Abstract
Restriction fragment length polymorphism (RFLP) analysis of IS6110 is commonly used to DNA fingerprint Mycobacterium tuberculosis. However, low-copy (≤5) IS6110 M. tuberculosis strains are poorly differentiated, requiring secondary typing. When spoligotyping was used as the secondary method, only 13% of Maryland culture-positive tuberculosis (TB) patients with low-copy IS6110-spoligotyped clustered strains had epidemiologic linkages to another patient, compared to 48% of those with high-copy strains clustered by IS6110 alone (P < 0.01). Spoligotyping did not improve a population-based molecular epidemiologic study of recent TB transmission.
IS6110 is a 1,361-bp insertion sequence specific for Mycobacterium tuberculosis complex. Genetic stability and method standardization have made restriction fragment length polymorphism (RFLP) analysis of IS6110 useful for molecular epidemiologic studies of tuberculosis (TB) (6, 9, 12, 14, 19, 20, 23). In particular, patient isolates with matching strains (clustering) are used to study recent TB transmission (1–4, 17). Epidemiologic investigations of clustered patients with strains having fewer than six copies of the IS6110 sequence yield few relationships between patients, however, and secondary typing methods are recommended to improve strain differentiation (4, 5, 21, 23).
In a study of M. tuberculosis isolates from all culture-positive patients reported to the Maryland Division of TB Control, Refugee and Migrant Health from January 1996 through December 1998, spoligotyping was used as the secondary typing method for clustered isolates having less than six IS6110 copies. It was of interest to determine whether (i) low-copy IS6110 M. tuberculosis strains were well differentiated by spoligotyping and whether (ii) the resulting IS6110-spoligotyping clusters were associated with recent TB transmission.
For each culture-positive patient, one M. tuberculosis isolate was retrieved from the Maryland State TB Laboratory (73%) or another laboratory where the culture was initially processed. All isolates were freshly grown at the state laboratory and sent to a regional DNA fingerprinting laboratory assigned by the Centers for Disease Prevention and Control. RFLP analysis was performed with chromosomal DNA extraction, PvuII digestion, and hybridization by Southern blot techniques with a digoxigenin-labeled 245-bp portion of the IS6110 repeat element (19). Spoligotyping for all matching strains with fewer than six IS6110 copies was performed with a commercially available kit, according to the manufacturer's instructions (Isogen Bioscience BV, Maarssen, The Netherlands). Patients whose M. tuberculosis strains were genetically related to the strain from another patient were considered clustered. For high-copy (≥6) IS6110 strains, patients whose isolate patterns matched exactly, plus or minus one band, were assigned a single cluster designation (5, 10, 14, 23). For low-copy (<6) strains, cluster designations were assigned to those patients whose isolates exactly matched by both RFLP analysis and spoligotyping.
An epidemiologic linkage was assigned to patients who named another clustered patient or were in the same place at the same time as another patient (e.g., a bar or club), even without being named. Local health department medical records were abstracted for all clustered patients to determine whether linkages existed. Data included medical history, country of origin, travel, workplace or school, social activities (e.g., church or club), known or suspected TB exposure, and contact investigation records. If record reviews revealed no linkages, patients were personally interviewed; proxy interviews were attempted when patients were not available. Trained personnel conducted record reviews and interviews by using standardized questionnaires. Consent forms were signed before each interview, and Maryland State Institutional Review Board approval was given for all project activities.
We compared the proportions of clustered patients with identified epidemiologic linkages among patients with low- and high-copy clustered strains, adjusting for the number of patients in a cluster and geographic dispersal of cluster members throughout the state. Countries of origin were also reviewed, since low-copy strains are sometimes associated with the foreign-born, particularly those originating from Asia (9, 10). Statistical analyses were conducted with the chi-square test. P values of <0.05 were considered significant.
