Abstract
Introduction
Krüppel-like factors (KLFs), which comprise 17 family members, exert important functions during the development of cancer. The role of KLF16 seems controversial in carcinogenesis because both tumor suppressive and promoting effects have been reported.
Methods
The expression level of KLF16 was analyzed based on public data sets from The Cancer Genome Atlas (TCGA) and evaluated by immunohistochemical (IHC) staining. CCK8 assay, colony formation analysis, transwell assays and the PI/Annexin V-APC assay kit were performed to detect cell growth, colony formation, cell migration and apoptosis of BC cells. Xenograft tumorigenesis assay was performed to detect the KLF16 expression on BC growth in vivo. Dual-luciferase reporter assay and chromatin immunoprecipitation (ChIP)-qPCR assay were performed to analyze the interaction between KLF16 and its target.
Results
In this study, we explored the role of KLF16 in bladder cancer (BC). We demonstrated that KLF16 was overexpressed in human BC tissues. The high expression of KLF16 was a potential predictor of a poor prognosis in patients with BC. Interference with KLF16 expression in 563 cells, having relatively higher levels of KLF16, repressed cell proliferation and migration. In contrast, upregulation of KLF16 in T24 cells enhanced cellular function, including cell growth and migration. KLF16 also suppressed the apoptosis of BC cells. Additionally, KLF16 inhibited the expression of the TGF-type III receptor (TGFBR3) by binding to its promoter sequence and reducing transcriptional activity. There was a negative correlation between KLF16 and TGFBR3 in human BC tissues. Furthermore, TGFBR3 was revealed to be a negative regulator of BC cell proliferation and migration. KLF16 also supported BC tumorigenesis by downregulating TGFBR3 expression in vivo.
Discussion
These results suggested that KLF16 acts as an oncogene in BC through transcriptional inactivation of TGFBR3. This study provides evidence that targeting the KLF16/TGFBR3 axis may be beneficial for BC patients.
Keywords: bladder carcinogenesis, KLF16, TGFBR3, cell proliferation
Introduction
Bladder cancer (BC) is a malignancy of the urinary system. More than 60% of cases occur in developing countries, and the incidence is much higher in men than in women.1,2 According to a statistical study evaluating tumor epidemiology in China, the total incidence and mortality of BC in Chinese men are 10.9/100,000 and 4.2/100,000, respectively. In general, BC represents the seventh most frequent and tenth most lethal malignancy in China.3,4 Currently, BC treatment is still based on surgical resection, accompanied by bladder perfusion chemotherapy, systemic chemotherapy, and radiotherapy.5–8 Although emerging targeted therapies and immunotherapy provide new treatments for BC, the high recurrence rate still leads to poor treatment effects for BC.9 Therefore, exploring new pathogenic genes will help us gain greater insight into the mechanism of BC occurrence and development, and will contribute to the development of effective molecular markers for early diagnosis, detection of metastasis, and recurrence of BC, and will identify potential BC treatment targets.
KLF16, a transcription factor that contains a conserved zinc finger domain, belongs to the Krüppel-like factors (KLFs) family. Recent studies have found that KLFs play an important role in the occurrence and development of different tumors. For example, KLF1 is highly expressed in cervical cancer. Downregulation of KLF1 can inhibit the invasion and migration ability of cervical cancer cells.10 KLF3 shows a high expression trend in lung cancer patients, and reducing KLF3 expression can inhibit the growth and migration of lung cancer cells.11 KLF16 also plays a role in tumor growth and metastasis. KLF16 promotes gastric cancer proliferation by regulating p21 and Cyclin-dependent kinases 4 (CDK4).12 However, in glioma, KLF16 acts as a tumor suppressor by targeting the expression of mitochondrial transcription factor A (TFAM).13 It appears that the role of KLF16 in cancer development may depend on the type of cancer. Although the involvement of KLFs members in BC development has been reported, such as KLF8,14 KLF4,15 KLF2,16 and KLF10,17 the clinical significance and function of KLF16 in BC remain to be determined.
In this study, we explored the importance of KLF16 in the clinical evaluation of BC using in vitro and in vivo experiments, in addition to exploring its downstream molecular effects. KLF16 was upregulated and conferred a worse prognosis in patients with BC. Ectopic expression of KLF16 promoted BC cell growth and migration, and opposite effects were presented when KLF16 expression was knocked down. We also determined that KLF16 transcriptionally repressed the TGF-type III receptor (TGFBR3), which has been proposed as a potential tumor suppressor gene. Furthermore, KLF16 downregulation of TGFBR3 was also critical for BC tumorigenesis in nude mice. The above results proposed that KLF16 promoted BC growth and migration through negative regulation of TGFBR3.
Materials and Methods
Analyzing KLF16 and TGFBR3 in BC from the Cancer Genome Atlas Datasets
The clinical relevance of KLF16 and TGFBR3 in BLCA was analyzed based on public data sets from The Cancer Genome Atlas (TCGA) by using the website: http://ualcan.path.uab.edu website, and The Cancer Genome Atlas (http://cancergenome.nih.gov) was performed for in-deep analysis, as previous studies indicated.18 A total of 404 cancer and 28 normal samples were included to examine the expression of KLF16 and TGFBR3. The patients were stratified according to high expression of KLF16 (n = 340) and low expression of KLF16 (n = 341) to analyze overall survival. The correlation between klf16 and TGFBR3 was examined in BC samples.
Immunohistochemical (IHC) Staining of KLF16 in BC Tissue Microarray
To detect the expression of KLF16 in BC samples, we obtained BC tissue microarray data from ServiceBio (Wuhan servicebio technologies co Ltd, Wuhan, China). The microarray contained 12 BC tissues and adjacent normal tissues. IHC staining of KLF16 was carried out according to the protocols described previously.19 The KLF16 antibody was purchased from Invitrogen. This study was approved by the Ethics Committee of Xuanwu Hospital Capital Medical University (No.2018-086)). All patients provided written informed consent for the information to be used in our hospital database. The study protocol and all related items followed the Declaration of Helsinki guidelines.
