Abstract
Objective
Intra-tumoral expression of the serine hydrolase carboxylesterase 2 (CES2) contributes to the activation of the pro-drug irinotecan in pancreatic ductal adenocarcinoma (PDAC). Given other potential roles of CES2, we assessed its regulation, downstream effects, and contribution to tumor development in PDAC.
Methods
Association between the mRNA expression of CES2 in pancreatic tumors and overall survival was assessed using The Cancer Genome Atlas. Cell viability, clonogenic, and anchorage-independent growth assays as well as an orthotopic mouse model of PDAC were used to evaluate the biological relevance of CES2 in pancreatic cancer. CES2-driven metabolic changes were determined by untargeted and targeted metabolomic analyses.
Results
Elevated tumoral CES2 mRNA expression was a statistically significant predictor of poor overall survival in PDAC patients. Knockdown of CES2 in PDAC cells reduced cell viability, clonogenic capacity, and anchorage-independent growth in vitro and attenuated tumor growth in an orthotopic mouse model of PDAC. Mechanistically, CES2 was found to promote the catabolism of phospholipids resulting in HNF4α activation through a soluble epoxide hydrolase (sEH)-dependent pathway. Targeting of CES2 via siRNA or small molecule inhibitors attenuated HNF4α protein expression and reduced gene expression of classical/progenitor markers and increased basal-like markers. Targeting of the CES2-sEH-HNF4α axis using small molecule inhibitors of CES2 or sEH reduced cell viability.
Conclusions
We establish a novel regulatory loop between CES2 and HNF4α to sustain the progenitor subtype and promote PDAC progression and highlight the potential utility of CES2 or sEH inhibitors for the treatment of PDAC as part of non-irinotecan-containing regimens.
Keywords: PDAC, HNF4α, CES2, Classical/progenitor subtype, Phospholipid catabolism
Abbreviations: CES2, serine hydrolase carboxylesterase 2; HNF4α, hepatocyte nuclear factor 4 alpha; sEH, soluble epoxide hydrolase; PDAC, pancreatic ductal adenocarcinoma; LC-MS, liquid chromatography mass spectrometry
1. Introduction
Serine hydrolase carboxylesterase 2 (CES2) is a member of the alpha/beta fold hydrolase family [1] that is commonly overexpressed in various malignances [[2], [3], [4], [5]] and plays a crucial role in the metabolism of endogenous esters, ester-containing drugs, and environmental toxicants [6]. The intrinsic role of CES2 in regulating lipid metabolism has been investigated in diabetes, liver steatosis, and nonalcoholic fatty liver disease [[7], [8], [9], [10]]. To date, the relevance of CES2 in cancer has primarily been considered with respect to its functional role in mediating the activation of pro-drugs irinotecan and LY2334737 into their respective active forms [4,5,[11], [12], [13], [14]], thereby providing a potential marker for predicting patient response to irinotecan- or LY2334737-containing regimens [2,5,15,16].
CES2 expression in tumor cells has been reported to be a prognostic indicator of poor overall survival in patients with metastatic colorectal cancer who do not respond to chemotherapy and in patients with neuroblastoma [3,17]. The mechanism(s) that mediate CES2 upregulation and the role that CES2 plays in cancer progression remain poorly understood. Here, we aimed to determine the biological role of CES2 in pancreatic cancer progression. We demonstrate a critical role for CES2 in pancreatic ductal adenocarcinoma (PDAC) progression and establish the occurrence of a novel regulatory loop of sustained CES2 expression, wherein CES2 activates its upstream transcriptional regulator HNF4α through a CES2-soluble epoxide hydrolase (sEH)-dependent pathway. Our findings further implicate CES2 and sEH as potential therapeutic targets for PDAC.
2. Materials and methods
Additional information regarding methodologies is provided in the Supplementary Methods.
2.1. Chemicals
The CES2 inhibitor fenofibrate was purchased from Sigma–Aldrich (Catalog #F6020). Trans-AUCB and GSK2256294, small molecular inhibitors of soluble epoxide hydrolase, were purchased from MedChemexpress LLC (Catalog #2220) and AOX Medchem LLC (Catalog #HY-113974), respectively.
2.2. Cell culture, transfection, and viral transduction
Pancreatic cancer cell lines used in this study (BxPC-3 [#CRL-1687), SW1990 [#CRL-2172], SU.86.86 [#CRL-1837], PANC-1 [#CRL-1469], Hs 766T [#HTB-134], CFPAC-1 [#CRL-1918], MIA PaCa-2 [#CRM-CRL-1420], AsPC-1 [#CRL-1682], Panc 03.27 [#CRL-2549], HPAF-II [#CRL-1997], and Capan-2 [#HTB-80]) were purchased from ATCC and maintained in RPMI-1640 with 10% fetal bovine serum (FBS). The identity of each cell line was confirmed by DNA fingerprinting via short tandem repeats at the time of mRNA and total protein lysate preparation using the PowerPlex 1.2 kit (Promega). Fingerprinting results were compared with reference fingerprints maintained by the primary source of the cell line.
Transient knockdown of CES2 was performed by transfecting the cells using the following siRNAs: siControl (Silencer Select Negative Control No. 1, Thermo Fisher Scientific) and siCES2 (s225041, s528; Thermo Fisher Scientific). Short-hairpin RNAs targeting human CES2 mRNA and cloned into the pLKO.1-puro vector were obtained from the human library MISSION® TRC-Hs 1.0 (Sigma–Aldrich, TRCN0000046965). For overexpression, CES2 was cloned into the pLenti-C-Myc-DDK-IRESPuro (OriGene) vector, and an empty vector was used as a control. Lentiviral infections were conducted using 293LTV cells (Cell Biolabs, Inc.).
2.3. Cell proliferation and viability assays
Effects of CES2 knockdown on cellular morphology and proliferation were assessed by live cell imaging confluency analysis (IncuCyte, Essen BioScience).
2.4. Soft agar assay for colony formation
Cells were mixed with 0.6% agarose solution (Corning) and seeded at a density of 1000 cells/well in 12-well plates pre-coated with 1% agarose. The cells were cultured for two weeks to allow the formation of colonies. The plates were then fixed and stained with 0.005% crystal violet solution (Sigma) for 30 min at 37 °C. After washig with PBS to remove excess staining solution, the plates were imaged with ChemiDoc scanner (Biorad), and ImageJ software was used to count the colonies.
2.5. Spheroid assay
Cells were added to the wells of 96-well round bottom ultralow attachment (ULA) plates (Corning) at a concentration of 500 cells per well. The microplates were then incubated at 37 °C/5% CO2 for 24 h to allow the cells to aggregate into spheroids. Bright-field images were captured every 2 days using the Cytation 3 (Biotek) instrument using a 10x objective. The visual differences were quantified using the cellular analysis capabilities in the Gen5 software (Biotek).
2.6. Animal studies
Animal experiment protocols were reviewed and approved by The University of Texas MD Anderson Cancer Center IRB and in accordance with the Guidelines for the Care and Use of Laboratory Animals published by the NIH (Bethesda, MD). Detailed information regarding patient-derived xenografts (PDXs) is described in a prior publication [15]. For orthotopic xenograft models of PDAC and in vivo imaging, an enhanced GFP/firefly luciferase double-expressing cassette FG12 was introduced into AsPC-1 shCES2 or scramble control cells by lentiviral infection, as described previously [18]. A total of 5 × 105 viable cells in 50 μl of complete growth medium with 50% growth factor-reduced matrigel were injected into the pancreas of 4- to 6-week-old female nude mice; five mice were used for each experimental group. The mice were monitored twice weekly over a period of 5 weeks for in vivo tumor growth real time using the Xenogeny In Vivo Imaging System (Alameda, CA). As the mice tumor burden increased, the mice were euthanized immediately upon becoming moribund, as required by our institutional animal care guidelines. After each mouse was killed, the tumor was harvested and processed for routine histological and immunohistochemical analyses.
