Abstract
To date, there has been no genomewide association study (GWAS) from the Middle East and North African (MENA) region to identify genetic variants associated with warfarin dose variability using this approach. In this study, we aimed to conduct the first GWAS of warfarin dose requirements in patients from the MENA region. A total of 132 Qatari (discovery) and 50 Egyptians (replication) were genotyped using Illumina Multi‐Ethnic Global BeadChip Array. A GWAS was performed on log‐transformed weekly warfarin dose in the studied population, adjusting for clinical characteristics and ancestry. The genomewide signals from the discovery cohort were tested in the Egyptian cohort. A GWAS meta‐analysis, including the Qatari and Egyptian cohorts, was also performed and the output from this analysis was used in a gene‐based analysis. The discovery analysis in Qatari identified five genomewide single‐nucleotide polymorphisms (SNPs) in chromosome 16. These signals were replicated in the Egyptian cohort. Combining the two data through a GWAS meta‐analysis strengthened the association in chromosome 16 with VKORC1 rs9934438 being the lead genomewide signal (β = −0.17, 6 × 10−15). Other SNPs were identified in chromosome 10 at a p value less than 1 × 10−5. The genetic variants within VKORC1 rs9934438 and CYP2C9 rs4086116 explained 39% and 27% of the variability in the weekly warfarin dose requirement in the Qatari and Egyptians, respectively. This is the first GWAS of warfarin dose variability in the MENA region. It confirms the importance of VKORC1 and CYP2C9 variants in warfarin dose variability among patients from the MENA region.
Keywords: anticoagulation, Arab, genome wide association, pharmacogenetics, warfarin
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Genomewide association study (GWAS) from different populations have shown that genetic polymorphisms in VKORC1 and CYP2C9 genes, both of which are involved in the pharmacodynamic and kinetic pathway of warfarin, are associated with warfarin dose variability. Variants in the CYP4F2, a gene coding for the vitamin K metabolizing enzyme, were also shown to provide a minor contribution to warfarin dose variability.
WHAT QUESTION DID THIS STUDY ADDRESS?
Which genetic mutations are associated with warfarin dose requirements in populations of Middle East and North African (MENA)? Are there any novel variants associated with warfarin dose in these populations?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This is the first GWAS in the MENA region to confirm the importance of VKORC1 (−1639G>A) and CYP2C9 rs4086116 variants in predicting warfarin dose in Arabs.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
This study is an important step to confirm the importance of population specific pharmacogenetic role in guiding warfarin dosing in the Arab population and support the initiation of clinical implementation precision medicine programs in the MENA region.
INTRODUCTION
For more than 6 decades, warfarin served as a cornerstone therapy for the treatment and prevention of a wide variety of thromboembolic conditions. 1 For many patients worldwide, warfarin remains a viable and effective option, given its low cost and generic availability. 2 Whereas direct oral anticoagulants use has increased over the past few years, warfarin is a widely used oral anticoagulant, especially in the Middle East and North Africa (MENA) region. 3 For example, in Qatar, warfarin use comprised 77% of all oral anticoagulants in 2015 compared to direct oral anticoagulants. 4 Even with its widespread use as a primary therapeutic option for many patients, warfarin’s narrow therapeutic index remains one of its disadvantages leading to an altered level of coagulation, resulting in bleeding and thrombotic adverse events, which may, in some situations, lead to death. 5 Additionally, warfarin regimens are usually associated with inter‐ and intrapatient variability, which makes it challenging to achieve optimal anticoagulation. For instance, warfarin maintenance dose requirements can vary from 0.5 to 20 mg per day. 6 , 7 Many factors may contribute to this variability, including age, body surface area, ethnicity, vitamin K intake, concomitant disease conditions, alcohol intake, diet, smoking, and genetics. 8
Studies have shown that pharmacogenetic algorithms can differ by race, with an improvement in anticoagulation control in certain populations versus others. Furthermore, genomewide association studies (GWAS) conducted in patients of different races have identified different genetic polymorphisms associated with warfarin dose variability. 9 , 10 , 11 , 12 , 13 , 14 However, to date, there have been no studies from the MENA region to identify genetic variants associated with warfarin dose variability using a GWAS approach. Thus, we aimed in this study to conduct the first GWAS to identify genetic variants associated with warfarin dose requirement and the extent to which genetic and nongenetic factors contribute to warfarin dose variability in patients from the MENA region, mainly from Qatar and Egypt.
METHODS
Study population
This study included a total of 182 warfarin‐treated participants (132 Qataris and 50 Egyptians), which were included in the discovery and replication phases, as explained below.
