Abstract
Background
Integrin α5 (ITGA5) was involved in a variety of cancers. However, the role of ITGA5 in laryngeal squamous cell carcinoma (LSCC) remains unknown.
Methods
The expression of ITGA5 and the corresponding clinicopathological parameters of LSCC patients the TCGA database. Five datasets (GSE51985, GSE59102, GSE84957, GSE27020, and GSE65858) were downloaded from the GEO database as validation sets. Kaplan–Meier plotter, Cox regression analysis, and nomogram were performed to determine the prognostic value of ITGA5 in LSCC. GO, KEGG, and GSEA were used to explore the underlying biological functions of ITGA5 in LSCC. The algorithms ESTIMATE and CIBERSORT were adopted to evaluate the association between ITGA5 and the infiltration of the immune cells. The algorithm pRRophetic was used to estimate the response to chemotherapeutic drugs.
Results
The expression of ITGA5 was higher in the LSCC samples and linked to poor overall survival and recurrence‐free survival. Further, the Cox regression analysis confirmed that high expression of ITGA5 was an independent unfavorable prognostic factor. The predictive performance of nomogram based on the expression of ITGA5 was accurate and practical. The functional enrichment analysis confirmed that ITGA5 was related to the construction of the components and structures of the extracellular matrix. Finally, patients with high ITGA5 expression were more likely to benefit from docetaxel and gemcitabine.
Conclusion
The expression of ITGA5 was elevated in the LSCC and was a predictor for prognosis and chemotherapeutic response in LSCC patients.
Keywords: chemotherapy, ITGA5, laryngeal squamous cell carcinoma, nomogram, prognosis
Integrin α5 (ITGA5) was involved in a variety of cancers. However, the role of ITGA5 in laryngeal squamous cell carcinoma (LSCC) remains unknown. TCGA database was used to download the expression of ITGA5 and the corresponding clinicopathological parameters of LSCC patients. The GEO database was used to download five datasets (GSE51985, GSE59102, GSE84957, GSE27020, and GSE65858) as validation sets. Kaplan‐Meier plotter, Cox regression analysis, and nomogram were performed to determine the prognostic value of ITGA5 in LSCC. Gene ontology, Kyoto Encyclopedia of Genes and Genomes, and Gene Set Enrichment Analysis were used to explore the underlying biological functions of ITGA5 in LSCC. The algorithms ESTIMATE and CIBERSORT were adopted to evaluate the association between ITGA5 and the infiltration of the immune cells. The algorithm pRRophetic was used to estimate the response to chemotherapeutic drugs. Our results showed that the expression of ITGA5 was higher in the LSCC samples and linked to poor overall survival and recurrence‐free survival. Further, the Cox regression analysis confirmed that high expression of ITGA5 was an independent unfavorable prognostic factor. The predictive performance of nomogram based on the expression of ITGA5 was accurate and practical. The functional enrichment analysis confirmed that ITGA5 was related to the construction of the components and structures of the extracellular matrix. Finally, patients with high ITGA5 expression were more likely to benefit from docetaxel and gemcitabine. The expression of ITGA5 was elevated in LSCC and was a predictor for prognosis and chemotherapeutic response in LSCC patients.
1. INTRODUCTION
Laryngeal cancer is one of the most common cancers in the respiratory system. Recent data reported that, in 2021, an estimated 12,620 new cases were diagnosed with about 3770 patients dying from laryngeal cancer in the United States. 1 Based on the pathological cases, approximately 98% of laryngeal cancer cases are laryngeal squamous cell carcinoma (LSCC). Although progress has been made in the comprehensive management of patients with LSCC, the 5‐year overall survival (OS) rate had reduced over the past 50 years, owing to the insidious symptoms and lack of effective diagnostic methods for early stages. 2 Approximately 60% of the patients with LSCC are diagnosed at an advanced stage or even with metastasis of the lymph node, and have had a poor prognosis. 3 Recently, more and more evidence has demonstrated that the prognosis and chemotherapeutic efficacy of patients with tumor was largely affected by the infiltration of the immune cells. 4 , 5 , 6 , 7 , 8 Therefore, the identification of novel potential biomarkers and their correlation with immune infiltration will potentially improve outcomes and direct individualized treatment for patients with LSCC.
Integrins, consisting of α and β subunits, are heterodimeric transmembrane proteins that function as surface adhesion receptors in different cells or cells and extracellular matrix communication. 9 Integrin α5 (ITGA5) is a member of the integrin alpha chain family and is located on chromosome 12q13.13 in Homo sapiens. 10 ITGA5 mainly recognizes its specific ligand through the formation of an α5β1 heterodimer and combines with integrin β1 (TGFβ1). 11 Increasing evidence has proved that ITGA5 is highly expressed in multiple cancers and is related to the progressions of tumor, such as cell proliferation, migration, and invasion. For instance, ITGA5 was upregulated in pancreatic ductal adenocarcinoma. Silencing ITGA5 could suppress the differentiation of human pancreatic stellate cells and reduce desmoplasia. 9 In oral cancer, ITGA5 facilitates cell growth, migration, and invasion by epithelial‐mesenchymal transition. 12 According to Yu et al., 11 ITGA5 was overexpressed in colorectal cancer and promoted cell proliferation and tumorigenesis and reduced cell apoptosis. Additionally, ITGA5 was shown to maintain chemotherapeutic resistance 9 , 13 and cancer cell stemness. 14 Russano et al. 15 reported that ITGA5 acts as a mediator of breast‐to‐bone metastasis and might improve the efficacy of volociximab/M200 in patients with breast cancer along with bone metastasis. These findings implied that ITGA5 may be a potential therapeutic target for tumor angiogenesis, lymphangiogenesis, and metastasis. However, the expression and potential functions of ITGA5 in the progression of LSCC remain largely unknown.
In this study, we explored the expression and prognostic value of ITGA5 in patients with LSCC, and then performed functional enrichment analysis. More importantly, we revealed the relationship between the expression of ITGA5 and tumor immune microenvironment (TME) as well as the infiltration of the immune cells in LSCC. Our results revealed that ITGA5 was highly expressed in LSCC and might appear as an independent prognostic factor and therapeutic target for LSCC.
