Abstract
Serine hydroxymethyltransferase (SHMT) is one of the most important enzyme families in one-carbon metabolic pathway and photorespiration within plant cells. Recently studies reported the active roles of plant SHMTs in defending abiotic stresses. However, genome-scale analysis of SHMT in tomato is currently unknown. In this study, seven SHMT genes were identified in the tomato genome using a genome-wide search approach. In addition, their physicochemical properties, protein secondary structure, subcellular localization, gene structure, conserved motifs, phylogenetic and collinear relationships were analyzed. Our results demonstrated that tomato SHMT members were divided into two group and four subgroups, and they were conserved with the orthologs of other plants. Analysis of cis-acting elements showed that each of the SlSHMT genes contained different kinds of hormones and stress-related cis-acting elements in their promoter regions. Finally, qRT-PCR analysis indicated that SlSHMTs were expressed at different levels in different tissues, and they responded to UV, cold, heat, NaCl, H2O2, ABA and PEG treatments. These results provided definite evidence that SlSHMTs might involve in growth, development and stress responses in tomato, which laid a foundation for future functional studies of SlSHMTs.
Keywords: Tomato, SHMT, Gene family, Photorespiration, Expression analysis, Abiotic stresses alleviation
Introduction
Serine hydroxymethyltransferase (SHMT, EC 2.1.2.1) was first discovered in rat and guinea pig in 1946 by Shemin (1946). With the exploration of protein purification and crystal structure, the property and function of this enzyme have been extensively identified in prokaryotic and eukaryotic organisms (Agrawal et al., 2004). SHMT is an α-class pyridoxal-5′-phosphate (PLP)-dependent enzyme which catalyzes the interconversion of serine and glycine. SHMT also can catalyze the generation of methylene tetrahydrofolate (THF), which carries one-carbon groups and takes part in the synthesis of thymidylate, purine and methionine (Zhang et al., 2010).
In mammals, the abnormal expression of SHMT may cause tumor growth (Pieroth et al., 2018; Barkla et al., 2014) and neurodegeneration (Troesch, Weber & Mohajeri, 2016; Escande-Beillard et al., 2020) in humans, thus SHMT is regarded as a target protein in genetic control of the corresponding diseases. SHMT genes have also been identified in many bacterial systems. More recently, the bacterial SHMTs has been confirmed to involve in alleviating salt and oxidative damage, and this function may be correlated with the photorespiration pathway (Srivastava et al., 2011; Waditee-Sirisattha et al., 2012; Shen et al., 2013; Waditee-Sirisattha et al., 2017; Nogués et al., 2020). Plant SHMT proteins, together with glycine decarboxylase (GDC), manifest an irreplaceable house-keeping function in the primary and secondary metabolisms (Rajinikanth, Harding & Tsai, 2007). In green leaves of plants, mitochondrial SHMT is important for chlorophyll biosynthesis by offering the synthetic precursors glycine and serine. The decreased enzyme activity of mitochondrial SHMT can cause photosynthesis deficiency and growth retardation (Bauwe & Kolukisaoglu, 2003). There also exists a photorespiration pathway in mitochondria of these photosynthetic tissues, which is regulated by phosphoglycolate phosphatase (PGLP), GDC and SHMT (Schwarte & Bauwe, 2007). Except for the release of carbon dioxide (CO2), the photorespiratory cycle can also remove the toxic metabolites of photosynthesis produced by ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) under high oxygen (O2) and low CO2 environment (Liu et al., 2019a; Liu et al., 2019b). In tomato (Solanum lycopersicum), a mitochondrial SHMT protein was found to regulate photorespiration as well as photosynthesis via interacting with chaperonin 60 α1 (SlCPN60 α1), confirming the vital role of SHMTs in adjusting plant metabolisms (Ye et al., 2020). Besides, SHMT activity was also detected in chloroplast, nucleus and cytosol in plants (Zhang et al., 2010; Lakhssassi et al., 2019). The plant-type SHMTs were found to be quite distant from the mammalian and the bacterial SHMTs, suggesting the occurrence of a gene duplication event during the divergence of animals, microorganisms and plants (Agrawal et al., 2004; Sodolescu et al., 2018; Batool et al., 2020).
Plant SHMTs can take part in the defense of pathogen infection. In soybean, GmSHMT08 was found to form a multi-protein complex with the soluble NSF attachment protein GmSNAP18 and the pathogenesis-related protein GmPR08-Bet VI (Lakhssassi et al., 2020). The multi-protein complex then modulated GmSHMT08 activity in maintaining the intracellular redox homeostasis in resistance to soybean cyst nematodes (SCN) (Lakhssassi et al., 2020). In rice, OsSHM1 participated in sheath blight resistance via a similar mechanism in soybean (Wang et al., 2021). In addition, a tomato SHMT protein regulated salicylic acid (SA) signaling-dependent basal defense against Pseudomonas syringae (Ahammed et al., 2018). Furthermore, adverse environmental conditions can stimulate the accumulation of SHMT in plant cells, such as salt (Mishra et al., 2016; Kito et al., 2017), drought (Liu et al., 2019a; Liu et al., 2019b), chilling (Fang et al., 2020), wound (Hourton-Cabassa et al., 1998) and heavy metal stress (Roth, Roepenack-Lahaye & Clemens, 2006). However, the restrained SHMT activity always comes along with a more severe growth deficiency under abiotic stresses (Moreno, Martín & Castresana, 2005; Liu et al., 2019a; Liu et al., 2019b). SHMT mainly decelerates abiotic stresses through modulating the level of ROS and recovering the impacted cellular metabolism (Ji et al., 2012; Zhou et al., 2012; Fang et al., 2020), which is also assumed to correlate with the photorespiration activity as in cyanobacteria. In Arabidopsis thaliana, AtSHMT1 was also found to adjust ABA-induced stomatal closure to reduce salt stress sensitivity (Liu et al., 2019a; Liu et al., 2019b), suggesting a crosstalk mechanism between plant hormones and SHMT proteins in defending abiotic stresses. Nevertheless, there also exists a negative role of SHMT in adapting to the changeable circumstance (Xu & Huang, 2010; Barkla et al., 2014; Fan et al., 2014; Chen et al., 2020a; Chen et al., 2020b), indicating a complex functional allocation of SHMT proteins in different plants. Yet, more explorations are still needed to uncover the accurate mechanism of SHMT proteins in enhancing plant resistance against abiotic stresses.
Tomato (Solanum lycopersicum) is a major horticultural crop widely cultivated in the world (Hu et al., 2021). Tomato is favoured by consumers worldwide due to its abundant nutrients (Wang & Liu., 2021). What’s more, tomato is an important model plant in horticultural research, including the study concerning the tolerance of adverse stress (Guo et al., 2021; Li et al., 2021). Hitherto, SHMT genes have been identified and characterized in pea, A. thaliana, soybean and poplar (Turner et al., 1992; McClung et al., 2000; Wu et al., 2016; Li & Cheng, 2020). As the biochemical properties and the environmental defense mechanism of tomato SHMTs are largely unknown, the genome-wide SHMT genes identification and characterization in tomato were investigated in the present study. Here, we analyzed the physicochemical properties, gene structures, evolutionary relationship, collinear relationship, conserved motifs, gene locations, cis-acting element distributions and tissue-specific expression patterns of SHMT genes in tomato plants. The expression patterns of the tomato SHMT genes under different abiotic stress conditions were also detected, and the potential function of SHMT in growth regulation and abiotic stresses alleviation was proposed.
Materials & Methods
Genome-wide identification of SHMT gene family members in tomato
The tomato genome sequence and annotation information of ITG3.2 were downloaded from the online database Phytozome v12.1 (https://phytozome.jgi.doe.gov/pz/) (Goodstein et al., 2012), and TBtools (Toolbox for ecological battle) v1.0985 software was used to organize and extract SHMT protein sequences by functions of ‘GXF Sequence Extract’ and ‘Batch Translate CDS’ to Protein in TBtools. Sequences of A. thaliana SHMT genes were downloaded from the online database SSWISS-PROT/Uniprot (https://www.uniprot.org). The HMM model of the conserved structural domain of SHMT (PF00464) was downloaded from the Pfam database. The preliminary SHMT candidate genes were obtained in comparison with the HMM search function of HMMER software. Then, single BLAST of the collected sequences between tomato and A. thaliana was performed using the function of ‘BLAST GUI Wrapper’ in TBtools software, and the corresponding bidirectional BLAST was performed using NCBI database (https://www.ncbi.nlm.nih.gov/). ‘Blast Xml to Table’ function in TBtools software was then used to obtain possible gene family members of tomato SHMTs. Additionally, the Swiss-Prot/Uniprot database in NCBI was used to further confirm sequences of tomato SHMT genes based on the corresponding conserved domains by the ‘Protein BLAST’ function of NCBI.
Characteristic analysis of tomato SHMT genes
For gene characteristic analysis, molecular weight and isoelectric point (pI) of different SHMT genes were conducted by use of the online website Expasy (https://web.expasy.org/compute_pi/). The corresponding gene structure was drawn based on the genome annotation file using the function of ‘Visualize Gene Structure (from GTF/GFF3 File)’ in TBtools software (Chen et al., 2020a; Chen et al., 2020b).
Phylogenetic analysis of tomato SHMT genes
The SHMT protein sequences of A. thaliana, soybean, poplar and tomato were downloaded from the online database phytozome. The multiple sequence alignment of proteins was performed by ClustalW with the Delay Divergent Cutoff value setting as 30 and other options as default. The Mega7.0 software was used to construct a phylogenetic tree of 41 SHMT protein sequences by applying the the Maximum Likelihood method (Kumar, Stecher & Tamura, 2016). In addition, the execution parameters were p-distance and Pairwise deletion, and the number of repeats of Bootstrap repetitions was set as 1000, and the remaining options were set as default. The online website evolview (https://www.evolgenius.info//evolview/#mytrees/clcle/123) was used to further embellish the evolutionary tree.
