Abstract
The aim of this study was to search for associations of genetic variants with celiprolol pharmacokinetics in a large set of pharmacokinetic genes, and, more specifically, in a set of previously identified candidate genes ABCB1, SLCO1A2, and SLCO2B1. To this end, we determined celiprolol single‐dose (200 mg) pharmacokinetics and sequenced 379 pharmacokinetic genes in 195 healthy volunteers. Analysis with 46,064 common sequence variants in the 379 genes did not identify any novel genes associated with celiprolol exposure. The candidate gene analysis showed that the ABCB1 c.3435T>C and c.2677T/G>A, and the SLCO1A2 c.516A>C variants were associated with reduced celiprolol area under the plasma concentration‐time curve (AUC0–∞). An alternative analysis with ABCB1 haplotypes showed that, in addition to SLCO1A2 c.516A>C, three ABCB1 haplotypes were associated with reduced celiprolol AUC0–∞. A genotype scoring system was developed based on these variants and applied to stratify the participants to low and high celiprolol exposure genotype groups. The mean AUC0–∞ of celiprolol in the low exposure genotype group was 55% of the mean AUC0–∞ in the high exposure group (p = 1.08 × 10−11). In addition, the results showed gene‐gene interactions in the effects of SLCO1A2 and ABCB1 variants on celiprolol AUC0–∞ (p < 5 × 10−6) suggesting an interplay between organic anion transporting polypeptide 1A2 and P‐glycoprotein in celiprolol absorption. Taken together, these data indicate that P‐glycoprotein and organic anion transporting polypeptide 1A2 play a role in celiprolol pharmacokinetics. Furthermore, patients with ABCB1 and SLCO1A2 genotypes associated with low celiprolol exposure may have an increased risk of poor blood‐pressure lowering response to celiprolol.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
High interindividual variability exists in the pharmacokinetics of celiprolol. There are no comprehensive studies evaluating how variability in pharmacokinetic genes associates with celiprolol exposure.
WHAT QUESTION DID THIS STUDY ADDRESS?
This study searched for associations of genetic variants with celiprolol pharmacokinetics in a large set of pharmacokinetic genes, and, more specifically, in a set of previously identified candidate genes ABCB1, SLCO1A2, and SLCO2B1.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This study showed that genetic variants in ABCB1 and SLCO1A2 are associated with celiprolol pharmacokinetics. Based on the results, a genotype scoring system was developed and applied to stratify the participants to low and high celiprolol exposure genotype groups.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
This knowledge might aid in identifying individuals with increased risk of insufficient celiprolol exposure and therapeutic failure. Furthermore, the data suggest an interplay between OATP1A2 and P‐gp in the small intestine, which may be relevant also for other drugs that are substrates of both of these transporters.
INTRODUCTION
Celiprolol is a selective β1‐adrenoceptor antagonist for treatment of hypertension. 1 , 2 It also mild partial β2‐agonist and vasodilator properties. Celiprolol is administered as a racemic mixture of two enantiomers: R‐celiprolol and S‐celiprolol. The plasma exposure to celiprolol exhibits considerable interindividual variability, with one study showing 33‐fold variability in the peak plasma concentration (Cmax) of celiprolol following a 200 mg oral dose in healthy volunteers. 3 The usual celiprolol dose is 200 mg once daily. The proportion of patients achieving target blood pressure increases along with the dose and a higher dose of 400 mg is recommended if the response is insufficient. 4 These data suggest that the blood pressure‐lowering effect of celiprolol is dose‐ and exposure‐dependent. In addition, although celiprolol is usually well‐tolerated, the risk of hypotension and other adverse effects may increase along with the exposure.
Celiprolol is a hydrophilic molecule with negligible metabolism in humans. 1 , 2 It has a dose‐dependent oral bioavailability ranging from 30% after a 100 mg dose to 70% after a 400 mg dose. This suggests there is a saturable efflux process in the absorption phase. Approximately 10% of a 200 mg oral celiprolol dose is excreted unchanged into urine, with the renal clearance indicating active excretion. 3 In vitro studies have suggested that celiprolol is a substrate of the efflux transporter P‐glycoprotein (P‐gp; encoded by ABCB1). 5 In addition, the influx transporters organic anion transporting polypeptide 1A2 (OATP1A2; encoded by SLCO1A2) and OATP2B1 (encoded by SLCO2B1) have transported celiprolol in vitro. 6 , 7 These transporters have been suggested to be expressed in various tissues, including intestine, liver, and/or kidneys, 8 , 9 indicating that they might play a role in the absorption and disposition of celiprolol.
Several studies have investigated the associations of ABCB1 variants, for example, the synonymous c.3435T>C single nucleotide variation (SNV), with the pharmacokinetics of P‐gp substrates but the results have been partly contradictory. 10 The mechanism underlying the associations of the c.3435T>C (rs1045642; p.Ile1145=) SNV with drug pharmacokinetics remains to be fully elucidated. The c.3435T allele has previously been associated with a reduced duodenal and hepatic expression of the P‐gp. 11 , 12 Other studies, however, failed to replicate these findings. 13 , 14 The associations of SLCO1A2 and SLCO2B1 variants on the pharmacokinetics of OATP1A2 and OATP2B1 substrates have not been as widely investigated as the associations of ABCB1 variants. Their clinical significance also remains to be clarified.
Few relatively small studies have investigated the association of genetic variants with celiprolol pharmacokinetics, but comprehensive studies are lacking. In one study, individuals homozygous for the SLCO2B1 c.1457C>T (rs2306168; p.Ser486Phe) SNV had 50% smaller area under the plasma concentration time curve (AUC) of celiprolol than individuals homozygous for the reference allele, but the difference was not statistically significant. 7 In addition, the ABCB1 c.3435T>C SNV was not associated with celiprolol plasma concentrations in one small study investigating the effects of rifampin on celiprolol pharmacokinetics. 15 The aim of this study was to search for associations of genetic variants with celiprolol pharmacokinetics in a large set of pharmacokinetic genes, and, more specifically, in a set of previously identified candidate genes ABCB1, SLCO1A2, and SLCO2B1.
METHODS
A total of 195 healthy unrelated White Finnish volunteers participated in the study after giving written informed consent. Their health was confirmed by medical history, clinical examination, and laboratory tests. Participants were not on any continuous medication nor were tobacco smokers. The intake of alcohol was prohibited 1 day before celiprolol administration, on the study day, and on the following blood sampling day. The study was approved by the Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa (record number 267/13/03/00/2011) and the Finnish Medicines Agency Fimea (EudraCT number 2011‐004645‐40). Of the participants, 96 were women and 99 men. Their mean ± SD age was 23 ± 4 years, height 174 ± 9 cm, body weight 69 ± 12 kg, body mass index 22.8 ± 2.5 kg/m2, lean body weight (LBW) 54 ± 9 kg, and body surface area (BSA) 1.83 ± 0.20 m2. LBW and BSA were calculated as described previously. 16 , 17
Celiprolol pharmacokinetics and pharmacodynamics
After an overnight fast, each participant ingested a 200 mg dose of racemic celiprolol (Selectol; Leiras, Helsinki, Finland) with 150 ml of water at 8 a.m. Standardized meals were served at 4, 7, and 10 h after celiprolol ingestion. Timed blood samples (4–9 ml each) were collected to light‐protected ethylenediaminetetraacetic acid tubes prior to and 0.5, 1, 1.5, 2, 3, 4, 5, 7, 9, 12, and 24 h after celiprolol administration. Tubes were immediately placed on ice. Plasma was separated within 30 min and stored at −70°C until analysis.