2.7. Western blot analysis
Cell pellets or tissues were lysed in RIPA buffer. Tumor tissues were cut into small pieces and homogenized using a Dounce homogenizer in RIPA buffer. The lysates were then separated by SDS-PAGE and subjected to Western blotting. The following antibodies were used: CES2 (Sigma Aldrich, #HPA018897), total HNF4α (Cell Signaling, #3113), β-actin (Sigma–Aldrich, #5316), Lamin A/C (Cell Signaling, #4777), and GAPDH (Cell Signaling, #5174).
2.8. Gene expression analysis
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN, #74104) following the manufacturer's instructions. Reverse transcription of total RNA into cDNA was performed using the High-Capacity Reverse Transcriptase Kit (Applied Biosystems, #4368813). The following probes were used: CES2 (Thermo Fisher Scientific, Hs00187279_m1), CLDN1 (Thermo Fisher Scientific, Hs00221623_m1), MUC5AC (Thermo Fisher Scientific, Hs01365616_m1), FOXA2 (Thermo Fisher Scientific, Hs00232764_m1), HNF1A (Thermo Fisher Scientific, Hs00167041_m1), HNF4A (Thermo Fisher Scientific, Hs00230853_m1), KRT5 (Thermo Fisher Scientific, Hs00361185_m1), KRT14 (Thermo Fisher Scientific, Hs00265033_m1), S100A2 (Thermo Fisher Scientific, Hs00195582_m1), and SIX4 (Thermo Fisher Scientific, Hs00213614_m1).
2.9. HNF4α activity assay
Cignal™ HNF4α (luc) reporter assay kit (Qiagen, #CCS-3039L) was used to determine the effect of CES2 knockdown or overexpression on HNF4α activity in PDAC cells according to the manufacturer's instructions. Briefly, ∼4 × 104 shControl or shCES2 AsPC-1 cells were seeded onto a 96-well plate and transfected on the following day with a non-inducible negative firefly luciferase reporter, HNF4α-responsive firefly luciferase reporter, or a constitutively expressed positive firefly luciferase reporter in combination with a constitutively expressed Renilla luciferase reporter. Negative controls were additionally included. After three days, the cells were washed twice with PBS, followed by sequential additions of Dual-Glo Luciferase Reagent and Dual-Glo Stop and Glo Reagent (Promega, Madison WI) to measure the activity of Firefly and Renilla luciferases, respectively. Luminescence (RLU) of firefly luciferase was normalized with that of Renilla luciferase; background signal was removed by subtracting the normalized luminescence (RLU) of negative firefly reporter control. CES2 activity was measured as described previously [5].
2.10. Gene expression datasets
RNA-seq data and clinical information for 162 pancreatic tumors from The Cancer Genome Atlas (TCGA) pancreatic cancer dataset was downloaded from cbioportal (https://www.cbioportal.org/) [19]. Gene expression for PDAC cell lines was derived from the Broad Institute Cancer Cell Line Encyclopedia (CCLE) database.
2.11. Mass spectrometry-based analyses
Detailed information regarding mass spectrometry-based analyses is provided in the Supplementary Methods.
2.12. Untargeted metabolomics analyses
Untargeted metabolomics was conducted on a Waters Acquity™ UPLC system coupled with a Xevo G2-XS quadrupole time-of-flight MS using standardized operating procedures as described previously [20,21].
2.13. Targeted analyses of oxylipins
Analyses for oxylipins were performed using a Waters ACQUITY i-Class LC system (Milford, MA) coupled with a Sciex 6500+ QTRAP (Redwood City, CA) and operated in the multiple reaction monitoring (MRM) mode. LC-MS/MS methods and parameters for oxylipins are detailed elsewhere [22].
2.14. MALDI imaging analyses
MALDI imaging was performed on a Waters MALDI SYNAPT G2-Si with Matrix-Assisted Laser Desorption Ionization MS as described previously [23].
2.15. Proteomic analyses
Proteomic analyses of cell line whole cell lysates were performed on a LTQ-ORBITRAP ELITE mass spectrometer (Thermo Scientific) coupled with a nanoflow chromatography system (Easy nanoLC 1000, Thermo Scientific) using a 25 cm separation column (75 μm ID, C18, 3um, Column Technology Inc) and a Symmetry C18 180 μm × 20 mm trap column (Waters) as described previously [15].
MS/MS spectra were searched against the Uniprot proteome database (Human, January 2017) using X!Tandem in the Trans-Proteomic pipeline (TPP-ver4.8). The mass error allowed was 20 ppm for parent monoisotopic and 0.5 Da for MS2 fragment monoisotopic ions. The searched result was filtered with FDR = 0.01.
2.16. Data analysis and statistics
For two-class comparisons, two-sided Student's t-tests or Wilcoxon rank-sum tests were used depending on data distribution. For orthotopic xenograft models of PDAC, we employed two-way repeated-measures ANOVA using group (control vs shCES2) and time as the variables of interest. Spearman correlation analyses were performed to evaluate the association between CES2 mRNA expression and lipid profiles of PDAC cell lines. Log rank statistic-based methods as described by Contal and O'Quigley [24] were used to determine the optimal cutoff point for tumoral CES2 mRNA expression for predicting survival in the TCGA-PDAC transcriptomic dataset [25]. Survival curves for the TCGA-PDAC transcriptomic dataset were generated using the Kaplan–Meier method and the statistical significance of the difference in survival was evaluated using the log-rank test. Univariable and multivariable Cox proportional hazard models were also used to evaluate the association between tumoral CES2 mRNA expression > or ≤ the cutoff value and overall survival in the TCGA-PDAC transcriptomic dataset. Sex, age (stratified by median), and stage were included as categorical co-variables in multivariable Cox proportional hazard models. To test for the proportionality of hazard assumption of a Cox regression test, we utilized the method of Patricia et al. [26].