Gene structure and chromosomal localization
The gene structure of each member of SHMT was analyzed using the ‘Visualize Gene Structure (from GTF/GFF3 File)’ function in TBtools software. The tomato GFF3 file downloaded from the online database Phytozome was prepared to visualize gene locations by Gene Location Visualize from GTF/GFF function in TBtools, Then chromosome location of each tomato SHMT gene members was mapped to the tomato genome (Chen et al., 2020a; Chen et al., 2020b). The SHMT gene members were renamed according to their chromosome distributions.
Cis-acting element analysis of tomato SHMT genes
The promoter regions of all the tomato SHMT genes were extracted by the function of ‘GXF Sequences Extract’ in TBtools software, and the corresponding promoter sequences of SHMT gene set were further extracted and then submitted to the online PlantCARE (Song et al., 2019) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) website to predict different kinds of cis-acting elements in SHMT gene promoter regions. The cis-acting element analysis results of PlantCARE website were collated and simplified, and were conducted to visualization processing by using ‘Simple BioSequence Viewer’ function in TBtools software.
Conserved motif analysis of tomato SHMT members
The online website MEME (http://meme-suite.org/tools/meme) was used to carry out the conserved motifs information of tomato SHMT gene family members (Liu et al., 2019a; Liu et al., 2019b). Then the corresponding motif information and the evolutionary tree information of tomato SHMT family derived from Mega7.0 were combined to be analyzed by the ‘gene structure view (Advances)’ function of TBtools software to visualize the conserved motifs of tomato SHMT members.
Protein second structure analysis of tomato SHMT genes
The online website prabi (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) was used to analyze the secondary structure of tomato SHMT family proteins and the corresponding information was exported and imaged.
Collinearity analysis
The online database phytozome was used to determine the genetic relationship between A. thaliana, soybean, poplar and tomato. The relative fasta and GFF3 files representing the genetic relationship were downloaded and used for collinearity analysis by ‘Text Merge for MCScanX’ function in TBtools software. Finally, the results were visualized by ‘Multiple Systeny Plot’ function in TBtools software (Chen et al., 2020a; Chen et al., 2020b).
Plant materials, growth conditions and stress treatments
Tomato (Lycopersicum esculentum L. ‘Micro-Tom’) seeds with full grains and consistent size were selected in a 50 mL centrifuge tube, and surface sterilized with 1% NaClO solution for 10 min. The sterilized seeds were put into a 250 mL conical flask filled with 100 mL sterile water, then placed in a HYG-C type shaker and cultured at a rotation speed of 180 r min−1 at 25 °C for 3 days. The sterile water was changed once a day. The germinated tomato seeds were planted in a hole dish containing culture soil and placed in a growth chamber for culture. The light intensity in the growth chamber was 250 mol photons m−2s−1, 26 ± 2 °C for 16 h during the day and 20 ± 2 °C for 8 h at night. The relative humidity was 60%. After 21 days, healthy seedlings of uniform size were selected for subsequent treatments.
As for salt, ABA, H2O2 and drought stress treatments, the selected seedlings were transferred to 1/2 nutrient solution containing 200 mM NaCl, 100 mM ABA, 10% (w/v) (2.94 M) hydrogen peroxide (H2O2) and 20% (w/v) PEG6000, respectively. Plants of the control group was grown in the 1/2 nutrient solution without adding other reagents. All the seedlings were grown in the same conditions of a growth chamber. For cold and heat treatments, the seedlings were transferred to the other growth chambers and put into 1/2 nutrient solution under 4 °C and 40 °C without adding other reagents, respectively. Some other selected seedlings were transferred to a growth chamber equipped with 253.7 nm UV-C radiation by a UV-C lamp (TUV PL-S 40 W/4P, Philips, Poland), and the other growth conditions were same as the control. After 0, 6, 12 and 24 h, the aboveground parts of the treated seedlings in each replication were harvested separately, then were frozen with liquid nitrogen and stored at −80 °C, respectively. Each treatment contained three biological replicates, and each replicate consisted of eight seedlings.
Also, the roots, stems and leaves of the 21-day old untreated seedlings were collected for SHMT genes expression analysis of the vegetative growth period. The roots, stems, leaves and flowers of the untreated plants were collected at flowering stage (55-day old seedlings), and the corresponding green fruits (25 days after pollination) and mature fruits (45 days after pollination) were also collected (one fruit selected in each plant) to analyze SHMT genes expression levels of the reproductive growth period. There were three biological replications with eight plants in each replication collected under the identical experimental condition.
RNA isolation and qRT-PCR
The methods of RNA isolation and qRT-PCR were done according to Dan et al. (2021) with some modifications. The plant samples in each replication were collected and homogenized in liquid nitrogen to powder. Total RNA was extracted from the homogenized powder by applying a MiniBEST PLANT RNA extraction Kit (TaKaRa). The concentration of the isolated RNA was measured by a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, USA). Then 4 µg RNA was used for producing the reverse-transcribed complementary DNA (cDNA) with Dnase I, oligo (dT) primers, dNTPs and M-MLV (TIANGEN) in a 20 µL reaction. A total of 2 µL cDNA from the above reaction was employed as a template to determine the transcript levels of the tested genes with a SuperReal PreMix Plus kit (TIANGEN) on a Roche LightCycler instrument. There were three biological replicates per treatment. The primers used for qRT-PCR were designed using primer premier 5 software, and were listed in File S1. The tomato ACTIN gene was used to normalize relative expression levels.
Results
Genome-wide identification of SHMT genes in tomato
In our study, seven SHMT sequences were screened out from the tomato genome database. Our results show that all the seven sequences contain a typical SHMT domain (Pfam: PF00464) and belong to the tomato SHMT family (File S2). According to the sequential positions of tomato SHMT genes identified on tomato chromosomes, the SHMT genes were named as SlSHMT1-SlSHMT7 (Table 1). Then, we analyzed physicochemical properties of the identified SlSHMT members. The full length of the open reading frame of the SlSHMT genes is between 1,416 bp (SlSHMT4, SlSHMT7) and 1,785 bp (SlSHMT1), and the corresponding amino acids of tomato SHMTs is between 471 (SlSHMT4, SlSHMT7) and 594 (SlSHMT1). As a result, SlSHMT1 has the longest protein sequence, and its molecular weight is 65521.19 Da, while the molecular weight of SlSHMT4 is lightest (52059.26 Da). Moreover, the isoelectric point (pI) of tomato SHMT members ranges from 6.62 (SlSHMT4) to 8.13 (SlSHMT2). SlSHMT4 is the only acidic (pI < 7) protein in the tomato SHMT family and the rest are alkalescent (pI > 7) (Table 1).
Table 1. Physical and chemical property of SHMT gene family in Solanum lycopersicum.
| Gene | Gene ID | Gene locus | ORF (bp) |
Amino acid | Molecular weight |
pI | SHMT domain location |
|---|---|---|---|---|---|---|---|
| SlSHMT1 | Solyc01g104000.3.1.ITAG3.2 | Chr01 | 1785 | 594 | 65521.19 | 7.13 | 137–542 |
| SlSHMT2 | Solyc02g091560.3.1.ITAG3.2 | Chr02 | 1557 | 518 | 57232.43 | 8.13 | 56–454 |
| SlSHMT3 | Solyc04g076790.3.1.ITAG3.2 | Chr04 | 1554 | 517 | 57246.39 | 8.11 | 55–453 |
| SlSHMT4 | Solyc05g053810.3.1.ITAG3.2 | Chr05 | 1416 | 471 | 52059.26 | 6.62 | 12–412 |
| SlSHMT5 | Solyc08g065490.3.1.ITAG3.2 | Chr08 | 1575 | 524 | 56994.42 | 7.17 | 78–468 |
| SlSHMT6 | Solyc12g095930.2.1.ITAG3.2 | Chr12 | 1584 | 527 | 57176.71 | 7.14 | 81–471 |
| SlSHMT7 | Solyc12g098490.2.1.ITAG3.2 | Chr12 | 1416 | 471 | 52276.55 | 7.16 | 12–412 |
Protein secondary structure and subcellular localization of SlSHMT members
The secondary structure of the proteins encoded by tomato SHMT was analyzed (Table 2). The results show that tomato SHMT family mainly includes alpha helix, random coil and beta turn, with the percentage of 40.07%∼46.91%, 33.01%∼38.72%, 5.89%∼7.02%, respectively.
Table 2. The secondary structure of SHMT gene family in Solanum lycopersicum.
Blue indicates alpha helix; Green indicates beta turn; Red indicates extended strand; Pink indicates random coil.
| Protein | Alpha helix (%) | Beta turn (%) | Random coil (%) | Distribution of secondary structure elements |
|---|---|---|---|---|
| SlSHMT1 | 40.07 | 5.89 | 38.72 |
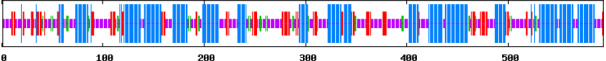
|
| SlSHMT2 | 46.91 | 6.95 | 33.01 |
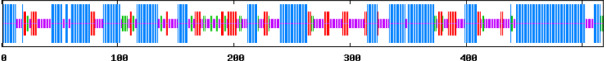
|
| SlSHMT3 | 45.26 | 6.58 | 35.01 |
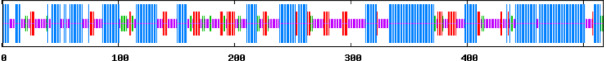
|
| SlSHMT4 | 44.80 | 6.16 | 35.24 |
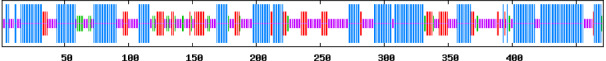
|
| SlSHMT5 | 42.75 | 6.49 | 36.26 |
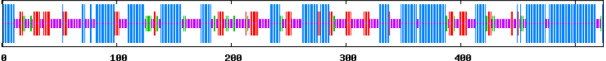
|
| SlSHMT6 | 42.13 | 7.02 | 36.05 |
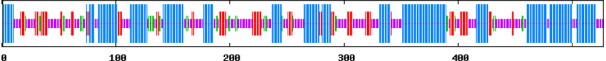
|
| SlSHMT7 | 44.59 | 5.94 | 36.94 |
|
Subcellular localization of tomato SHMTs showed that SHMT genes are mainly distributed in chloroplasts, mitochondria, cytoskeletons and cytoplasms (Table 3). However, the distribution of each member in different cellular parts is different. SlSHMT1, SlSHMT3, SlSHMT5 and SlSHMT6 in tomato SHMT family were found to be chloroplast-localized. Only SlSHMT2 and SlSHMT3 are distributed in mitochondria. SlSHMT4 and SlSHMT7 were found to mainly localize in cytoskeletons, and the cytoplasms-localized proteins are SlSHMT1, SlSHMT2 and SlSHMT4. The localizations of tomato SHMTs in nucleus and cytoplasms are relatively low.