2. MATERIALS AND METHODS
2.1. Data preparation
The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) was used to download the ITGA5 mRNA expression (FPKM form) and the corresponding clinicopathological parameters of 111 LSCC tissues and 12 normal tissues. GSE51985 (including 10 LSCC tissues and 10 adjacent normal tissues), GSE59102 (including 29 LSCC tissues and 13 normal tissues), and GSE84957 (including 9 LSCC tissues and 9 adjacent normal tissues), and the corresponding platform annotation files were downloaded from the Gene Expression Omnibus (GEO) database to validate the expression levels of ITGA5 in LSCC. Additionally, although there were no normal tissues in the GSE65858 (including 48 LSCC samples with OS and recurrence‐free survival [RFS] information) and GSE27020 (including 109 LSCC samples with RFS information), these two datasets with survival data were also downloaded to validate the prognostic value of ITGA5 in LSCC. The clinicopathological features of the patients in this study are listed in Table 1.
TABLE 1.
Table listing the clinical characteristics of patients with LSCC in this study
| Characteristics | TCGA | GSE65858 | GSE27020 | GSE51985 | GSE84957 | GSE59102 |
|---|---|---|---|---|---|---|
| Gender | ||||||
| Female | 20 | 5 | — | 0 | 0 | 1 |
| Male | 91 | 43 | — | 10 | 9 | 28 |
| Age | ||||||
| ≤60 | 47 | 18 | 43 | 6 | — | — |
| >60 | 64 | 30 | 66 | 4 | — | — |
| Smoking history | ||||||
| Yes | 102 | 41 | — | — | — | — |
| No | 6 | 7 | — | — | — | — |
| Unknown | 3 | |||||
| Alcohol history | ||||||
| Yes | 70 | 43 | — | — | — | — |
| No | 39 | 5 | — | — | — | — |
| Unknown | 2 | |||||
| Histologic grade | ||||||
| G1 | 8 | — | 42 | — | — | — |
| G2‐3 | 99 | — | 65 | — | — | — |
| Unknown | 4 | 2 | ||||
| Pathologic T | ||||||
| T1‐2 | 19 | 20 | — | 3 | — | — |
| T3‐4 | 79 | 28 | — | 7 | — | — |
| Unknown | 13 | |||||
| Pathologic N | ||||||
| N0 | 39 | 29 | — | 5 | — | — |
| N1‐3 | 53 | 19 | — | 5 | — | — |
| Unknown | 19 | |||||
| Pathologic M | ||||||
| M0 | 40 | 46 | — | 10 | — | — |
| M1 | 9 | 2 | — | 0 | — | — |
| Unknown | 62 | |||||
| Clinical stage | ||||||
| Stage I‐II | 13 | 17 | — | 3 | 0 | 14 |
| Stage III‐IV | 85 | 31 | — | 7 | 9 | 15 |
| Unknown | 13 | |||||
| Radiation therapy | ||||||
| Yes | 65 | — | — | — | — | — |
| No | 23 | — | — | — | — | — |
| Unknown | 23 | |||||
| Targeted therapy | ||||||
| Yes | 31 | — | — | — | — | — |
| No | 51 | — | — | — | — | — |
| Unknown | 29 | |||||
| Survival status | ||||||
| Dead | 50 | 13 | — | — | — | — |
| Alive | 61 | 35 | — | — | — | — |
| Recurrence | ||||||
| Yes | 42 | 19 | 34 | — | — | — |
| No | 69 | 29 | 75 | — | — | — |
2.2. The expression of ITGA5 and its association with the clinical characteristics of patients with LSCC
“Limma” R package was applied to estimate the expression of ITGA5 between LSCC and non‐tumor samples in the TCGA cohort. GSE51985, GSE59102, and GSE84957 were the validation cohorts. Further, the association between the expression of ITGA5 and the clinical characteristics was also explored in patients with LSCC.
2.3. Prognostic analysis of ITGA5 in patients with LSCC
According to the optimal cut‐off point by routing algorithms, the OS and the RFS curves were plotted by the Kaplan–Meier (K‐M) method, and the difference in the survival rates between high‐ITGA5 and low‐ITGA5 groups was assessed using the log‐rank test. Univariate and multivariate Cox regression analyses were used to evaluate the prognostic value of ITGA5 in LSCC. Further, a nomogram was constructed based on the expression of ITGA5 and the clinical features to predict the survival probability. The 1‐, 3‐, and 5‐years calibration curves and receiver operating characteristic (ROC) curves were used to assess the prediction performance of the nomogram.
2.4. Functional enrichment analysis
Patients with LSCC from the TCGA were separated into the high‐ITGA5 and the low‐ITGA5 groups according to their median value. The Gene Set Enrichment Analysis (GSEA, version 4.1.0) was performed in our previous study. 16 The c2.cp.kegg.v7.2.symbols.gmt was utilized as reference gene sets. Pathways with false discovery rate (FDR) p < 0.05 were considered statistically enriched. Differentially expressed genes (DEGs) between the high‐ITGA5 and the low‐ITGA5 groups were identified with FDR p < 0.05 and |Log2FC| > 1. The Gene ontology (GO) analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed based on the DEGs to explore the ITGA5‐related functional pathways in LSCC.
2.5. TME and the infiltration of the immune cells
The ESTIMATE algorithm was used to calculate the immuneScore, stromalScore, and estimate score of the LSCC samples. 17 The CIBERSORT algorithm was adopted to quantify the absolute abundance of 22 tumor‐infiltrating immune cell types in the LSCC samples. 18 Then, the difference in the TME score and the infiltration of the immune cells between the high‐ITGA5 and the low‐ITGA5 groups were analyzed.
2.6. Chemotherapeutic drug sensitivity analysis
In this study, we demonstrated whether ITGA5 could influence the clinical response to treatment and could be considered as a potential biomarker for drug screening. The half‐maximum inhibitory concentration (IC50) was estimated using the “pRRophetic” package 19 to predict the chemotherapeutic response of four commonly used chemotherapeutic drugs (paclitaxel, gemcitabine, docetaxel, and cisplatin) in the high‐ITGA5 and the low‐ITGA5 groups.