Cell Culture
The human normal bladder epithelial cell line SV-HUC-1 and the BC cell lines T24, 5637, and RT-4 were obtained from ATCC. Cells were cultured and grown in Dulbecco-modified Eagle medium, following the protocols described previously.19
KLF16 and TGFBR3 Knockdown
The expression of KLF16 and TGFBR3 was knocked down using small interfering RNAs (siRNAs), which were obtained from Huzhou Hippo Biotechnology Co., Ltd. The siRNAs were transfected using RNAiMAX (Invitrogen), following the manufacturer’s instructions. Targeted sequences for siCtrl, siKLF16#1, siKLF16#2, and siTGFBR3 were as follows: 5′-UUCUCCGAACGUGUCACGU-3′, 5′-GGGUUCUUCCAAAGAACAU-3′, 5′-GGUAUCACGUGACAAUCAA-3′, and 5′-TAATGGATTTCCGGGAGATAT-3′, respectively.
Overexpression of KLF16 and TGFBR3
KLF16 and TGFBR3 were overexpressed by infecting BC cells with lentiviral vectors, which were packaged in 293FT cells and enriched by ultracentrifugation. After infecting BC cells twice, cells were collected for overexpression detection and subsequent studies, such as CCK8, colony formation, apoptosis, Transwell assays, and effects on in vivo tumorigenesis.
Immunoblotting
Immunoblotting was carried out according the protocols described previously.19 Antibody information was described as follows: anti-KLF16 (Invitrogen, 1:1000), anti-TGFBR3 (ThermoFisher, 1:800), anti-GAPDH (Proteintech, 1:3000), anti-β-actin (Proteintech, 1:3000), secondary antibodies (Proteintech, 1:8000).
Quantitative Real-Time PCR (qRT-PCR)
RNA extraction, reverse transcription, and qPCR assays were performed according to protocols described previously.19 The qPCR primer sequences used were as follows: KLF16-F 5′-CAAGTCCTCGCACCTAAAGTC-3′ and KLF16-R 5′-AGCGGGCGAACTTCTTGTC-3′; TGFBR3-F 5′-GTGTTCCCTCCAAAGTGCAAC-3′ and TGFBR3-R 5′-AGCTCGATGATGTGTACTTCCT-3′; PCNA-F 5′-CCTGCTGGGATATTAGCTCCA-3′ and PCNA-R 5′-CAGCGGTAGGTGTCGAAGC-3′; and β-actin-F 5′-CATGTACGTTGCTATCCAGGC-3′ and β-actin-R 5′-CTCCTTAATGTCACGCACGAT-3′.
Cell Proliferation
Cell proliferation was assessed using the CCK8 assay. Briefly, an equal number of T24 and 5637 cells with KLF16/TGFBR3 knockdown or overexpression were seeded in 96-well plates. Cells were incubated with 10% of CCK8 solution for 2 to 4 hours, as indicated by the experiment. After detecting OD450 values, cell proliferation was adjusted to the OD450 values on day 1.
Colony Formation Assay
To perform the colony formation assay, we seeded an equal number of indicated cells in 6-well plates and supplied each well with 3 mL of complete medium. Nine days later, the medium was removed and the colonies were stained with Giemsa stain and washed with clean water. The images of the colonies were obtained using a camera.
Apoptosis Assay
To perform the apoptosis assay, the PI/Annexin V-APC assay kit (Thermo) was performed. The equal number of BC cells were collected and detected using a FACScan flow cytometer (BD Bioscience) as the manufacturers’ protocols indicated.
Transwell Assays
Cell migration was determined by the Transwell assay. We first seeded an equal number of BC cells on the upper surface of the Transwell chamber, which contained 200 μL of FBS-free culture medium. The chamber was then inserted into a 24-well plate containing 500 μL complete medium. After 24 hours, the cells on the upper surface were removed and the cells on the lower surface were stained with GIMSA buffer. Cell migration pictures were taken under the microscope.
Tumorigenesis of T24 Cells
An equal number of T24 cells (106/mice) were injected subcutaneously into the right flank region of female nude mice; 4–6-week-old Balb/c mice were obtained from Charles River and maintained in a standard animal room (25°C, 12-h light/12-h dark cycle). Fifteen mice were randomly divided into three groups. Tumors developed 14 days after implantation, and tumor volume was calculated by measuring the longest and shortest diameter. The mice were sacrificed by peritoneal injection of chloral hydrate. Animal experiments were carried out following the animal experimentation guidelines and were approved by the Ethics Committee of the Xuanwu Hospital of Capital Medical University.
Dual-Luciferase Reporter Assay
The coding sequence of KLF16 and a Flag sequence were inserted into pCDNA3.1 plasmids. The TGFBR3 promoter sequence was inserted into the pGL3-Basic vectors. To verify dual luciferase activity, pCDNA3.1 (pCDNA3.1-empty or pCDNA3.1-KLF16) plasmids, as well as the pGL3. Basic-TGFBR3 and TK plasmids, were transfected in HEK293 cells. After 48 hours, dual luciferase activity was measured with TK acting as an internal control.
Chromatin Immunoprecipitation (ChIP)-qPCR Assay
The ChIP-qPCR assay was carried out according to the manufacturer’s protocols (Cell Signaling, 9005S). The results were normalized to IgG expression. The qPCR primer sequences TGFBR3 used were forward 5′-GGGAGCCCTGGAAAGTTCAA-3′ and reverse 5′-GAGAGAGAAGGGGCTAGGGG-3′, respectively.