Table 3. Subcellular localization prediction of SHMT gene family in Solanum lycopersicum.
| Gene | Gene ID | Chloroplast | Nucleus | Plasma membrane | Mitochondria | Cytoskeleton | Cytoplasm |
|---|---|---|---|---|---|---|---|
| SlSHMT1 | Solyc01g104000.3.1.ITAG3.2 | 7 | 2 | – | – | 1 | 4 |
| SlSHMT2 | Solyc02g091560.3.1.ITAG3.2 | 1 | – | – | 6 | – | 7 |
| SlSHMT3 | Solyc04g076790.3.1.ITAG3.2 | 8 | – | – | 6 | – | – |
| SlSHMT4 | Solyc05g053810.3.1.ITAG3.2 | 2 | 2 | – | – | 6 | 4 |
| SlSHMT5 | Solyc08g065490.3.1.ITAG3.2 | 11 | 1 | 1 | – | 1 | – |
| SlSHMT6 | Solyc12g095930.2.1.ITAG3.2 | 13 | – | – | – | – | 1 |
| SlSHMT7 | Solyc12g098490.2.1.ITAG3.2 | – | 3 | – | – | 8 | 3 |
Gene structure analysis of SlSHMT genes
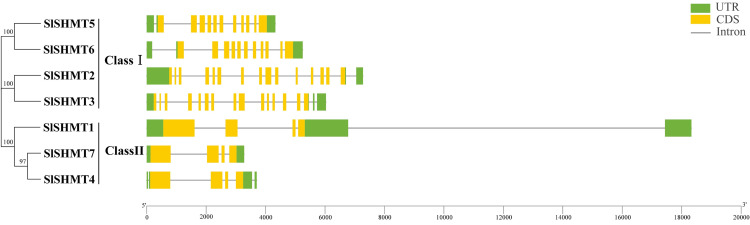
According to the information of phylogenetic analysis, tomato SHMT proteins are divided into two groups (Fig. 1). In detail, SlSHMT2, SlSHMT3, SlSHMT5 and SlSHMT6 are classified into Class I, and SlSHMT1, SlSHMT4 and SlSHMT7 are classified into Class II. Gene structure analysis is regarded as a very effective method to determine gene function and reflect the phylogenetic relationship between genes. Therefore, the gene structure of tomato SHMT family members were evaluated by using a TBtools software (Fig. 1). Our results show that SlSHMT5 and SlSHMT6 (Class I) both contain 11 exons and 11 introns. SlSHMT2 and SlSHMT3 (Class I) possess 15 exons and 15-16 introns. Thus, SlSHMT2 and SlSHMT3 were more similar in their gene structures than SlSHMT5 and SlSHMT6 in Class I. In Class II, all the members contain 4 exons, and SlSHMT1, SlSHMT4 and SlSHMT7 have 4, 4, and 3 introns, respectively. Gene structures of SlSHMT4 and SlSHMT7 were quite similar with each other, and SlSHMT1 gene possessed relative longer exons, introns and UTR regions. In all, the distributions of exon-intron in the same protein group are similar, and the sequence lengths of SHMT genes vary between different subfamilies. This difference is mainly caused by the sequence length differences of non-coding regions and the amounts of introns. Thus, the two subfamilies may have undergone functional differentiation during evolution.
Figure 1. Exon-intron structure of SHMT gene family in tomato.
The evolutionary tree was constructed based on the full length of tomato SHMT protein sequences using MEGA7.0. The exon-intron graph of tomato SHMT genes was drawn using TBtools software.
Phylogeny and Collinearity analysis of SlSHMT members
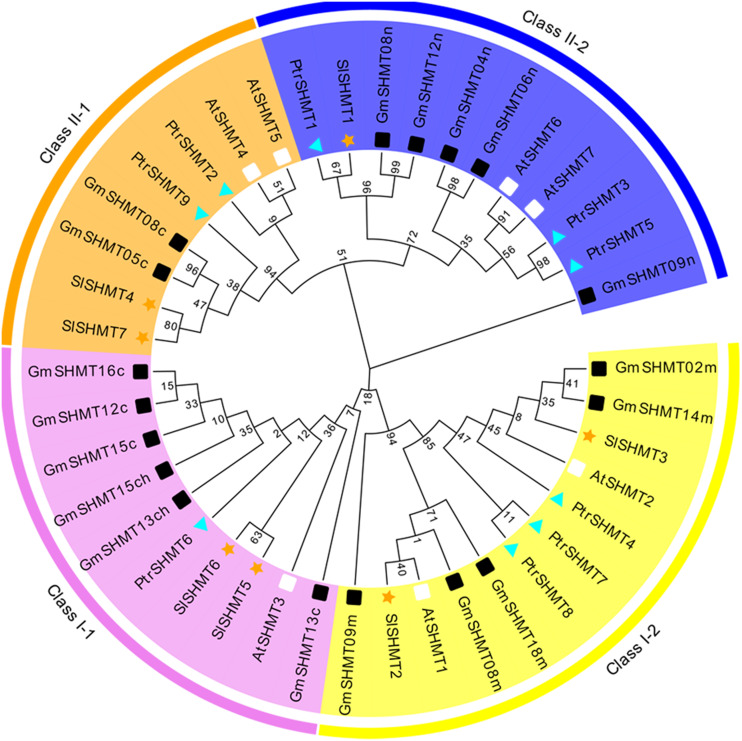
In order to further understand the functions and characteristics of tomato SHMT family genes, we compared the full-length sequences of 41 SHMT proteins in A. thaliana, soybean (Glycine max), poplar (Populus trichocarpa) and tomato (Fig. 2 and File S3). The 41 SHMT proteins of these plants are divided into two groups: Class I and Class II, which is parallel with the classification in Fig. 1. The two groups are further divided into four subgroups: Class I-1, Class I-2, Class II-1 and Class II-2. There are two members of tomato SHMT family (SlSHMT5 and SlSHMT6) in Class I-1, two members (SlSHMT2 and SlSHMT3) in Class I-2, two members (SlSHMT4 and SlSHMT7) in Class II-1, and one member (SlSHMT1) in Class II-2. In addition, the homology coefficient of SlSHMT6 and SlSHMT7 is very high, and they are located on the same chromosome, indicating that gene replication may take place between them.
Figure 2. The unrooted phylogenetic tree of SHMT gene family in Solanum lycopersicum, Glycine max, Populus trichocarpa and A. thaliana.
The maximum likelihood method was used to construct phylogenetic tree containing seven tomato, seven A. thaliana (At), nine poplar (Ptr), and 18 soybean (Gm) SHMT proteins. The four subgroups are colored differently. The four differently-colored shapes represent SHMT proteins from four species. The orange pentacle, white rectangle, blue triangle, black rectangle represent tomato, A. thaliana, poplar, and soybean SHMT proteins, respectively. The number on the node in the phylogenetic tree represents the percentage of trustworthiness of the branch in the bootstrap validation.
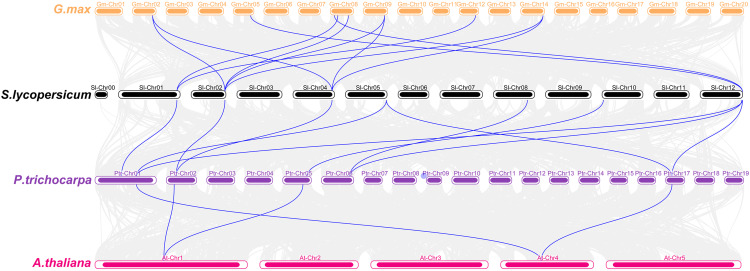
Then, we also explored the collinear relationship between tomato SHMT genes and the related genes of the three typical plants A. thaliana, soybean and poplar based on the evolutionary relationship among different species (Fig. 3). The results of collinearity analysis showed that seven tomato genes are collinear with nine soybean genes, six poplar genes and two A. thaliana genes. There are 11 homologous pairs between tomato and soybean, 10 homologous pairs between tomato and poplar, and four homologous pairs between poplar and A. thaliana. The above results indicated that tomato SHMTs are closely related to other SHMT members in the above three plant species.
Figure 3. Collinearity analysis of SHMT gene family in S. lycopersicum, G. max, P. trichocarpa and A. thaliana.
The blue lines delineate the syntenic SHMT gene pairs.
