication [14]. The effect of ASTEX on mTOR was further validated by an mTOR-focused protein array which showed that ASTEX increased inhibitory phosphorylation sites on mTOR (Thr2446) and reduced phosphorylation of the AKT1 activation site (Thr72; Fig. 2C,D). Moreover, the mTOR downstream signaling kinase to P70S6K was hypo-phosphorylated at the activation residues Thr421 and Ser423 (Fig. 2E,F)). Finally, the mTOR regulator PP2a was hyperphosphorylated in ASTEX-treated cells (Fig. S3E). In conclusion, following SARS-CoV-2 infection, mTOR signaling is activated, and ASTEX attenuates such activation.
Fig. 2.
ASTEX represses SARS-CoV-2-mediated activation of the mTOR pathway. (A) Heatmap of transcriptomic analysis of uninfected Calu-3 cells (Mock) or those pre-treated with vehicle (Veh), ASTEX, or remdesivir (Rem) followed by SARS-CoV-2 (CoV-2) exposure (B). Sequencing data demonstrating that ASTEX repress the mTOR pathway activators in infected cells down to levels comparable to uninfected cells (C). Phospho-array (n = 6 replicates) of cell lysates showing post-translational repression of the mTOR in ASTEX-pretreated cells. ASTEX represses mTOR (through hypophosphorylation of Thr2446; D), AKT1 (through hyperphosphorylation of Thr72; (D), and P70S6K (through hypophosphorylation of Thr421 and Ser423; E, F) in ASTEX-pretreated infected cells compared to vehicle. Analysis done using Student's T-test; *p < 0.05, **p < 0.01, ***p < 0.001 using a 95% CI.
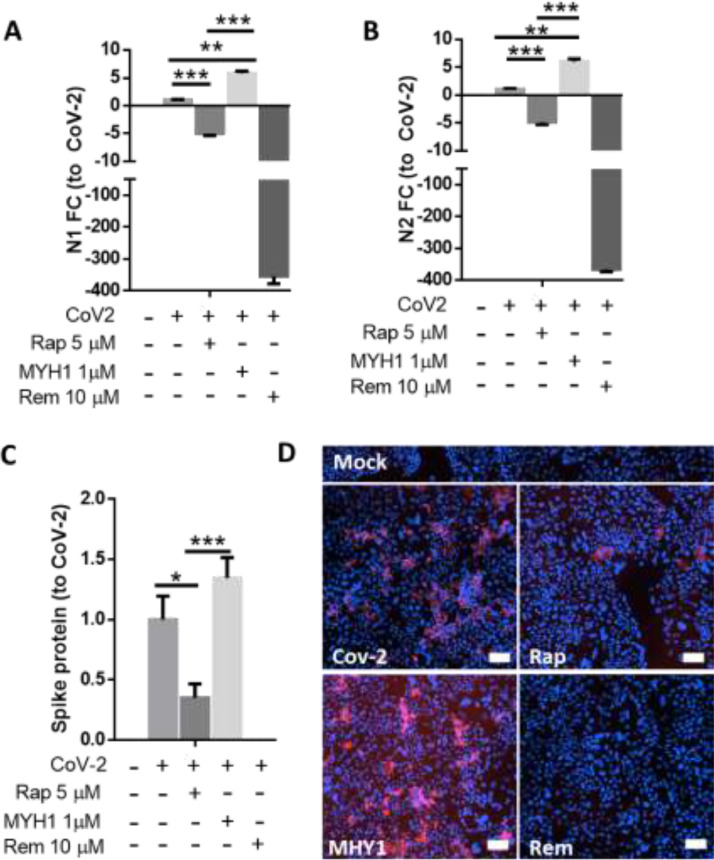
mTOR activation exacerbates infection and mTOR inhibition attenuates it
To confirm the role of mTOR in SARS-CoV-2 infection, we investigated the effect of mTOR modulation on viral replication using the mTOR inhibitor rapamycin and activator MY1485. Indeed inhibition of mTOR attenuated viral replication as shown by reduction in copies of N1 and N2 (Fig. 3A,B). This was further confirmed by reductions in spike protein levels (Fig. 3C,D).
Fig. 3.
Inhibition of the mTOR pathway inhibits viral replication and its activation enhances it. Rapamycin attenuated viral replication as shown by reduced copies of SARS-CoV-2 N1 and N2 copies (A,B) and Spike protein; pooled data in (C) and representative image in (D). (A–D; n = 3 biological replicates). Analysis was done using One Way ANOVA with Tukey's test for multiple comparisons: *p < 0.05, **p < 0.01, ***p < 0.001 using a 95% CI. Scale Bar: 200 μm.