Plasma samples were processed as previously described with minor modifications. 18 Briefly, an aliquot of 0.3 ml of plasma, 0.1 ml of internal standard (50 ng/ml deuterium labeled celiprolol in water) and 50 µl of 1 M sodium hydroxide were mixed prior to extraction with 3 ml of methyl‐tert‐butyl ether. The organic phase was separated and evaporated to dryness, and celiprolol was derivatized using 0.1 ml of (R)‐1‐(1‐naphthyl)ethyl isocyanate (0.005% in hexane‐isopropanol 95:5 v/v). The sample extract was then incubated for 30 min at room temperature and reconstituted in 0.1 ml of 45% acetonitrile.
Quantification of celiprolol enantiomers was carried out on an API 3000 liquid chromatography‐tandem mass spectrometer interfaced with an electrospray ion source (AB Sciex).
The separation of R‐celiprolol and S‐celiprolol was performed on a Kinetex 2.1 × 100 mm, 2.6 μm (Phenomenex) analytical column protected by a KrudKatcher Ultra in‐line filter (Phenomenex). The mobile phase consisted of a mixture of aqueous solution of 10 mM ammonium formate adjusted to pH 4 with 98% formic acid (solvent A) and acetonitrile (solvent B). Initial eluent composition of solvent B was 30% followed by a linear ramp over 8 min to 80% B, which was maintained for 1 min prior to the equilibration step back to 30% B. The column temperature and the mobile phase flow rate were maintained at 30°C and 0.2 ml/min. The mass spectrometer was operated in positive multiple reaction monitoring (MRM) mode using the mass‐to‐charge (m/z) transitions 577 to 380 (celiprolol) and 586 to 389 (celiprolol‐D9) for the quantification. The lower limit of quantification for R‐celiprolol and S‐celiprolol was 0.25 ng/ml and the day‐to‐day coefficient of variation was below 10% at relevant concentrations. The AUC from 0 h to infinity (AUC0–∞), Cmax, and elimination half‐life (t ½) values were calculated for R‐celiprolol, S‐celiprolol, and total celiprolol (calculated from the sum of R‐celiprolol and S‐celiprolol concentration) with standard noncompartmental methods using Phoenix WinNonlin, version 8.2 (Certara).
Systolic and diastolic blood pressures, and heart rate were measured in a sitting position with an automatic oscillometric blood pressure monitor (Omron Healthcare Europe BV) before and at 4, 12, and 24 h after celiprolol ingestion. The average change in diastolic and systolic blood pressure, were calculated by dividing the incremental area under the blood pressure‐time curve from time 0 to 24 h with 24 h. The maximum decrease in heart rate was calculated by subtracting the initial heart rate at 0 h from the minimum heart rate value at 4, 12, or 24 h.
DNA sequencing and genotyping
Genomic DNA was extracted from EDTA blood samples using the Maxwell 16 LEV Blood DNA Kit on a Maxwell 16 Research automated nucleic acid extraction system (Promega). A total of 379 pharmacokinetic genes ±20 kb were completely sequenced in the study participants using targeted massive parallel sequencing at the Technology Centre at Institute for Molecular Medicine Finland (Helsinki, Finland) as described previously. 19 , 20 , 21 , 22 The pharmacokinetic genes included phase I and II metabolizing enzymes, influx and efflux drug transporters, and regulatory proteins. 23 , 24 Coverage depth greater than or equal to 10×, Hardy‐Weinberg equilibrium p < 3.15 × 10−7 (Bonferroni‐correction), and proportion missing less than or equal to 0.05 were used as quality thresholds for the sequencing data. A total of 46,064 SNVs with minor allele frequency (MAF) greater than or equal to 0.05 passed these criteria and were included in the statistical analysis. TaqMan genotyping assays on a QuantStudio 12K Flex Real‐Time PCR System were used to supplement individual missing genotypes for SLCO2B1 c.1457C>T and SLCO2B1 c.601G>A (rs35199625, p.Val201Met) sequence variations (Thermo Fisher Scientific).
Statistical analysis
The number of participants was estimated to be sufficient to detect an effect size of f2 larger than 0.2 with two predictors in multiple linear regression analysis, with a power greater than 80% (Bonferroni corrected α level 1.09 × 10−6). The data were analyzed with the statistical programs JMP Genomics 8.2 (SAS Institute Inc.) and IBM SPSS 22.0 for Windows. The pharmacokinetic variables were logarithmically transformed before analysis. 25 Sex, and logarithmically transformed body weight, LBW, 16 BSA, 17 and estimated glomerular filtration rate, 26 were tested as demographic covariates for pharmacokinetic data using stepwise linear regression analysis, with p value thresholds of 0.05 for entry and 0.10 for removal. Possible associations of genetic variants with pharmacokinetic variables were investigated using linear regression analysis fixed for significant demographic covariates with a stepwise approach. A Bonferroni‐corrected p value threshold of 1.09 × 10−6 was used for the 379 gene and thresholds of 0.05 for entry and 0.10 for removal for the candidate gene analysis. Additive coding was used for the genetic variants. The pharmacokinetic variables of total celiprolol were compared between genotype score groups using analysis of variance adjusting for significant demographic covariates and pairwise comparisons with the Fisher’s Least Significant Difference method. A p value below 0.05 was considered statistically significant. Haplotype computations for the ABCB1 gene were performed with PHASE version 2.1.1. 27 , 28 SNV‐SNV and SNV‐haplotype interaction testing was performed with a regression testing for a linear trend of alleles, with a p value below 0.05 considered statistically significant. Comparison of the maximum decrease in heart rate and average change in diastolic and systolic blood pressure between genotype score groups were investigated with analysis of variance, with baseline values (0 h) as covariates and p value below 0.05 considered statistically significant. Pharmacokinetic data are given as geometric means with geometric coefficients of variation (CV), 90% confidence intervals (CIs), ranges, or geometric SDs, or medians with ranges.
RESULTS
Celiprolol pharmacogenomics
Among the 195 healthy volunteers, the AUC0–∞ and Cmax of R‐celiprolol varied 37‐fold and 90‐fold, those of S‐celiprolol 34‐fold and 79‐fold, and those of total celiprolol 35‐fold and 85‐fold, respectively (Table 1). The elimination t ½ of R‐celiprolol, S‐celiprolol, and total celiprolol ranged from 3 to 10 h. LBW was a significant covariate for all AUC0–∞ and Cmax values and sex and estimated glomerular filtration rate for t ½ values. These covariates were used in all subsequent analyses.
TABLE 1.
Pharmacokinetic variables of R‐celiprolol, S‐celiprolol, and total celiprolol in 195 healthy volunteers and the effects of significant demographic covariates on these variables
| Variable | Geometric mean (90% CI) | CV | Range | Demographic covariate | Effect (90% CI) b | p value |
|---|---|---|---|---|---|---|
| R‐celiprolol | ||||||
| Cmax (ng/ml) | 317 (296, 340) | 64% | 9.3–828 | LBW | −9.6% (−12.8%, −6.2%) | 1.02 × 10−5 |
| Tmax (h) a | 4 | – | 0.5–7 | – | ||
| AUC0‐∞ (ng·h/ml) | 1619 (1531, 1713) | 50% | 99–3625 | LBW | −8.6% (−11.2%, −5.9%) | 1.03 × 10−6 |
| t ½ (h) | 4.3 (4.2, 4.4) | 16% | 3.2–10 | Sex | −10.5% (−13.5%, −7.4%) | 2.57 × 10−7 |
| eGFR | 2.5% (4.0%, 1.0%) | 6.69 × 10−3 | ||||
| S‐celiprolol | ||||||
| Cmax (ng/ml) | 307 (287, 329) | 64% | 10–802 | LBW | −9.3% (−12.5%, −5.9%) | 2.13 × 10−5 |
| Tmax (h) a | 4 | – | 0.5–7 | – | ||
| AUC0‐∞ (ng·h/ml) | 1642 (1552, 1737) | 51% | 108–3705 | LBW | −8.3% (−11.0%, −5.6%) | 2.69 × 10−6 |
| t ½ (h) | 4.7 (4.7, 4.8) | 16% | 3.4–10 | Sex | −11.9% (−14.8%, −9.0%) | 1.51 × 10−9 |
| eGFR | 3.0% (4.4%, 1.5%) | 9.27 × 10−4 | ||||
| Total celiprolol | ||||||
| Cmax (ng/ml) | 624 (582, 669) | 64% | 19–1630 | LBW | −9.4% (−12.7%, −6.0%) | 1.48 × 10−5 |
| Tmax (h) a | 4 | – | 0.5–7 | – | ||
| AUC0‐∞ (ng·h/ml) | 3262 (3084, 3450) | 51% | 207–7330 | LBW | −8.5% (−11.1%, −5.7%) | 1.64 × 10−6 |
| t ½ (h) | 4.5 (4.5, 4.6) | 16% | 3.3–10 | Sex | −11.4% (−14.2%, −8.4%) | 8.25 × 10−9 |
| eGFR | 2.7% (4.2%, 1.3%) | 2.22 × 10−3 | ||||
Abbreviations: AUC0‐∞, area under the plasma concentration‐time curve from 0 h to infinity; CI, confidence interval; Cmax, peak plasma concentration; CV, geometric coefficient of variation; eGFR, estimated glomerular filtration rate; LBW, lean body weight; Tmax, concentration peak time; t ½, elimination half‐life.