2.7. Statistical analysis
The R statistical language (version 4.1.0) was used for all statistical analysis and visualizations. To compare the expression of ITGA5 among different groups, we performed Wilcoxon signed rank test or Chi square test where required. Spearman correlation analysis was used to examine the correlation analysis. Unless specified otherwise, p < 0.05 was considered statistically significant.
3. RESULTS
3.1. Expression of ITGA5 in LSCC
By data mining using the TCGA database, ITGA5 was determined to be elevated in LSCC compared with the normal tissues (Figure 1A, p < 0.001). Further, the differential expression of ITGA5 in LSCC was verified based on three independent datasets from the GEO database, which revealed that the expression of ITGA5 in LSCC was significantly higher than in the normal tissues (GSE51985 [Figure 1B, p = 0.011], GSE59102 [Figure 1C, p < 0.001], and GSE84957 [Figure 1D, p = 0.001]). Subsequently, we analyzed the correlations between the expression of ITGA5 and the pathological parameters in LSCC (Figure 2A). Both Chi square test (Figure 2B, p = 0.017) and Wilcoxon signed rank test (Figure 2C, p = 0.005) showed that the dead patients had an elevated expression of ITGA5 than the alive patients. However, no correlation between the expression of ITGA5 and other clinical characteristics was found.
FIGURE 1.
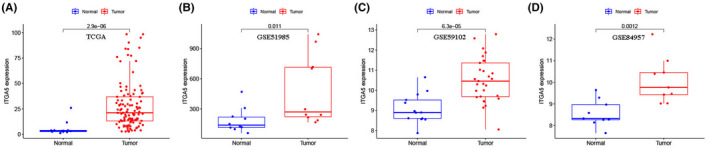
Expression of ITGA5 in laryngeal squamous cell carcinoma (LSCC) and normal tissues from the Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) databases. (A) Expression of ITGA5 in 111 LSCC and 12 normal tissues from TCGA cohort; (B) Expression of ITGA5 in 10 LSCC and 10 normal tissues from GSE51985 cohort; (C) Expression of ITGA5 in 29 LSCC and 13 normal tissues from GSE59102 cohort; (D) Expression of ITGA5 in 9 LSCC and 9 normal tissues from GSE84957 cohort
FIGURE 2.
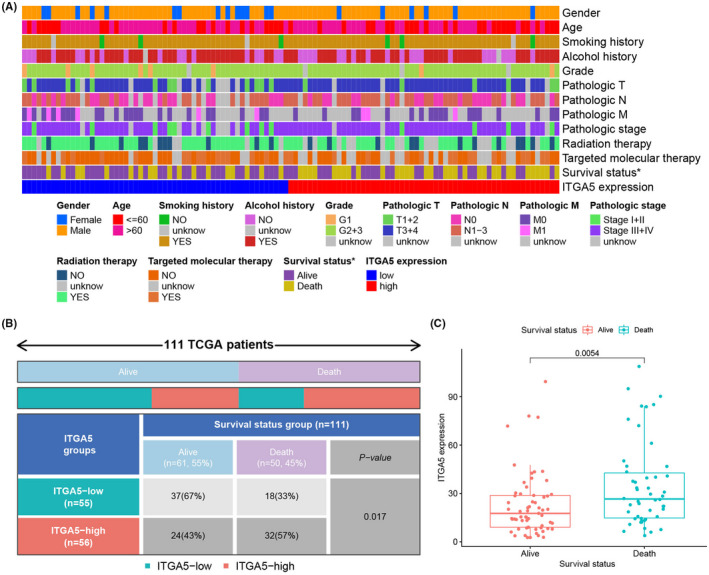
The association between the expression of ITGA5 and the clinical characteristics in patients with laryngeal squamous cell carcinoma (LSCC). (A) Heatmap showing the distribution of the clinical characteristics of patients with LSCC between the high‐ITGA5 and the low‐ITGA5 expression groups; (B) The survival status in the high‐ ITGA5 and the low‐ITGA5 expression groups were compared using Chi square test; (C) Significant statistical differences between the high‐ITGA5 and the low‐ITGA5 expression groups were assessed using Wilcoxon signed‐rank test
3.2. ITGA5 is associated with poor prognosis of patients with LSCC
The K‐M curves and the log‐rank test showed that patients in the high‐ITGA5 group had worse OS than the patients in the low‐ITGA5 group in the TCGA cohort (Figure 3A, p < 0.001). The patients with LSCC in the GSE65858 dataset were used as validation cohort and confirmed that the high‐ITGA5 group exhibited shorter OS than the low‐ITGA5 group (Figure 3B, p = 0.031). The univariate (Figure 3C, HR = 3.247, p < 0.001) and multivariate Cox regression analyses (Figure 3D, HR = 4.272, p < 0.001) demonstrated that ITGA5 was an independent prognostic factor for OS. We also assessed the ability of ITGA5 to predict the relapse in patients with LSCC. In the TCGA cohort (Figure 4A, p < 0.001), the K‐M survival curves indicated that patients with LSCC and high expression of ITGA5 had significantly worse RFS compared with patients with low expression of ITGA5. Additionally, GSE27020 (Figure 4B, p < 0.001) and GSE65858 (Figure 4C, p = 0.009) datasets as validation cohorts confirmed this result. Further, the univariate (Figure 4D, HR = 4.442, p < 0.001) and multivariate Cox regression analyses (Figure 4E, HR = 4.741, p < 0.001) demonstrated that ITGA5 was an independent predictive factor for recurrence in patients with LSCC.
FIGURE 3.