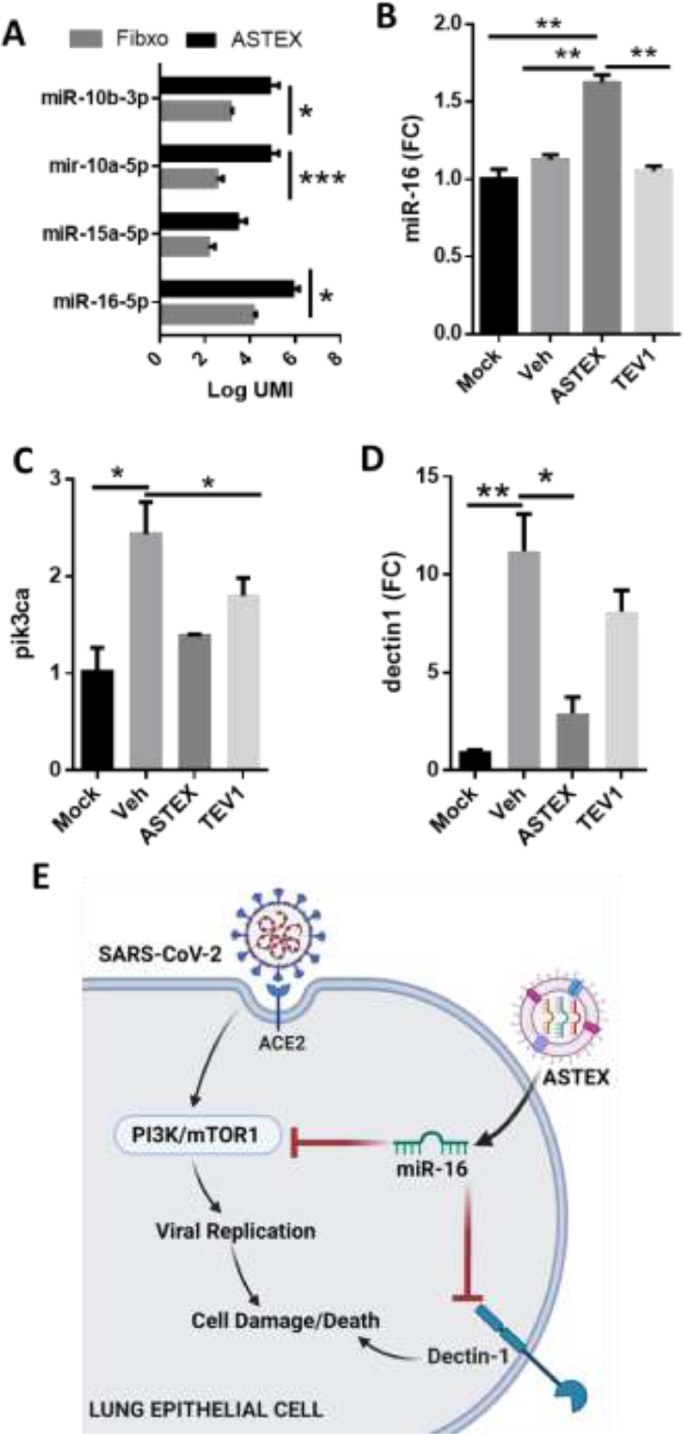
MiR-16 contributes to the ASTEX-mediated anti-viral effect
To identify constituents of ASTEX payload that may account for the antiviral and anti-inflammatory effects, we surveyed the most highly abundant and most differentially expressed miRNAs in ASTEX compared to EVs derived from non-transduced fibroblast parent cells. Two miR clusters were uniquely enriched in ASTEX: miR-10a/b and miR-16/15a (Fig. 4A). Furthermore, miR-16 is upregulated in cells pre-treated with ASTEX compared to other groups (Fig. 4B). Given the observed post-translational regulation of mTOR (Fig. 2C,D, Fig. S3E-G), we investigated the expression of PI3K, the principal kinase involved in the modulation of mTOR. Cells exposed to ASTEX had suppressed levels of pi3k, comparable to those in uninfected cells (Fig. 4C). Indeed, both mirs −10a and −16 has been shown to target pik3ca. In addition miR-16 also targets dectin 1, a pattern recognition receptor implicated in the inflammatory response post-SARS-CoV-2 infection (Fig. 4D). Therefore, miR-16 may mediate the anti-viral and anti-inflammatory effects of ASTEX against SARS-CoV-2 infection through targeting pik3ca and dectin 1 (Fig. 4E), but the association remains conjectural.