Discovery
The discovery cohort consisted of 132 self‐identified Qatari national patients recruited from the anticoagulation clinics at Al‐Wakra Hospital, Heart Hospital, and Hamad General Hospital. The three hospitals are part of the Hamad Medical Corporation, the main governmental healthcare entity in Qatar. Recruitment started in September 2016 and was completed in March 2017. DNA samples were processed and analyzed at the laboratories of Qatar University (QU) between November 2016 and June 2017. Detailed inclusion and exclusion criteria have been previously explained by our group. 15
Replication cohort
The replication cohort consisted of 50 Egyptian patients recruited from four different clinical sites in Cairo, including Nasser Institute Hospital, Cardiology Clinic at Al Azhar University, Demerdash Hospital, and an Egyptian Cardiology Clinic. Eligible patients were defined as those taking stable weekly doses of warfarin for three consecutive clinic visits, occurring over a minimum of 2 months. We defined a stable weekly dose of warfarin as a dose that did not change by more than 10% across clinic visits, with international normalized ratio (INR) values at each of the three visits being within the patient’s INR target range. We excluded patients with liver cirrhosis, advanced malignancy, a recent hospitalization within the previous 4 weeks, and patients who had diarrhea and any febrile illness within the past 2 weeks from the visit.
The research ethics committee approved the protocols for each study at each institution. For the Qatari study, approvals were obtained from the Institutional Review Board of Hamad Medical Corporation and QU. For the Egyptian study, approval was obtained from the Research Ethics Committee at the Faculty of Medicine, Ain Shams University, Cairo, Egypt.
Sample collection and DNA extraction
For the Qatari samples, genomic DNA was extracted from either whole blood or saliva using the PureLink Genomic DNA mini kits, Invitrogen or the prepIT L2P manual protocol, respectively. 15 For the Egyptian samples, genomic DNA was collected from peripheral blood using QIAamp DNA Blood Mini Kit or automated QIAcube device (QIAGEN), following the manufacturer’s protocols and guidelines. 16
Genotyping and quality control procedures
Both Qatari and Egyptian samples were genotyped together using the Illumina Multi‐Ethnic Global BeadChip Array platform. Quality control procedures were performed in PLINK version 1.9. Single‐nucleotide polymorphisms (SNPs) were included if they had a genotyping call rate greater than 95%. Sample missingness rate was set at 95%, and samples were removed if they had a missingness rate higher than this threshold. Samples with a mismatch between genetic and demographic sex were removed. Deviation from Hardy‐Weinberg Equilibrium was tested using Fisher’s exact test. We conducted Identity By Descent/Identity By Sample, Heterozygosity, and Principal Component of Ancestry (PCA) analyses using linkage disequilibrium (LD)‐pruned, high‐quality SNPs with minor allele frequency greater than 10%.
Assessed phenotype
The study phenotype was defined as a stable, total weekly, therapeutic warfarin dose. Weekly warfarin doses were log‐transformed to limit heteroscedasticity and improve model fit.
Statistical analysis
Genomewide association analysis
First, we performed a linear regression analysis in PLINK for a total of 1,132,000 genotyped SNPs, using an additive genetic model (Figure 1). This analysis was performed separately in both discovery (Qatari samples, N = 132) and replication (Egyptian samples, N = 50), adjusting for age, body surface area, smoking status, first four PCAs for ancestry, and clinical comorbidities, such as hypertension and diabetes. Genomewide significant signals were defined as those with association p value less than 5 × 10−8. Using Haploview, 17 we assessed the LD between signals in the top associated regions from the GWAS. The GWAS significant SNPs, at p < 5 × 10−8, in Qatari (discovery cohort) were tested in Egyptians (replication cohort). SNPs were considered replicated if the direction of association in the replication cohort was similar to that observed in the discovery cohort, with a statistically significant Bonferroni corrected p value (0.05/number of independent SNPs tested for replication). Furthermore, we performed a conditional analysis adjusting for the top GWAS‐associated signal to identify additional independent signals that may be masked by the effect of the top GWAS signal.
FIGURE 1.
