Abstract
Aims
Genome-wide association studies (GWASs) have discovered hundreds of common genetic variants for atherosclerotic disease and cardiovascular risk factors. The translation of susceptibility loci into biological mechanisms and targets for drug discovery remains challenging. Intersecting genetic and gene expression data has led to the identification of candidate genes. However, previously studied tissues are often non-diseased and heterogeneous in cell composition, hindering accurate candidate prioritization. Therefore, we analysed single-cell transcriptomics from atherosclerotic plaques for cell-type-specific expression to identify atherosclerosis-associated candidate gene–cell pairs.
Methods and results
We applied gene-based analyses using GWAS summary statistics from 46 atherosclerotic and cardiovascular disease, risk factors, and other traits. We then intersected these candidates with single-cell RNA sequencing (scRNA-seq) data to identify genes specific for individual cell (sub)populations in atherosclerotic plaques. The coronary artery disease (CAD) loci demonstrated a prominent signal in plaque smooth muscle cells (SMCs) (SKI, KANK2, and SORT1) P-adj. = 0.0012, and endothelial cells (ECs) (SLC44A1, ATP2B1) P-adj. = 0.0011. Finally, we used liver-derived scRNA-seq data and showed hepatocyte-specific enrichment of genes involved in serum lipid levels.
Conclusion
We discovered novel and known gene–cell pairs pointing to new biological mechanisms of atherosclerotic disease. We highlight that loci associated with CAD reveal prominent association levels in mainly plaque SMC and EC populations. We present an intuitive single-cell transcriptomics-driven workflow rooted in human large-scale genetic studies to identify putative candidate genes and affected cells associated with cardiovascular traits. Collectively, our workflow allows for the identification of cell-specific targets relevant for atherosclerosis and can be universally applied to other complex genetic diseases and traits.
Keywords: Atherosclerosis, Cardiovascular disease, Genome-wide association study, Single-cell analysis
Graphical Abstract
Graphical Abstract.
Translational perspective .
Genome-wide association studies (GWAS) identified a large number of genomic loci associated with atherosclerotic disease. The translation of these results into drug development and faster diagnostics remains challenging. With our approach, we cross-reference the GWAS findings for atherosclerotic disease with single-cell RNA sequencing data of disease-relevant tissue and bring the GWAS findings closer to the functional and mechanistic studies.
Introduction
Since the first publication in 2005 on age-related macular degeneration,1 genome-wide association studies (GWAS) soared, and to date, the GWAS Catalogue2 registered over 5000 publications covering thousands of genetic associations. These efforts also identified hundreds of common genetic risk loci for cardiovascular diseases and traits, including coronary artery disease (CAD),3 ischaemic stroke subtypes,4 coronary artery calcification,5 and carotid intima and media thickness.6 Despite tremendous socio-economic and medical progress, cardiovascular disease remains the leading cause of death,7 highlighting the need for new biomarkers and pharmaceutical targets. Previous studies convincingly show that GWAS offer a solid foundation for innovative drug development8 as they agnostically identify genetic relationships with diseases and point to potential causal mechanisms.9
Identifying the causal genes and underlying pathological mechanisms that drive the signal in genetic risk loci is not straightforward. One possible approach to narrow down the lists of candidate genes is to utilize gene expression data, by identifying expression quantitative trait loci (eQTLs) that colocalize with genetic risk loci with expression changes in relevant tissues.10 Alternatively, transcriptome-wide association studies estimate genetic correlation between tissue-specific gene expression and complex traits to predict causal relations between traits.11 However, these powerful methods take advantage of whole-tissues collected in large biobanks and the variable cell composition confounds the resulting signal. Because tissues are composed of many cell types and tissue expression is averaged across all those cells, the signal from less prevalent but important cell populations is obscured. In order to test potential therapeutic targets, one requires suitable functional assays. This in turn requires an idea of the function and cellular expression of targets, which is complicated by tissue heterogeneity. Single-cell transcriptomics enables measurement of cell-specific expression, cell-to-cell expression variability, transcriptional noise, or temporal dynamics.12 Intersecting GWAS data with human transcriptomic single-cell data would increase the resolution both in terms of candidate gene and more importantly, it would identify the cell type most relevant for the given disease.