Fig. 4.
ASTEX are enriched in mTOR-targeting miRs. ASTEX are enriched in miRs that regulate the mTOR pathway compared to EVs derived from fibroblasts (A; n = 3 biological replicates). Analysis was done using Student's T test. Higher levels of miR-16 in cells treated with ASTEX at 48 h post-treatment (B).Validated targets of miR-10 (pik3ca; C) and miR-16 (dectin-1; D). Analysis was done using One Way ANOVA with Tukey's test for multiple comparisons. *p < 0.05, **p < 0.01, ***p < 0.001 using a 95% CI. Conceptual figure of the mechanism of action of ASTEX in promoting cell survival and inhibiting SARS-CoV-2 viral infection (E).
Discussion
The therapeutic bioactivity of ASTEX has been demonstrated in a number of models relevant to COVID-19 pathology including cardiac, skeletal muscle, and lung injury. This prior work motivated us to develop ASTEX as a potential therapeutic candidate for COVID-19. In a cellular model of SARS-CoV-2 infection, we not only verified cytoprotective effects but also discovered an unanticipated ability of ASTEX to inhibit viral replication. The effects were specific to ASTEX as they were not fully mimicked by other types of therapeutic EVs. ASTEX attenuated viral replication and virus-mediated cytotoxicity, while also blunting IL6 levels and NFκB translocation. Furthermore, ASTEX attenuated the shedding of the viral host receptor ACE2 potentially inhibiting renin-angiotensin-aldosterone system (RAAS) dysregulation (a common consequence in COVID-19). Transcriptomic analysis of infected lung epithelial cells exposed to ASTEX revealed suppression of the mTOR pathway.
Viruses exploit the cellular machinery for self-replication through the modulation of a myriad of pathways. One of these strategies involves dysregulation of cap-dependent vs. cap-independent protein synthesis [15]. A central pathway of protein synthesis is the mTOR pathway, a keystone regulator driving cap-dependent protein synthesis (via mTORC1) and cellular metabolism and stress response (via mTORC2). Given this central role, it is a frequent target of viruses during infection. Viruses that utilize cap-independent protein synthesis including picornaviruses (e.g. poliovirus) enhance the cap-independent pathway through inhibition of mTOR. Conversely, viruses that translate their proteins through a cap-dependent mechanism including influenza [16] and coronavirus suppress cap-independent translation through suppression of the mTOR [11]. While mTOR hyperactivation has been confirmed in Middle East Respiratory Syndrome (MERS) [10] it has not yet been demonstrated in SARS-CoV-2. However, the use of mTOR inhibitors is currently being considered as a possible therapeutic approach [12,14,17].
Future studies will validate specific ASTEX payloads and identify the specific targets in the mTOR pathway that they affect. Furthermore, EV therapies function as a mosaic; several defined factors work in concert to exert therapeutic efficacy. This necessitates the use of in vivo models which will interrogate the full capacity of ASTEX therapy on different compartments including immune and epithelial tissue. Other effects seen with ASTEX in SARS-CoV-2 infection in vivo may be mTOR dependent. The work shown here represents a promising proof of concept for an EV therapeutic against COVID-19 and describes a mechanism of action by which viral replication and cell death are attenuated. Indeed, this represents a promising next step in the evolution of therapies not only against COVID-19 but potential other viruses (including influenza) where mTOR is a critical pathway for viral infection. It is important to note that this study remains preliminary and includes a number of key limitations. For instance, in this study EVs were isolated using ultrafiltration. While the large cut-off size of the filter preferentially concentrates EVs and filters out soluble proteins, investigations using purer isolation methods like size-exclusion chromatography will be necessary to definitively ascribe these effects to the vesicles alone. Another limitation is that all of the studies were done in a cell line. Therefore, validation will need to be recapitulated in primary in vitro models, namely primary lung epithelial cells. Since Covid-19 is a pathology that involves a complex interplay between organ tissue and the immune response, it will be also necessary to fully validate the effects of ASTEX in an animal model of Covid-19. A clear mechanism of action remains to be elucidated. ASTEX, like other EVs, contain a diversity of molecular signals, it will be important to further mine and validate ASTEX payload, especially those that target the mTOR pathway. Finally, this study was designed to evaluate the protective capacity of ASTEX in SARS-CoV-2 infection (as cells were pre-treated to ASTEX prior to infection). Therefore from a clinical utility standpoint, ASTEX may be therapeutically relevant in the early stages of Covid-19 to slow or halt disease progression by limiting viral spread and attenuating the damaging effects of hyperinflammation.. In conclusion, this work represents promising preliminary findings that warrant further investigation of the therapeutic bioactivity of ASTEX in Covid-19.