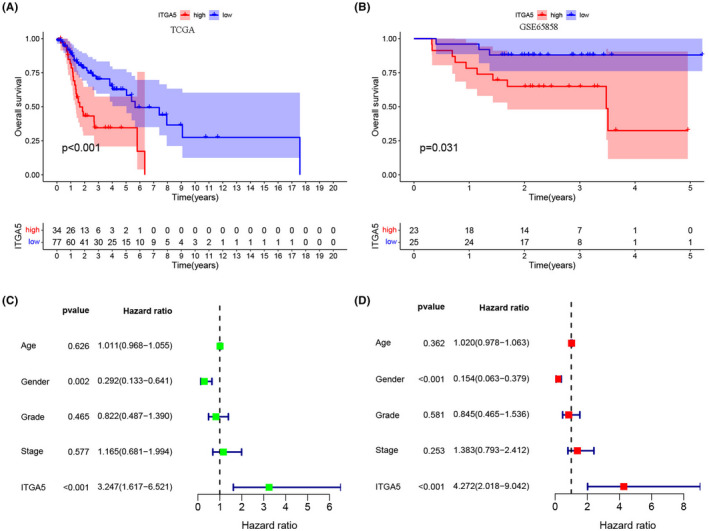
The expression of ITGA5 for prediction of overall survival (OS) in patients with laryngeal squamous cell carcinoma (LSCC). (A) Survival curves of OS from the Cancer Genome Atlas (TCGA) cohort (n = 111); (B) Survival curves of OS from the GSE65858 cohort (n = 48); The univariate (C) and multivariate Cox regression analyses (D) of OS according to the expression of ITGA5 after adjusting the clinical characteristics in the TCGA cohort
FIGURE 4.
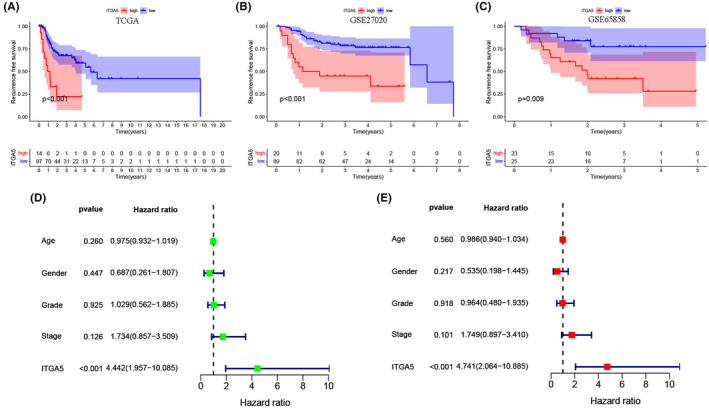
The expression of ITGA5 for the prediction of recurrence‐free survival (RFS) in patients with laryngeal squamous cell carcinoma (LSCC). (A) Survival curves of RFS from the Cancer Genome Atlas (TCGA) cohort (n = 111); (B) Survival curves of RFS from the GSE27020 cohort (n = 109); (C) Survival curves of RFS from the GSE65858 cohort (n = 48); The univariate (D) and multivariate Cox regression analyses (E) of RFS according to the expression of ITGA5 after adjusting the clinical characteristics in the TCGA cohort
3.3. Establishment and validation of a prognostic nomogram
A prognostic nomogram was constructed based on age, gender, grade, stage, and expression of ITGA5 for predicting 1‐, 3‐, and 5‐years survival rates of patients with LSCC (Figure 5A). The calibration curves of 1‐, 3‐, and 5‐years (Figure 5B) were all close to the reference line (at 45°), which represents the perfect prediction performance of the nomogram. The time‐dependent ROC curve analysis evaluated the accuracy of the nomogram and the area under the curve at 1‐, 3‐, and 5‐years which were 0.715, 0.726, and 0.649, respectively (Figure 5C), confirming that the predictive value of the nomogram was superior to any of the clinical factors (Figure 5D, age [0.425], gender [0.370], grade [0.475], and stage [0.502]).
FIGURE 5.
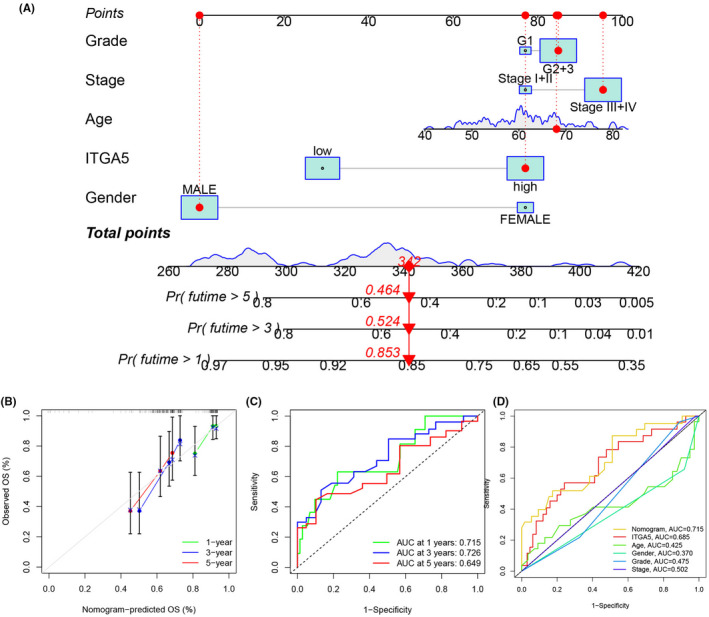
Construction and validation of a nomogram for patients with laryngeal squamous cell carcinoma (LSCC). (A) A nomogram was constructed to predict the 1‐, 3‐, and 5‐years of OS for patients with LSCC; (B) Calibration curves of 1‐, 3‐, and 5‐years of the nomogram; (C) Receiver operating characteristic (ROC) curves of 1‐, 3‐, and 5‐years of the nomogram; (D) ROC curves of the nomogram and other clinical characteristics
3.4. Functional enrichment analysis of ITGA5 in LSCC
To clarify the molecular mechanism of ITGA5 in LSCC, we performed the GSEA analysis based on the high‐ITGA5 and the low‐ITGA5 expressions. There were six KEGG pathways significantly enriched in the high‐ITGA5 group (Figure 6A), including extracellular matrix (ECM)‐receptor interaction, focal adhesion, glycosaminoglycan biosynthesis‐chondroitin sulfate, glycosaminoglycan biosynthesis‐keratan sulfate, leukocyte transendothelial migration, and regulation of the actin cytoskeleton. These significantly enriched the KEGG pathways that participated in a variety of cellular activities by regulating the construction of the ECM, such as adhesion, migration, differentiation, proliferation, and apoptosis. The GO and the KEGG enrichment analysis were performed using 415 DEGs (Figure 6B, Table S1) between the high‐ITGA5 and the low‐ITGA5 groups. The results of the GO analysis (Figure 6C, D) and the KEGG analysis (Figure 6E, F) confirmed that the DEGs were related to the construction of the ECM components and structures.