Statistical Analysis
The statistical data were presented as mean ± standard error of mean (SEM) with three independent results. GraphPad prism software was used for statistical analysis. Student’s t tests were used to compare differences between two groups. One-way ANOVA was used to compare differences among groups. A p-value <0.05 was considered statistically significant.
Results
Increased KLF16 Expression in Human Bladder Cancer Specimens
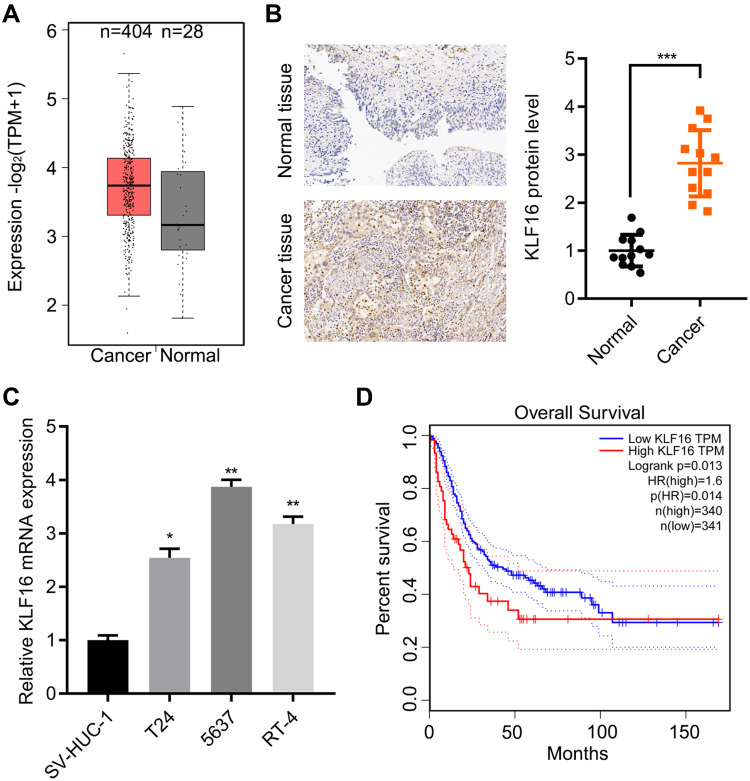
To examine the expression of KLF16 in human BC patients, we first analyzed datasets obtained from TCGA database, in which 404 cancer and 28 normal specimens were available. The expression of KLF16 mRNA was increased in cancer specimens (Figure 1A). Next, we detected the expression of the KLF16 protein expression by IHC staining and found a marked nuclear expression of KLF16 in cancer tissues compared to normal tissues (Figure 1B). The expression of KLF16 was also evaluated in normal bladder cells SV-HUC-1 and a panel of BC cell lines. An increased expression of KLF16 mRNA was observed in T24, 5637 and RT-4 cells (Figure 1C). To provide stronger evidence supporting the clinical importance of KLF16, we analyzed the correlation between KLF16 expression and the prognosis of BC patients from TCGA. The results showed that patients with high expression of KLF16 had shorter overall survival than those with low expression of KLF16 (Figure 1D). Taken together, KLF16 overexpression was likely associated with the malignant progression of patients with BC.
Figure 1.
KLF16 is highly expressed in BC specimens. (A) The levels of KLF16 transcript were evaluated in 404 of bladder urothelial carcinoma (BLCA) and 28 normal samples from the TCGA database, p<0.05. (B) Immunohistochemical staining of KLF16 in 12 pairs of BC and normal samples. The quantitative result is presented. ***p<0.001. (C) qRT-PCR detection of KLF16 in SV-HUC-1, T24, 5637, and RT-4 cells. The expression of KLF16 was adjusted to β-actin expression. *p<0.05. **p<0.01. (D) BLCA patients were grouped into high- and low-expression KLF16 groups according to TCGA database. Overall survival was analyzed in the patients. p=0.013.
KLF16 is Critical for BC Cell Growth and Migration
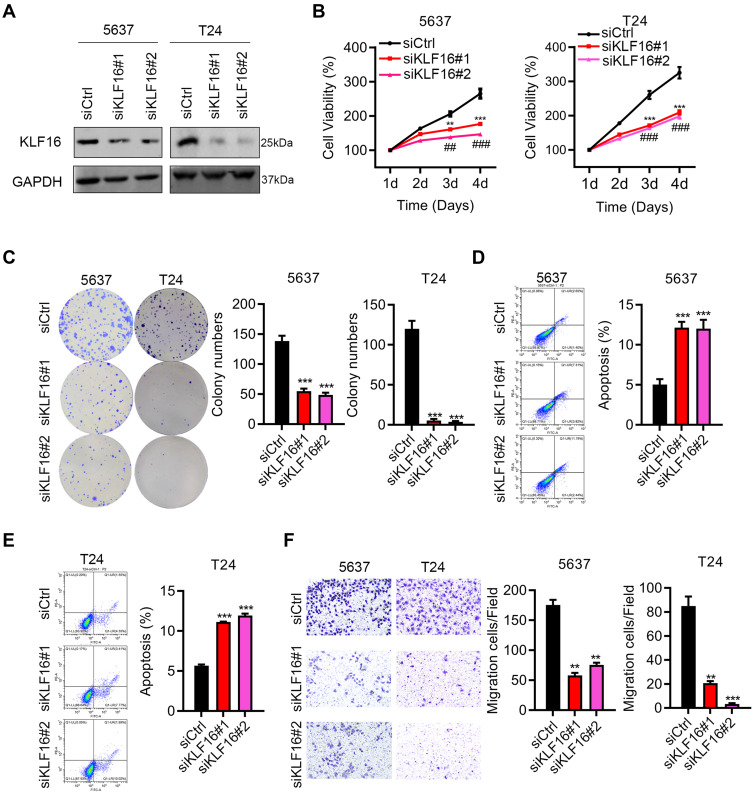
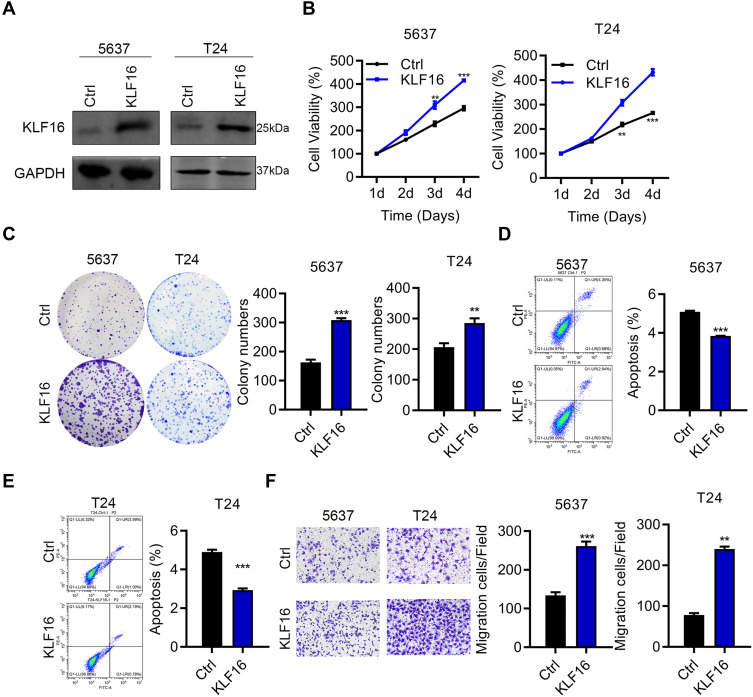