Tmax data given as median.
Per 10% increase in LBW and per 10% decrease in eGFR; Sex: women vs. men.
To identify novel genes affecting celiprolol pharmacokinetics, we first tested the associations of 46,064 SNVs with MAF of at least 0.05 in the whole set of 379 pharmacokinetic genes. In a stepwise linear regression analysis, none of these SNVs was significantly associated with R‐celiprolol, S‐celiprolol, or total celiprolol AUC0–∞ at the Bonferroni corrected significance level (p = 1.09 × 10−6; Figure 1).
FIGURE 1.
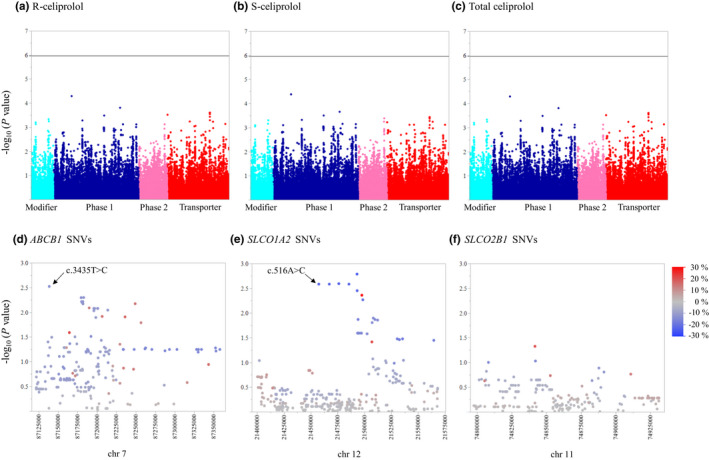
The associations of 46,064 SNVs in 379 pharmacokinetic genes with R‐celiprolol (a), S‐celiprolol (b), and total celiprolol (c) AUC0–∞ adjusting for LBW. The Y‐axes describe the negative logarithm of the p value for each SNV and the horizontal lines indicate the Bonferroni‐corrected significance level of 1.09 × 10−6. The X‐axes show individual SNVs grouped by protein function. The associations of SNVs (MAF ≥ 0.05) in SLCO1A2 (d), ABCB1 (e), and SLCO2B1 (f) with total celiprolol AUC0–∞. The Y‐axes describe the negative logarithm of the p value for each SNV and the X‐axes show the chromosomal positions (GRCh37/hg19 assembly). The blue to red scale shows the effect size (%) of each SNV per copy of the minor allele. AUC0–∞, area under the plasma concentration‐time curve from 0 to infinity; LBW, lean body weight; MAF, minor allele frequency; SNV, single nucleotide variation
Candidate gene analysis with SNVs
We next carried out a candidate gene analysis for celiprolol AUC0–∞ without correction for multiple testing. In this analysis, we included missense and other potentially functional variants with MAF of greater than or equal to 0.01 in genes suggested to be involved in celiprolol pharmacokinetics (i.e., ABCB1, SLCO1A2, and SLCO2B1; Table S1, Figure 1). In the analysis, SLCO1A2 c.516A>C (rs11568563, p.Glu172Asp), ABCB1 c.3435T>C, and ABCB1 c.2677T/G>A (rs2032582, p.Ser/Ala893Thr) were associated with reduced AUC0–∞ of R‐celiprolol, S‐celiprolol, and total celiprolol (Table 2). According to the linear regression models, the AUC0–∞ values were 25% smaller (p < 5 × 10−4) per copy of the SLCO1A2 c.516A>C minor allele, 13% smaller (p < 5 × 10−3) per copy of the ABCB1 c.3435T>C minor allele, and 21–22% smaller per copy of the ABCB1 c.2677T/G>A minor allele (p < 0.05). The association of the c.2677A allele was not, however, significant after correction for multiple testing. The effects of SLCO1A2 c.516A>C and ABCB1 c.3435T>C SNVs on celiprolol AUC0–∞ showed a statistically significant interaction (p = 1.18 × 10−6) in the SNV‐SNV interaction test.
TABLE 2.
Results of the candidate gene analysis with SNVs or SNVs and ABCB1 haplotypes on R‐celiprolol, S‐celiprolol, and total celiprolol AUC0–∞
| Pharmacokinetic variable | Covariate/SNV/haplotype | Effect a | p value | Bonferroni adjusted p value | Adjusted R 2 for each step | |
|---|---|---|---|---|---|---|
| Average | 90% CI | |||||
| R‐celiprolol AUC0–∞ | LBW | −8.4% | −11.0%, −5.7% | 1.14 × 10−6 | 0.11 | |
| (ABCB1 SNVs) | ABCB1 c.3435T>C (rs1045642) | −13.4% | −19.7%, −6.6% | 1.84 × 10−3 | 0.0257 | 0.15 |
| SLCO1A2 c.516A>C (rs11568563) | −24.9% | −34.3%, −14.2% | 4.84 × 10−4 | 6.77 × 10−3 | 0.19 | |
| ABCB1 c.2677T>A (rs2032582) | −21.2% | −35.0%, −4.5% | 0.0421 | 0.590 | 0.21 | |
| S‐celiprolol AUC0–∞ | LBW | −8.2% | −10.8%, −5.4% | 2.77 × 10−6 | 0.10 | |
| (ABCB1 SNVs) | SLCO1A2 c.516A>C (rs11568563) | −25.4% | −34.8%, −14.7% | 4.10 × 10−4 | 5.74 × 10−3 | 0.14 |
| ABCB1 c.3435T>C (rs1045642) | −13.2% | −19.5%, −6.3% | 2.50 × 10−3 | 0.0351 | 0.18 | |
| ABCB1 c.2677T>A (rs2032582) | −22.2% | −35.9%, −5.4% | 0.0347 | 0.486 | 0.20 | |
| Total celiprolol AUC0–∞ | LBW | −8.3% | −10.9%, −5.6% | 1.75 × 10−6 | 0.11 | |
| (ABCB1 SNVs) | SLCO1A2 c.516A>C (rs11568563) | −25.2% | −34.6%, −14.4% | 4.42 × 10−4 | 6.19 × 10−3 | 0.14 |
| ABCB1 c.3435T>C (rs1045642) | −13.3% | −19.6%, −6.5% | 2.14 × 10−3 | 0.0300 | 0.19 | |
| ABCB1 c.2677T>A (rs2032582) | −21.7% | −35.5%, −5.0% | 0.0381 | 0.533 | 0.20 | |
| R‐celiprolol AUC0–∞ | LBW | −8.7% | −11.3%, −6.2% | 2.05 × 10−7 | 0.11 | |
| (ABCB1 haplotypes) | ABCB1 H8 (AGCAAC) | −41.0% | −53.4%, −25.2% | 3.02 × 10−4 | 5.14 × 10−3 | 0.15 |