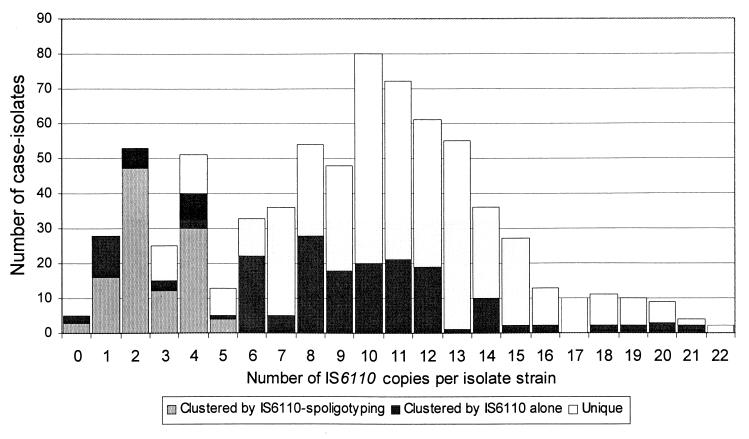
Of 742 culture-positive patients, two with Mycobactarium bovis were excluded. Four (0.5%) M. tuberculosis patient isolates without available DNA fingerprints were also excluded. Of the remaining 736, 175 (24%) had low-copy strains, including 5 with no IS6110 copies. RFLP analysis of IS6110 alone produced 13 groups with identical patterns among 145 patients with low-copy strains (median, 7 patients/group; range, 2 to 52). These groups were further differentiated by spoligotyping into 24 clusters of 112 patients with low-copy strains (median, 6 patients/cluster; range, 2 to 29). (The remaining 33 patient isolates with matching IS6110 patterns produced unique spoligotypes.) The 112 patients plus 157 patients with high-copy strains accounted for all clustered patients (269 of 736 [37%]) in 72 total clusters (median, 3 patients/cluster; range, 2 to 29). Seventy-two percent (405 of 561) of high-copy isolates had unique DNA fingerprints. However, only 36% (63 of 175) of low-copy isolates were unique even with secondary typing (P < 0.01) (Fig. 1). All 269 medical records were reviewed; 84% of eligible, locatable patients were interviewed in person (n = 69) or by proxy (n = 3).
FIG. 1.
Distributions of M. tuberculosis patient isolates clustered by IS6110 alone and by IS6110 spoligotyping, by copy number, in Maryland 1996 to 1998. Note that secondary typing was conducted by the spoligotyping method for all isolates with ≤5 copies of IS6110 that were clustered by RFLP analysis of IS6110 alone.
Nearly one-quarter of clustered patients were 65 years of age or older, although elderly patients are more likely to have acquired their TB infection in the distant past and consequently to have unique M. tuberculosis strains (1, 15, 17). Further investigation revealed that our elderly clustered patients were significantly more likely than younger ones to have low-copy M. tuberculosis strains (53% versus 41%; P = 0.04).
Epidemiologic linkages were identified for 34% of the 269 clustered patients compared with 20 to 25% reported in most studies (1–3, 17). The likelihood of finding linkages was not related to cluster size (≤5 patients/cluster versus >5 patients/cluster) (Table 1). Linkages were identified four times more frequently among high-copy than low-copy clustered patients, regardless of the number of jurisdictions encompassed by a single cluster or the countries of origin of cluster members.
TABLE 1.
Identification of epidemiologic linkages among DNA clustered patients by number of patients in the cluster and number of IS6110 copies in clustered M. tuberculosis strains in Maryland 1996 to 1998
| Cluster characteristics | No. (%) of clustered patients:
|
P value | |
|---|---|---|---|
| With identified linkages | Without linkages | ||
| Large (>5 patients) | 41 (34.2) | 79 (65.8) | 0.92 |
| Small (≤5 patients) | 50 (33.5) | 99 (66.5) | |
| Low-copy strain (≤5 copies) | 15 (13.3) | 97 (86.7) | <0.01 |
| High-copy strain (>5 copies) | 76 (48.4) | 81 (51.6) | |
Low-copy clustered strains were not significantly associated with foreign birth (P = 0.26) and were more common among patients in mixed ethnicity clusters than foreign-born-only clusters (P = 0.046). Approximately one-half of clustered patients originating from Asia (18 of 32) and Africa (11 of 22) had low-copy strains. U.S.-born patients with low-copy strains were significantly more likely than foreign-born patients to be 65 years or older (37% versus 15%, respectively; P = 0.01). No foreign-born Hispanic clustered patients (n = 8) had a low-copy M. tuberculosis strain.
Secondary typing to improve strain differentation among low-copy IS6110 isolates is often conducted by using the highly specific polymorphic GC-rich repetitive sequence (PGRS) found in the plasmid pTBN12 (4, 5, 7). However, the process is costly and labor intensive, and its usefulness has been questioned for population-based studies of TB transmission (16). Spoligotyping (spacer oligonucleotide typing) detects unique spacer sequences in the M. tuberculosis genome, independent from the IS6110 sequence, that can be amplified and hybridized (13). Spoligotyping is rapid, easily performed, and inexpensive and seemed a practical approach to increase strain differentiation.