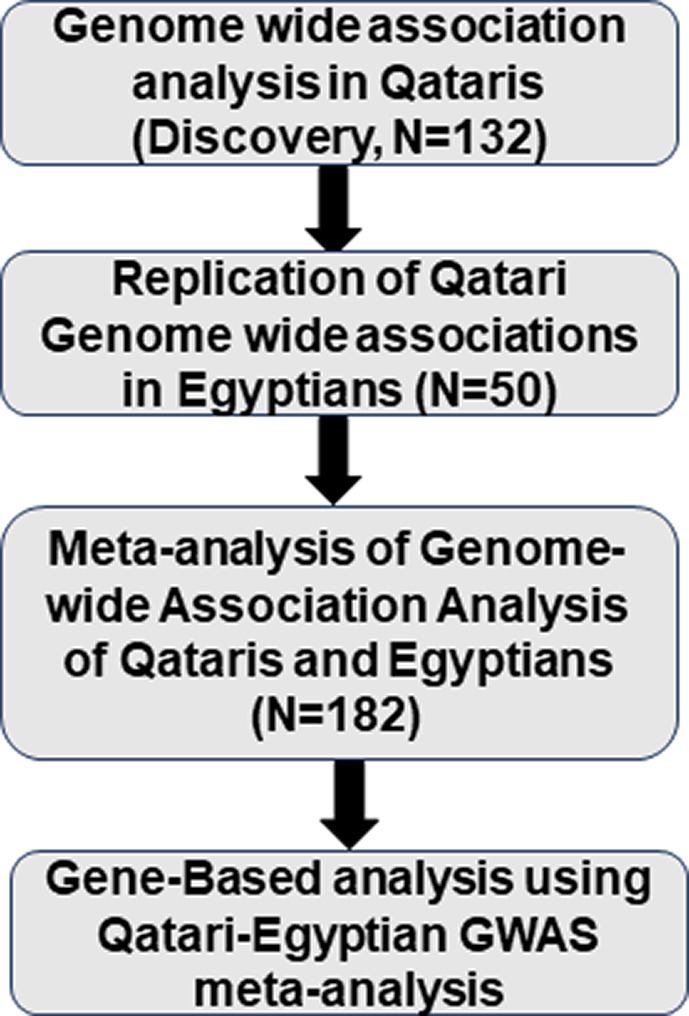
Overall description of the study analysis steps. A total of 1,132,000 SNPs were genotyped using an additive genetic model. This analysis was performed separately in both discovery (Qatari samples, N = 132) and replication (Egyptian samples, N = 50). This was followed by gene‐based analysis using a Qatari‐Egyptian combined cohort. GWAS, genomewide association study; SNPs, single‐nucleotide polymorphisms
In an attempt to detect novel warfarin associations that were not discovered in either of the cohorts alone, we meta‐analyzed the summary statistics from both the discovery and replication cohorts (Qatari and Egyptian samples) to increase the sample size and power of our analysis. The meta‐analysis was performed in METAL using fixed effect, inverse‐variance method. 18
Gene‐based analysis
Previous studies have shown that running GWAS using a gene‐based approach rather than an SNP‐based approach provides more power due to the aggregate effect of multiple SNPs being larger than that of individual SNPs. 19 Thus, we performed a gene‐based analysis relying on the input from the Qatari‐Egyptians meta‐analysis results. This analysis was performed using MAGMA (Multi‐marker Analysis of GenoMic Annotation), 19 a tool that is broadly used to run gene‐based analysis. For this analysis, SNPs were mapped to genes based on their physical distances (within 10 kb) from protein‐coding genes in the human reference assembly (hg 19). Based on this gene mapping strategy, a total of 14,075 genes were defined and served as the basis for the gene‐based analysis in MAGMA. A Bonferroni corrected p value was used to adjust for multiple testing, and according to the number of tested genes, the level of gene‐based GWAS significance was set at 0.05/14,075 = 3.55 × 10−6.
Contribution of associated SNPs to warfarin dose variability
A stepwise linear regression analysis was performed, using R (version 3.6.2), to evaluate the percent of variability in warfarin dose requirements that could be explained by significant covariates. We performed the regression analysis in the derivation cohort (Qatari) and variable selection was executed using a forward selection to include variables with p value less than 0.20, and backward elimination to only retain those with p values less than 0.05 in the model. The My.stepwise.lm function from the “My.stepwise” library was used to perform both forward selection and backward elimination method. Independent Qatari genetic signals that were replicated in the Egyptian cohort were included in the final regression model in addition to the lead SNPs within the top genes that passed the gene‐based significance threshold. Partial r 2 of the clinical and genetic factors and adjusted r 2 were reported in the final model.
RESULTS
A total of 182 warfarin‐treated patients from Qatar (N = 132) and Egypt (N = 50) were included in this study. The average age of patients in years was 62 ± 13 and 41 ± 12 in the Qatari and Egyptian cohorts, respectively. The median and interquartile range for the weekly warfarin dose in Qataris and Egyptians was 32 mg (24.5) and 37 mg (42.0), respectively (Table 1). The indication for warfarin varied between the two cohorts, with atrial fibrillation being the most common indication for warfarin in Qatari (n = 87, 66%) and heart valve replacement in Egyptians (n = 33, 66%).
TABLE 1.
Clinical characteristics of Egyptian and Qatari cohorts (N = 182)
| Clinical characteristics or demographics |
Qataris N = 132 |
Egyptians N = 50 |
p value |
|---|---|---|---|
| Age, years, mean ± SD | 6262 ± 13 | 41 ± 12 | 2 × 10−16 |
| Female, n (%) | 75 (57%) | 50 (50%) | 0.4 |
| BSA mean ± SD | 1.9 ± 0.2 | 1.9 ± 0.2 | 0.5 |
| Smoker, n (%) | 11 (8%) | 10 (20%) | 0.03 |
| Mean weekly, mg (SD) | 35 (16) | 44 (28) | 0.04 |
| Median weekly dose, mg (IQR) | 32 (24.5) | 37 (42.0) | 0.29 |
| Indication for warfarin | |||
| Atrial Fibrillation, n (%) | 87 (66%) | 0 | 1 × 10−15 |
| Venous thromboembolism, n (%) | 18 (14%) | 11 (22%) | 0.2 |
| Valve replacement, n (%) | 28 (21%) | 33 (66%) | 1 × 10−8 |
| Cardiovascular/metabolic comorbidities | |||
| Hypertension, n (%) | 89 (67%) | 6 (12%) | 2.3 × 10−11 |
| Diabetes, n (%) | 75 (57%) | 2 (4%) | 1.2 × 10−10 |
Categorical variables were expressed as frequency (%). Numerical variables were expressed as mean and standard deviation (SD) as in age and BSA, or median and IQR as in weekly warfarin doses.