Next, we investigated the function of KLF16 by modulating its expression in BC cells. Because KLF16 levels were relatively higher in 5637 cells, we used siRNAs to interfere with KLF16 expression. Knockdown efficiency was demonstrated by immunoblotting results (Figure 2A). Downregulation of KLF16 suppressed the proliferation and colony growth of 5637 and T24 cells (Figure 2B and C). Increased apoptosis was detected in 5637 and T24 cells after knocking down KLF16 expression (Figure 2D and E). Furthermore, KLF16 downregulation reduced the migration capacity of 5637 and T24 cells (Figure 2F). To determine whether KLF16 expression is sufficient to induce cell growth and migration, we overexpressed KLF16 in 5637 and T24 cells, which had relatively lower expression of KLF16. The immunoblot results suggested that KLF16 was ectopically expressed by lentivirus infection (Figure 3A). The CCK8 and colony formation assays showed that KLF16 overexpression enhanced the growth ability of T24 cells (Figure 3B and C). Apoptosis was reduced by upregulation of KLF16 (Figure 3D and E). KLF16 also promoted T24 cell migration (Figure 3F). Therefore, KLF16 acts as an oncogene in BC.
Figure 2.
KLF16 is essential for BC cell growth and migration. (A) Immunoblotting analysis of KLF16 in siCtrl, siKLF16#1 and siKLF16#2-transfected 5637 and T24 cells. Expression of KLF16 was adjusted to GAPDH expression. (B) Cell proliferation was examined in siCtrl, siKLF16#1, and siKLF16#2-treated 5637 and T24 cells. siCtrl vs siKLF16#1 **p<0.01 and siCtrl vs siKLF16#1 ***p<0.001, siCtrl vs siKLF16#2 ##p<0.01, siCtrl vs siKLF16#2 ###p<0.001. (C) Colony growth was evaluated in siCtrl, siKLF16#1, and siKLF16#2-treated 5637 and T24 cells. **p<0.01, ***p<0.001. (D and E) Cell apoptosis was detected by PI/Annexin V staining in siCtrl, siKLF16#1 and siKLF16#2-treated 5637 and T24 cells, respectively. ***p<0.001. (F) Cell migration was assessed using Transwell assays using 5637 and T24 cells exposed to siCtrl, siKLF16#1, and siKLF16#2, respectively. **p<0.01, ***p<0.001.
Figure 3.
Overexpression of KLF16 promoted BC cell growth and migration. (A) Immunoblot analysis of KLF16 in 5637 and T24 cells overexpressing Ctrl and KLF16. Expression of KLF16 was adjusted to GAPDH expression. (B) Cell proliferation was examined in 5637 and T24 cells overexpressing Ctrl and KLF16. **p<0.01, ***p<0.001. (C) Colony growth was evaluated in 5637 and T24 cells overexpressing Ctrl and KLF16. **p<0.01, ***p<0.001. (D and E) Cell apoptosis was detected by PI/Annexin V staining in Ctrl and KLF16 overexpressing 5637 and T24 cells. ***p<0.001. (F) Cell migration was detected by the Transwell assay in 5637 and T24 cells overexpressing Ctrl and KLF16. **p<0.01, ***p<0.001.