FIGURE 6.
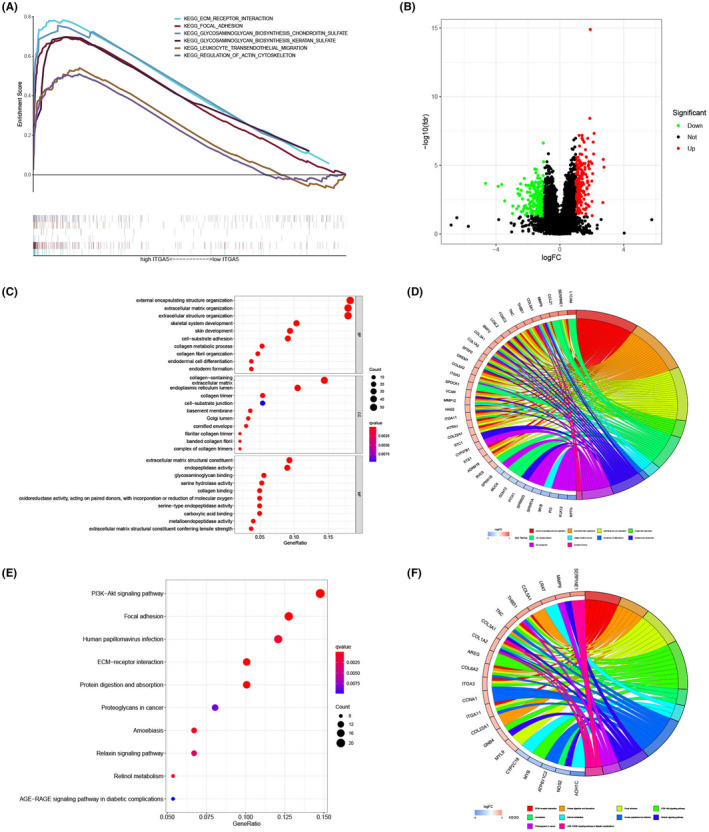
Functional enrichment analysis of ITGA5 in laryngeal squamous cell carcinoma (LSCC). (A) Gene Set Enrichment Analysis (GSEA) between the high‐ITGA5 and the low‐ITGA5 expression groups; (B) Volcano plot of differential expression genes between the high‐ITGA5 and the low‐ITGA5 expression groups; (C) Bubble plot of the GO enrichment analysis; (D) Circos plot of the gene ontology (GO) enrichment analysis; (E) bubble plot of the KEGG enrichment analysis; (F) Circos plot of the KEGG enrichment analysis
3.5. Association between ITGA5 and TME as well as the infiltration of the immune cells in LSCC
According to the TME analysis, using the ESTIMATE algorithm, the results showed that patients with high‐ITGA5 had elevated stromalScore than patients with low‐ITGA (Figure 7, p < 0.01), indicating that ITGA5 might play important roles in the components of the ECM. Further, we evaluated the correlation between the expression of ITGA5 and 22 immune cells. As shown in the violin plot (Figure 8A), patients with high‐ITGA5 had a higher abundance of M0 macrophages and resting CD4 memory T cells and a lower abundance of CD8 T cells, naive B cells, plasma cells, and follicular helper T cells. Pearson correlation analysis revealed that high expression levels of ITGA5 were directly proportional to M0 macrophages (Figure 8B) and resting CD4 memory T cells (Figure 8C) as well as resting natural killer (NK) cells (Figure 8D), and inversely proportional to CD8 T cells (Figure 8E), follicular helper T cells (Figure 8F) as well as plasma cells (Figure 8G).
FIGURE 7.
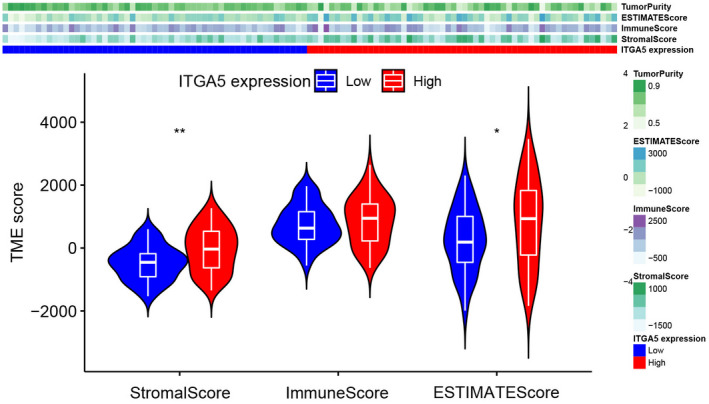
The stromal, immune, and ESTIMATE scores between the high ITGA5 and the low ITGA5 expression groups using the ESTIMATE algorithm
FIGURE 8.