3. Results
3.1. CES2 gene expression is associated with poor overall survival in PDAC
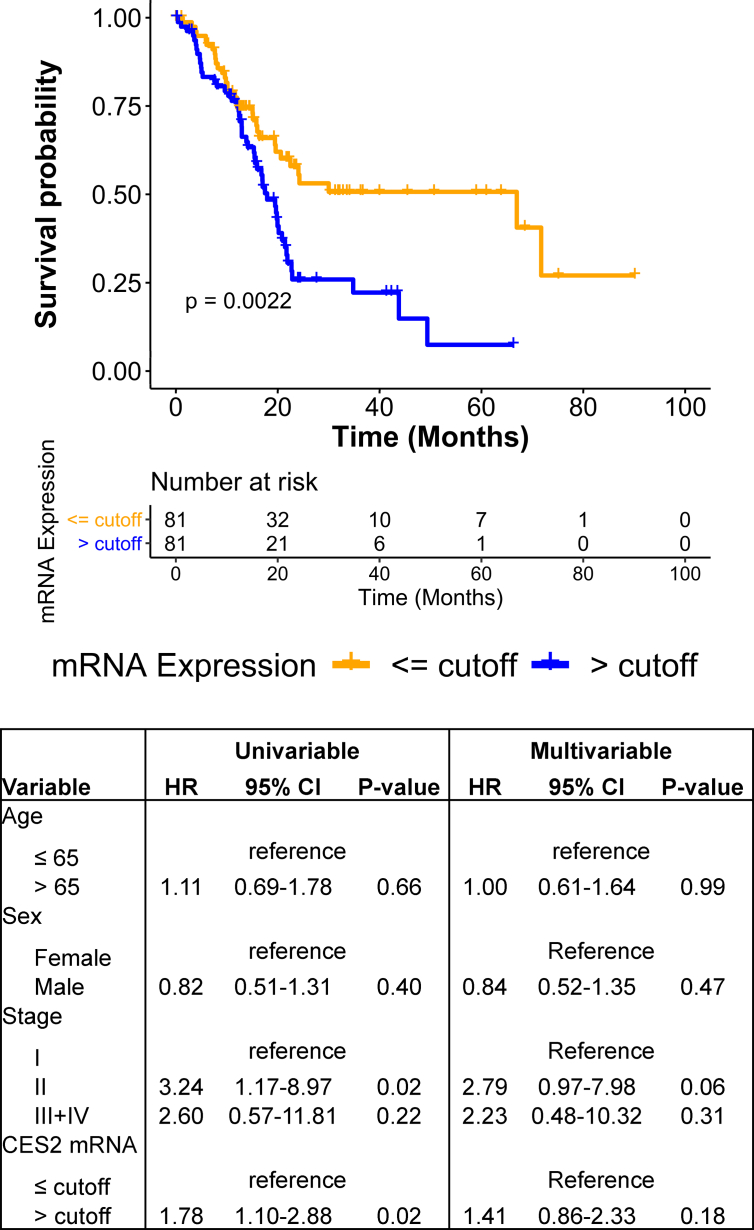
Using Cox proportional hazard models, we evaluated whether CES2 mRNA levels were prognostic of overall survival within the TCGA-PDAC transcriptomic dataset. In univariable analyses, compared with low tumor CES2 levels, elevated tumor CES2 mRNA levels were associated with statistically significantly worse overall survival (hazard ratio: 1.78 (95% CI: 1.10–2.88), two-sided p-value: 0.02); the Kaplan–Meier survival curve is shown in Figure 1.
Figure 1.
CES2 expression predicts poor prognosis in PDAC patients. Kaplan–Meier survival curves depict the relationship between tumoral mRNA expression of CES2 > or ≤ an optimal cutoff value and overall survival in the TCGA-PDAC transcriptomic dataset. The cutoff value was determined using log rank statistics-based methods (see Materials and methods). Results of the univariable and multivariable Cox proportional hazard model analyses are provided. For multivariable analyses, age (stratified by median), sex, and stage were included as co-variables.
3.2. Loss of CES2 attenuates PDAC tumor growth
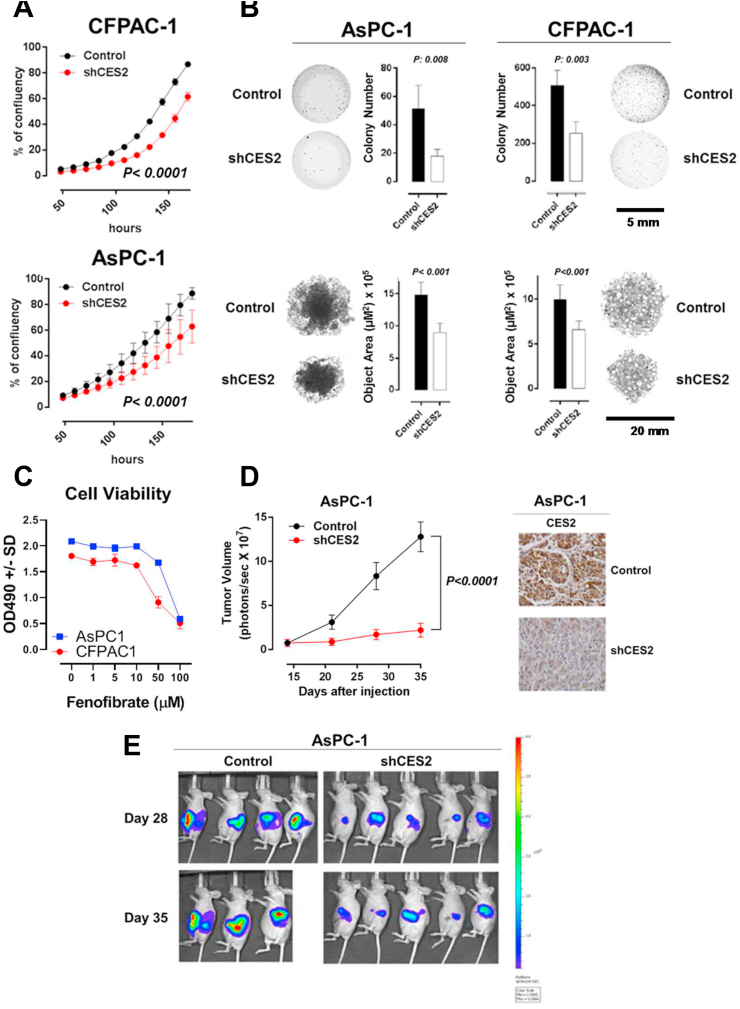
We performed stable knockdown of CES2 in PDAC cell lines CFPAC-1 and AsPC-1 and then assessed the differences in cell viability, clonogenic capacity, and anchorage-independent growth. Loss of CES2 in AsPC-1 and CFPAC-1 cells markedly reduced proliferation as well as clonogenic capability and anchorage-independent growth in vitro (two-sided Student's t-test p-value < 0.05) (Figure 2A–B). Consistent with these findings, treatment of AsPC-1 and CFPAC-1 cells with fenofibrate, a small molecule inhibitor of CES2 [27], resulted in dose-dependent reductions in cell viability (Figure 2C).
Figure 2.
Loss of CES2 attenuates PDAC growth in vitro and in vivo. A) Cell confluency of CFPAC-1 and AsPC-1 was checked every 12 h starting 48 h after the cells were seeded. B) Colony formation in soft agar or anchorage-independent growth in 96-well round bottom ultralow attachment (ULA) plate of CFPAC-1 and AsPC-1 cells. C) Growth of AsPC1 and CFPAC1 was measured by MTS assay following 96-h treatment with either DMSO or the indicated concentrations of fenofibrate. D) The growth of AsPC-1 cells expressing high (control) or low (shCES2) level of CES2 in nude mice was monitored every week. The effect of CES2 knockdown in xenograft tumor tissue was confirmed by immunohistochemistry using α-CES2 antibody. E) Luciferase-based noninvasive bioluminescence imaging of nude mice inoculated with control and shCES2 AsPC-1 cells at day 28 and day 35.
To test whether CES2 is essential for tumor growth in vivo, we implanted control or shCES2 AsPC1 PDAC cells labeled with luciferase into the pancreas of nude mice. Tumor growth, based on tumor size and luciferase-based bioluminescence imaging, was statistically significantly attenuated in shCES2-tumor-bearing mice, compared with controls (two-way repeated measures ANOVA group effect, p-value < 0.0001) (Figure 2D–E).