Conserved motifs analysis of tomato SHMT proteins
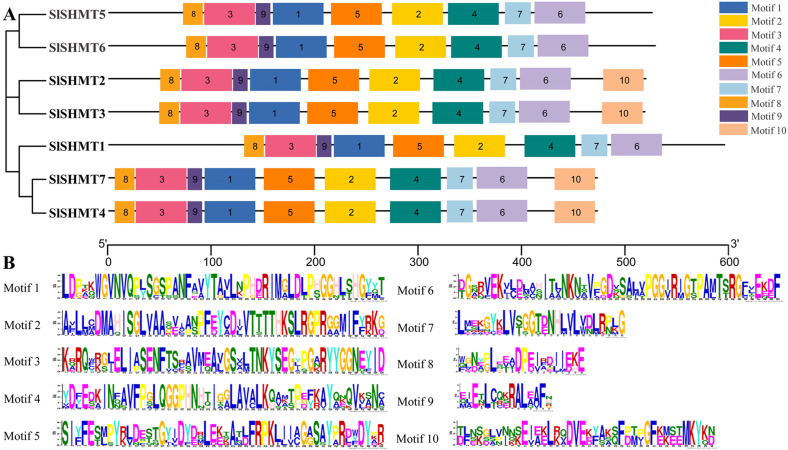
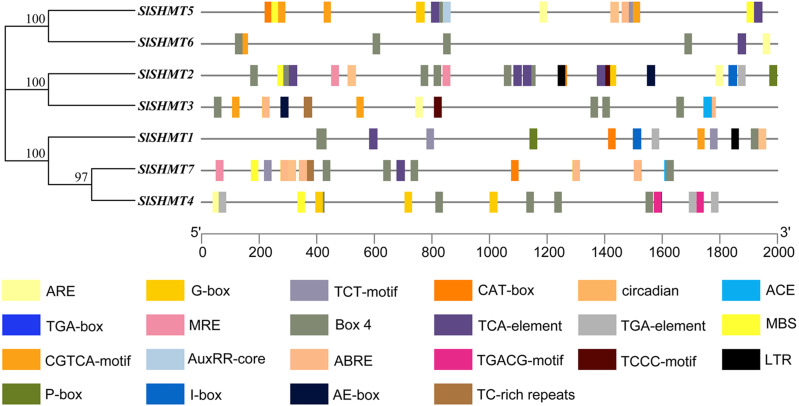
The online site MEME was used to study the conserved region of tomato SHMT proteins. In the recent studies, 10 conserved motifs were found in tomato SHMT proteins (Fig. 4). The sequence information of the identified conserved motifs was listed in Table 4, and the length of each motif ranges from 15 to 50 amino acids. Using TBtools software, we found that the tomato SHMT proteins belonging to the same subfamily in the evolutionary tree contain the similar or identical composition of motifs. For example, all the members of Class I-1 (SlSHMT5 and SlSHMT6) lack the tenth conserved motif, while the members of Class I-2 (SlSHMT2 and SlSHMT3) contain this motif. The members of Class II-2 (SlSHMT1) contain nine motifs as that of class I-2, and the motif locations are significant different with that in Class II-1. SlSHMT4 and SlSHMT7 in Class II-1 contain all the 10 conserved motifs. Furthermore, the sequence of motifs in each subfamily is almost the same. These results are in agreement with the phylogenetic analysis of the tomato SHMT proteins, which further suggest that these 4 subgroups may have developed functional difference during evolution.
Figure 4. The motif composition and distribution of tomato SHMT proteins.
(A) Colored boxes represent different conserved motifs, and motifs 1-10 are shown in (B). (B) Amino acid sequences of different conserved motifs displayed by stacks of letters at each position. The total height of the stack represents the information content of the relative amino acid in the position of each letter in the motif in bits. The height of the individual letter in a stack is calculated by the probability of the letter at that position times the total information content of the stack. X- and Y-axis represents the width and the bits of each letter, respectively.
Table 4. Details of the 10 conserved motifs of Solanum lycopersicum SHMT proteins.
| Motif | Width (aa) |
Motif Sequence |
|---|---|---|
| Motif 1 | 50 | LDPKKWGVNVQPLSGSPANFAVYTAVLNPHDRIMGLD LPHGGHLSHGYYT |
| Motif 2 | 50 | ALLLCDMAHISGLVAASVIANPFEYCDIVTTTTHKS LRGPRGGMIFYRKG |
| Motif 3 | 50 | KQRQFRGJELIASENFTSRAVMEAVGSALTNKYSEG LPGARYYGGNEYID |
| Motif 4 | 50 | YDFEDKINFAVFPGLQGGPHNHTIGGLAVALKQAKT PEFKAYQEQVKANA |
| Motif 5 | 50 | SIYFESMPYRLDESTGYIDYDRLEKSATLFRPKLIIA GASAYPRDWDYPR |
| Motif 6 | 50 | DGSRVEKVLDLAHITLNKNSVPGDKSALVPGGIRIGT PAMTSRGFVEKDF |
| Motif 7 | 26 | LMEKGYKLVSGGTDNHLVLVDLRPLG |
| Motif 8 | 20 | WGNEPLEEADPEIADIIEKE |
| Motif 9 | 15 | ZIETLCQKRALEAFH |
| Motif 10 | 40 | DFNKGLVNNKEIEELKQDVEKYAKQFPTPGFEKEEMKYKD |
Chromosomal location of tomato SHMT genes
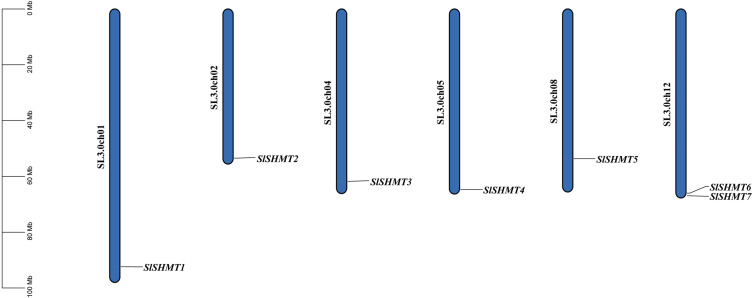
Using tomato genome annotation information and TBtools software, we visualized the chromosomal distributions of tomato SHMT gene family members (Fig. 5). As could be seen from Figs. 5, seven genes of tomato SHMT family are unevenly distributed on six chromosomes (Chr.1, Chr.2, Chr.4, Chr.5, Chr.8 and Chr.12), and the number of genes on each chromosome is unrelated to the chromosome size. Among the tomato SHMTs, SlSHMT1 is distributed on chromosome 1, SlSHMT2 is distributed on chromosome 2, SlSHMT3 is distributed on chromosome 4, SlSHMT4 is distributed on chromosome 5, SlSHMT5 is distributed on chromosome 8, SlSHMT6 and SlSHMT7 are both distributed on chromosome 12. There are no SHMT members on the chromosome 3, 6, 7, 9, 10 and 11. Most SlSHMT genes are located on the proximal or distal end of the tomato chromosomes.
Figure 5. The distribution of SHMT gene family members of chromosomes in Solanum lycopersicum.
Cis-acting element analysis of tomato SHMT genes
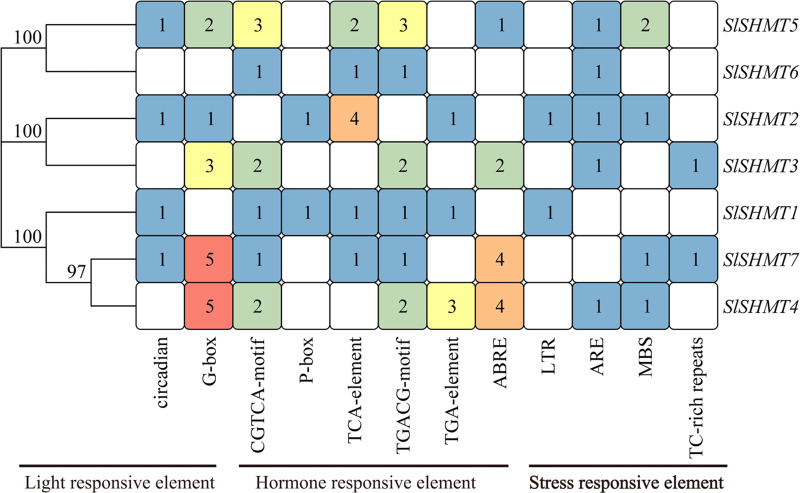
In the current study, 46 types of elements were analyzed in the promoter region of SlSHMT genes. The predicted cis-acting elements were classified as stress responsive elements, light responsive elements, and hormone responsive elements according to the function and action of these elements (Table 5). The results show that SlSHMT members contain 7-23 cis-acting elements in their promoter region (Fig. 6 and File S4). The stress responsive elements include LTR, ARE, MBS and TC-rich repeats. The light responsive elements include Circadian and G-box. Some hormone responsive elements are also found in most of the SlSHMT genes, including CGTCA-Motif, P-box, TCA-Element, TGACG-Motif, TGA-Element, and ABRE (Fig. 7).
Table 5. Summary of cis-acting elements of Solanum lycopersicum SHMT genes.
| Element | Sequence | Description |
|---|---|---|
| Circadian | CAAAGATATC | cis-acting regulatory element involved in circadian control |
| G-box | CACGAC | cis-acting regulatory element involved in light responsiveness |
| CGTCA-motif | CGTCA | cis-acting regulatory element involved in the MeJA-responsiveness |
| P-box | CCTTTTG | gibberellin-responsive element |
| TCA-element | CCATCTTTTT | cis-acting element involved in salicylic acid responsiveness |
| TGACG-motif | TGACG | cis-acting regulatory element involved in the MeJA-responsiveness |
| TGA-element | AACGAC | auxin-responsive element |
| ABRE | ACGTG | cis-acting element involved in the abscis ic acid responsiveness |
| LTR | CCGAAA | cis-acting element involved in low-temperature responsiveness |
| ARE | AAACCA | cis-acting regulatory element essential for the anaerobic induction |
| MBS | CAACTG | MYB binding site involved in drought-inducibility |
| TC-rich repeats | ATTCTCTAAC | cis-acting element involved in defense and stress responsiveness |
Figure 6. The distribution of cis-acting elements in Solanum lycopersicum SHMT genes.
Figure 7. The number of cis-acting elements in Solanum lycopersicum SHMT genes.
As can be seen from Fig. 7, G-box elements are mostly distributed in SlSHMT3, SlSHMT4, SlSHMT5 and SlSHMT7. CGTCA-Motif elements are mostly distributed in SlSHMT3, SlSHMT4 and SlSHMT5. TCA-element elements are mostly distributed in SlSHMT2 and SlSHMT5. TGACG-motif elements are mostly distributed in SlSHMT4. ABRE elements are more distributed in SlSHMT3, SlSHMT4 and SlSHMT7. Finally, MBS elements are mostly distributed in SlSHMT5. Our analysis of cis-acting elements shows that most of the SlSHMT genes may play an important role in stress, light and hormone responses.