Recently, Soini et al. (18) reported that the use of spoligotyping as a secondary typing method for low-copy IS6110 M. tuberculosis strains could provide a “reliable index of disease transmission.” Their finding of 14.5% epidemiologic linkages was nearly identical to our 13.3%, even though Houston's (Tex.) case of infection rate was three times Maryland's (19.7 versus 6.4 per 100,000), and their clustered population differed ethnically from ours. One might expect that high disease incidence would be associated with increased transmission, producing a greater proportion of linkages between patients. Unlike their study, however, we reported no foreign-born Hispanics with low-copy clustered strains and little association with other countries of origin. In both population-based studies, intensive study of clustered patients with low-copy strains yielded no linkages for 85% of patients. By the same methodology, however, we were able to identify linkages for nearly one-half of clustered patients with high-copy strains. We concluded that spoliogotyping did not substantially differentiate between related and unrelated patients with low-copy IS6110 strains.
We did not subtype our low-copy M. tuberculosis isolates by the PGRS method. However, Yang et al.(22) reported that PGRS fingerprinting was more discriminating than spoligotyping for determining related strains among low-copy IS6110 isolates and that IS6110-PGRS clusters were more closely associated with finding linkages between patients than were IS6110- spoligotyping clusters.
Spoligotyping alone or in tandem with RFLP analysis of IS6110 appears to be useful for timely confirmation of laboratory contamination and outbreak evaluation (8, 11). We recently identified a cluster of eight TB patients who had a common single-copy M. tuberculosis strain with a spoligotype that first appeared in our state in 1999. Five (63%) of the patients had epidemiologic linkages. Confining investigation of low-copy clusters to those with rare spoligotypes may also be helpful in population-based epidemiologic studies.
In conclusion, spoligotyping did not appreciably improve strain differentiation among isolates with few IS6110 copies. Although this method may be useful for timely study of suspected outbreaks, more discriminating methods for secondary typing of low-copy M. tuberculosis strains should be utilized to evaluate recent TB transmission in population-based studies.
REFERENCES
- 1.Alland D, Kalkut G E, Moss A R, McAdam R A, Hahn J A, Bosworth W, Drucker E, Bloom B R. Transmission of tuberculosis in New York City: an analysis by DNA fingerprinting and conventional epidemiological methods. N Engl J Med. 1994;300:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 2.Barnes P F, Yang Z, Preston-Martin S, Pogoda J M, Jones B E, Otaya M, Eisenach K D, Knowles L, Harvey S, Cave M D. Patterns of tuberculosis transmission in Central Los Angeles. JAMA. 1997;278:1159–1163. [PubMed] [Google Scholar]
- 3.Bishai W R, Graham N M H, Harrington S, Pope D S, Hooper N, Astemborski J, Sheely L, Vlahov D, Glass G E, Chaisson R E. Molecular and geographic patterns of tuberculosis transmission after 15 years of directly observed therapy. JAMA. 1998;280:1679–1684. doi: 10.1001/jama.280.19.1679. [DOI] [PubMed] [Google Scholar]
- 4.Braden C R, Templeton G L, Cave M D, Valway S, Onorato I M, Castro K G, Moers D, Yang Z, Stead W W, Bates J H. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J Infect Dis. 1997;175:1446–1452. doi: 10.1086/516478. [DOI] [PubMed] [Google Scholar]
- 5.Burman W J, Reves R R, Hawkes A P. DNA fingerprinting with two probes decreases clustering of Mycobacterium tuberculosis. Am J Respir Crit Care Med. 1997;155:1140–1146. doi: 10.1164/ajrccm.155.3.9117000. [DOI] [PubMed] [Google Scholar]