Abbreviations: BSA, body surface area; IQR, interquartile range.
Warfarin association results in Qatari with validation in Egyptians
A total of five SNPs on chromosome 16 reached genomewide significance at p < 5 × 10−8, with the top signal being the VKORC1 rs9934438 (β = −0.16, p = 2.1 × 10−12; Table 2 and Figure S1). Qatari patients carrying the rs9934438 A‐allele had lower weekly warfarin dose (AA genotypes = 27 mg/week, AG = 39 mg/week, and GG = 52 mg/week; Figure S2). Consistent with this association, Egyptian patients carrying two copies of the A‐allele of SNP rs9934438 had lower warfarin dose requirements, 19 mg/week, compared to heterozygous carriers, 41 mg/week, and noncarriers, 46 mg/week. The association of the other four genomewide significant SNPs was evaluated in Egyptians, which showed a consistent association with warfarin dose as in Qatari at a corrected Bonferroni p value (Table 2). A conditional analysis adjusting for the VKORC1 rs9934438 SNP led to nonsignificant associations of the other genomewide significant SNPs, suggesting that VKORC1 rs9934438 SNP is the main driver of the association observed in this region. Evaluating LD for the five genomewide significant signals in Qatari and Egyptians, separately, confirmed that these five SNPs are in high LD (Figure 2).
TABLE 2.
Top signals in the Qatari cohort (N = 132) that were replicated in Egyptian cohort (N = 50)
| Top associated signals | Qatari, N = 132 | Egyptians, N = 50 | Meta‐analysis (N = 182) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Gene | A1 | β | p value | β | p value | β | p value |
| rs9934438 | 16 | VKORC1 | A | −0.16 | 2.1E−12 | −0.21 | 0.0006 | −0.16 | 7.6E−15 |
| rs10871454 | 16 | STX4 | T | −0.16 | 9.1E−12 | −0.21 | 0.0006 | −0.16 | 4.76E−14 |
| rs2359612 | 16 | VKORC1 | A | −0.15 | 1.1E−10 | −0.22 | 0.0005 | −0.16 | 2.0E−14 |
| rs2303222 | 16 | 5’ UTR‐ ZNF668 | C | −0.15 | 4.1E−10 | −0.21 | 0.0003 | −0.16 | 2.33E−14 |
| rs4889603 | 16 | SETD1A | G | −0.15 | 9.5E−10 | −0.19 | 0.001 | −0.15 | 3.45E−11 |
Abbreviation: SNP, single‐nucleotide polymorphism.
FIGURE 2.
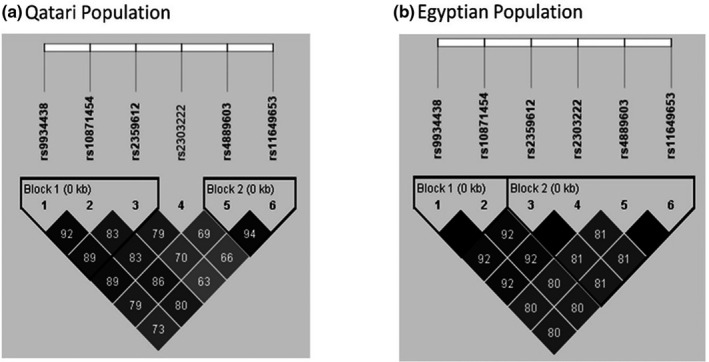
Linkage disequilibrium for top genomewide association study signals on chromosome 16 in the Qatari population (a) and the Egyptian population (b)
Warfarin association results in the Qatari and Egyptian GWAS meta‐analysis
The meta‐analysis of the Qatari‐Egyptian cohort strengthened the observed association in chromosome 16, with VKORC1 rs9934438 significantly impacting warfarin dose requirements and reaching the highest association in this region (p = 7.6 × 10−15; Table 2, Table S1, Figure 3). Besides chromosome 16 signals, no other genetic signals passed the genomewide significance level (p < 5 × 10−8).
FIGURE 3.