3.3. CES2 sustains HNF4α expression through a CES2-soluble epoxide hydrolase (sEH)-dependent pathway
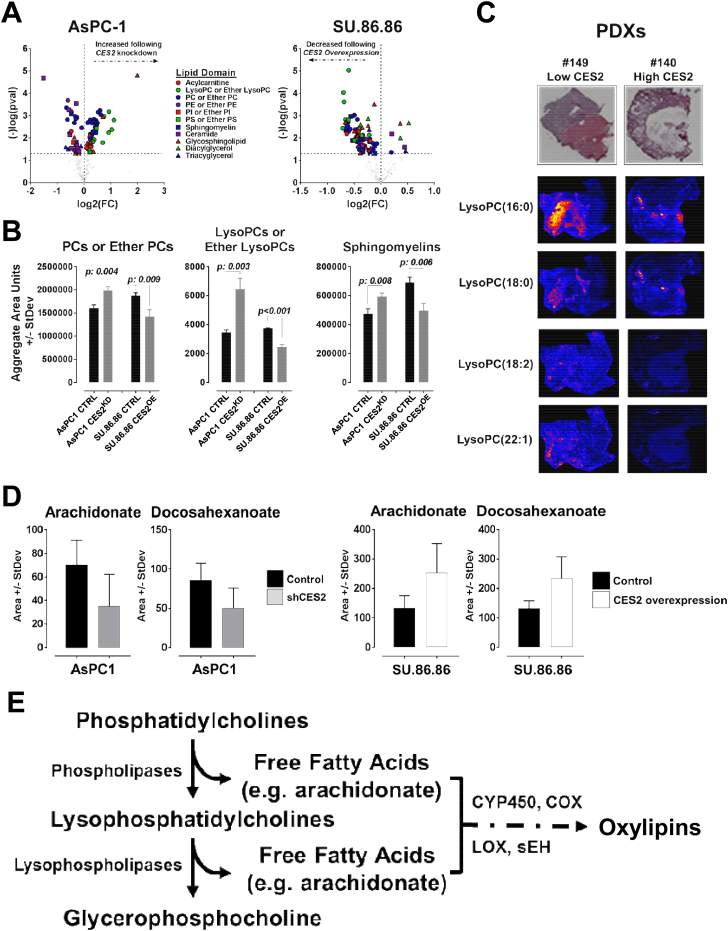
CES2 is known to play a role in the metabolism of endogenous esters including cholesteryl esters, triacylglycerols, and other lipid types [7,28]. To elucidate the mechanism(s) by which CES2 promotes PDAC tumor growth, we compared the lipidome of 11 PDAC cell lines profiled by liquid chromatography mass spectrometry (LC-MS) with mRNA expression of CES2. Spearman correlation analyses revealed negative associations between CES2 mRNA levels and several choline-containing lipid species (Supplementary Fig. S1). We further compared the lipid profiles of SU.86.86 (CES2 low) and AsPC-1 (CES2 high) PDAC cells following the overexpression or shRNA-mediated knockdown of CES2. Overexpression of CES2 in SU.86.86 PDAC cells resulted in reduced intracellular levels of choline-containing lipids, whereas knockdown of CES2 in AsPC-1 resulted in elevations in choline-containing lipid species (two-sided Student's t-tests, p < 0.05) (Figure 3A–B). Matrix-assisted laser desorption/ionization imaging (MALDI) of tumors derived from PDXs with different levels of CES2 protein expression that were engrafted subcutaneously into nude mice also revealed high CES2 tumor expression to be associated with reductions in tumor levels of choline-containing lysophopholipids (Figure 3C).
Figure 3.
CES2 promotes the catabolism of choline-containing lipids. A) Volcano plot of individual lipid changes classified by their lipid domain upon CES2 overexpression or knockdown in SU.86.86 and AsPC-1 cell lines, respectively. The x-axis specifies the fold changes (FC) and the y-axis specifies the negative logarithm to the base 10 of the t-test p-values relative to the respective control cell line. The black dots represent lipids showing a p-value < 0.05. B) Aggregate area counts for choline-containing phospholipids (left panel), lysophospholipids (center panel), or sphingomyelins (right panel) following CES2 overexpression (CES2OE) or knockdown (CES2KD) in SU.86.86 and AsPC-1 cell lines, respectively. Aggregate area counts were based on the sum of individual lipids belonging to the respective lipid class. Abbrev. PC, phosphatidylcholine; LysoPC, lysophosphatidylcholine. Significance was determined using the two-sided Student t-test. C) MALDI imaging analysis of lysophosphatidylcholines in PDX tumors with different levels of CES2 expression. The top panels show an H&E image of the analyzed tissues. D) Untargeted metabolomics analysis of the free fatty acids arachidonate and docosahexaenoate levels in AsPC-1 and SU.86.86 cell lines after CES2 knockdown (shCES2) and overexpression (CES2 overexpression), respectively. Values represent the average metabolite peak area ± SD. E) Schematic of the phosphatidylcholine catabolism pathway.
CES2-mediated reductions in choline-containing phospholipids were associated with increased levels of free fatty acids arachidonate (AA) and docosahexanoate (DHA) (Figure 3D). Free fatty acids, particularly AA, serve as precursors for a variety of downstream bioactive enzymatically oxidized lipids (Figure 3E) [29]. Complementary proteomic analyses of SU.86.86 and AsPC-1 following the overexpression or knockdown of CES2 revealed positive associations of CES2 with protein signatures of elevated lipid hydrolysis, lipid oxidation, and eicosanoid metabolism (Supplementary Table S1). To this end, knockdown of CES2 in AsPC-1 cells was associated with reductions in protein expression of CYP3A5 and bifunctional epoxide hydrolase 2 (EPHX2, sEH) (Supplementary Table S1 and Supplementary Figs. S2A–B). Collectively, our proteomic analyses support our metabolomics findings.
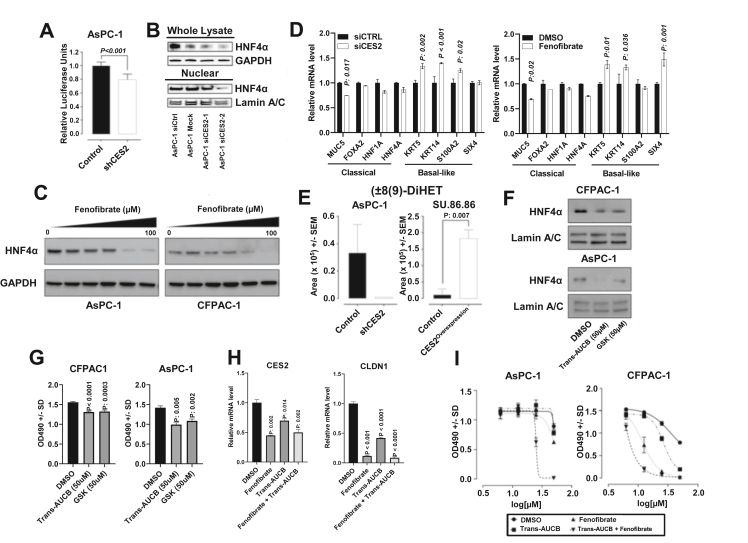
Essential fatty acids can serve as endogenous, reversible ligands for HNF4α, which have been shown to be essential for tumor growth [30,31]. We previously demonstrated that HNF4α is the upstream transcriptional regulator of CES2 [15]. We next assessed whether CES2 may regulate the activation of HNF4α. Loss of CES2 in AsPC-1 was associated with statistically significantly reduced HNF4α transcriptional activity as compared with the control (two-sided Student t-test, p < 0.001) (Figure 4A); siRNA-mediated knockdown of CES2 in AsPC-1 cells similarly attenuated nuclear protein expression of HNF4α (Figure 4B) without a change at the mRNA level (Supplementary Fig. S3A). Treatment of CES2-high AsPC-1 and CFPAC-1 PDAC cell lines with the CES2 inhibitor fenofibrate attenuated HNF4α protein levels as well as mRNA levels in a dose-dependent manner (Figure 4C and Supplementary Fig. S3B), although we acknowledge that the reduction in HNF4α protein and mRNA levels may be attributed to alternative mechanisms of fenofibrates [32,33].