| SLCO1A2 c.516A>C (rs11568563) | −23.0% | −32.3%, −12.3% | 1.03 × 10−3 | 0.0176 | 0.20 | |
| ABCB1 H2 (AGCGAC) | −13.2% | −19.7%, −6.1% | 3.36 × 10−3 | 0.0571 | 0.22 | |
| ABCB1 H7 (AGCTAT) | −22.9% | −37.3%, −5.2% | 0.0390 | 0.662 | 0.24 | |
| S‐celiprolol AUC0–∞ | LBW | −8.5% | −11.1%, −5.9% | 4.97 × 10−7 | 0.10 | |
| (ABCB1 haplotypes) | ABCB1 H8 (AGCAAC) | −42.1% | −54.4%, −26.4% | 2.14 × 10−4 | 3.64 × 10−3 | 0.15 |
| SLCO1A2 c.516A>C (rs11568563) | −23.5% | −32.9%, −12.9% | 8.43 × 10−4 | 0.0143 | 0.19 | |
| ABCB1 H2 (AGCGAC) | −12.8% | −19.4%, −5.6% | 4.79 × 10−3 | 0.0814 | 0.22 | |
| ABCB1 H7 (AGCTAT) | −23.1% | −37.6%, −5.3% | 0.0387 | 0.657 | 0.23 | |
| Total celiprolol AUC0–∞ | LBW | −8.6% | −11.2%, −6.0% | 3.13 × 10−7 | 0.11 | |
| (ABCB1 haplotypes) | ABCB1 H8 (AGCAAC) | −41.5% | −53.9%, −25.9% | 2.52 × 10−4 | 4.28 × 10−3 | 0.15 |
| SLCO1A2 c.516A>C (rs11568563) | −23.3% | −32.6%, −12.6% | 9.26 × 10−4 | 0.0157 | 0.20 | |
| ABCB1 H2 (AGCGAC) | −12.9% | −19.6%, −5.8% | 4.00 × 10−3 | 0.0679 | 0.22 | |
| ABCB1 H7 (AGCTAT) | −23.0% | −37.4%, −5.2% | 0.0387 | 0.658 | 0.24 | |
Abbreviations: AUC0–∞, area under the plasma concentration‐time curve from 0 h to infinity; CI, confidence interval; LBW, lean body weight; SNV, single nucleotide variation.
ABCB1 haplotypes are presented in Figure 2 and are based on the c.61A>G, c.1199G>A, c.1236T>C, c.2677T>G/A, c.3320A>C, and c.3435T>C SNVs.
Per minor allele or haplotype copy; per 10% increase in LBW.
ABCB1 linkage disequilibrium and haplotype analysis
We next investigated the linkage disequilibrium profile of the ABCB1 SNVs included in the candidate gene analysis (Figure 2). The c.3435T>C synonymous variant was in a relatively strong linkage disequilibrium with the synonymous variant c.1236T>C (r2 = 0.40, p = 9.88 × 10−19) and with the G allele of the tri‐allelic missense variant c.2677T>G/A (r2 = 0.54, p = 9.18 × 10−25). The A allele of c.2677T>G/A was in a significant linkage disequilibrium with the c.1236T>C SNV, but their correlation was weak because the A allele is rare (r2 = 0.04, p = 4.35 × 10−3).
FIGURE 2.
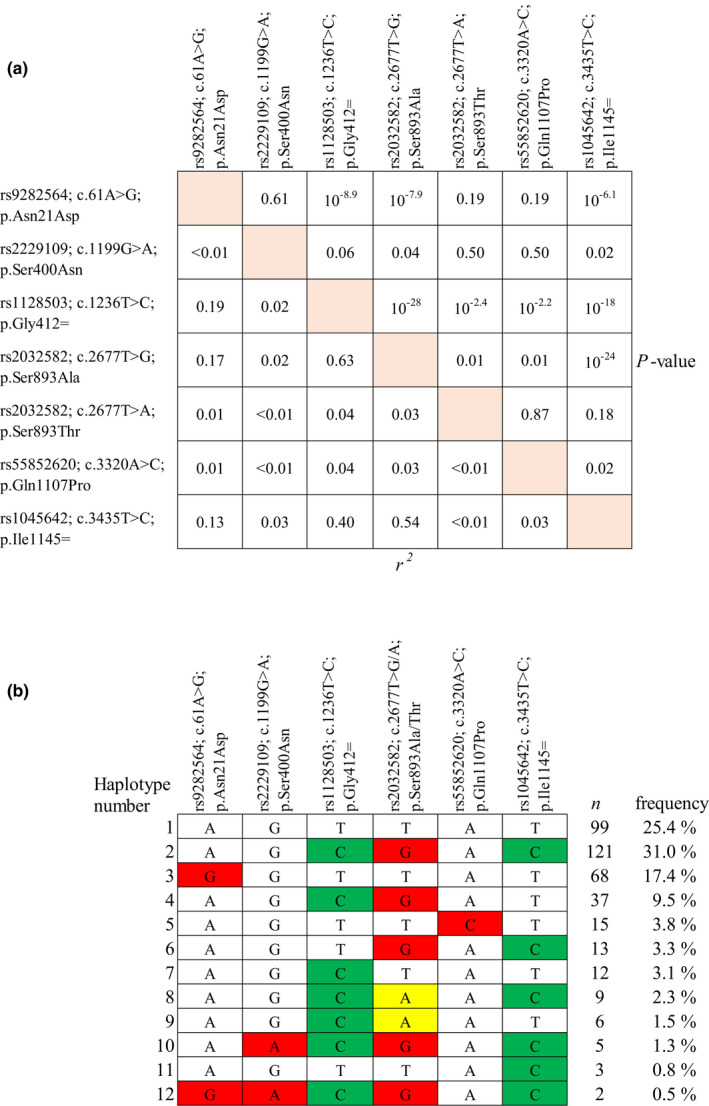
(a) Linkage disequilibrium of missense and potentially functional variants of the ABCB1 gene. (b) ABCB1 haplotypes inferred with missense and potentially functional variants. Synonymous variants are depicted in green and missense variants in red or yellow
Previous studies have suggested that ABCB1 haplotypes rather than SNVs affect P‐gp function. 10 Therefore, we computed haplotypes using ABCB1 SNVs included in the candidate gene analysis. A total of 12 ABCB1 haplotypes were inferred in the analysis (Figure 2). The c.3435C minor allele was present in six haplotypes, for example, in the most frequent haplotype that included also the minor alleles c.1236C and c.2677G (n = 121; H2). The c.2677A allele was present in two haplotypes; one of them included c.2677A with minor alleles c.1236C and c.3435C (n = 9; H8) and the other one included c.2677A with minor allele c.1236C (n = 6; H9).
Candidate gene analysis with ABCB1 haplotypes
To investigate the effects of ABCB1 haplotypes on celiprolol AUC0–∞ values, we repeated the candidate gene analysis using the 10 inferred haplotypes with MAF greater than or equal to 0.01 (Figure 2). In addition to SLCO1A2 c.516A>C, the ABCB1 H8 (including minor alleles c.1236C‐c.2677A‐c.3435C), H2 (c.1236C‐c.2677G‐c.3435C), and H7 (c.1236C) haplotypes were associated with reduced AUC of R‐celiprolol, S‐celiprolol, and total celiprolol (Table 2). Of these, the H8 haplotype had the largest effect size and the strongest association; the haplotype was associated with 41–42% reduced AUC values per copy (p < 5 × 10−4). The associations of the H2 and H7 haplotypes were not significant after correction for multiple testing. The effects of the SLCO1A2 c.516A>C SNV and the ABCB1 H2 haplotype on celiprolol AUC0–∞ showed a statistically significant interaction (p = 1.68 × 10−7) in the interaction test. In addition, the ABCB1 H7 haplotype showed a statistically significant interaction with SLCO1A2 c.516A>C (p = 3.5 × 10−6), but only three participants carried these alleles concomitantly (Figure 3).