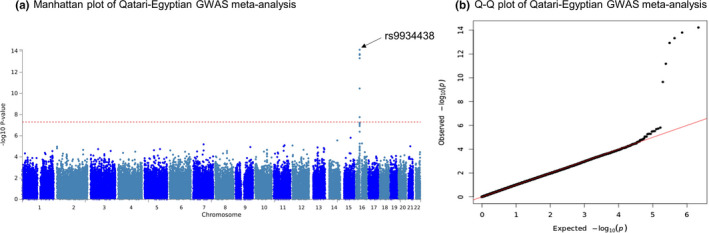
Association with log‐transformed weekly warfarin dose in Qatari‐Egyptian GWAS meta‐analysis. Panel a: The Manhattan plot of the GWAS association shows the SNPs on chromosome 16 that reached genome‐wide significance at p < 5 × 10−8, for association with warfarin dose. Top signal included the VKORC1 rs9934438 as well as rs10871454, rs2359612, rs2303222, rs4889603, and rs11649653. Panel b: Quantile‐quantile(Q‐Q) plot of the data shown in the Manhattan plot. GWAS, genomewide association study; SNPs, single‐nucleotide polymorphisms
Gene‐based analysis
The results of the gene‐based GWAS identified six significantly associated genes that passed the Bonferroni corrected significance threshold, as explained in the methods. Of these six genes, five are located on chromosome 16, and one is located on chromosome 10, with VKORC1 and CYP2C9 being among the top associated genes. The rs9934438 and rs4086116 SNPs were the lead signals in chromosomes 16 and 10, respectively (Figure 4 and Table S2). The rs4086116 T‐allele was significantly associated with lower warfarin weekly dose requirement in Qatari’s (β = −0.11, p = 3.6 × 10−4) and Egyptians (β = −0.18, p = 0.02; Figure S3). Patients carrying the rs4086116 T‐allele had lower weekly warfarin dose compared to patients with homozygous C‐allele in Qataris and Egyptians (Qataris: [TC/TT] = 34 mg, CC = 43 mg; Egyptians: [TC/TT] = 33 mg, CC = 51 mg; Figure S3). Of note, according to the 1000 Genomes Project data, the rs4086116 SNP is in moderate LD (r 2 = 0.62, D’ = 1) with CYP2C9*2 SNP (rs1799853).
FIGURE 4.
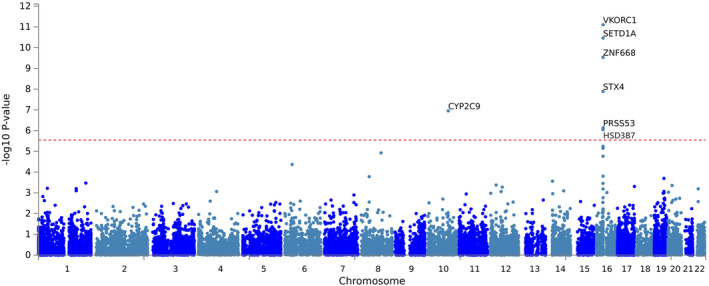
Manhattan plot showing association results of gene‐based Qatari‐Egyptian GWAS meta‐analysis. Significance level was set at p = 3.55 × 10−6 (0.05/#genes). Top signal being VKORC1, SETD1A, ZNF668, STX4, CYP2C9, PRSS53, and HSD3B7. GWAS, genomewide association study
Contribution of identified genetic variants on warfarin dose variability
A linear regression model was performed in Qatari (derivation cohort) to identify the percent of variability explained by the replicated genetic polymorphisms. This model included the replicated independent signals from Qatari GWAS analysis (VKORC1 rs9934438) and the lead SNP within the CYP2C9 gene (rs4086116), which passed the significant threshold of the gene‐based analysis. As explained in the methods section, several clinical variables were also included to evaluate the percent of variability in warfarin dose requirements that can be explained using both genetic and clinical variables in Qatari. Collectively, the clinical and genetic predictors explained a total of 53% of the variability in weekly warfarin dose requirement (Table 3). Whereas the clinical predictors, including age, smoking, valve replacement, diabetes, and hypertension explained ~12% of warfarin dose variability, the genetic signals alone (CYP2C9 rs4086116 and VKORC1 rs9934438) explained ~39% variability in Qatari’s and ~27% in Egyptians.
TABLE 3.
Contribution of clinical characteristics and VKORC1 and CYP2C9 SNPs to warfarin dose requirement in Qatari (N = 132)
| Variables | β (SE) | p value | Partial r 2 |
|---|---|---|---|
| Age, years | −0.002 (0.001) | 0.02 | 0.05 |
| Diabetes | 0.06 (0.03) | 0.04 | 0.04 |
| Hypertension | −0.08 (0.03) | 0.008 | 0.05 |
| Smoking | 0.13 (0.05) | 0.006 | 0.06 |
| VKORC1‐rs9934438 | −0.17 (0.02) | <2 × 10−16 | 0.42 |
| CYP2C9‐rs4086116 a | −0.10 (0.02) | 1.1 × 10−5 | 0.14 |
| Adjusted R 2 | 53% | ||
According to 1000 Genomes Project data, the rs4086116 SNP is in moderate linkage disequilibrium (r 2 = 0.62, D’ = 1) with CYP2C9*2 (rs1799853).