KLF16 Downregulated TGFBR3 by Directly Inactivating Its Transcriptional Activity
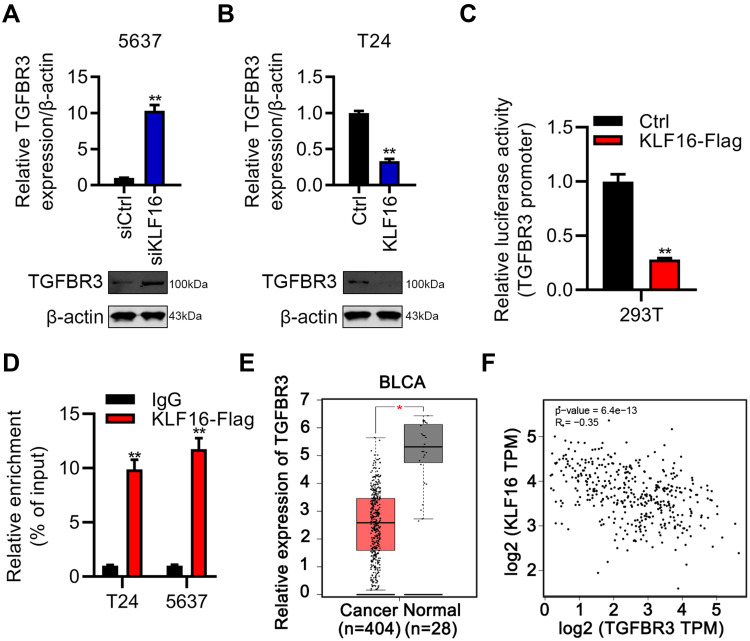
After demonstrating the oncogenic function of KLF16, we then attempted to dissect the molecular mechanism downstream of KLF16 in BC. Based on the results of qRT-PCR and immunoblotting, we showed that KLF16 interference resulted in upregulation of TGFBR3 in 5637 cells. Conversely, KLF16 overexpression decreased TGFBR3 mRNA and protein expression in 5637 and T24 cells, respectively (Figure 4A and B). To verify whether KLF16 regulated the transcription activity of TGFBR3, we inserted a KLF16-flag sequence into pCDNA3.1 vector and the promoter sequence of TGFBR3 into the pGL3. Basic vector. Subsequently, a dual-luciferase activity reporter assay was carried out by co-transfection of the vectors into HEK293 cells. KLF16 overexpression reduced luciferase activity, indicating that KLF16 negatively regulated the transcriptional activity of TGFBR3 (Figure 4C). To identify the interaction between KLF16 and TGFBR3, a ChIP-qPCR assay was performed and the results showed that KLF16 bound to the promoter sequence of TGFBR3 (Figure 4D). Finally, we analyzed the clinical importance of TGFBR3 in BC by analyzing the TCGA database. We found that TGFBR3 was downregulated in BC samples (Figure 4E). Importantly, KLF16 expression exhibited an inverse correlation with TGFBR3 expression in BC samples (Figure 4F). These results suggested that KLF16 may downregulate TGFBR3 expression in BC patients.
Figure 4.
KLF16 negatively regulates TGFBR3 by inhibiting its transcriptional activity. (A and B) mRNA and protein were determined by qRT-PCR and immunoblotting and normalized to β-actin in KLF16 silencing and overexpressing cells. (C) 293T cells were transfected with pCDNA3.1-Ctrl or pCDNA3.1-KLF16, pGL3. Basic-TGFBR3 promoter, and TK vectors. 48 hours later, dual luciferase activity was assessed and adjusted for TK. **p<0.01. (D) The ChIP-qPCR assay was performed in T24 and 5637 cells overexpressing KLF16-flag. Relative enrichment was adjusted to the IgG group. **p<0.01. (E) TGFBR3 transcript levels in 404 BC and 28 normal samples were assessed using datasets obtained from TCGA database. *p<0.05. (F) Spearman correlation between KLF16 and TGFBR3 was analyzed in BLCA samples from TCGA database. p<0.0001.
KLF16 Downregulation of TGFBR3 Supported BC Growth and Migration
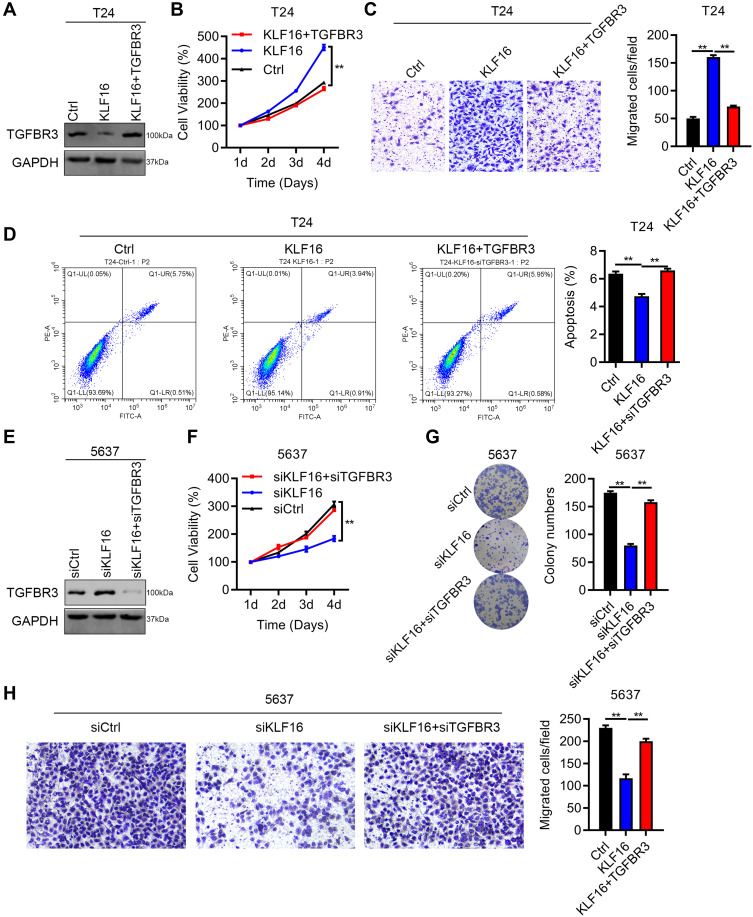
Previous studies have reported that TGFBR3 acted as a tumor suppressor gene in the development of various cancers, including lung cancer and head and neck cancer. However, the role of TGFBR3 in BC remains to be investigated. As TGFBR3 is downregulated by KLF16, we upregulated TGFBR3 in cells overexpressing KLF16 (Figure 5A). We showed that TGFBR3 overexpression reduced the growth capacity of T24 cells (Figure 6B). As expected, TGFBR3 overexpression also inhibited the migration of T24 cells overexpressing KLF16 (Figure 6C). Consistently, the apoptosis of T24 cells suppressed by KLF16 overexpression was increased after overexpressing TGFBR3 (Figure 6D). Next, we silenced TGFBR3 expression in 5637 cells with KLF16 knockdown (Figure 6E). The results of the CCK8 and colony formation assays showed that TGFBR3 downregulation restored the growth ability of 5637 cells, which was suppressed by KLF16 knock-down (Figure 6F and G). Similar results were observed in the migration capacity of 5637 cells (Figure 6H). Therefore, our findings revealed that downregulation of TGFBR3 contributed to the oncogenic role of KLF16 in BC cell growth and migration.