FIGURE 3.
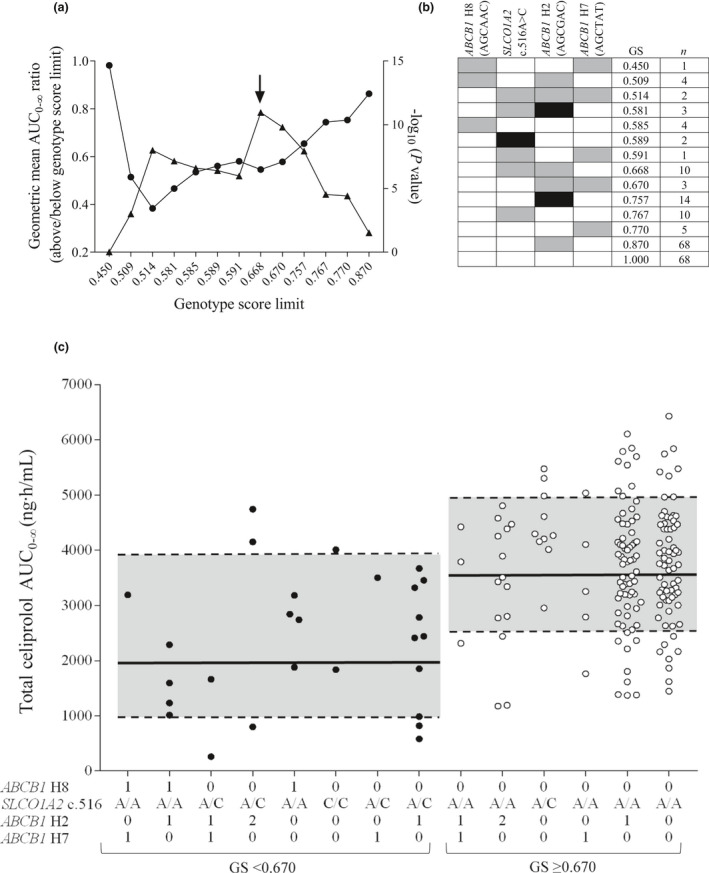
Associations of total celiprolol AUC0–∞ with genotype scores calculated with SLCO1A2 c.516A>C and ABCB1 haplotypes. (a) The total celiprolol geometric mean AUC0–∞ ratios between groups below and above each genotype score limit (circles) and respective p values (triangles). The arrow depicts the optimal cutoff value. (b) Genotype scores (GS) for individuals with different genotype combinations. ABCB1 and SLCO1A2 reference genotypes are depicted with white, heterozygous with gray, and homozygous variant genotypes with black rectangles. (c) The LBW‐adjusted AUC0–∞ values of total celiprolol grouped by genotype scores. The 0, 1, and 2 indicate the number of ABCB1 haplotype copies. The black lines in the gray areas depict the geometric mean and dashed lines the ± geometric SD AUC0–∞ values for genotype score groups below and above the cutoff limit. AUC0–∞, area under the plasma concentration‐time curve from 0 to infinity; LBW, lean body weight
Genotype score
The effects of the SNVs and haplotypes were nearly identical for celiprolol enantiomers and the pharmacokinetic variables of the enantiomers did not significantly differ from each other. Thus, for clarity, all further analyses were carried out for total celiprolol only. To predict total celiprolol AUC0–∞ in individuals with different combinations of the SLCO1A2 c.516A>C SNV and ABCB1 haplotypes, we calculated genotype scores (GS) using the candidate gene linear regression model using ABCB1 haplotypes with the following equation:
where n is the number of variant alleles (0, 1, or 2) of SLCO1A2 c.516A>C or number of ABCB1 haplotype (0, 1, or 2) (Figure 3). Genotype score is 1.00 in individuals who carry neither SLCO1A2 c.516C allele nor any of the associated ABCB1 haplotypes. For others, the score shows the predicted fold difference in celiprolol AUC0–∞ compared to 1.00.
Next, we determined a genotype score cutoff value for optimal differentiation, defined as the strongest statistical significance, between individuals with a lower and higher exposure to celiprolol. The optimal cutoff value was 0.670 (Figure 3). The geometric mean Cmax of celiprolol was 49% lower (p = 1.07 × 10−9) and AUC 45% smaller (p = 1.08 × 10−11) in the group of individuals with a genotype score less than 0.670 than in the group with a genotype score greater than or equal to 0.670 (Table 3, Figures 3, 4). A genotype score calculated with ABCB1 SNVs instead of haplotypes performed similarly well in identifying individuals with low celiprolol exposure (Table S2, Figure S1). The maximum decrease in heart rate or average change in diastolic or systolic blood pressure were not significantly different between the low and high exposure groups defined either by the haplotype or the SNV‐based genotype score (data not shown).
TABLE 3.
Pharmacokinetic variables of total celiprolol in subjects with SLCO1A2 c.516A>C and ABCB1 haplotype‐based genotype scores <0.670 and ≥0.670
| Variable | Geometric mean (90% CI) | Geometric mean ratio (90% CI) | p value | |
|---|---|---|---|---|
| Genotype score <0.670; n = 27 | Genotype score ≥0.670; n = 168 | |||
| Cmax (ng/ml) | 347 (294, 408) | 686 (643, 732) | 0.51 (0.42, 0.60) | 1.07 × 10−9 |
| Tmax (h) a | 4 (0.5–5) | 4 (0.5–7) | 0.470 | |
| AUC0–∞ (ng·h/ml) | 1937 (1704, 2202) | 3548 (3370, 3733) | 0.55 (0.48, 0.63) | 1.08 × 10−11 |
| t ½ (h) | 4.8 (4.6, 5.0) | 4.5 (4.4, 4.6) | 1.07 (1.02, 1.12) | 0.013 |
The Cmax and AUC0–∞ data are adjusted for lean body weight and t ½ data for sex and estimated glomerular filtration rate.
Abbreviations: AUC0–∞, area under the plasma concentration‐time curve from 0 h to infinity; CI, confidence interval; Cmax, peak plasma concentration; Tmax concentration peak time; t ½, elimination half‐life.
Tmax is given as median with range.
FIGURE 4.
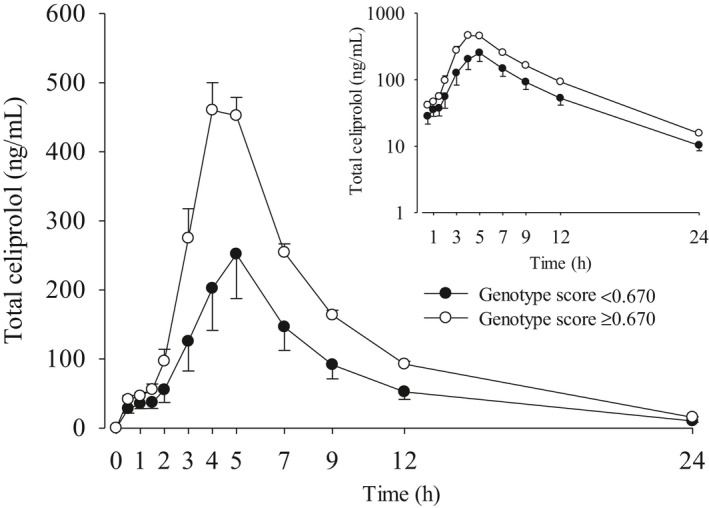
Geometric mean (90% CI) LBW‐adjusted plasma concentrations of celiprolol after a single 200 mg oral dose of celiprolol in 195 healthy volunteers with different combinations of SLCO1A2 c.516A>C and ABCB1 haplotypes. The inset depicts the same data on a semilogarithmic scale. The volunteers were grouped according to the genotype score (GS) limit 0.670: GS < 0.670 (n = 27) and GS ≥ 0.670 (n = 168). CI, confidence interval; LBW, lean body weight
DISCUSSION
This study searched for associations of genetic variants with celiprolol pharmacokinetics in a large set of pharmacokinetic genes, and, more specifically, in a set of previously identified candidate genes in 195 healthy volunteers. No variants in novel genes affecting celiprolol exposure were identified, but the candidate gene analysis indicates that SNVs and haplotypes in the ABCB1 and SLCO1A2 genes are associated with a reduced AUC of celiprolol. A scoring system was developed on the basis of the associated ABCB1 and SLCO1A2 variants and applied to determine a genotype score cutoff value to stratify the participants to low and high celiprolol exposure genotype groups. The mean AUC of celiprolol in the low exposure genotype group was about half of that in the high exposure genotype group.