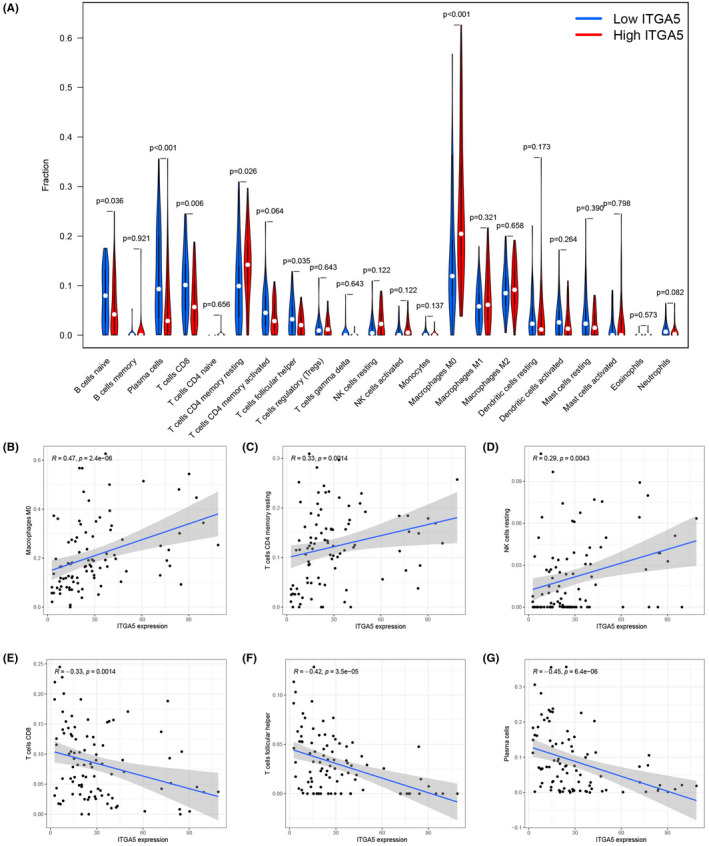
The correlation between the expression of ITGA5 and tumor immune cell infiltrating using the CIBERSORT algorithm. (A) the fraction of immune cells between the high‐ITGA5 and the low‐ITGA5 expression groups are presented in the violin plot; A significant positive correlation was observed between M0 macrophages (B), resting CD4 memory T cells (C), and resting natural killer (NK) cells (D) with ITGA5 expression; A significant negative correlation was observed between CD8 T cells (E), follicular helper T cells (F), and plasma cells (G) with ITGA5 expression
3.6. Chemosensitivity analysis
To explore the potential role of ITGA5 in the clinical treatment strategy, we estimated the IC50 of four common chemotherapeutic drugs recommended for laryngeal cancer therapy. Patients with low expression of ITGA5 had higher IC50 of docetaxel (Figure 9A) and gemcitabine (Figure 9B), indicating that patients with high expression of ITGA5 were more likely to respond to these two chemotherapeutic drugs. However, there was no difference in IC50 between the high‐ITGA5 and the low‐ITGA5 groups for cisplatin (Figure 9C) and paclitaxel (Figure 9D) therapies.
FIGURE 9.
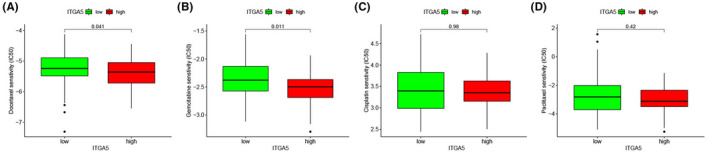
The correlation between the expression of ITGA5 and chemosensitivity in laryngeal squamous cell carcinoma (LSCC). The IC50 values of docetaxel (A) and gemcitabine (B) decreased significantly in the high‐ITGA5 expression group; The IC50 values of cisplatin (C) and paclitaxel (D) were not significantly different between the high‐ITGA5 and the low‐ITGA5 groups
4. DISCUSSION
Laryngeal squamous cell carcinoma is the second most common malignant tumor of the upper respiratory system as well as the head and neck. 20 The current treatment for LSCC is a comprehensive protocol that is based on surgical operation combined with chemotherapy, radiotherapy, targeted therapy, and immunotherapy. 21 However, LSCC is a heterogeneous disease, resulting in the treatment outcome to be significantly different. Therefore, more biomarkers for prediction and risk stratification are needed urgently for patients with LSCC. ITGA5 is an important member of the integrin family, which acts as an adhesive receptor for the ECM proteins. Recent research indicates that ITGA5 is overexpressed in various types of tumors and promotes proliferation, migration, invasion, and metastasis of cancer cells. 14 , 22 , 23 , 24 However, the role of ITGA5 in LSCC remains unclear. Hence, we investigated the expression profile and clinical significance of ITGA5 in LSCC by analyzing the TCGA and the GEO databases.
In this study, we combined results from the TCGA database with three independent datasets from the GEO database as validation cohorts, and confirmed that the expression levels of ITGA5 were upregulated in LSCC, which was consistent with the findings of previous studies in other cancers. 12 , 24 After the analysis of the clinicopathological data, it was found that the expression level of ITGA5 was correlated with the survival status of patients with LSCC. Breuksch et al. also indicated ITGA5 as a potent biomarker to discriminate the prognosis of patients with renal cell carcinoma. 25 Further, we evaluated the prognostic value of ITGA5 in LSCC. Both TCGA cohort and GSE65858 dataset showed that high expression of ITGA5 was strongly associated with worse OS in patients with LSCC. Regional recurrence was an important factor associated with the prognosis of patients with LSCC. 26 Interestingly, we found the expression level of ITGA5 was significantly associated with poor RFS in the TCGA cohort, which was validated by GSE27020 and GSE65858 datasets. Moreover, the univariate and multivariate Cox regression analyses demonstrated that ITGA5 was an independent predictive factor for death and recurrence in patients with LSCC. Collectively, all the above results strongly supported that the expression of ITGA5 is a perfect prognostic biomarker for LSCC. Therefore, we developed a nomogram based on the expression of ITGA5, which can be used to precisely predict the survival rate of patients with LSCC. Calibration curves and ROC curves confirmed the accuracy of the nomogram prediction, which was superior to any single factor. However, in the near future, further studies will be needed to validate the nomogram using a larger sample size.