Figure 5.
Downregulation of TGFBR3 by KLF16 contributes to BC growth and migration. (A–D) T24 cells were transfected with lentivirus overexpressing Ctrl, KLF16, and KLF16+TGFBR3. The cells were subjected to immunoblotting analysis of TGFBR3 (A), CCK8 assay (B), Transwell assay (C), and PI/Annexin V-FITC staining of apoptosis (D). Quantification results were presented. **p<0.01. (E–H) 5637 cells were transfected with siRNAs against Ctrl, KLF16 and KLF16+TGFBR3. The cells were subjected to immunoblotting analysis of TGFBR3 (E), CCK8 assay (F), colony formation (G), and Transwell assay (H). Quantification results were presented. **p<0.01.
Figure 6.
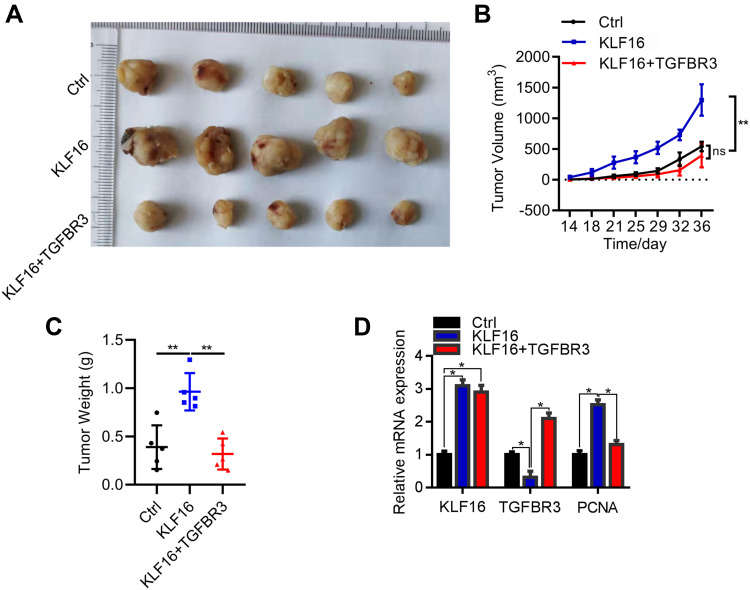
Downregulation of TGFBR3 by KLF16 promotes BC tumorigenesis. (A) Macroscopic images of xenografted tumors of T24 cells transfected with the lentivirus Ctrl (negative control), KLF16 (KLF16 overexpression), and KLF16+TGFBR3 (KLF16 and TGFBR3 overexpression) lentivirus. (B) Tumor growth was evaluated on day 14 after implantation. ns, not significant. **p<0.01. (C) Tumors were weighed on day 36 after implantation. **p<0.01. (D) qRT-PCR analysis of KLF16, TGFBR3, and PCNA in tumors. *p<0.05.
Downregulation of TGFBR3 by KLF16 Contributed to BC Tumorigenesis
Finally, we explored the in vivo function of the KLF16/TGFBR3 axis in BC development. To this end, we injected an equal amount of T24 cells, which were transfected with the lentivirus Ctrl (negative control), KLF16 (KLF16 overexpression), or KLF16+TGFBR3 (KLF16 and TGFBR3 overexpression) lentivirus, into the right upper limb of 5-week-old nude mice. Each group contained 5 mice. Tumorigenesis was monitored after implantation. The results showed that upregulation of KLF16 promoted T24 tumorigenesis in nude mice, which could be suppressed by TGFBR3 overexpression (Figure 6A–C). The tumor tissues were then subjected to qRT-PCR analysis of components of the KLF16/TGFBR3 axis. The results showed that KLF16 negatively regulated TGFBR3 during BC tumorigenesis (Figure 6D). We also found that KLF16 overexpression increased proliferating cell nuclear antigen (PCNA) levels, which could be reversed by ectopic expression of TGFBR3 (Figure 6D). Collectively, our findings suggested that the downregulation of TGFBR3 by KLF16 promotes BC tumorigenesis in vivo.
Discussion
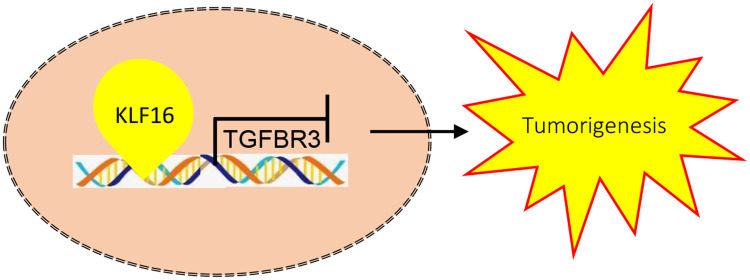
KLFs have been shown to participate in the progression of a wide variety of cancers, while the function of KLF16 in BC is poorly understood. In this study, we demonstrated that KLF16 served as an oncoprotein in BC. KLF16 was not only overexpressed in BC samples, but also predicted a poor prognosis in BC patients. In vitro, the upregulation of KLF16 promoted tumor formation, while its downregulation suppressed BC cell growth and migration. We also found that KLF16 expression supported tumorigenesis of BC cells in nude mice. Mechanistically, KLF16 protein could interact with the promoter sequence of TGFBR3. Through this interaction, KLF16 reduced the transcription activity of the TGFBR3 gene. In vitro and in vivo studies demonstrated that the negative regulation of TGFBR3 by KLF16 was responsible for cancer growth and migration. Thus, KLF16 may contribute to BC progression by downregulating TGFBR3 (Figure 7).