Celiprolol is a substrate of the influx transporters OATP1A2 and OATP2B1 as well as the efflux transporter P‐gp in vitro. 5 , 6 , 7 In vivo, the P‐gp inhibitor itraconazole has increased and the P‐gp inducer rifampin (INN, rifampicin) has reduced celiprolol plasma concentrations. 15 , 29 In addition, grapefruit juice, orange juice, and green tea have significantly reduced celiprolol plasma concentrations possibly by inhibiting intestinal OATP1A2 and/or OATP2B1 transporters. 7 , 29 , 30 , 31 , 32 , 33 To investigate the roles of P‐gp, OATP1A2, and OATP2B1 in celiprolol pharmacokinetics, we carried out a candidate gene analysis with their missense variants and other potentially functional variants. The results showed that the SLCO1A2 c.516A>C, ABCB1 c.3435T>C, and ABCB1 c.2677T/G>A SNVs were associated with a reduced AUC of celiprolol. No associations were found with SLCO2B1 SNVs. Thus, the candidate gene analysis corroborates the previous in vitro and in vivo studies indicating that P‐gp and OATP1A2 play a role in celiprolol pharmacokinetics. The data therefore also support celiprolol as a potential index substrate for P‐gp‐ and/or OATP1A2‐mediated drug interactions.
The ABCB1 c.3435T>C SNV is a synonymous variant with a high minor allele frequency, 0.39 in this study. Previously, studies on this variant have commonly reported the effects of the c.3435T allele on the pharmacokinetics of P‐gp substrates. 10 In the present study, the c.3435C allele was, however, the minor allele and thus the results of the regression analyses relate to the effects of the C allele. Accordingly, the c.3435C allele was associated with a modestly reduced exposure to celiprolol. In accordance with our results, the c.3435T allele has been associated with increased exposure to P‐gp substrates, such as digoxin, cyclosporine, and fexofenadine, although the results have not always been reproducible. 10 , 11 , 34 , 35 Moreover, one study reported a genomewide significant association of the intronic ABCB1 rs4148738 SNV with dabigatran pharmacokinetics, with 12% increase in Cmax per copy of each minor allele. 36 The minor allele of this SNV is in a relatively strong linkage disequilibrium with the c.3435T allele. Altogether, the balance of evidence suggests that the c.3435T>C SNV associates with the pharmacokinetics of P‐gp substrates, although the magnitude of the effect is relatively small.
In addition to the c.3435T>C SNV, the A allele of the tri‐allelic ABCB1 c.2677T>G/A missense SNV was associated with 21% reduced celiprolol AUC in the candidate gene SNV analysis. The frequency of the A allele was relatively low, 0.04, and association of the allele with celiprolol AUC was not significant after correction for multiple testing. In vitro, the c.2677A allele, has significantly increased the transport of the P‐gp substrate vincristine. 37 Due to the rarity of the A allele, its effects have not been widely characterized in vivo. Nevertheless, the plasma concentrations of fexofenadine have been significantly lower in carriers of the c.2677A/A‐c.3435C/C genotype than in noncarriers. 34 Furthermore, paclitaxel clearance has been significantly higher in patients carrying the c.2677G/A genotype than in noncarriers. 38
Previous studies have suggested that ABCB1 haplotypes rather than SNVs affect P‐gp function. 10 Therefore, to investigate whether the effects of ABCB1 haplotypes differ from the effects of SNVs, we carried out an additional candidate gene analysis with the ABCB1 haplotypes. Three haplotypes were associated with reduced AUC of celiprolol. Of these, the association of the H7 haplotype, containing the minor allele c.1236C alone, and H2 haplotype, containing minor alleles c.1236C‐c.2677G‐c.3435C, were not significant after correction for multiple testing and should be interpreted with caution. Nevertheless, the c.1236C‐c.2677G‐c.3435C haplotype has been associated with reduced exposure to some P‐gp substrates, for example, simvastatin acid, atorvastatin, and cyclosporine. 39 , 40 Interestingly, the association of the H8 haplotype, which contains both the c.2677A and the c.3435C alleles, showed the strongest association of all the associated SNVs and haplotypes. In the regression model, this haplotype reduced celiprolol AUC by 42% per its copy. Previously, one study reported that this haplotype was associated with increased instead of decreased trough concentrations of cyclosporine in renal transplant recipients. 41 Despite this, the results of our study suggest that c.2677A and c.3435C together in the same haplotype produce a larger effect on celiprolol AUC than either of these alleles individually or in haplotypes with other ABCB1 SNVs.
In addition to the ABCB1 variants, the SLCO1A2 c.516A>C missense SNV was associated with reduced celiprolol AUC. In vitro, the SNV has consistently decreased the function of OATP1A2. 42 , 43 , 44 , 45 However, the SNV has not been associated with the plasma exposure to the OATP1A2 substrates imatinib or lopinavir. 44 , 46 OATP1A2 has been found, for example, in the kidneys, liver, and blood‐brain barrier. 42 Studies regarding the intestinal expression of OATP1A2 have, however, been contradictory. 47 , 48 Some studies have found SLCO1A2 mRNA and OATP1A2 protein in intestine samples 9 , 44 and one study suggested that OATP1A2, like P‐gp, is expressed on the apical membrane of small intestinal enterocytes. 49 The present results would be compatible with reduced absorption of celiprolol in the small intestine due to the SLCO1A2 c.516A>C SNV.
The association of the SLCO1A2 c.516A>C SNV with celiprolol AUC seemed to exist only in individuals who concomitantly carried the ABCB1 c.3435C allele (Figure 3, Figure S1). Accordingly, the c.516A>C SNV showed a significant interaction with the c.3435T>C SNV and with the ABCB1 H2 haplotype containing the minor alleles c.1236C‐c.2677G‐c.3435C. These statistical interactions suggest an influx‐efflux transporter interplay between enterocyte OATP1A2 and P‐gp in celiprolol absorption. This interplay might also affect the absorption of other drugs which are substrates of both OATP1A2 and P‐gp, such as fexofenadine and nadolol. 31 , 49 Further studies are warranted on the potential OATP1A2‐P‐gp interplay and its mechanisms.
This study was carried out in healthy young individuals, whereas patients using celiprolol are usually elderly. The pharmacokinetic profile of celiprolol is similar in elderly and younger healthy individuals and the steady‐state concentrations are predictable from the single‐dose pharmacokinetics. 4 , 50 Therefore, the effects of genetic variants on the steady‐state plasma concentrations of celiprolol in the elderly should be similar to the effects on the AUC0–∞ of celiprolol observed in our study after a single dose. Moreover, elderly patients often have several concomitant medications and they may therefore be susceptible to drug‐drug interactions. These interactions may vary depending on the ABCB1 and SLCO1A2 genotypes.