- 6.Cave M D, Eisenbach K D, Templeton G, Salfinger M, Mazurek G, Bates J H, Crawford J T. Stability of DNA genotype pattern produced with IS6110 in strains of Mycobacterium tuberculosis. J Clin Microbiol. 1994;32:262–266. doi: 10.1128/jcm.32.1.262-266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaves F, Yang Z, el Hajj H, Alonso M, Burman W J, Eisenach K D, Dronda F, Bates J H, Cave M D. Usefulness of the secondary probe pTBN12 in DNA fingerprinting of Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:1118–1123. doi: 10.1128/jcm.34.5.1118-1123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis A B, Ridzon R, Novick L F, Driscoll J, Blair D, Oxtoby M, McGarry M, Hiscox B, Faulkner C, Taber H, Valway S, Onorato I M. Analysis of Mycobacterium tuberculosis transmission patterns in a homeless shelter. Int J Tuber Lung Dis. 2000;4:308–313. [PubMed] [Google Scholar]
- 9.Das S, Paramativan C N, Lowrie D B, Prabhakar R, Narayanan P R. IS6110 restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, South India. Tuber Lung Dis. 1995;76:550–554. doi: 10.1016/0962-8479(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 10.Das S, Chan S L, Allen B W, Mitchison D A, Lowrie D B. Application of DNA fingerprinting with IS986 [IS6110] to sequential mycobacterial isolates obtained from pulmonary tuberculosis patients in Hong Kong before, during and after short-course therapy. Tuber Lung Dis. 1993;74:47–51. doi: 10.1016/0962-8479(93)90068-9. [DOI] [PubMed] [Google Scholar]
- 11.de C. Ramos M, Soini H, Roscanni G C, Jaques M, Villares M C, Musser J M. Extensive cross-contamination with Mycobacterium tuberculosis in a reference laboratory. J Clin Microbiol. 1999;37:916–919. doi: 10.1128/jcm.37.4.916-919.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermans P W M, Messadi F, Guebrexabher H, van Sooligan D, Haas P E W, Heersman H, de Neeling H, Ayoub A, Portaels F, Frommel D, Zribi M, van Embden J D A. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and the Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J Infect Dis. 1995;171:1504–1513. doi: 10.1093/infdis/171.6.1504. [DOI] [PubMed] [Google Scholar]
- 13.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Sooligen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazurek G H, Cave M D, Eisenach K D, Wallace R J, Jr, Bates J H, Crawford J T. Chromosomal DNA fingerprint patterns produced with IS6110 as strain-specific markers for epidemiologic study of tuberculosis. J Clin Microbiol. 1991;29:2030–2033. doi: 10.1128/jcm.29.9.2030-2033.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powell K E, Farer L S. The rising age of the tuberculosis patient: a sign of success and failure. J Infect Dis. 1980;142:946–948. doi: 10.1093/infdis/142.6.946. [DOI] [PubMed] [Google Scholar]
- 16.Rhee J T, Tanaka M M, Behr M A, Agasino C B, Paz E A, Hopewell P C, Small P M. Use of multiple markers in population-based molecular epidemiologic studies of tuberculosis. Int J Tuber Lung Dis. 2000;4:1111–1119. [PubMed] [Google Scholar]
- 17.Small P M, Hopewell P C, Singhe S P, Paz A, Parsonnet J, Ruston D C, Schechter G F, Daley C L, Schoolnik G K. The epidemiology of tuberculosis in San Francisco: a population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 18.Soini H, Pan X, Teeter L, Musser J M, Graviss E A. Transmission dynamics and molecular characterization of Mycobacterium tuberculosis isolates with low copy numbers of IS6110. J Clin Microbiol. 2001;39:217–221. doi: 10.1128/JCM.39.1.217-221.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Soolingen D, Hermans P W M, de Haas P E W, Soll D R, van Embden J D A. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren R, Richardson M, Sampson S, Hauman J H, Beyers N, Donald P R, van Helden P D. Genotyping of Mycobacterium tuberculosis with additional markers enhances accuracy in epidemiological studies. J Clin Microbiol. 1996;34:2219–2224. doi: 10.1128/jcm.34.9.2219-2224.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z H, Ijaz K, Bates J H, Eisenach K D, Cave M D. Spoligotyping and polymorphic GC-rich repetitive sequence fingerprinting of Mycobacterium tuberculosis strains having few copies of IS6110. J Clin Microbiol. 2000;38:3572–3576. doi: 10.1128/jcm.38.10.3572-3576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh R W, Ponce de Leon A, Agasino C B, Hahn J A, Daley C L, Hopewell P C, Small P M. Stability of Mycobacterium tuberculosis DNA genotypes. J Infect Dis. 1998;177:1107–1111. doi: 10.1086/517406. [DOI] [PubMed] [Google Scholar]