Figure 7.
The schematic mechanisms of KLF16 promotes BC proliferation and migration via downregulating TGFBR3.
The well-known transcription factors include specificity proteins (SPs) and Krüppel-like factors (KLFs), both of which harbor the conserved zinc finger.20 Similar to SPs, KLFs recognize the DNA sequence that contains enriched 5′-CACCC-3′ elements.21 Unlike SPs, KLFs have different downstream effectors and exhibit distinct physiological and pathological roles. For the past decades, scientists have paid greater attention to the role of transcription factors in cancer development. As a member of the KLF family, KLF16 can be overexpressed or downregulated in different types of cancer. For example, KLF16 was upregulated in samples of gastric cancer, prostate cancer, retinoblastoma, and breast cancer. KLF16 overexpression is correlated with the stage of the disease, the metastasis status, and the prognosis of cancer patients. Upregulation of KLF16 also contributed to tumor growth and metastasis in these cancers by modulating cyclin-dependent kinase inhibitor 1A (p21), CDK4, and Bcl-2-like protein 15 (BCL2L15).12,22–24 In contrast, KLF16 was downregulated in patients with glioma and had tumor suppressive function by regulating transcription factor A, mitochondrial (TFAM).13 These results indicated that KLF16 had both oncogenic and tumor suppressive function based on cancer type and downstream effector diversities. Consistent with the oncogenic function of KLF16, herein we demonstrated that KLF16 was highly expressed in BC specimens and was inversely associated with the survival of BC patients. By altering the expression of KLF16, we revealed that the expression of KLF16 was essential for the growth and migration ability of BC cells in vitro. KLF16 also repressed apoptosis in vitro and promoted tumorigenesis in vivo. These results suggested that KLF16 was likely an oncogene in BC.
Furthermore, we also found that KLF16 suppressed TGFBR3 expression at both the mRNA and protein level. Consistent with previous studies that confirmed that KLF16 negatively regulated the expression of BLC2L15 and TFAM, we show here that KLF16 reduced the transcription activity of the TGFBR3 gene by direct binding to its promoter sequence. This implied that KLF16 had a transcription repressor function in regulating TGFBR3. Importantly, KLF16 expression was inversely correlated with TGFBR3 expression in human BC samples, suggesting that KLF16 may downregulate TGFBR3 during human BC progression. Previously studies have reported that TGFBR3 exhibited tumor suppressive roles in all kinds of cancers. Both a mothers against DPP homolog 4 (SMAD4)-dependent or independent mechanisms are involved in TGFBR3-induced suppression of head and neck cancer cell migration and invasion.25 In addition, TGFBR3 blocks lung cancer tumorigenesis by regulating epithelial-mesenchymal transition.26 A tumor suppressive function of TGFBR3 has also been described in other types of cancer, such as renal cell carcinoma,27 tongue squamous cell carcinoma,28 and hepatocellular carcinoma.29 However, TGFBR3 is elevated in esophageal squamous cell carcinoma and may serve as an oncogene. In this study, based on loss-of-function and gain-of-function results, we showed that TGFBR3 overexpression reduced the promoting effect of KLF16 overexpression, while TGFBR3 knockdown enhanced the suppressive effect of KLF16 silencing on BC cell growth, migration, and tumorigenesis. Our study suggested that TGFBR3 was a tumor suppressor in BC.
Conclusions
In summary, our findings showed that KLF16 is a potential oncogene in BC. Upregulation of KLF16 was observed in human BC samples or in patients with shorter overall survival. In vitro and in vivo, KLF16 transcriptionally suppressed TGFBR3 expression to promote BC cell growth, migration, and tumorigenesis. Our study provides evidence that the negative regulation of TGFBR3 by KLF16 contributes to the progression of BC. Targeting this axis may become a promising treatment approach for patients with BC.
Acknowledgments
We thank Chunsong Jia for his assistance on experiments performance during this study.
Funding Statement
The research was supported by Beijing Municipal Hospital Scientific Research and Cultivation Project (XWJL-2019025). The funders provided assistance for materials purchasing but not for study design and experiments performance.
Data Sharing Statement
Research Data are included in the manuscript.
Author Contributions
XC and PW contributed to the study design, experiments performance and data analysis. TO and JL provided assistance for experiments performance and data analysis. XC and PW wrote the manuscript. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declared no conflicts of interest in this work.