Based on significant ABCB1 haplotypes and SLCO1A2 c.516A>C we calculated genotype scores to predict the exposure to celiprolol. In addition, we determined a genotype score cutoff value, which optimally differentiated individuals with lower and higher exposure to celiprolol. The significantly lower celiprolol AUC and Cmax but not t ½ in individuals below the cutoff value as compared with individuals above the cutoff value suggest that the ABCB1 and SLCO1A2 variants affect the absorption of celiprolol. Although in this study with normotensive healthy volunteers, the blood pressure or heart rate responses did not differ significantly between the groups, patients with a genotype score below the cutoff value may have an increased risk of insufficient celiprolol exposure and therapeutic failure.
The participants of this study were White Finnish individuals. The ABCB1 c.3435C allele is generally very common in European (MAF 0.48) and South‐Asian (0.42) populations and it is the major allele in East‐Asian (0.60) and Sub‐Saharan African (0.85) populations (Table S1). The c.2677A allele, on the other hand, is very rare in Sub‐Saharan African population (<0.001) and relatively rare in Europeans (0.018). However, it is more common in South‐Asian (0.050) and especially in East‐Asian (0.13) populations. The different allele frequencies naturally result also in different ABCB1 haplotype frequencies. 10 Moreover, the SLCO1A2 c.516A>C SNV is rare in Asians and Africans (0–0.02) as compared with Europeans (0.07), making combinations of the ABCB1 c.3435C and the SLCO1A2 c.516C alleles more frequent in Europeans. Therefore, the extent to which the ABCB1 and SLCO1A2 variants and their combinations explain population variability in celiprolol pharmacokinetics likely differs markedly between ethnic groups.
In conclusion, the results of this study indicate that genetic variants in ABCB1 and SLCO1A2 are associated with celiprolol pharmacokinetics. Celiprolol could be a useful index substrate for P‐gp‐ and/or OATP1A2‐mediated drug interactions. Furthermore, especially individuals carrying the ABCB1 c.2677A or the SLCO1A2 c.516C allele together with the ABCB1 c.3435C allele may have a risk of low celiprolol exposure. These individuals may thus be at an increased risk of poor blood pressure‐lowering efficacy of celiprolol. Moreover, the data suggest an interplay of OATP1A2 and P‐gp in the small intestine.
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS
P.H. and M.Ni. wrote the manuscript. P.H., A.T., J.T.B., and M.Ni. designed the research. P.H., A.T., T.T., M.P.‐H., M.Ne., J.T.B., and M.Ni. performed the research. P.H. and M.Ni. analyzed the data.
Supporting information
Table S1
Table S2
Figure S1
ACKNOWLEDGEMENTS
The authors thank Ms. Katja Halme, Ms. Hanna Hyvärinen, Ms. Satu Karjalainen, Mr. Jouko Laitila, Ms. Eija Mäkinen‐Pulli, Ms. Raija Nevala, and Ms. Lisbet Partanen for skillful assistance in conducting the clinical pharmacokinetic study, and Mr. Pekka Ellonen and Ms. Maija Lepistö, M. Sc., for the massive parallel sequencing.
Hirvensalo P, Tornio A, Tapaninen T, et al. Pharmacogenomics of celiprolol – evidence for a role of P‐glycoprotein and organic anion transporting polypeptide 1A2 in celiprolol pharmacokinetics. Clin Transl Sci. 2022;15:409–421. 10.1111/cts.13159
Funding information
This study was supported by grants from the European Research Council (Grant agreement 282106), State funding for university‐level health research, the Sigrid Jusélius Foundation (Helsinki, Finland), the Biomedicum Helsinki Foundation (Helsinki, Finland), and the Orion Research Foundation sr (Espoo, Finland).
REFERENCES
- 1. Riddell JG, Shanks RG, Brogden RN. Celiprolol. A preliminary review of its pharmacodynamic and pharmacokinetic properties and its therapeutic use in hypertension and angina pectoris. Drugs. 1987;34:438‐458. [DOI] [PubMed] [Google Scholar]
- 2. Milne RJ, Buckley MM. Celiprolol. An updated review of its pharmacodynamic and pharmacokinetic properties, and therapeutic efficacy in cardiovascular disease. Drugs. 1991;41:941‐969. [DOI] [PubMed] [Google Scholar]
- 3. Hartmann C, Frölich M, Krauss D, Spahn‐Langguth H, Knauf H, Mutschler E. Comparative enantioselective pharmacokinetic studies of celiprolol in healthy volunteers and patients with impaired renal function. Eur J Clin Pharmacol. 1990;39:573‐576. [DOI] [PubMed] [Google Scholar]
- 4. Dunn CJ, Spencer CM. Celiprolol. An evaluation of its pharmacological properties and clinical efficacy in the management of hypertension and angina pectoris. Drugs Aging. 1995;7:394‐411. [DOI] [PubMed] [Google Scholar]
- 5. Karlsson J, Kuo SM, Ziemniak J, Artursson P. Transport of celiprolol across human intestinal epithelial (Caco‐2) cells: mediation of secretion by multiple transporters including P‐glycoprotein. Br J Pharmacol. 1993;110:1009‐1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kato Y, Miyazaki T, Kano T, Sugiura T, Kubo Y, Tsuji A. Involvement of influx and efflux transport systems in gastrointestinal absorption of celiprolol. J Pharm Sci. 2009;98:2529‐2539. [DOI] [PubMed] [Google Scholar]
- 7. Ieiri I, Doi Y, Maeda K, et al. Microdosing clinical study: pharmacokinetic, pharmacogenomic (SLCO2B1), and interaction (grapefruit juice) profiles of celiprolol following the oral microdose and therapeutic dose. J Clin Pharmacol. 2012;52:1078‐1089. [DOI] [PubMed] [Google Scholar]
- 8. International Transporter Consortium , Giacomini KM, Huang SM, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Couto N, Al‐Majdoub ZM, Gibson S, et al. Quantitative proteomics of clinically relevant drug‐metabolizing enzymes and drug transporters and their intercorrelations in the human small intestine. Drug Metab Dispos. 2020;48:245‐254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolking S, Schaeffeler E, Lerche H, Schwab M, Nies AT. Impact of genetic polymorphisms of ABCB1 (MDR1, P‐Glycoprotein) on drug disposition and potential clinical implications: update of the literature. Clin Pharmacokinet. 2015;54:709‐735. [DOI] [PubMed] [Google Scholar]
- 11. Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug‐resistance gene: multiple sequence variations and correlation of one allele with P‐glycoprotein expression and activity in vivo. Proc Natl Acad Sci USA. 2000;28:3473‐3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang D, Johnson AD, Papp AC, Kroetz DL, Sadée W. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet Genomics. 2005;15:693‐704. [PubMed] [Google Scholar]
- 13. Siegmund W, Ludwig K, Engel G, et al. Variability of intestinal expression of P‐glycoprotein in healthy volunteers as described by absorption of talinolol from four bioequivalent tablets. J Pharm Sci. 2003;92:604‐610. [DOI] [PubMed] [Google Scholar]
- 14. Meier Y, Pauli‐Magnus C, Zanger UM, et al. Interindividual variability of canalicular ATP‐binding‐cassette (ABC) transporter expression in human liver. Hepatology. 2006;44:62‐74. [DOI] [PubMed] [Google Scholar]
- 15. Lilja JJ, Niemi M, Neuvonen PJ. Rifampicin reduces plasma concentrations of celiprolol. Eur J Clin Pharmacol. 2004;59:819‐824. [DOI] [PubMed] [Google Scholar]
- 16. Hallynck TH, Soep HH, Thomis JA, Boelaert J, Daneels R, Dettli L. Should clearance be normalised to body surface or to lean body mass? Br J Clin Pharmacol. 1981;11:523‐526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haycock GB, Schwartz GJ, Wisotsky DH. Geometric method for measuring body surface area: a height‐weight formula validated in infants, children, and adults. J Pediatr. 1978;93:62‐66. [DOI] [PubMed] [Google Scholar]
- 18. Verbesselt R, Zugravu A, Tjandramaga TB, De Schepper PJ. Liquid chromatographic determination of total celiprolol or (S)‐celiprolol and (R)‐celiprolol simultaneously in human plasma. J Chromatogr B Biomed Appl. 1996;683:231‐236. [DOI] [PubMed] [Google Scholar]
- 19. Sulonen A‐M, Ellonen P, Almusa H, et al. Comparison of solution‐based exome capture methods for next generation sequencing. Genome Biol. 2011;12:R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hirvensalo P, Tornio A, Neuvonen M, et al. Comprehensive pharmacogenomic study reveals an important role of UGT1A3 in montelukast pharmacokinetics. Clin Pharmacol Ther. 2018;104:158‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hirvensalo P, Tornio A, Neuvonen M, et al. Enantiospecific pharmacogenomics of fluvastatin. Clin Pharmacol Ther. 2019;106:668‐680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hirvensalo P, Tornio A, Launiainen T, et al. UGT1A3 and sex are major determinants of telmisartan pharmacokinetics‐a comprehensive pharmacogenomic study. Clin Pharmacol Ther. 2020;108:885‐895. [DOI] [PubMed] [Google Scholar]
- 23. PharmaADME website. Available from: http://pharmaadme.org. Accessed March 26, 2021.