DISCUSSION
The geographical location of the Middle East at the crossroads of Africa, Europe, and Asia has enabled populations within this region to be the first to migrate out of Africa, which makes them particularly interesting for genomics research. 20 Qatar is a peninsula in the Persian/Arabian Gulf. It resides at the eastern edge of the Arabian Peninsula, making it the center of migration patterns that took part in the region over the era of human history. Various migrations and historical events have created a genomic admixed Qatari population consisting mainly of Arabic (mainly Bedouins), Persian (Iran, Pakistan, and Afghanistan), and African (Sub‐Saharan Africa) ancestry. A high rate of consanguinity still exists among Qatari (35% in 2010), with first cousins’ marriages being widely accepted and predominant. 21 Such high levels of consanguinity may have had a profound influence on the genetic make‐up of this population. 22 Egyptians are also characterized by having strong Arabic (mainly Bedouins) and African (Sub‐Saharan Africa) ancestry. 16 There has been limited genetic research in Arabs despite the unique genetic admixture of that population. Additionally, warfarin is still widely used in the region. 4 Thus, it was imperative to conduct GWAS for warfarin dosing in Arabs, which, to the best of our knowledge, is the first in this population.
Our GWAS indicates that VKORC1 rs9934438 is the main SNP associated with warfarin dose variability in a cohort of Qatari and was replicated in an independent cohort of Egyptians (Figures S1 and S2). There were four more genomewide significant SNPs on chromosome 16 (Figure 3a), but the association turned to be nonsignificant after the conditional adjustment for the VKORC1 rs9934438 genotype (data not shown). This suggests that the association in this region is driven primarily by the VKORC1 rs9934438 signal. VKORC1 is the gene coding for the vitamin K epoxide reductase (VKOR), an essential enzyme for the vitamin K cycle activity and the activation of the coagulation factors. By inhibiting this enzyme, warfarin prevents the activation of the vitamin K‐dependent coagulation factors and mediates its anticoagulant activity. 23 , 24
Apart from VKORC1, there were associated signals in CYP2C9 when the Qatari and Egyptian cohorts were combined together in the gene‐based analysis. CYP2C9 was one of the signals that passed the gene‐based significant threshold, with SNP rs4086116 being the lead SNP in this gene. The lead SNP, rs4086116, was an intronic SNP, which, according to data from the 1000 Genomes Project, it is in moderate LD (r 2 = 0.62, D’ = 1) with CYP2C9*2 variant (rs1799853). Warfarin S isomer is mainly metabolized by the cytochrome P450 2C9 (CYP2C9) enzyme and is 3–5 times more potent than the R isomer, but has faster clearance. 25 Findings from this work are in alignment with previous results from warfarin GWAS in other ethnicities, which identified VKORC1 and CYP2C9 to be the strongest genetic determinants of warfarin dosing variability. 9 , 10 , 11 , 12 , 13 , 14 SNPs with the following genomic regions/genes, STX4, ZNF668, SETD1A, and CTF1 also showed GWAS significant association with warfarin dose requirement, but these signals are likely mediated by the VKORC1.
In an effort to demonstrate the clinical utility of genetic‐guided dosing of warfarin, multiple randomized controlled trials (RCTs) have been conducted over the past decade. In 2013, the first two landmark trials—the Clarification of Optimal Anticoagulation through Genetics (COAG) and the European Pharmacogenetics of Anticoagulant Therapy (EU‐PACT) were published. 7 , 26 Whereas the EU‐PACT trial showed benefit of genetic‐guided dosing of warfarin, results from COAG were negative. Furthermore, COAG found that the percent time in therapeutic range (PTTR) was significantly lower in Black patients in the genetic‐guided arm compared to the clinical dosing arm. 26 This is possibly due to the fact that Black patients may have other less common variants affecting warfarin dose that were not well‐represented in the genetic algorithm used in the COAG trial. 27 This point also highlights the importance of examining the genetic variation affecting the warfarin dose across populations instead of relying on a generic warfarin dosing algorithm predominantly derived in patients with European ancestry. Recently, a third landmark trial—Genetics‐InFormatics Trial (GIFT) also tested the utility of warfarin pharmacogenetic‐guided dosing. 28 The pharmacogenetic dosing algorithm used included genotypes for CYP2C9*2 and *3, CYP4F2*3, and VKORC1‐1639. The primary end point was composite of major bleeding, INR greater than or equal to 4, venous thromboembolism, or death. GIFT indicated that genotype‐guided dosing could improve the composite outcome of efficacy and safety. Almost 11% of the participants had at least one composite end point in the genotype‐guided arm, compared to 14.7% in the clinical arm, resulting in an absolute risk difference of 3.9% (95% confidence interval, 0.7% to 7.2%, p = 0.02). There was an improvement in the mean PTTR (54.7% vs. 51.3%, p = 0.003) in the genotype‐guided group compared to the clinical group. Based on the results from these RCTs, it is still not recommended to perform genetic testing prior to warfarin initiation, as per the evidence‐based guidelines. In this paper, we documented that variants in VKORC1 (rs9934438) and CYP2C9 (rs4086116) were associated with warfarin dose requirements in the population from the MENA region and, together with age, presence of valve replacement, diabetes, hypertension, and smoking, 53% of the variability in warfarin dose requirements could be explained in Qatari. The use of genetic information alone (CYP2C9 rs4086116 and VKORC1 rs9934438) explained ~39% and ~27% of the variability in Qatari and Egyptians, respectively, suggesting that these genetic variants are important warfarin dose determinants in this population.