Expression analysis of tomato SHMT genes in different organs of different growth stages
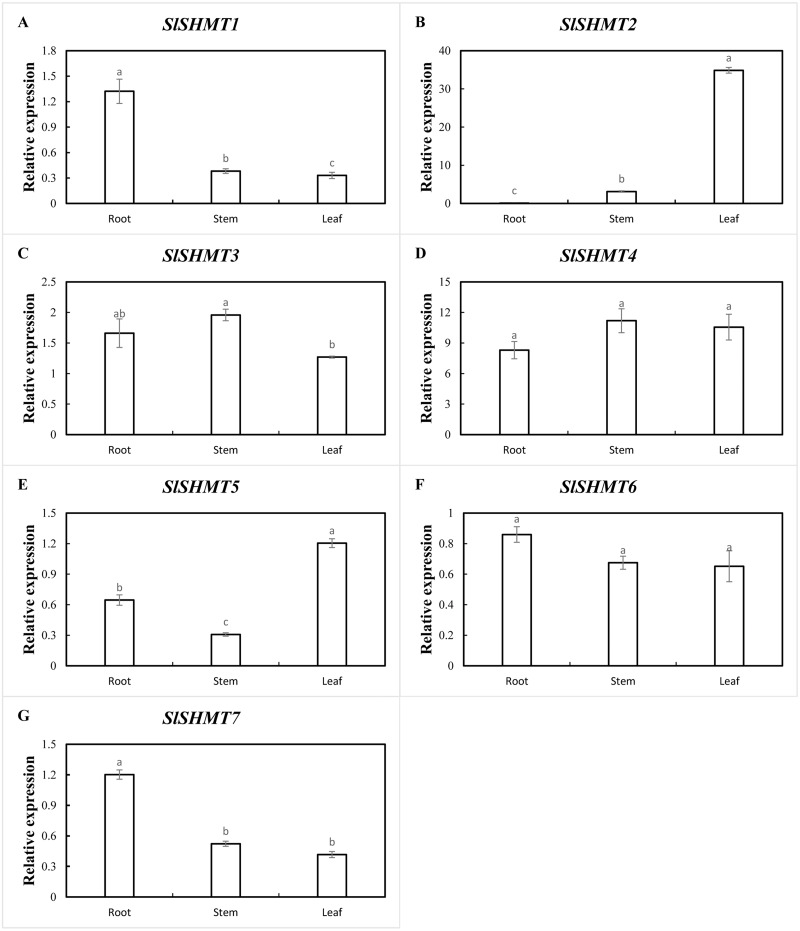
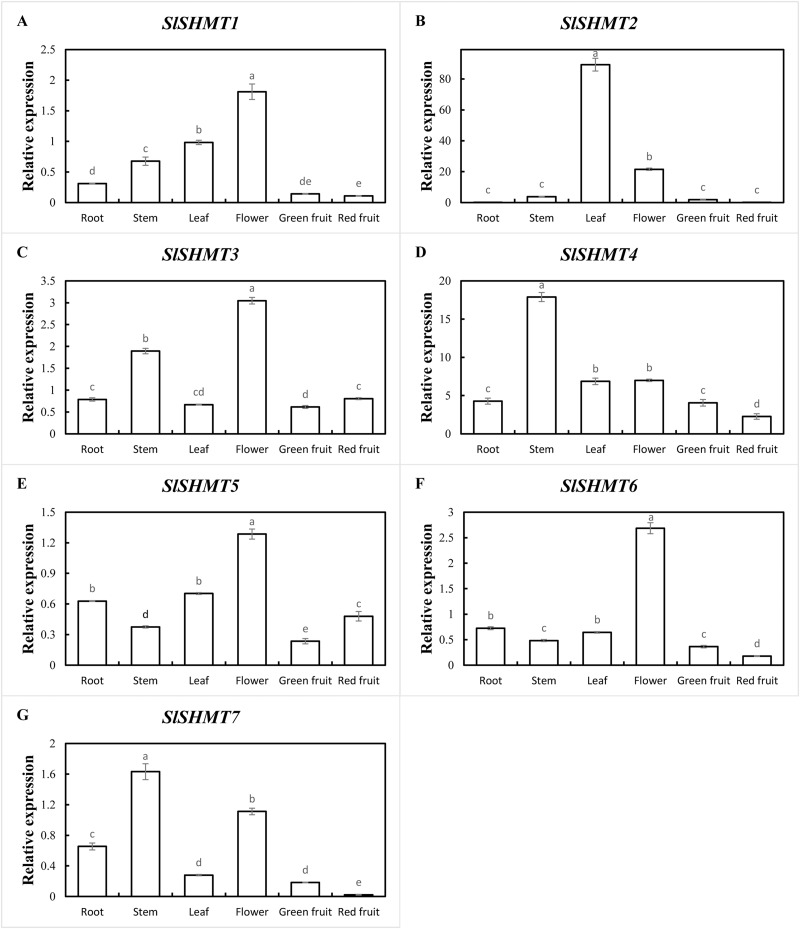
In order to determine the expression specificity of tomato SHMT genes in different growth stages and organs, qRT-PCR was used to detect the expression levels of SlSHMT genes in different organs of tomato during vegetative and reproductive growth stages. In vegetative growth stage, SlSHMT1 and SlSHMT7 are highly expressed in root. The expressions of SlSHMT2 and SlSHMT5 are most abundant in leaf. However, the expression levels of SlSHMT4 and SlSHMT6 in root, stem and leaf are similar. The expression level of SlSHMT3 gene in stem is relatively high (Fig. 8). In reproductive growth stage, SlSHMT1, SlSHMT3, SlSHMT5 and SlSHMT6 are highly expressed in flower. SlSHMT4 and SlSHMT7 are mainly expressed in stem. SlSHMT2 gene is highly expressed in leaf (Fig. 9). The results show that SlSHMT genes might play special roles in different growth stages and different plant organs.
Figure 8. Expression levels of SlSHMT genes of different plant organs in vegetative growth period.
The expression patterns of SlSHMT1-SlSHMT7 in different tissues are shown in A–G, respectively. Error bars represent the standard error (SE) of three replicates. The relative expression of each gene in different tissues is expressed as mean ± SE (n = 3). Bars with different lowercase letters were significantly different by Duncan’s multiple range tests (p < 0.05).
Figure 9. Expression levels of SlSHMT genes of different organs in reproductive growth period.
The expression patterns of SlSHMT1-SlSHMT7 in different tissues are shown in A–G, respectively. Error bars represent the standard error (SE) of three replicates. The relative expression of each gene in different tissues is expressed as mean ± SE (n = 3). Bars with different lowercase letters were significantly different by Duncan’s multiple range tests (p < 0.05).
Expression analysis of tomato SHMT genes under different treatments
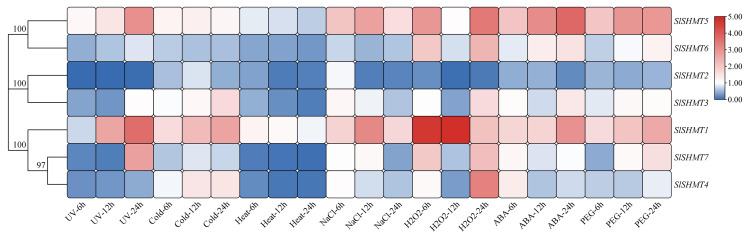
The correlation of plant SHMTs with biotic and abiotic stress defenses has long been discovered. For further analyze the response of tomato SHMT members to abiotic stresses and plant to hormones, qRT-PCR experiments were carried out and TBtools software was used to draw clustering heat map of SlSHMT gene expression levels under different treatments (Fig. 10 and File S5). The results show that expression levels of SlSHMT genes are different under different treatments. In terms of SlSHMT1, there is almost no change of expression level under heat treatment, but it is significantly up-regulated under UV radiation, cold, ABA and PEG treatments, reaching the highest expression level at 24 h. SlSHMT1 is also significantly up-regulated under NaCl and H2O2 treatments, obtaining the highest expression level at 12 h. For SlSHMT2, its transcription level exhibits a downward trend under all treatments. SlSHMT3 is slightly down-regulated under UV radiation, heat and NaCl treatments, and slightly up-regulated under cold, H2O2 and ABA treatments, but almost unchanged under PEG treatment. SlSHMT4 is mildly down-regulated by UV radiation, heat, NaCl and PEG treatments, and slightly up-regulated under cold treatment, while the expression level of SlSHMT4 under H2O2 and ABA are complicated. SlSHMT5 is up-regulated under UV radiation, cold, NaCl, H2O2, PEG and ABA treatments but down-regulated under heat treatment. SlSHMT6 is mildly down-regulated by UV radiation, cold, heat, NaCl and PEG treatments, but slightly up-regulated under ABA and H2O2 treatments. SlSHMT7 is down-regulated and then up-regulated under UV radiation and PEG treatments, reaching the highest expression level at 24 h. Meanwhile, SlSHMT7 is slightly down-regulated by heat treatment, and its expression does not change under other treatments.
Figure 10. Expression levels of SlSHMT genes under UV radiation, cold, heat, NaCl, ABA, H2O2 and drought (PEG) treatments.
Seedlings were treated with 253.7 nm UV radiation, 4 °C Cold, 40 °C Heat, 200 mM NaCl, 100 mM ABA, 10% (w/v) H2O2 and 20% (w/v) PEG. The color scale represents the folding changes normalized by log2 transformed data. Blue represents downregulated genes and red represents upregulated genes.