Figure 4.
CES2-sEH pathway sustains HNF4α activation. A) Luciferase reporter activity for HNF4α in AsPC-1 cells following CES2 knockdown (shCES2). Significance was determined using the two-sided Student t-test. B) Western blots for HNF4α in whole lysate and nuclear fraction following siRNA-mediated knockdown of CES2 in AsPC-1 cells; GAPDH was used as a loading control for whole lysate; Lamin A/C was used as a loading control for nuclear isolates. C) Western blots for HNF4α in AsPC-1 and CFPAC-1 cells treated with variable concentrations of CES2 inhibitor fenofibrate for 48 h. D) Relative mRNA expression (2−Δ/ΔCT) of classical/progenitor-associated markers and basal-like markers in CFPAC pancreatic cancer cells following siRNA-mediated knockdown of CES2 or treatment with the CES2 inhibitor fenofibrate. The statistical significance was determined using two-sided Student t-tests. E) Targeted metabolomics analysis of the dihydroxyeicosatrienoic acid (±)8 (9)-DiHET levels in the conditioned media of AsPC-1 and SU.86.86 cell lines after CES2 knockdown (shCES2) and overexpression (CES2overexpression), respectively. Values represent area units; p-values were calculated using the two-sided Student t-test. F) Western blots for HNF4α in nuclear fraction following the sEH inhibitor Trans-AUCB (50 μM) or GSK2256294A (50 μM) treatment for 48 h in CFPAC-1 and AsPC-1 cells. Lamin A/C was used as a loading control for nuclear isolates. G) MTS cell proliferation assay for CFPAC-1 and AsPC-1 cells treated with the sEH inhibitors Trans-AUCB (50 μM) and GSK2256294A (50 μM). P-values were calculated using the two-sided Student t-test. H) mRNA levels of CES2 and CLDN1 in AsPC-1 cells following treatment with fenofibrate (20 μM), Trans-AUCB (20 μM), or fenofibrate plus Trans-AUCB (20 μM + 20 μM) for 48 h. Statistical significance was determined by Dunnet's multiple comparison tests and adjusted two-sided p-values reported in comparison with DMSO control. I) MTS cell proliferation assay for AsPC-1 and CFPAC-1 cells following 48 h treatment with fenofibrate, Trans-AUCB, or the combination of fenofibrate plus Trans-AUCB at the indicated concentration.
HNF4α is a pivotal transcription factor associated with the classical/progenitor subtypes of pancreatic cancer [34]. Loss of HNF4α, in addition to GATA6, has been reported to drive PDAC cells to acquire a basal-like phenotype [34]. Using endogenous CES2-high CFPAC-1 PDAC cells, which exhibit the characteristics of the classical subtype [35], we performed siRNA-mediated knockdown of CES2 and assessed the gene expression profiles associated with the classical (MUC5, FOXA2, HNF1A, and HNF4A) and basal-like subtypes (KRT5, KRT14, S100A2, and SIX4). Our analyses revealed that loss of CES2 was associated with a statistically significant (two-sided student t-test p < 0.05) reduction in MUC5 mRNA expression and increases in several basal-like markers including KRT5, KRT14, and S100A2 (Figure 4D). Inhibition of CES2 using fenofibrate yielded comparable findings (Figure 4D). Taken together, the results provided in Figure 4D suggest a role of CES2 in maintaining the classical subtype of PDAC in a HNF4α-dependent manner.
Proteomic analyses of SU.86.86 and AsPC-1 following the overexpression or knockdown of CES2 revealed a positive association of CES2 with CYP3A5 and EPHX2. Targeted analysis of oxylipins in conditioned media of SU.86.86 and AsPC-1 cell lines following the overexpression or knockdown of CES2 identified dihydroxyeicosatrienoic acid (±)8 (9)-DiHET, a diol generated from the hydrolysis of arachidonate-derived 8,9-EET via epoxide hydrolase (EH), to be positively associated with CES2 expression (Figure 4E). To determine whether EPHX2 is essential for HNF4α expression, we treated AsPC-1 and CFPAC-1 with the EPHX2 inhibitors Trans-AUCB and GSK 2256294A (GSK) and found that the inhibition of EPHX2 resulted in dose-dependent reductions in nuclear protein levels of HNF4α with concomitant loss of cell viability (Figure 4F–G). Of relevance, targeting of CES2 and/or sEH in AsPC-1 cells with fenofibrate, Trans-AUCB, or the combination of fenofibrate plus Trans-AUCB resulted in statistically significant (Dunn's multiple comparison test adjusted 2-sided P < 0.05) reductions in mRNA levels of HNF4α and its transcriptional targets CES2 and CLDN1 (Figure 4H and Supplementary Fig. S3B) [15,36,37]. Moreover, treatment of AsPC-1 and CFPAC-1 PDAC cells with the combination of Trans-AUCB (EPHX2 inhibitor) and fenofibrate (CES2 inhibitor) yielded improved anticancer effects compared with either drug alone (Figure 4I).
4. Discussion
We demonstrate here that CES2 is critical for PDAC progression and that elevated tumoral CES2 mRNA expression is predictive of poor overall survival. We further establish a novel mechanism by which PDAC cells sustain elevated CES2 expression through a CES2-sEH-HNF4α axis to promote PDAC growth (Figure 5). Moreover, we demonstrate that targeting of the CES2-sEH-HNF4α axis using small-molecule inhibitors of CES2 or sEH leads to a potent anticancer effect.
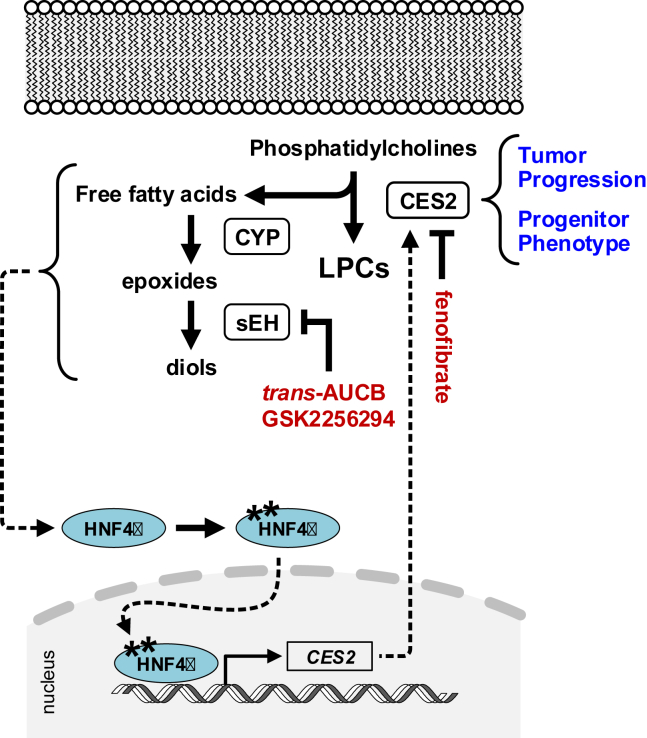
Figure 5.