References
- 1.Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71(1):96–108. doi: 10.1016/j.eururo.2016.06.010 [DOI] [PubMed] [Google Scholar]
- 2.Kamat A, Hahn N, Efstathiou J, et al. Bladder cancer. Lancet. 2016;388(10061):2796–2810. doi: 10.1016/S0140-6736(16)30512-8 [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Miller K, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade P, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 5.Ashrafizadeh M, Hushmandi K, Hashemi M, et al. Role of microRNA/epithelial-to-mesenchymal transition axis in the metastasis of bladder cancer. Biomolecules. 2020;10(8):8. doi: 10.3390/biom10081159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirzaei S, Gholami MH, Mahabady MK, et al. Pre-clinical investigation of STAT3 pathway in bladder cancer: paving the way for clinical translation. Biomed Pharmacother. 2021;133:111077. [DOI] [PubMed] [Google Scholar]
- 7.Ashrafizadeh M, Yaribeygi H, Sahebkar A. Therapeutic effects of curcumin against bladder cancer: a review of possible molecular pathways. Anticancer Agents Med Chem. 2020;20(6):667–677. doi: 10.2174/1871520620666200203143803 [DOI] [PubMed] [Google Scholar]
- 8.Mirzaei S, Paskeh MDA, Hashemi F, et al. Long non-coding RNAs as new players in bladder cancer: lessons from pre-clinical and clinical studies. Life Sci. 2021;288:119948. doi: 10.1016/j.lfs.2021.119948 [DOI] [PubMed] [Google Scholar]
- 9.Ashrafizadeh M, Zarrabi A, Samarghandian S, Najafi M. PTEN: what we know of the function and regulation of this onco-suppressor factor in bladder cancer? Eur J Pharmacol. 2020;881:173226. doi: 10.1016/j.ejphar.2020.173226 [DOI] [PubMed] [Google Scholar]
- 10.Zhu B, Liu Q, Han Q, Zeng B, Chen J, Xiao Q. Downregulation of Krüppel‑like factor 1 inhibits the metastasis and invasion of cervical cancer cells. Mol Med Rep. 2018;18(4):3932–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R, Xu J, Xu J, et al. MiR-326/Sp1/KLF3: a novel regulatory axis in lung cancer progression. Cell Prolif. 2019;52(2):e12551. doi: 10.1111/cpr.12551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma P, Sun C, Wang Y, et al. KLF16 promotes proliferation in gastric cancer cells via regulating p21 and CDK4. Am J Transl Res. 2017;9(6):3027–3036. [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Li S, Ke Y, et al. KLF16 suppresses human glioma cell proliferation and tumourigenicity by targeting TFAM. Artif Cells Nanomed Biotechnol. 2018;46(sup1):608–615. doi: 10.1080/21691401.2018.1431654 [DOI] [PubMed] [Google Scholar]
- 14.Liang K, Liu T, Chu N, et al. KLF8 is required for bladder cancer cell proliferation and migration. Biotechnol Appl Biochem. 2015;62(5):628–633. doi: 10.1002/bab.1310 [DOI] [PubMed] [Google Scholar]
- 15.Ohnishi S, Ohnami S, Laub F, et al. Downregulation and growth inhibitory effect of epithelial-type Krüppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun. 2003;308(2):251–256. doi: 10.1016/S0006-291X(03)01356-1 [DOI] [PubMed] [Google Scholar]
- 16.Yang H, Li G, Cheng B, Jiang R. ZFAS1 functions as an oncogenic long non-coding RNA in bladder cancer. Biosci Rep. 2018;38(3):3. doi: 10.1042/BSR20180475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Q, Yan D, Dong W, et al. circRNA circFUT8 upregulates Krüpple-like factor 10 to inhibit the metastasis of bladder cancer via sponging miR-570-3p. Mol Ther Oncolytics. 2020;16:172–187. doi: 10.1016/j.omto.2019.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Wang P, Wang S, Li J, Ou T, Zeng X. CIZ1 knockdown suppresses the proliferation of bladder cancer cells by inducing apoptosis. Gene. 2019;719:143946. doi: 10.1016/j.gene.2019.143946 [DOI] [PubMed] [Google Scholar]
- 20.Kim C, He P, Bialkowska A, Yang V. SP and KLF transcription factors in digestive physiology and diseases. Gastroenterology. 2017;152(8):1845–1875. doi: 10.1053/j.gastro.2017.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaczynski J, Cook T, Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003;4(2):206. doi: 10.1186/gb-2003-4-2-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Yu W, Wang X, Hu B, Wu D, Shi G. KLF16 affects the MYC signature and tumor growth in prostate cancer. Onco Targets Ther. 2020;13:1303–1310. doi: 10.2147/OTT.S233495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Yan X, Wu S, Liu Q, Ma J. KLF16 overexpression deleteriously affects the proliferation and migration of retinoblastoma by transcriptionally repressing BCL2L15. Biochem Biophys Res Commun. 2020;529(4):977–983. doi: 10.1016/j.bbrc.2020.06.027 [DOI] [PubMed] [Google Scholar]
- 24.Bang S, Li J, Zhang M, et al. The clinical relevance and function of Krüppel-like factor 16 in breast cancer. Cancer Manag Res. 2020;12:6373–6383. doi: 10.2147/CMAR.S256490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang W, Kuo Y, Chang J, et al. The tumor suppressor TGFBR3 blocks lymph node metastasis in head and neck cancer. Cancers. 2020;12(6):1375. doi: 10.3390/cancers12061375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang JJ, Corona AL, Dunn BP, Cai EM, Prakken JN, Blobe GC. Increased type III TGF-β receptor shedding decreases tumorigenesis through induction of epithelial-to-mesenchymal transition. Oncogene. 2019;38(18):3402–3414. doi: 10.1038/s41388-018-0672-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishida J, Miyazono K, Ehata S. Decreased TGFBR3/betaglycan expression enhances the metastatic abilities of renal cell carcinoma cells through TGF-β-dependent and -independent mechanisms. Oncogene. 2018;37(16):2197–2212. doi: 10.1038/s41388-017-0084-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, Liu K, Li Z, Wang J, Wang X. miR-19a and miR-424 target TGFBR3 to promote epithelial-to-mesenchymal transition and migration of tongue squamous cell carcinoma cells. Cell Adh Migr. 2018;12(3):236–246. doi: 10.1080/19336918.2017.1365992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S, Sun W, Wu J, Gu Y, Wei W. Decreased expression of the type III TGF-β receptor enhances metastasis and invasion in hepatocellullar carcinoma progression. Oncol Rep. 2016;35(4):2373–2381. doi: 10.3892/or.2016.4615 [DOI] [PubMed] [Google Scholar]