To further explore the potential mechanisms that might be mediated by ITGA5 in LSCC, we performed the GSEA, GO, and KEGG enrichment analysis. The functional enrichment analysis showed that ITGA5 was involved in pathways related to the construction of the components of the ECM and intercellular signaling within the matrix. Additionally, ESTIMATE analysis also confirmed that stromalScore increased significantly in the high‐ITGA5 expression group than in the low‐ITGA5 expression group. The components of the ECM have been gaining attention due to their extremely important functions in carcinogenesis and tumor progression. 27 Increasing evidence has confirmed the crucial role of ECM in tumor metastasis. 28 Furthermore, ITGA5 is an important member of the integrin family, which can provide a bridge for the mechanical adhesion of cells and increase the ability of tumor cells to metastasize and invade. 29 Therefore, we have a reason to believe that ITGA5 might play a pivotal role in tumorigenesis, progression, and metastasis of LSCC by supporting the formation of the ECM and facilitating the tumor microenvironment.
The CIBERSORT algorithm was used to examine the relative proportions of infiltrating immune cells in each LSCC sample. Our results revealed that patients in the high‐ITGA5 group had higher proportions of immunosuppressive cells such as M0 macrophages and resting CD4 memory T cells than the low‐ITGA5 group, whereas the fraction of immune active cells such as CD8 T cells and follicular helper T cells were more abundant in the low‐ITGA5 group than the high‐ITGA5 group. Tian et al. showed that the high infiltration of M0 macrophages and resting CD4 memory T cells are associated with an unfavorable prognosis in glioma. 30 Conversely, previous studies have suggested that the infiltration of the CD8 T cells was positively correlated to improve prognosis and survival in several cancers, including melanoma, 31 lung adenocarcinoma, 32 and cutaneous angiosarcoma. 33 In line with these results, this might partly explain that the high‐ITGA5 group had a poorer prognosis than that of the low‐ITGA5 group in patients with LSCC.
Chemotherapy is one of the important treatments for patients with advanced LSCC, whereas the development of chemoresistance is rising and the efficacy of chemotherapy varies from person to person. 34 , 35 Hou et al. reported that ITGA5 was associated with chemoresistance in esophageal squamous cell carcinoma. 36 To evaluate the predictive value of ITGA5 in clinical treatment for LSCC, we calculated the sensitivity of four common chemotherapeutic drugs based on the pRRophetic algorithm. Interestingly, our results indicated that LSCC patients with high expression levels of ITGA5 tended to be more sensitive to docetaxel and gemcitabine, which supported more clues for the reasonable choice of a chemotherapeutic drug for patients with LSCC in the clinic.
However, there are some limitations in our study, and additional research is required to be carried out in the future. First, our findings are based entirely on the public databases using bioinformatics analysis. Although we also used the GEO database to verify the reliability of our results, further studies with a large sample size across different populations are needed to validate the current results. Second, we used the GO, KEGG, and GSEA analyses to detect the biological functions of ITGA5 in LSCC. Functional experiments are needed to verify these results. Finally, the association between ITGA5 and chemotherapy still needs to be validated in clinical trials.
In conclusion, our research was the first to identify ITGA5 as an independent potential of OS and an RFS predictor for patients with LSCC. We also revealed the potential biological functions and pathways of ITGA5 in LSCC. Additionally, LSCC patients with high expression levels of ITGA5 might benefit from chemotherapy. The comprehensive analysis of ITGA5 in patients with LSCC provided the possibility to improve the prognostic prediction and risk stratification for patients with LSCC and provided novel insights into the development of individual treatment strategies.
CONFLICT OF INTEREST
The authors declare that there are no financial or other conflicts of interest associated with this study.
AUTHOR CONTRIBUTIONS
The research was designed by CZ and HD. The data analysis from the public database was made by CZ, ZW, MT, JZ, and YS. The data were analyzed by CZ, ZS, and YS. CZ supervised the writing of the draft. The manuscript was reviewed by all the authors.
Supporting information
Table S1
ACKNOWLEDGMENTS
The Zhejiang Provincial Natural Science Foundation of China (No. LQ21H130001, No. LY20H130001, and No.LY19H160014), National Natural Science Foundation of China (No. 81670920), Ningbo Medical and Health Brand Discipline (No. PPXK2018‐02), Zhejiang Province Medical and Health Science Research Foundation(No. 2021KY307, No. 2020KY274, No. 2020RC107 and No. 2022KY1086), Ningbo Natural Science Foundation (No. 2019A610319, and No. 202003N4239), Ningbo Public Science Research Foundation(No. 2021S171 and No. 2021S170 ), and Ningbo “Technology Innovation 2025” Major Special Project (No. 2020Z097) supported this work.
Zhou C, Shen Y, Wei Z, et al. ITGA5 is an independent prognostic biomarker and potential therapeutic target for laryngeal squamous cell carcinoma. J Clin Lab Anal. 2022;36:e24228. doi: 10.1002/jcla.24228
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available from The Cancer Genome Atlas database (https://portal.gdc.cancer.gov/) and the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo).