Conclusions
In summary, we demonstrate that EVs derived from therapeutically engineered cells (ASTEX) inhibit SARS-CoV-2 replication, attenuate RAAS dysregulation and induce cytoprotective effects in vitro. These cytoprotective effects are driven, at least in part, by suppression of infection-mediated activation of the Pi3k/mTOR pathway. Specific targeting or promotion of mTOR signaling attenuated and potentiatied viral replication, respectively. Therefore, ASTEX merit further investigation as promising therapeutics for Covid-19.
CRediT authorship contribution statement
A.G. Ibrahim: Conceptualization, Methodology, Formal analysis, Writing – original draft. A. Ciullo: Methodology, Formal analysis. C. Li: Methodology, Formal analysis. G. Garcia: Methodology, Formal analysis. K. Peck: Methodology, Formal analysis. K. Miyamoto: Methodology, Formal analysis. V. Arumugaswami: Conceptualization, Writing – original draft. E. Marbán: Conceptualization, Writing – original draft.
Declaration of Competing Interest
EM owns founder stock in Capricor Therapeutics. All other authors declare no competing financial interests.
Acknowledgments
We thank the Cedars Sinai Genomics Core for RNA sequencing. Work in the Marbán lab was supported by NIH R01124074. Work in the Arumugaswami lab was supported by the UCLA DGSOM and Broad Stem Cell Research Center institutional (OCRC #20–15), and the National Eye Institute of NIH (1R01EY032149) awards as well as the California Institute for Regenerative Medicine Discovery Award (TRAN1COVID19–11975).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbiosy.2022.100042.
Appendix. Supplementary materials
References
- 1.Beigel J.H., et al. Remdesivir for the treatment of Covid-19 - final report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez M.A. Lack of effectiveness of repurposed drugs for COVID-19 treatment. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.635371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinkowski J. Drug-drug interactions could imperil COVID-19 treatment. Medscape. 2020 [Google Scholar]
- 4.Lee S.H. The advantages and limitations of mesenchymal stem cells in clinical application for treating human diseases. Osteoporos Sarcopenia. 2018;4:150. doi: 10.1016/j.afos.2018.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boukouris S., Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteom Clin Appl. 2015;9:358–367. doi: 10.1002/prca.201400114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibrahim A., Marban E. Exosomes: fundamental biology and roles in cardiovascular physiology. Annu Rev Physiol. 2016;78:67–83. doi: 10.1146/annurev-physiol-021115-104929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibrahim A.G.E., et al. Augmenting canonical Wnt signalling in therapeutically inert cells converts them into therapeutically potent exosome factories. Nat Biomed Eng. 2019;3:695–705. doi: 10.1038/s41551-019-0448-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibrahim A., et al. Engineered fibroblast extracellular vesicles attenuate pulmonary inflammation and fibrosis in bleomycin-induced lung injury. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.733158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dittmar M., et al. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kindrachuk J., et al. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob Agents Chemother. 2015;59:1088–1099. doi: 10.1128/AAC.03659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramaiah M.J. mTOR inhibition and p53 activation, microRNAs: the possible therapy against pandemic COVID-19. Gene Rep. 2020;20 doi: 10.1016/j.genrep.2020.100765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullen P.J., et al. SARS-CoV-2 infection rewires host cell metabolism and is potentially susceptible to mTORC1 inhibition. Nat Commun. 2021;12:1876. doi: 10.1038/s41467-021-22166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Appelberg S., et al. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg Microbes Infect. 2020;9:1748–1760. doi: 10.1080/22221751.2020.1799723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia G., et al. Antiviral drug screen identifies DNA-damage response inhibitor as potent blocker of SARS-CoV-2 replication. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.108940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson P.J., et al. IRES-dependent translational control during virus-induced endoplasmic reticulum stress and apoptosis. Front Microbiol. 2012;3:92. doi: 10.3389/fmicb.2012.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuss-Duerkop S.K., et al. Influenza virus differentially activates mTORC1 and mTORC2 signaling to maximize late stage replication. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karam B.S., et al. mTOR inhibition in COVID-19: a commentary and review of efficacy in RNA viruses. J Med Virol. 2020 doi: 10.1002/jmv.26728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Y.N., et al. Extracellular vesicles from immortalized cardiosphere-derived cells attenuate arrhythmogenic cardiomyopathy in desmoglein-2 mutant mice. Eur Heart J. 2021;42:3558–3571. doi: 10.1093/eurheartj/ehab419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.