Discussion
SHMT catalyzes the conversion of serine to glycine and take part in cellular metabolism through providing one-carbon units for many biosynthetic reactions in many organisms (Bauwe & Kolukisaoglu, 2003). SHMT also acts as an indispensable role in the photorespiratory pathway of aerobic photosynthetic organisms (Waditee-Sirisattha et al., 2017). In addition, SHMT is widely distributed in plants and plays an active role in regulating plant stress resistance (Fang et al., 2020). Genes encoding SHMT have been identified in many higher plants. It has been shown that 18, 7 and nine SHMT genes are present in soybean (Lakhssassi et al., 2019), A. thaliana (Zhang et al., 2010) and P. trichocarpa (Li & Cheng, 2020), respectively. However, the SHMT gene family in tomato has not been studied in detail. In the study, we identified 7 SHMT genes in the tomato genome and they are randomly distributed on six chromosomes of tomato (Table 1, Fig. 5). Thus, the difference in genome size may lead to diverse amounts of SHMT family members (Xie et al., 2018). Soybean is the only tetraploid among the above plants, and its genome size is the largest. Soybean contains the most abundant SHMT members in comparison with the diploid tomato, A. thaliana and Populus trichocarpa, indicating a difference replication pattern of each kind of plants during the evolution.
We analyzed the gene structure of SlSHMT genes and found that the intron numbers of different SlSHMT genes are quite distant, ranging from three to 16 (Fig. 1). In parallel, the average exon length of SlSHMT2, SlSHMT3, SlSHMT5 and SlSHMT6 with multiple introns is relatively shorter than that of SlSHMT1, SlSHMT4 and SlSHMT7 with fewer introns, and the length of coding sequence in each SlSHMT gene does not show too much difference (Table 1). This phenomenon is similar with the SHMT genes in A. thaliana and soybean (Lakhssassi et al., 2019; Li & Cheng, 2020), demonstrating that SHMT genes may evolve from parts of members through alternative splicing. Here, the conserved regions of tomato SHMT proteins was also analyzed (Fig. 4). There are 10 conserved motifs found in SlSHMT proteins, and all SlSHMT proteins contain the typical SHMT (Pfam: PF00464) domain. Nevertheless, only four family members contain the tenth motif, suggesting different protein properties and functions between these SHMT proteins.
In order to further explore the relationship between tomato SHMT members and other species, a phylogenetic tree was constructed including SHMT members of tomato, A. thaliana, soybean and poplar. All the SHMTs members are divided into two groups and four subgroups namely ClassI-1, ClassI-2, ClassII-1 and ClassII-2, which is similar with previous studies in A. thaliana, soybean and poplar (Lakhssassi et al., 2019; Li & Cheng, 2020). Each subgroup contained at least one member of tomato SHMT (Fig. 2). In addition, the 7 predicted tomato SHMT genes are collinear with SHMTs of A. thaliana, soybean and poplar, and there are more than 4 homologous pairs between every two of these species (Fig. 3), indicating a conserved relationship within SHMTs in A. thaliana, soybean and poplar during evolution (Rajinikanth, Harding & Tsai, 2007). Further analysis of molecular weight, subcellular localization, motif composition and distribution of SlSHMTs shows that the SlSHMT members within each subgroup of ClassI-1, ClassI-2 and ClassII-1 are quite similar (Tables 1, 3 and Fig. 4). For example, the length of SlSHMT proteins in the same subgroup differs by no more than three amino acids (Table 1), suggesting that gene duplication events may occur at the species or lineage levels, and the proteins in each subgroup may exhibit a redundant function in plant growth and development (Wu et al., 2016).
In the current study, a part of the tomato SHMTs distribute in mitochondria, chloroplasts, and cytosol (Table 3), similar with that in A. thaliana and soybean (Bauwe & Kolukisaoglu, 2003; Lakhssassi et al., 2019). In plants, the serine produced by SHMT and GDC in mitochondria enters the cytosol and then serves as the primary one carbon donor to generate nucleotide, vitamin and amino acid, which is directed by the cytosolic SHMT (Rajinikanth, Harding & Tsai, 2007). The chloroplastic SHMT also presents a vital role for photoreception and for the biosynthesis concerning one carbon metabolism (Zhang et al., 2010). Thus, tomato SHMTs may also provide outstanding contributions on plant cellular metabolisms, which could be partially certified by the extensive expressions in root, stem, leaf and flower in tomato seedlings (Figs. 8, 9). Moreover, many mitochondrial SHMTs, such as AtSHMT1, OsSHMT1 and GmSHMT08, have been shown to participate in defense activities against biotic and abiotic stresses (Moreno, Martín & Castresana, 2005; Wang et al., 2015; Lakhssassi et al., 2020). We speculate that the mitochondrion-localized SlSHMT2 and SlSHMT3 may have similar functions with them, which was confirmed by a recent study in tomato (Ahammed et al., 2018). Unpredictably, the class II-1 members SlSHMT4 and SlSHMT7 are shown to localize in cytoskeletons (Table 3), which has not been reported in other plants so far, and their accurate function remains to be discovered. Meanwhile, the tomato SHMT members rarely distribute in nucleus.
In plants, SHMT genes have been identified in many tissues (McClung et al., 2000; Zhang et al., 2010; Lakhssassi et al., 2019), but the expression levels vary according to different SHMT members in different growth and development stages. In A. thaliana, AtSHMT1 mainly distributes in leaf, stem and flower, and AtSHMT1 mutation induces aberrant regulation of cell death and enhances susceptibility to pathogens and abiotic stress (Moreno, Martín & Castresana, 2005). AtSHMT3 is mainly expressed in germinated seed, which potentially regulates metabolic fluxes in plastids (Zhang et al., 2010), while AtSHMT4 transcripts accumulation is restricted to root in young seedlings (McClung et al., 2000). BvSHMTa is expressed in the leaf and root in sugar beet, and its expression level could be strengthened by salt stress (Kito et al., 2017). In soybean, most of the GmSHMT genes present a ubiquitous expression in all the tissues with some exceptions, while some of these members display irredundant responses during SCN infection (Lakhssassi et al., 2019). In the present study, all the tomato SHMT genes could be expressed in root, stem, leaf and flower, except for a very low expression of SlSHMT2 in root, indicating that the enzyme activities of tomato SHMTs are essential for the growth and development of various growth stages. In addition, SlSHMT2 transcripts is the most abundant ones in all the analyzed tissues, and SlSHMT2 is specifically expressed in leaf, which is similar with the tissue localization of AtSHMT1 and OsSHMT1 (Moreno, Martín & Castresana, 2005; Wang et al., 2015). Thus, the molecular function of SlSHMT2 in green leaves may be conserved in tomato, rice and A. thaliana. Further, SlSHMT1, SlSHMT3, SlSHMT5 and SlSHMT6 exhibit the most abundant expression levels in flower, while expressions of SlSHMT4 and SlSHMT7 are relatively higher in stem in reproductive growth stage. Thus, SHMT members may function redundantly in leave, stem and flower to support the metabolism activities concerning fruit development, and the specific mechanism needs to be explored in detail. Simultaneously, we detected a mildly and redundantly expression of tomato SHMT genes in the fruit tissues, which was divergent with the reported expressions of SHMT members in seeds (Zhang et al., 2010; Lakhssassi et al., 2019), suggesting a different regulation mechanism of SHMTs in fruits and the mature seeds.
The cis-acting elements in the promoter region of genes have long been studied as the binding sites of specific transcription factors, which help to modulate the initiation of genes transcription (Carrier et al., 2020). Up to now, many of the cis-acting elements have been well characterized and classified into different groups (Hadiarto & Tran, 2011). We have identified a number of cis-acting elements in the promoter regions of tomato SHMT genes, including abiotic stresses-related LTR, ARE, MBS and TC-rich repeats; hormones-related CGTCA-motif, P-box, TCA-element, TGACG-motif, TGA-element and ABRE; and light-related Circadian and G-box (Table 5). The widely present of these cis-acting elements suggests a crucial role of SlSHMT members in response to abiotic stresses and hormone stimulations. For example, expression of SlSHMT5 is up-regulated under PEG and ABA treatments, which was consistent with the distributions of drought and ABA response elements (MBS, ABRE) in SlSHMT5 (Fig. 10). However, some conflicting results also appeared in our further study. Both SlSHMT4 and SlSHMT7 contain AREB binding site in their promoter regions (Fig. 7), but neither of them responds to ABA treatment. SlSHMT3 has been found to mediate plant basal defence against Pseudomonas syringae in a salicylic acid (SA)-dependent manner (Ahammed et al., 2018), but it does not have a SA response element (TCA-element). This phenomenon was also found in other gene families previously (Gao et al., 2021). In addition, the restricted analyzed tissue, growth stage and treated period may influence the judgment of their relationships. Thus, more explorations are yet to be done to uncover the detailed relationships between different responses of SlSHMTs and the corresponding cis-acting elements.
As an important house-keeping regulator of plant growth and development, the response of SHMT gene family members to adverse environmental conditions has been reported in many species. In our study, SlSHMT genes could respond to variety kinds of circumstance stimuli. In detail, SlSHMT1 gene expression is increased by UV, cold, salt, H2O2 and PEG stress. Expression level of SlSHMT5 is significantly enhanced by UV, salt, H2O2 and PEG treatment. The alleviation of salt stress by SHMT members have been discovered in many higher plants as well as in the procaryotic organism cyanobacterium. In A. thaliana, a ubiquitin-specific protease UBP16 interacts with AtSHMT1 to stabilize it, and they cooperate to increase Na+/H+ antiport activity under salt stress (Zhou et al., 2012). In rice, the overexpression level of OsSHMT3 was found to be up-regulated in a salt-tolerant line ‘CSR27′(Mishra et al., 2016; Mishra et al., 2019). Overexpression of ApSHMT (a gene isolated from cyanobacterium) in Escherichia coli. could increase the accumulation of glycine betaine, which leads to an intensive tolerance of the transgenic cells (Srivastava et al., 2011; Waditee-Sirisattha et al., 2012). Plant SHMTs were also found to relieve damages of chilling and drought through maintaining redox homeostasis and regulating stomatal closure (Liu et al., 2019a; Liu et al., 2019b; Fang et al., 2020). Interestingly, heat stress reduces the expression levels of all SlSHMTs except for SlSHMT1. Moreover, SlSHMT2 is down-regulated by all the treatments analyzed. The similar phenomenon was also reported in pitaya (Hylocereus undatus), A. thaliana and rice (Xu & Huang, 2010; Barkla et al., 2014; Fan et al., 2014; Chen et al., 2020a; Chen et al., 2020b). Plant SHMTs influence gene replication through regulating nucleotide synthesis, and the decrease of SHMT activities under abiotic stresses may reduce the probability of inaccurate replication events (Xu & Huang, 2010). The responses of other SlSHMT members varied in different conditions, indicating a redundant function of tomato SHMTs in resisting environmental stresses. Also, the diverse responses to abiotic stresses may due to the evolutionary discordance within SHMT gene members in different species.