REFERENCES
- 1. Trekova NA. [Effect of thymalin on the motor activity of the progeny of neurosensitized rats]. Zh Vyssh Nerv Deiat Im I P Pavlova. 1987;37(5):987‐988. [PubMed] [Google Scholar]
- 2. Lauwerends LJ, Galema HA, Hardillo JAU, et al. Current intraoperative imaging techniques to improve surgical resection of laryngeal cancer. a systematic review. Cancers. 2021;13(8):1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Steuer CE, El‐Deiry M, Parks JR, Higgins KA, Saba NF. An update on larynx cancer. CA Cancer J Clin. 2017;67(1):31‐50. [DOI] [PubMed] [Google Scholar]
- 4. Byrne A, Savas P, Sant S, et al. Tissue‐resident memory T cells in breast cancer control and immunotherapy responses. Nat Rev Clin Oncol. 2020;17(6):341‐348. [DOI] [PubMed] [Google Scholar]
- 5. Tran Janco JM, Lamichhane P, Karyampudi L, Knutson KL. Tumor‐infiltrating dendritic cells in cancer pathogenesis. J Immunol. 2015;194(7):2985‐2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang H, Liu H, Shen Z, et al. Tumor‐infiltrating neutrophils is prognostic and predictive for postoperative adjuvant chemotherapy benefit in patients with gastric cancer. Ann Surg. 2018;267(2):311‐318. [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor‐infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 2020;17(8):807‐821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zuo S, Wei M, Wang S, Dong J, Wei J. Pan‐cancer analysis of immune cell infiltration identifies a prognostic Immune‐Cell Characteristic Score (ICCS) in lung adenocarcinoma. Front Immunol. 2020;11:1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuninty PR, Bansal R, De Geus SWL, et al. ITGA5 inhibition in pancreatic stellate cells attenuates desmoplasia and potentiates efficacy of chemotherapy in pancreatic cancer. Sci Adv. 2019;5(9):eaax2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schaefer KL, Wai DH, Poremba C, et al. Characterization of the malignant melanoma of soft‐parts cell line GG‐62 by expression analysis using DNA microarrays. Virchows Arch. 2002;440(5):476‐484. [DOI] [PubMed] [Google Scholar]
- 11. Yu M, Chu S, Fei B, Fang X, Liu Z. O‐GlcNAcylation of ITGA5 facilitates the occurrence and development of colorectal cancer. Exp Cell Res. 2019;382(2):111464. [DOI] [PubMed] [Google Scholar]
- 12. Deng Y, Wan Q, Yan W. Integrin alpha5/ITGA5 promotes the proliferation, migration, invasion and progression of oral squamous carcinoma by epithelial‐mesenchymal transition. Cancer Manag Res. 2019;11:9609‐9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saatci O, Kaymak A, Raza U, et al. Targeting lysyl oxidase (LOX) overcomes chemotherapy resistance in triple negative breast cancer. Nat Commun. 2020;11(1):2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xiao Y, Li Y, Tao H, et al. Integrin alpha5 down‐regulation by miR‐205 suppresses triple negative breast cancer stemness and metastasis by inhibiting the Src/Vav2/Rac1 pathway. Cancer Lett. 2018;433:199‐209. [DOI] [PubMed] [Google Scholar]
- 15. Russano M, Cortellini A, Giusti R, et al. Clinical outcomes of NSCLC patients experiencing early immune‐related adverse events to PD‐1/PD‐L1 checkpoint inhibitors leading to treatment discontinuation. Cancer Immunol Immunother. 2021. https://doi.org/10.1007/s00262‐021‐03045‐9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qiu S, Li D, Shen Z, et al. Diagnostic and prognostic value of FOXD1 expression in head and neck squamous cell carcinoma. J Cancer. 2021;12(3):693‐702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9):e107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu F, Zhu Y, Qian Y, et al. Effects of gold nanorods modified with antiepidermal growth factor receptor monoclonal antibody on laryngeal cancer cells. Turk J Biol. 2018;42(2):144‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dyckhoff G, Warta R, Herold‐Mende C, Rudolph E, Plinkert PK, Ramroth H. Could primary chemoradiotherapy in T2 glottic cancers yield results comparable to primary radiotherapy in T1? Considerations from 531 German early stage patients. Cancers. 2021;13(7):1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Etienne‐Selloum N, Prades J, Bello‐Roufai D, et al. Expression analysis of alpha5 integrin subunit reveals its upregulation as a negative prognostic biomarker for glioblastoma. Pharmaceuticals. 2021;14(9):882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pantano F, Croset M, Driouch K, et al. Integrin alpha5 in human breast cancer is a mediator of bone metastasis and a therapeutic target for the treatment of osteolytic lesions. Oncogene. 2021;40(7):1284‐1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Y, Wang Y, Che X, et al. Integrin alpha5 promotes migration and invasion through the FAK/STAT3/AKT signaling pathway in icotinib‐resistant non‐small cell lung cancer cells. Oncol Lett. 2021;22(1):556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Breuksch I, Prosinger F, Baehr F, et al. Integrin alpha5 triggers the metastatic potential in renal cell carcinoma. Oncotarget. 2017;8(64):107530‐107542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen H, Song S, Zhang L, Dong W, Chen X, Zhou H. Preoperative platelet‐lymphocyte ratio predicts recurrence of laryngeal squamous cell carcinoma. Future Oncol. 2020;16(6):209‐217. [DOI] [PubMed] [Google Scholar]
- 27. Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196(4):395‐406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326(5957):1216‐1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kechagia JZ, Ivaska J, Roca‐Cusachs P. Integrins as biomechanical sensors of the microenvironment. Nat Rev Mol Cell Biol. 2019;20(8):457‐473. [DOI] [PubMed] [Google Scholar]
- 30. Tian Y, Ke Y, Ma Y. High expression of stromal signatures correlated with macrophage infiltration, angiogenesis and poor prognosis in glioma microenvironment. PeerJ. 2020;8:e9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ladanyi A, Kiss J, Somlai B, et al. Density of DC‐LAMP(+) mature dendritic cells in combination with activated T lymphocytes infiltrating primary cutaneous melanoma is a strong independent prognostic factor. Cancer Immunol Immunother. 2007;56(9):1459‐1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Du M, Liang Y, Liu Z, et al. Identification of key genes related to CD8+ T‐cell infiltration as prognostic biomarkers for lung adenocarcinoma. Front Oncol. 2021;11:693353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fujii H, Arakawa A, Utsumi D, et al. CD8(+) tumor‐infiltrating lymphocytes at primary sites as a possible prognostic factor of cutaneous angiosarcoma. Int J Cancer. 2014;134(10):2393‐2402. [DOI] [PubMed] [Google Scholar]
- 34. Magnes T, Wagner SM, Melchardt T, et al. Postoperative chemoradiotherapy with cisplatin is superior to radioimmunotherapy with cetuximab and radiotherapy alone: analysis of the Austrian head and neck cancer registry of the AGMT. Wien Klin Wochenschr. 2021;133(21–22):1131‐1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marcu LG, Marcu DC. Current omics trends in personalised head and neck cancer chemoradiotherapy. J Pers Med. 2021;11(11):1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hou S, Jin W, Xiao W, et al. Integrin alpha5 promotes migration and cisplatin resistance in esophageal squamous cell carcinoma cells. Am J Cancer Res. 2019;9(12):2774‐2788. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Data Availability Statement
The data that support the findings of this study are openly available from The Cancer Genome Atlas database (https://portal.gdc.cancer.gov/) and the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo).