Diagram of reciprocal regulation between CES2 and HNF4α. Proposed regulatory pathway wherein CES2 promotes the catabolism of choline-containing phospholipids to yield lipid species that sustain HNF4α protein expression and activation in a soluble epoxide hydrolase (sEH)-dependent manner; activated HNF4α in turn increases the transcription of CES2. Elevated CES2 enables tumor progression and maintains the progenitor phenotype.
We note that while increased tumoral CES2 expression was associated with poor overall survival in PDAC, no association was found between HNF4α mRNA expression and survival outcomes. This may be attributed to our findings that knockdown of CES2 reduced HNF4α protein levels but not mRNA levels in pancreatic cancer cells.
Our prior study established HNF4α as the upstream transcriptional regulator of CES2, which is enriched in the progenitor subtype of PDAC [15]. Aberrant expression of HNF4α is characteristic of several malignancies and its altered expression is predictive of clinical outcome [[38], [39], [40], [41]]. The regulation of HNF4α in PDAC is multifaceted, with both pro- or anti-tumor effects occurring in a context-dependent manner [40,41]. This paradox is likely to be attributed to the different splice variants that are generated by transcription from two alternative promoters (P1 and P2) and by two different ‘3’ splicing events that play discrete physiological roles in development and transcriptional regulation of target genes [42,43]. In the adult pancreas, the P2 promoter-driven HNF4α is predominantly expressed [44].
Here, we demonstrate that CES2 promotes the catabolism of phospholipids, particularly choline-containing phospholipids, in PDAC, resulting in the accumulation of free fatty acids that are further enzymatically oxidized to their downstream lipid diol derivatives. Fatty acids are established reversible endogenous ligands of HNF4α [30,31], thereby providing an explanation for the attenuation of HNF4α activation following knockdown or chemical inhibition of CES2. Interestingly, inhibition of epoxide hydrolase similarly decreased HNF4α activation, implying the relevance of diols in maintaining activated HNF4α and establishing the CES2-sEH-HNF4α axis.
HNF4α is a key driver of endodermal differentiation and is associated with the classical/progenitor subtypes of pancreatic cancer [34,45]. HNF4α is typically absent in normal pancreatic ducts, undifferentiated or poorly differentiated PDAC, but shows high expression in well-differentiated PDAC [46]. Loss of HNF4α, in addition to GATA6, has been reported to drive PDAC cells toward a basal-like phenotype [34]. Silencing of HNF4A due to hypermethylation is well characterized in poorly differentiated basal-like PDAC [45]. Our findings demonstrate that loss of CES2 through siRNA-mediated knockdown or small-molecule inhibition attenuates HNF4α activation, which is met with a concordant loss in gene expression of classical/progenitor markers and elevations in basal-like markers. These findings suggest a novel role of CES2 in maintaining the classical/progenitor subtype of PDAC.
Targeting of CES2 using fenofibrate reduced the cell viability of PDAC cells in our study. Fenofibrate is an FDA-approved drug used for patients with hypertriglyceridemia, primary hypercholesterolemia, or mixed dyslipidemia [47,48]. Prior studies on other cancer types have demonstrated the anticancer effects of fenofibrate [49]. Small-molecule inhibition of sEH via Trans-AUCB similarly reduced PDAC cell viability, consistent with prior studies on other cancer types [[50], [51], [52], [53]]. Notably, in our study, the combination of fenofibrate plus trans-AUCB yielded improved anticancer effects compared with either treatment alone. CES2 converts irinotecan into its active form, and CES2 expression levels in pancreatic tumors are predictive of response to irinotecan-containing therapies including FOLFIRINOX [5,15]. While outside the scope of the current study, the potential utility of CES2 or sEH inhibitors for the treatment of PDAC should be evaluated in the context of non-irinotecan-containing regimens.
In summary, we demonstrate that CES2 is an essential mediator of PDAC progression and reveal a novel sEH-dependent mechanism by which CES2 promotes HNF4α protein expression to maintain the classical/progenitor subtype of PDAC. Importantly, we show that small molecule inhibition of CES2 or sEH may have relevance for the treatment of PDAC.
Author contributions
Y.C. – performed the experiments, data analysis, and generation of figures and tables; involved in the conception and construction of the study; and did manuscript writing
M.C. – performed the experiments, data analysis, and generation of figures and tables; involved in the conception and construction of the study; and did manuscript writing
M.V.R. – provided the materials for the in vivo investigations and performed manuscript editing
J.V. – provided general guidance throughout the study, provided key insights, and performed manuscript editing
D.R. – provided key materials for the in vivo investigations and performed manuscript editing
Y.K. – provided key materials for the in vivo investigations and performed manuscript editing
L.R.P. – provided key materials for the in vivo investigations, and performed manuscript editing
H.K. – performed proteomic analyses and manuscript editing
E.I. – performed statistical analyses and manuscript editing
A.F. – performed experiments and manuscript editing
S.F.B. – provided key materials for in vivo investigations and performed manuscript editing
J.B.D. –performed metabolomic analyses and manuscript editing
O.F. –- performed targeted metabolomic analyses and manuscript editing
A.M. – provided general guidance throughout the study, provided key insights, and performed manuscript editing
H.W. – provided key materials for in vivo investigations and performed manuscript editing
P.J.C. –- provided key materials for in vivo investigations and performed manuscript editing
M.H.G.K. – provided key materials for in vivo investigations and performed manuscript editing
J.B.F. – provided key materials for in vivo investigations and performed manuscript editing
S.M.H. – provided funding support, general guidance throughout study, and key insights and performed manuscript editing
J.F.F. – performed data analysis and generation of figures and tables, involved in the conception and construction of the study, provided critical review of the manuscript, provided guidance and mentorship throughout the study, and did manuscript writing
Conflict of interest
None declared.