Conclusions
In general, seven SHMT genes in tomato genome were identified in the present study. Physicochemical properties, gene and protein structures, and evolutionary relationships may predict structure and function similarity as well as divergence among tomato SHMTs and their orthologs in other dicotyledonous members. What’s more, subcellular location and tissue-specific analysis shows that SlSHMT members conduct a pivotal house-keeping function through regulating cellular metabolisms during plant growth and development. Last but not the least, we provide evidence that tomato SHMTs participate in alleviating various abiotic stresses and hormone responses. In summary, this study attempts to support further studies on the involvement of the SlSHMT gene family in growth regulation and stress response in tomato, and provide theoretical foundation for further exploration of the function of plant SHMT members.
Supplemental Information
Funding Statement
This work was supported by the National Natural Science Foundation of China (Nos. 32072559, 31860568, 31560563, and 31160398); the Key Research and Development Program of Gansu Province, China (No. 21YF5WA096); the Research Fund of Higher Education of Gansu, China (No. 2018C-14 and 2019B-082); and the Natural Science Foundation of Gansu Province, China (Nos. 1606RJZA073, 1606RJZA077, and 1606RJYA252). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Zesheng Liu conceived and designed the experiments, performed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Xuejuan Pan performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Chunlei Wang conceived and designed the experiments, analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Fahong Yun and Fujin Ye performed the experiments, prepared figures and/or tables, and approved the final draft.
Dengjing Huang analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Yandong Yao, Rong Gao and Xingjuan Liu analyzed the data, prepared figures and/or tables, and approved the final draft.
Weibiao Liao conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplementary Files.
References
- Agrawal et al. (2004).Agrawal S, Kumar A, Srivastava V, Mishra BN. Cloning, expression, activity and folding studies of serine hydroxymethyltransferase: a target enzyme for cancer chemotherapy. Journal of Molecular Microbiology and Biotechnology. 2004;6(2):67–75. doi: 10.1159/000076737. [DOI] [PubMed] [Google Scholar]
- Ahammed et al. (2018).Ahammed GJ, Li X, Zhang G, Zhang H, Shi J, Pan C, Yu J, Shi K. Tomato photorespiratory glycolate-oxidase-derived H2O2 production contributes to basal defence against Pseudomonas syringae. Plant, Cell & Environment. 2018;41(5):1126–1138. doi: 10.1111/pce.12932. [DOI] [PubMed] [Google Scholar]
- Barkla et al. (2014).Barkla BJ, Vera-Estrella R, Miranda-Vergara MC, Pantoja O. Quantitative proteomics of heavy metal exposure in Arabidopsis thaliana reveals alterations in one-carbon metabolism enzymes upon exposure to zinc. Journal of Proteomics. 2014;111:128–138. doi: 10.1016/j.jprot.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Batool et al. (2020).Batool N, Ko KS, Chaurasia AK, Kim KK. Functional identification of serine hydroxymethyltransferase as a key gene involved in lysostaphin resistance and virulence potential of Staphylococcus aureus strains. International Journal of Molecular Sciences. 2020;21(23):9135. doi: 10.3390/ijms21239135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwe & Kolukisaoglu (2003).Bauwe H, Kolukisaoglu U. Genetic manipulation of glycine decarboxylation. Journal of Experimental Botany. 2003;54(387):1523–1535. doi: 10.1093/jxb/erg171. [DOI] [PubMed] [Google Scholar]
- Carrier et al. (2020).Carrier MC, Ng Kwan Lim E, Jeannotte G, Massé E. Trans-acting effectors versus RNA cis-elements: a tightly knit regulatory mesh. Frontiers in Microbiology. 2020;11:609237. doi: 10.3389/fmicb.2020.609237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2020a).Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant. 2020a;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2020b).Chen J, Huang XY, Salt DE, Zhao FJ. Mutation in OsCADT1 enhances cadmium tolerance and enriches selenium in rice grain. New Phytologist. 2020b;226(3):838–850. doi: 10.1111/nph.16404. [DOI] [PubMed] [Google Scholar]
- Dan et al. (2021).Dan Y, Niu Y, Wang C, Yan M, Liao W. Genome-wide identification andexpression analysis of the trehalose-6-phosphate synthase (TPS) gene family incucumber (Cucumis sativus L.) PeerJ. 2021;9:e11398. doi: 10.7717/peerj.11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escande-Beillard et al. (2020).Escande-Beillard N, Loh A, Saleem SN, Kanata K, Hashimoto Y, Altunoglu U, Metoska A, Grandjean J, Ng FM, Pomp O, Baburajendran N, Wong J, Hill J, Beillard E, Cozzone P, Zaki M, Kayserili H, Hamada H, Shiratori H, Reversade B. Loss of PYCR2 causes neurodegeneration by increasing cerebral glycine levels via SHMT2. Neuron. 2020;107(1):82–94. doi: 10.1016/j.neuron.2020.03.028. [DOI] [PubMed] [Google Scholar]
- Fan et al. (2014).Fan QJ, Yan FX, Qiao G, Zhang BX, Wen XP. Identification of differentially-expressed genes potentially implicated in drought response in pitaya (Hylocereus undatus) by suppression subtractive hybridization and cDNA microarray analysis. Gene. 2014;533(1):322–331. doi: 10.1016/j.gene.2013.08.098. [DOI] [PubMed] [Google Scholar]
- Fang et al. (2020).Fang C, Zhang P, Li L, Yang L, Mu D, Yan X, Li Z, Lin W. Serine hydroxymethyltransferase localised in the endoplasmic reticulum plays a role in scavenging H2O2 to enhance rice chilling tolerance. BMC Plant Biology. 2020;20(1):236. doi: 10.1186/s12870-020-02446-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao et al. (2021).Gao R, Luo Y, Yun F, Wu X, Wang P, Liao W. Genome-wide identification, expression profile, and alternative splicing analysis of CAMTA family genes in cucumber (Cucumis sativus L.) Agronomy. 2021;11:1827. doi: 10.3390/agronomy11091827. [DOI] [Google Scholar]
- Goodstein et al. (2012).Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo et al. (2021).Guo L, Yu H, Kharbach M, Zhang W, Wang J, Niu W. Biochar improves soil-tomato plant, tomato production, and economic benefits under reduced nitrogen application in northwestern China. Plants. 2021;10(4):759. doi: 10.3390/plants10040759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadiarto & Tran (2011).Hadiarto T, Tran LS. Progress studies of drought-responsive genes in rice. Plant Cell Reports. 2011;30(3):297–310. doi: 10.1007/s00299-010-0956-z. [DOI] [PubMed] [Google Scholar]
- Hourton-Cabassa et al. (1998).Hourton-Cabassa C, Ambard-Bretteville F, Moreau F, Davy de Virville J, Rémy R, Francs-Small CC. Stress induction of mitochondrial formate dehydrogenase in potato leaves. Plant Physiology. 1998;116(2):627–635. doi: 10.1104/pp.116.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2021).Hu J, Cai J, Umme A, Chen Y, Xu T, Kang H. Unique features of mRNA m6A methylomes during expansion of tomato (Solanum lycopersicum) fruits. Plant Physiology. 2021:kiab509. doi: 10.1093/plphys/kiab509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji et al. (2012).Ji K, Wang Y, Sun W, Lou Q, Mei H, Shen S, Chen H. Drought-responsive mechanisms in rice genotypes with contrasting drought tolerance during reproductive stage. Journal of Plant Physiology. 2012;169(4):336–344. doi: 10.1016/j.jplph.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Kito et al. (2017).Kito K, Tsutsumi K, Rai V, Theerawitaya C, Cha-Um S, Yamada-Kato N, Sakakibara S, Tanaka Y, Takabe T. Isolation and functional characterization of 3-phosphoglycerate dehydrogenase involved in salt responses in sugar beet. Protoplasma. 2017;254(6):2305–2313. doi: 10.1007/s00709-017-1127-7. [DOI] [PubMed] [Google Scholar]
- Kumar, Stecher & Tamura (2016).Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhssassi et al. (2019).Lakhssassi N, Patil G, Piya S, Zhou Z, Baharlouei A, Kassem MA, Lightfoot DA, Hewezi T, Barakat A, Nguyen HT, Meksem K. Genome reorganization of the GmSHMT gene family in soybean showed a lack of functional redundancy in resistance to soybean cyst nematode. Scientific Reports. 2019;9(1):1506. doi: 10.1038/s41598-018-37815-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhssassi et al. (2020).Lakhssassi N, Piya S, Knizia D, El Baze A, Cullen MA, Meksem J, Lakhssassi A, Hewezi T, Meksem K. Mutations at the serine hydroxymethyltransferase impact its interaction with a soluble NSF attachment protein and a pathogenesis-related protein in soybean. Vaccine. 2020;8(3):349. doi: 10.3390/vaccines8030349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li & Cheng (2020).Li B, Cheng Y. Analysis of SHMT gene family and production of PtrSHMT9 mutant in Populus trichocarpa. Bulletin of Botanical Research. 2020;40(6):906–912. doi: 10.7525/j.issn.1673-5102.2020.06.013. [DOI] [Google Scholar]