With the advancement in whole genome sequencing and the tremendous drop in its cost, genetic data are expected to be present in all healthcare records. In the state of Qatar, initiatives from the Qatar Genome and Qatar Biobank already started in 2015 with a primary goal to use the latest DNA sequencing technologies to establish a genomic map of the Qatari population. 29 , 30 One of the long‐term goals of these projects is to integrate this genetic data into the electronic health record system used by government hospitals. Therefore, evidence from our GWAS and other studies may provide some guidance on how to use this genetic data once available. This could be through the use of an exact algorithm to predict the warfarin maintenance dose of the patient or to guide the use of direct oral anticoagulants versus warfarin in patients carrying risk variants, such as CYP2C9*2, *3, or other variation leading to a very high or very low warfarin dose. Having confirmed the effect of VKORC1 (rs9934438) and CYP2C9 (rs4086116) on warfarin dose and their prevalence in the Arab population, it would be important as a next step to compare the performance of internationally validated warfarin dosing algorithms to those developed specifically in Arabs. This would be followed by a clinical utility study to compare the effectiveness of genetically guided dosing approach using the best algorithm to the conventional dosing before proceeding toward clinical implementation.
Although this study provides the first GWAS evidence on warfarin dosing in the Arab population, it was limited by the small sample size. This is likely the reason why CYP2C9 genetic polymorphisms did not show a genomewide significant association in GWAS discovery analysis. Nevertheless, with the sample size used, we were able to confirm the importance of VKORC1 rs9934438 SNP on warfarin dose requirements in the Qatari and Egyptian populations. Additionally, using gene‐based analysis, both VKORC1 and CYP2C9 were among the top genes influencing warfarin dose requirements, consistent with GWAS results previously reported in other global populations. CYP2C9*2 has been shown to be an important predictor of warfarin dose variability in previously conducted warfarin GWAS; however, CYP2C9*2 was not present within the GWAS chip used in this study. Despite the absence of CYP2C9*2 genotyping data, the top intronic CYP2C9 SNP identified in the Qatari‐Egyptians gene‐based analysis was in moderate LD (r 2 = 0.62, D’ = 1) with CYP2C9*2, according to data from the 1000 Genomes Project, which indicates that this association may be mediated via the CYP2C9*2 genetic polymorphism.
In conclusion, the results of this study identified VKORC1 rs9934438 and CYP2C9 rs4086116 variants as important genetic variants that impact warfarin dose requirements in populations from the MENA region, which emphasize the substantial role of these variants in guiding warfarin dosing in these populations.
With the limited resources in many countries in the MENA region, warfarin remains a cornerstone treatment for many patients who cannot afford direct oral anticoagulants. Warfarin dose prediction based on the variants identified in VKORC1 and CYP2C9 could be valuable, especially if confirmed in other patients from the same region. This study is an important step to confirm the importance of population specific pharmacogenetic role in guiding warfarin dosing in the Arab population and support the initiation of clinical implementation precision medicine programs in the MENA region.
CONFLICTS OF INTEREST
M.H.S. is an employee of Pfizer, Inc. All other authors have no competing interests for this work.
AUTHORS CONTRIBUTIONS
All authors wrote the manuscript. M.H.S., S.I.K., and H.E. designed the research. N.E.R., M.H.S., L.B., and H.E. performed the research. N.E.R., M.H.S., L.B., and H.E. analyzed the data.
Supporting information
Supplementary Material
El Rouby N, Shahin MH, Bader L, Khalifa SI, Elewa H. Genomewide association analysis of warfarin dose requirements in Middle Eastern and North African populations. Clin Transl Sci. 2022;15:558–566. doi: 10.1111/cts.13176
Nihal El Rouby and Mohamed H. Shahin contributed equally to this work.
Funding information
This study was funded by GCC grant through Qatar University (Grant #GCC‐2017‐011).