Acknowledgements
This study was supported by the National Pancreas Foundation (M.C.), MD Anderson's Moonshot Program, 2 faculty fellowships from The University of Texas MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment (J.F.F. and M.C.), 2 National Institutes of Health T32 fellowships (T32 CA009599 and P30 CA016672; M.V.R.P. and D.R.), and the Viragh Family Foundation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101426.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Imai T. Human carboxylesterase isozymes: catalytic properties and rational drug design. Drug Metabolism and Pharmacokinetics. 2006;21(3):173–185. doi: 10.2133/dmpk.21.173. [DOI] [PubMed] [Google Scholar]
- 2.Xu G., Zhang W., Ma M.K., McLeod H.L. Human carboxylesterase 2 is commonly expressed in tumor tissue and is correlated with activation of irinotecan. Clinical Cancer Research. 2002;8(8):2605–2611. [PubMed] [Google Scholar]
- 3.Zhang Y., Sun L., Sun Y., Chen Y., Wang X., Xu M., et al. Overexpressed CES2 has prognostic value in CRC and knockdown CES2 reverses L-OHP-resistance in CRC cells by inhibition of the PI3K signaling pathway. Experimental Cell Research. 2020;389(1):111856. doi: 10.1016/j.yexcr.2020.111856. [DOI] [PubMed] [Google Scholar]
- 4.Sanghani S.P., Quinney S.K., Fredenburg T.B., Sun Z., Davis W.I., Murry D.J., et al. Carboxylesterases expressed in human colon tumor tissue and their role in CPT-11 hydrolysis. Clinical Cancer Research. 2003;9(13):4983–4991. [PubMed] [Google Scholar]
- 5.Capello M., Lee M., Wang H., Babel I., Katz M.H., Fleming J.B., et al. Carboxylesterase 2 as a determinant of response to irinotecan and neoadjuvant FOLFIRINOX therapy in pancreatic ductal adenocarcinoma. Journal of the National Cancer Institute. 2015;107(8):djv132. doi: 10.1093/jnci/djv132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D., Zou L., Jin Q., Hou J., Ge G., Yang L. Human carboxylesterases: a comprehensive review. Acta Pharmaceutica Sinica B. 2018;8(5):699–712. doi: 10.1016/j.apsb.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Zalzala M., Jadhav K., Xu Y., Kasumov T., Yin L., et al. Carboxylesterase 2 prevents liver steatosis by modulating lipolysis, endoplasmic reticulum stress, and lipogenesis and is regulated by hepatocyte nuclear factor 4 alpha in mice. Hepatology. 2016;63(6):1860–1874. doi: 10.1002/hep.28472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruby M.A., Massart J., Hunerdosse D.M., Schönke M., Correia J.C., Louie S.M., et al. Human carboxylesterase 2 reverses obesity-induced diacylglycerol accumulation and glucose intolerance. Cell Reports. 2017;18(3):636–646. doi: 10.1016/j.celrep.2016.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R., Wang Y., Ning R., Hu J., Liu W., Xiong J., et al. Decreased carboxylesterases expression and hydrolytic activity in type 2 diabetic mice through Akt/mTOR/HIF-1alpha/Stra13 pathway. Xenobiotica. 2015;45(9):782–793. doi: 10.3109/00498254.2015.1020353. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y., Pan X., Hu S., Zhu Y., Cassim Bawa F., Li Y., et al. Hepatocyte-specific expression of human carboxylesterase 2 attenuates nonalcoholic steatohepatitis in mice. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2021;320(2):G166–G174. doi: 10.1152/ajpgi.00315.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khanna R., Morton C.L., Danks M.K., Potter P.M. Proficient metabolism of irinotecan by a human intestinal carboxylesterase. Cancer Research. 2000;60(17):4725–4728. [PubMed] [Google Scholar]
- 12.Cecchin E., Corona G., Masier S., Biason P., Cattarossi G., Frustaci S., et al. Carboxylesterase isoform 2 mRNA expression in peripheral blood mononuclear cells is a predictive marker of the irinotecan to SN38 activation step in colorectal cancer patients. Clinical Cancer Research. 2005;11(19 Pt 1):6901–6907. doi: 10.1158/1078-0432.CCR-05-0602. [DOI] [PubMed] [Google Scholar]
- 13.Pratt S.E., Durland-Busbice S., Shepard R.L., Heinz-Taheny K., Iversen P.W., Dantzig A.H. Human carboxylesterase-2 hydrolyzes the prodrug of gemcitabine (LY2334737) and confers prodrug sensitivity to cancer cells. Clinical Cancer Research. 2013;19(5):1159–1168. doi: 10.1158/1078-0432.CCR-12-1184. [DOI] [PubMed] [Google Scholar]
- 14.Sanghani S.P., Quinney S.K., Fredenburg T.B., Davis W.I., Murry D.J., Bosron W.F. Hydrolysis of irinotecan and its oxidative metabolites, 7-ethyl-10-[4-N-(5-aminopentanoic acid)-1-piperidino] carbonyloxycamptothecin and 7-ethyl-10-[4-(1-piperidino)-1-amino]-carbonyloxycamptothecin, by human carboxylesterases CES1A1, CES2, and a newly expressed carboxylesterase isoenzyme, CES3. Drug Metabolism & Disposition. 2004;32(5):505–511. doi: 10.1124/dmd.32.5.505. [DOI] [PubMed] [Google Scholar]
- 15.Capello M., Fahrmann J.F., Rios Perez M.Y., Vykoukal J.V., Irajizad E., Tripathi S.C., et al. CES2 expression in PAAD is predictive of response to irinotecan and is associated with type 2 diabetes. JCO Precision Oncology. 2020;4(2020):426–436. doi: 10.1200/PO.19.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones R.P., Sutton P., Greensmith R.M., Santoyo-Castelazo A., Carr D.F., Jenkins R., et al. Hepatic activation of irinotecan predicts tumour response in patients with colorectal liver metastases treated with DEBIRI: exploratory findings from a phase II study. Cancer Chemotherapy and Pharmacology. 2013;72(2):359–368. doi: 10.1007/s00280-013-2199-5. [DOI] [PubMed] [Google Scholar]
- 17.Uchida K., Otake K., Tanaka K., Hashimoto K., Saigusa S., Matsushita K., et al. Clinical implications of CES2 RNA expression in neuroblastoma. Journal of Pediatric Surgery. 2013;48(3):502–509. doi: 10.1016/j.jpedsurg.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Chang Z., Ju H., Ling J., Zhuang Z., Li Z., Wang H., et al. Cooperativity of oncogenic K-ras and downregulated p16/INK4A in human pancreatic tumorigenesis. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vykoukal J., Fahrmann J.F., Gregg J.R., Tang Z., Basourakos S., Irajizad E., et al. Caveolin-1-mediated sphingolipid oncometabolism underlies a metabolic vulnerability of prostate cancer. Nature Communications. 2020;11(1):4279. doi: 10.1038/s41467-020-17645-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang T., Fahrmann J.F., Lee H., Li Y.J., Tripathi S.C., Yue C., et al. JAK/STAT3-Regulated fatty acid β-oxidation is critical for breast cancer stem cell self-renewal and chemoresistance. Cell Metabolism. 2018;27(1):136–150. doi: 10.1016/j.cmet.2017.11.001. e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedersen T.L., Newman J.W. In: Clinical metabolomics: methods and protocols. Giera M., editor. Springer New York; New York, NY: 2018. Establishing and performing targeted multi-residue analysis for lipid mediators and fatty acids in small clinical plasma samples; pp. 175–212. [DOI] [PubMed] [Google Scholar]
- 23.Kandel P., Semerci F., Bajic A., Baluya D., Ma L., Chen K., et al. 2020. Oleic acid triggers hippocampal neurogenesis by binding to TLX/NR2E1. bioRxiv:2020.2010.2028.359810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cecile Contal J.O.Q. An application of changepoint methods in studying the effect of age on survival in breast cancer. Computational Statistics & Data Analysis. 1999;30(3):253–270. [Google Scholar]
- 25.Fahrmann J.F., Vykoukal J., Fleury A., Tripathi S., Dennison J.B., Murage E., et al. Association between plasma diacetylspermine and tumor spermine synthase with outcome in triple negative breast cancer. Journal of National Cancer Institute. 2019 doi: 10.1093/jnci/djz182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patricia M., Grambsch T.M.T. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81(3):12. [Google Scholar]