- Li et al. (2021).Li N, Xu R, Wang B, Wang J, Huang S, Yu Q, Gao J. Genome-wide identification and evolutionary analysis of the SRO gene family in tomato. Frontiers in Genetics. 2021;12:753638. doi: 10.3389/fgene.2021.753638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2019a).Liu Y, Mauve C, Lamothe-Sibold M, Guérard F, Glab N, Hodges M, Jossier M. Photorespiratory serine hydroxymethyltransferase 1 activity impacts abiotic stress tolerance and stomatal closure. Plant, Cell & Environment. 2019a;42(9):2567–2583. doi: 10.1111/pce.13595. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2019b).Liu Y, Wen H, Qi X, Zhang X, Zhang K, Fan H, Tian Y, Hu Y, Li Y. Genome-wide identification of the Na+/H+ exchanger gene family in Lateolabrax maculatus and its involvement in salinity regulation. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics. 2019b;29:286–298. doi: 10.1016/j.cbd.2019.01.001. [DOI] [PubMed] [Google Scholar]
- McClung et al. (2000).McClung CR, Hsu M, Painter JE, Gagne JM, Karlsberg SD, Salomé PA. Integrated temporal regulation of the photorespiratory pathway, Circadian regulation of two Arabidopsis genes encoding serine hydroxymethyltransferase. Plant Physiology. 2000;123(1):381–392. doi: 10.1104/pp.123.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra et al. (2019).Mishra P, Jain A, Takabe T, Tanaka Y, Negi M, Singh N, Jain N, Mishra V, Maniraj R, Krishnamurthy SL, Sreevathsa R, Singh NK, Rai V. Heterologous expression of serine hydroxymethyltransferase-3 from rice confers tolerance to salinity stress in E. coli and Arabidopsis. Frontiers in Plant Science. 2019;10:217. doi: 10.3389/fpls.2019.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra et al. (2016).Mishra P, Mishra V, Takabe T, Rai V, Singh NK. Elucidation of salt-tolerance metabolic pathways in contrasting rice genotypes and their segregating progenies. Plant Cell Reports. 2016;35(6):1273–1286. doi: 10.1007/s00299-016-1959-1. [DOI] [PubMed] [Google Scholar]
- Moreno, Martín & Castresana (2005).Moreno JI, Martín R, Castresana C. Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. The Plant Journal. 2005;41(3):451–463. doi: 10.1111/j.1365-313X.2004.02311.x. [DOI] [PubMed] [Google Scholar]
- Nogués et al. (2020).Nogués I, Tramonti A, Angelaccio S, Ruszkowski M, Sekula B, Contestabile R. Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance. International Journal of Biological Macromolecules. 2020;159:517–529. doi: 10.1016/j.ijbiomac.2020.05.081. [DOI] [PubMed] [Google Scholar]
- Pieroth et al. (2018).Pieroth R, Paver S, Day S, Lammersfeld C. Folate and its impact on cancer risk. Current Nutrition Reports. 2018;7(3):70–84. doi: 10.1007/s13668-018-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajinikanth, Harding & Tsai (2007).Rajinikanth M, Harding SA, Tsai CJ. The glycine decarboxylase complex multienzyme family in Populus. Journal of Experimental Botany. 2007;58(7):1761–1770. doi: 10.1093/jxb/erm034. [DOI] [PubMed] [Google Scholar]
- Roth, Roepenack-Lahaye & Clemens (2006).Roth U, Roepenack-Lahaye Evon, Clemens S. Proteome changes in Arabidopsis thaliana roots upon exposure to Cd2+ Journal of Experimental Botany. 2006;57(15):4003–4013. doi: 10.1093/jxb/erl170. [DOI] [PubMed] [Google Scholar]
- Schwarte & Bauwe (2007).Schwarte S, Bauwe H. Identification of the photorespiratory 2-phosphoglycolate phosphatase, PGLP1, in Arabidopsis. Plant Physiology. 2007;144(3):1580–1586. doi: 10.1104/pp.107.099192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemin (1946).Shemin D. The biological conversion of 1-serine to glycine. The Journal of Biological Chemistry. 1946;162:297–307. [PubMed] [Google Scholar]
- Shen et al. (2013).Shen T, Rui B, Zhou H, Zhang X, Yi Y, Wen H, Zheng H, Wu , J, Shi Y. Metabolic flux ratio analysis and multi-objective optimization revealed a globally conserved and coordinated metabolic response of E. coli to paraquat-induced oxidative stress. Molecular bioSystems. 2013;9(1):121–132. doi: 10.1039/c2mb25285f. [DOI] [PubMed] [Google Scholar]
- Sodolescu et al. (2018).Sodolescu A, Dian C, Terradot L, Bouzhir-Sima L, Lestini R, Myllykallio H, Skouloubris S, Liebl U. Structural and functional insight into serine hydroxymethyltransferase from Helicobacter pylori. PLOS ONE. 2018;13(12):e208850. doi: 10.1371/journal.pone.0208850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song et al. (2019).Song Z, Pan F, Lou X, Wang D, Yang C, Zhang B, Zhang H. Genome-wide identification and characterization of Hsp70 gene family in Nicotiana tabacum. Molecular Biology Reports. 2019;46(2):1941–1954. doi: 10.1007/s11033-019-04644-7. [DOI] [PubMed] [Google Scholar]
- Srivastava et al. (2011).Srivastava AK, Alexova R, Jeon YJ, Kohli GS, Neilan BA. Assessment of salinity-induced photorespiratory glycolate metabolism in Anabaena sp. PCC 7120. Microbiology. 2011;157(Pt 3):911–917. doi: 10.1099/mic.0.045682-0. [DOI] [PubMed] [Google Scholar]
- Troesch, Weber & Mohajeri (2016).Troesch B, Weber P, Mohajeri MH. Potential links between impaired one-carbon metabolism due to polymorphisms, inadequate B-vitamin status, and the development of Alzheimer’s disease. Nutrients. 2016;8(12):803. doi: 10.3390/nu8120803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner et al. (1992).Turner SR, Ireland R, Morgan C, Rawsthorne S. Identification and localization of multiple forms of serine hydroxymethyltransferase in pea (Pisum sativum) and characterization of a cDNA encoding a mitochondrial isoform. The Journal of biological chemistry. 1992;267(19):13528–13534. [PubMed] [Google Scholar]
- Waditee-Sirisattha et al. (2012).Waditee-Sirisattha R, Sittipol D, Tanaka Y, Takabe T. verexpression of serine hydroxymethyltransferase from halotolerant cyanobacterium in Escherichia coli results in increased accumulation of choline precursors and enhanced salinity tolerance. FEMS Microbiology Letters. 2012;333(1):46–53. doi: 10.1111/j.1574-6968.2012.02597.x. [DOI] [PubMed] [Google Scholar]
- Waditee-Sirisattha et al. (2017).Waditee-Sirisattha R, Kageyama H, Tanaka Y, Fukaya M, Takabe T. Overexpression of halophilic serine hydroxymethyltransferase in fresh water cyanobacterium Synechococcus elongatus PCC7942 results in increased enzyme activities of serine biosynthetic pathways and enhanced salinity tolerance. Archives of Microbiology. 2017;199(1):29–35. doi: 10.1007/s00203-016-1271-z. [DOI] [PubMed] [Google Scholar]
- Wang & Liu (2021).Wang X, Liu J. Tomato anomalies detection in greenhouse scenarios based on YOLO-Dense. Frontiers in Plant Science. 2021;12:634103. doi: 10.3389/fpls.2021.634103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2015).Wang D, Liu H, Li S, Zhai G, Shao J, Tao Y. Characterization and molecular cloning of a serine hydroxymethyltransferase 1 (OsSHM1) in rice. Journal of Integrative Plant Biology. 2015;57(9):745–756. doi: 10.1111/jipb.12336. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2021).Wang A, Shu X, Jing X, Jiao C, Chen L, Zhang J, Ma L, Jiang Y, Yamamoto N, Li S, Deng Q, Wang S, Zhu J, Liang Y, Zou T, Liu H, Wang L, Huang Y, Li P, Zheng A. Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome-wide association study. Plant Biotechnology Journal. 2021;19(8):1553–1566. doi: 10.1111/pbi.13569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu et al. (2016).Wu XY, Zhou GC, Chen YX, Wu P, Liu LW, Ma FF, Wu M, Liu CC, Zeng YJ, Chu AE, Hang YY, Chen JQ, Wang B. Soybean cyst nematode resistance emerged via artificial selection of duplicated serine hydroxymethyltransferase genes. Frontiers in Plant Science. 2016;7:998. doi: 10.3389/fpls.2016.00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie et al. (2018).Xie T, Chen C, Li C, Liu J, Liu C, He Y. Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics. 2018;19(1):490. doi: 10.1186/s12864-018-4880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu & Huang (2010).Xu C, Huang B. Differential proteomic response to heat stress in thermal Agrostis scabra and heat-sensitive Agrostis stolonifera. Physiologia Plantarum. 2010;139(2):192–204. doi: 10.1111/j.1399-3054.2010.01357.x. [DOI] [PubMed] [Google Scholar]
- Ye et al. (2020).Ye J, Chen W, Feng L, Liu G, Wang Y, Li H, Ye Z, Zhang Y. The chaperonin 60 protein SlCpn60 α1 modulates photosynthesis and photorespiration in tomato. Journal of Experimental Botany. 2020;71(22):7224–7240. doi: 10.1093/jxb/eraa418. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2010).Zhang Y, Sun K, Sandoval FJ, Santiago K, Roje S. One-carbon metabolism in plants: characterization of a plastid serine hydroxymethyltransferase. Biochemical Journal. 2010;430(1):97–105. doi: 10.1042/BJ20100566. [DOI] [PubMed] [Google Scholar]
- Zhou et al. (2012).Zhou H, Zhao J, Yang Y, Chen C, Liu Y, Jin X, Chen L, Li X, Deng XW, Schumaker KS, Guo Y. Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na(+)/H(+) antiport activity and serine hydroxymethyltransferase stability. Plant Cell. 2012;24(12):5106–5122. doi: 10.1105/tpc.112.106393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplementary Files.