REFERENCES
- 1. Ageno W, Gallus AS, Wittkowsky A, Crowther M, Hylek EM, Palareti G. Oral anticoagulant therapy: antithrombotic therapy and prevention of thrombosis, 9th ed: American College of Chest Physicians Evidence‐Based Clinical Practice Guidelines. Chest. 2012;141:e44S‐e88S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pengo V, Pegoraro C, Cucchini U, Iliceto S. Worldwide management of oral anticoagulant therapy: the ISAM study. J Thromb Thrombolysis. 2006;21:73‐77. [DOI] [PubMed] [Google Scholar]
- 3. Barnes GD, Lucas E, Alexander GC, Goldberger ZD. National trends in ambulatory oral anticoagulant use. Am J Med. 2015;128:1300‐1305.e1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elewa H, Alhaddad A, Al‐Rawi S, Nounou A, Mahmoud H, Singh R. Trends in oral anticoagulant use in Qatar: a 5‐year experience. J Thromb Thrombolysis. 2017;43:411‐416. [DOI] [PubMed] [Google Scholar]
- 5. Landefeld CS, Beyth RJ. Anticoagulant‐related bleeding: clinical epidemiology, prediction, and prevention. J Am Coll Cardiol. 1993;95:315‐328. [DOI] [PubMed] [Google Scholar]
- 6. Nunnelee JD, Review of an Article: The International Warfarin Pharmacogenetics Consortium . Estimation of the warfarin dose with clinical and pharmacogenetic data. NEJM. 2009;360(8):753‐764. J Vasc Nurs. 27, 109 (2009). [DOI] [PubMed] [Google Scholar]
- 7. Pirmohamed M, Burnside G, Eriksson N, et al. A randomized trial of genotype‐guided dosing of warfarin. NEJM. 2013;369:2294‐2303. [DOI] [PubMed] [Google Scholar]
- 8. Yuan HY, Chen J‐J, Lee MTM, et al. A novel functional VKORC1 promoter polymorphism is associated with inter‐individual and inter‐ethnic differences in warfarin sensitivity. Hum Mol Genet. 2005;14:1745‐1751. [DOI] [PubMed] [Google Scholar]
- 9. Cha P‐C, Mushiroda T, Takahashi A, et al. Genome‐wide association study identifies genetic determinants of warfarin responsiveness for Japanese. Hum Mol Genet. 2010;19:4735‐4744. [DOI] [PubMed] [Google Scholar]
- 10. Cooper GM, Johnson JA, Langaee TY, et al. A genome‐wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022‐1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parra EJ, Botton MR, Perini JA, et al. Genome‐wide association study of warfarin maintenance dose in a Brazilian sample. Pharmacogenomics. 2015;16:1253‐1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perera MA, Cavallari LH, Limdi NA, et al. Genetic variants associated with warfarin dose in African‐American individuals: a genome‐wide association study. Lancet. 2013;382:790‐796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takeuchi F, McGinnis R, Bourgeois S, et al. A genome‐wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teichert M, Eijgelsheim M, Rivadeneira F, et al. A genome‐wide association study of acenocoumarol maintenance dosage. Hum Mol Genet. 2009;18:3758‐3768. [DOI] [PubMed] [Google Scholar]
- 15. Bader L, Mahfouz A, Kasem M, et al. The effect of genetic and nongenetic factors on warfarin dose variability in Qatari population. Pharmacogenomics J. 2020;20:277‐284. [DOI] [PubMed] [Google Scholar]
- 16. Shahin MH, Khalifa SI, Gong Y, et al. Genetic and nongenetic factors associated with warfarin dose requirements in Egyptian patients. Pharmacogenet Genomics. 2011;21:130‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263‐265. [DOI] [PubMed] [Google Scholar]
- 18. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta‐analysis of genomewide association scans. Bioinformatics. 2010;26:2190‐2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene‐set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rodriguez‐Flores JL, Fakhro K, Agosto‐Perez F, et al. Indigenous Arabs are descendants of the earliest split from ancient Eurasian populations. Genome Res. 2016;26:151‐162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sandridge AL, Takeddin J, Al‐kaabi E, Frances Y. Consanguinity in Qatar: knowledge, attitude and practice in a population born between 1946 and 1991. J Biosoc Sci. 2010;42:59‐82. [DOI] [PubMed] [Google Scholar]
- 22. Hunter‐Zinck H, Musharoff S, Salit J, et al. Population genetic structure of the people of Qatar. Am J Hum Genet. 2010;87:17‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gong X, Gutala R, Jaiswal AK. Quinone oxidoreductases and vitamin K metabolism. Vitam Horm. 2008;78:85‐101. [DOI] [PubMed] [Google Scholar]
- 24. Oldenburg J, Marinova M, Muller‐Reible C, Watzka M. The vitamin K cycle. Vitam Horm. 2008;78:35‐62. [DOI] [PubMed] [Google Scholar]
- 25. Hirsh J, Fuster V, Ansell J, Halperin JL. American Heart Association/American College of Cardiology Foundation guide to warfarin therapy. J Am Coll Cardiol. 2003;41:1633‐1652. [DOI] [PubMed] [Google Scholar]
- 26. Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369:2283‐2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson JA, Cavallari LH. Warfarin pharmacogenetics. Trends Cardiovasc Med. 2015;25:33‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gage BF, Bass AR, Lin H, et al. Effect of genotype‐guided warfarin dosing on clinical events and anticoagulation control among patients undergoing hip or knee arthroplasty: the GIFT randomized clinical trial. JAMA. 2017;318:1115‐1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zayed H. The Qatar genome project: translation of whole‐genome sequencing into clinical practice. Int J Clin Pract. 2016;70:832‐834. [DOI] [PubMed] [Google Scholar]
- 30. Fakhro KA, Staudt MR, Ramstetter MD, et al. The Qatar genome: a population‐specific tool for precision medicine in the Middle East. Hum Genome Var. 2016;3:16016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