- 27.Fukami T., Takahashi S., Nakagawa N., Maruichi T., Nakajima M., Yokoi T. In vitro evaluation of inhibitory effects of antidiabetic and antihyperlipidemic drugs on human carboxylesterase activities. Drug Metabolism & Disposition. 2010;38(12):2173–2178. doi: 10.1124/dmd.110.034454. [DOI] [PubMed] [Google Scholar]
- 28.Ruby M.A., Massart J., Hunerdosse D.M., Schonke M., Correia J.C., Louie S.M., et al. Human carboxylesterase 2 reverses obesity-induced diacylglycerol accumulation and glucose intolerance. Cell Reports. 2017;18(3):636–646. doi: 10.1016/j.celrep.2016.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajeyah A.A., Griffiths W.J., Wang Y., Finch A.J., O'Donnell V.B. The biosynthesis of enzymatically oxidized lipids. Frontiers in Endocrinology. 2020;11:591819. doi: 10.3389/fendo.2020.591819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan X., Ta T.C., Lin M., Evans J.R., Dong Y., Bolotin E., et al. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS One. 2009;4(5) doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wisely G.B., Miller A.B., Davis R.G., Thornquest A.D., Jr., Johnson R., Spitzer T., et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10(9):1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- 32.Frazier-Wood A.C., Ordovas J.M., Straka R.J., Hixson J.E., Borecki I.B., Tiwari H.K., et al. The PPAR alpha gene is associated with triglyceride, low-density cholesterol and inflammation marker response to fenofibrate intervention: the GOLDN study. The Pharmacogenomics Journal. 2013;13(4):312–317. doi: 10.1038/tpj.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viswakarma N., Jia Y., Bai L., Vluggens A., Borensztajn J., Xu J., et al. Coactivators in PPAR-regulated gene expression. PPAR Research. 2010;2010:250126. doi: 10.1155/2010/250126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunton H., Caligiuri G., Cunningham R., Upstill-Goddard R., Bailey U.M., Garner I.M., et al. HNF4A and GATA6 loss reveals therapeutically actionable subtypes in pancreatic cancer. Cell Reports. 2020;31(6):107625. doi: 10.1016/j.celrep.2020.107625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collisson E.A., Sadanandam A., Olson P., Gibb W.J., Truitt M., Gu S., et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nature Medicine. 2011;17(4):500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Battle M.A., Konopka G., Parviz F., Gaggl A.L., Yang C., Sladek F.M., et al. Hepatocyte nuclear factor 4 orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(22):8419–8424. doi: 10.1073/pnas.0600246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walesky C., Edwards G., Borude P., Gunewardena S., O'Neil M., Yoo B., et al. Hepatocyte nuclear factor 4 alpha deletion promotes diethylnitrosamine-induced hepatocellular carcinoma in rodents. Hepatology. 2013;57(6):2480–2490. doi: 10.1002/hep.26251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oshima T., Kawasaki T., Ohashi R., Hasegawa G., Jiang S., Umezu H., et al. Downregulated P1 promoter-driven hepatocyte nuclear factor-4alpha expression in human colorectal carcinoma is a new prognostic factor against liver metastasis. Pathology International. 2007;57(2):82–90. doi: 10.1111/j.1440-1827.2006.02061.x. [DOI] [PubMed] [Google Scholar]
- 39.Ehehalt F., Rümmele P., Kersting S., Lang-Schwarz C., Rückert F., Hartmann A., et al. Hepatocyte nuclear factor (HNF) 4α expression distinguishes ampullary cancer subtypes and prognosis after resection. Annals of Surgery. 2011;254(2):302–310. doi: 10.1097/SLA.0b013e31821994a8. [DOI] [PubMed] [Google Scholar]
- 40.Sun Q., Xu W., Ji S., Qin Y., Liu W., Hu Q., et al. Role of hepatocyte nuclear factor 4 alpha in cell proliferation and gemcitabine resistance in pancreatic adenocarcinoma. Cancer Cell International. 2019;19(1):49. doi: 10.1186/s12935-019-0767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camolotto S.A., Belova V.K., Torre-Healy L., Vahrenkamp J.M., Berrett K.C., Conway H., et al. Reciprocal regulation of pancreatic ductal adenocarcinoma growth and molecular subtype by HNF4α and SIX1/4. Gut. 2020;70(5):900–914. doi: 10.1136/gutjnl-2020-321316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang S., Tanaka T., Iwanari H., Hotta H., Yamashita H., Kumakura J., et al. Expression and localization of P1 promoter-driven hepatocyte nuclear factor-4α (HNF4α) isoforms in human and rats. Nuclear Receptor. 2003;1(1):5. doi: 10.1186/1478-1336-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yusuf D., Butland S.L., Swanson M.I., Bolotin E., Ticoll A., Cheung W.A., et al. The transcription factor encyclopedia. Genome Biology. 2012;13(3):R24. doi: 10.1186/gb-2012-13-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka T., Jiang S., Hotta H., Takano K., Iwanari H., Sumi K., et al. Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha in the pathogenesis of human cancer. The Journal of Pathology. 2006;208(5):662–672. doi: 10.1002/path.1928. [DOI] [PubMed] [Google Scholar]
- 45.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 46.Kim J., Hoffman J.P., Alpaugh R.K., Rhim A.D., Reichert M., Stanger B.Z., et al. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Reports. 2013;3(6):2088–2099. doi: 10.1016/j.celrep.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balakumar P., Sambathkumar R., Mahadevan N., Muhsinah A.B., Alsayari A., Venkateswaramurthy N., et al. Molecular targets of fenofibrate in the cardiovascular-renal axis: a unifying perspective of its pleiotropic benefits. Pharmacological Research. 2019;144:132–141. doi: 10.1016/j.phrs.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 48.Tarantino N., Santoro F., Correale M., De Gennaro L., Romano S., Di Biase M., et al. Fenofibrate and dyslipidemia: still a place in therapy? Drugs. 2018;78(13):1289–1296. doi: 10.1007/s40265-018-0965-8. [DOI] [PubMed] [Google Scholar]
- 49.Lian X., Wang G., Zhou H., Zheng Z., Fu Y., Cai L. Anticancer properties of fenofibrate: a repurposing use. Journal of Cancer. 2018;9(9):1527–1537. doi: 10.7150/jca.24488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J., Sanidad K.Z., Zhang G. Cytochrome P450 monooxygenase/soluble epoxide hydrolasemediated eicosanoid pathway in colorectal cancer and obesityassociated colorectal cancer. Oncoscience. 2019;6(9–10):371–375. doi: 10.18632/oncoscience.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang G., Panigrahy D., Hwang S.H., Yang J., Mahakian L.M., Wettersten H.I., et al. Dual inhibition of cyclooxygenase-2 and soluble epoxide hydrolase synergistically suppresses primary tumor growth and metastasis. Proceedings of the National Academy of Sciences of the U S A. 2014;111(30):11127–11132. doi: 10.1073/pnas.1410432111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho H.J., Switzer C.H., Kamynina A., Charles R., Rudyk O., Ng T., et al. Complex interrelationships between nitro-alkene-dependent inhibition of soluble epoxide hydrolase, inflammation and tumor growth. Redox Biology. 2020;29:101405. doi: 10.1016/j.redox.2019.101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Das Mahapatra A., Choubey R., Datta B. Small molecule soluble epoxide hydrolase inhibitors in multitarget and combination therapies for inflammation and cancer. Molecules. 2020;25(23) doi: 10.3390/molecules25235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.