- 24. Morrissey KM, Wen CC, Johns SJ, Zhang L, Huang SM, Giacomini KM. The UCSF‐FDA TransPortal: a public drug transporter database. Clin Pharmacol Ther. 2012;92:545‐546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clinical Pharmacology & Therapeutics Editorial Team . Statistical guide for Clinical Pharmacology & Therapeutics. Clin Pharmacol Ther. 2010;88:150‐152. [DOI] [PubMed] [Google Scholar]
- 26. Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604‐612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978‐989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162‐1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lilja JJ, Backman JT, Laitila J, Luurila H, Neuvonen PJ. Itraconazole increases but grapefruit juice greatly decreases plasma concentrations of celiprolol. Clin Pharmacol Ther. 2003;73:192‐198. [DOI] [PubMed] [Google Scholar]
- 30. Lilja JJ, Juntti‐Patinen L, Neuvonen PJ. Orange juice substantially reduces the bioavailability of the beta‐adrenergic‐blocking agent celiprolol. Clin Pharmacol Ther. 2004;75:184‐190. [DOI] [PubMed] [Google Scholar]
- 31. Misaka S, Yatabe J, Müller F, et al. Green tea ingestion greatly reduces plasma concentrations of nadolol in healthy subjects. Clin Pharmacol Ther. 2014;95:432‐438. [DOI] [PubMed] [Google Scholar]
- 32. Sonoda J, Ogata K, Yoshikawa N, Sato K, Ikeda R, Shimodozono Y. Impact of green tea intake on the pharmacokinetics of celiprolol in healthy subjects. Int J Clin Pharmacol Ther. 2021;59:198‐201. [DOI] [PubMed] [Google Scholar]
- 33. Bailey DG. Fruit juice inhibition of uptake transport: a new type of food‐drug interaction. Br J Clin Pharmacol. 2010;70:645‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yi SY, Hong KS, Lim HS, et al. A variant 2677A allele of the MDR1 gene affects fexofenadine disposition. Clin Pharmacol Ther. 2004;76:418‐427. [DOI] [PubMed] [Google Scholar]
- 35. Hu Y‐F, Qiu W, Liu Z‐Q, et al. Effects of genetic polymorphisms of CYP3A4, CYP3A5 and MDR1 on cyclosporine pharmacokinetics after renal transplantation. Clin Exp Pharmacol Physiol. 2006;33:1093‐1098. [DOI] [PubMed] [Google Scholar]
- 36. Paré G, Eriksson N, Lehr T, et al. Genetic determinants of dabigatran plasma levels and their relation to bleeding. Circulation. 2013;127:1404‐1412. [DOI] [PubMed] [Google Scholar]
- 37. Schaefer M, Roots I, Gerloff T. In‐vitro transport characteristics discriminate wild‐type ABCB1 (MDR1) from ALA893SER and ALA893THR polymorphisms. Pharmacogenet Genomics. 2006;16:855‐861. [DOI] [PubMed] [Google Scholar]
- 38. Gréen H, Söderkvist P, Rosenberg P, et al. Pharmacogenetic studies of Paclitaxel in the treatment of ovarian cancer. Basic Clin Pharmacol Toxicol. 2009;104:130‐137. [DOI] [PubMed] [Google Scholar]
- 39. Keskitalo JE, Kurkinen KJ, Neuvonen PJ, Niemi M. ABCB1 haplotypes differentially affect the pharmacokinetics of the acid and lactone forms of simvastatin and atorvastatin. Clin Pharmacol Ther. 2008;84:457‐461. [DOI] [PubMed] [Google Scholar]
- 40. Fanta S, Niemi M, Jönsson S, et al. Pharmacogenetics of cyclosporine in children suggests an age‐dependent influence of ABCB1 polymorphisms. Pharmacogenet Genomics. 2008;18:77‐90. [DOI] [PubMed] [Google Scholar]
- 41. Qiu X‐Y, Jiao Z, Zhang M, et al. Association of MDR1, CYP3A4*18B, and CYP3A5*3 polymorphisms with cyclosporine pharmacokinetics in Chinese renal transplant recipients. Eur J Clin Pharmacol. 2008;64:1069‐1084. [DOI] [PubMed] [Google Scholar]
- 42. Lee W, Glaeser H, Smith LH, et al. Polymorphisms in human organic anion‐transporting polypeptide 1A2 (OATP1A2): implications for altered drug disposition and central nervous system drug entry. J Biol Chem. 2005;280:9610‐9617. [DOI] [PubMed] [Google Scholar]
- 43. Badagnani I, Castro RA, Taylor TR, et al. Interaction of methotrexate with organic‐anion transporting polypeptide 1A2 and its genetic variants. J Pharmacol Exp Ther. 2006;318:521‐529. [DOI] [PubMed] [Google Scholar]
- 44. Eechoute K, Franke RM, Loos WJ, et al. Environmental and genetic factors affecting transport of imatinib by OATP1A2. Clin Pharmacol Ther. 2011;89:816‐820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee HH, Leake BF, Kim RB, Ho RH. Contribution of organic anion‐transporting polypeptides 1A/1B to doxorubicin uptake and clearance. Mol Pharmacol. 2017;91:14‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hartkoorn RC, Kwan WS, Shallcross V, et al. HIV protease inhibitors are substrates for OATP1A2, OATP1B1 and OATP1B3 and lopinavir plasma concentrations are influenced by SLCO1B1 polymorphisms. Pharmacogenet Genomics. 2010;20:112‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Drozdzik M, Gröer C, Penski J, et al. Protein abundance of clinically relevant multidrug transporters along the entire length of the human intestine. Mol Pharm. 2014;11:3547‐3555. [DOI] [PubMed] [Google Scholar]
- 48. Franke RM, Scherkenbach LA, Sparreboom A. Pharmacogenetics of the organic anion transporting polypeptide 1A2. Pharmacogenomics. 2009;10:339‐344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Glaeser H, Bailey DG, Dresser GK, et al. Intestinal drug transporter expression and the impact of grapefruit juice in humans. Clin Pharmacol Ther. 2007;81:362‐370. [DOI] [PubMed] [Google Scholar]
- 50. Norris RJ, Lee EH, Muirhead D, Sanders SW. A pharmacokinetic evaluation of celiprolol in healthy elderly volunteers. J Cardiovasc Pharmacol. 1986;8(Suppl 4):S91‐S92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Figure S1