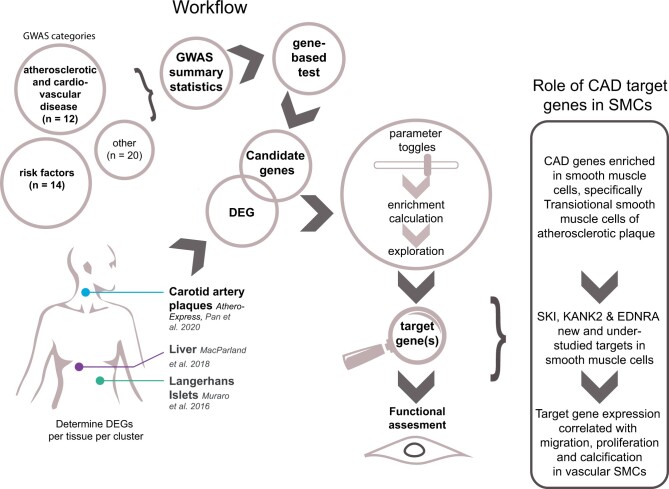
We hypothesized that genetic drivers for atherosclerotic disease can be found in plaque tissue. We aimed to project GWAS signals for 46 traits directly into a cell-type-specific expression using single-cell RNA sequencing (scRNA-seq) data derived from atherosclerotic plaques. To this end, we devised a workflow that utilized gene-based testing to identify candidate genes from GWAS summary statistics and calculated an enrichment score for each candidate list using expression profiles from scRNA-seq driven cell populations. This approach allows us to find the enrichment in expression patterns that point to specific, and potentially causal, cell populations. We report that loci associated with CAD reveal prominent association in mainly plaque smooth muscle cells (SMCs) and endothelial cells (ECs).
Methods
Single-cell sequencing: carotid artery plaques
Atherosclerotic lesions were collected from 12 female and 26 male patients undergoing a carotid endarterectomy procedure. All pathological tissue was included in the Athero-Express Biobank Study biobank (AE, www.atheroexpress.nl) at the University Medical Centre Utrecht (UMCU).13 Plaque processing for single-cell sequencing is described elsewhere.14 After sequencing, data were processed in an R 3.5 and 4.0 environment15 using Seurat version 3.2.2.16 Mitochondrial genes were excluded and doublets were omitted by gating for unique reads per cell (between 400 and 10 000) and total reads per cell (between 700 and 30 000). Data were corrected for sequencing batches using the function SCTransform. Clusters were created with 30 principal components at resolution 0.8. Multiple iterations of clustering were used to determine optimal clustering parameters. Population identities were assigned by evaluating gene expression per individual cell clusters. Sub-populations of SMCs were determined by isolating the SMC and EC populations from the complete population of cells mentioned above and re-assigned clusters. Clusters were created with 10 principal components at resolution 1.4. New subpopulation identities were assigned by evaluating gene expression per individual cell clusters. The newly defined SMC subpopulation identities were projected onto the SMC populations in the complete scRNA-seq dataset of atherosclerotic lesions for downstream processing.
Genome-wide association studies summary statistics
Genome-wide association studies summary statistics were leveraged from publicly available datasets. Mining these datasets uncovered a plethora of challenges. Not all GWAS summary statistics are available for download, can be requested by the authors, and/or only provide the lead single nucleotide nucleotide polymorphisms (SNPs). Other issues included ambiguous reports of sample size, genome build, or missing information such as standard errors and effect sizes. We were able to partly resolve these issues by using a custom pipeline, which automates tasks such as genome build liftover and SNP alignment and provides a uniform file format (Supplementary material online, Figure S2A). This significantly decreased workload and improved downstream applications.
Genome-wide association studies summary statistics were selected by means of relevance, ‘latest and the largest’ and publicly available, easily accessible and/or open-source data was given priority. Incomplete summary statistics were omitted during this phase (Supplementary material online, Figure S2A). A custom Python17 pipeline (see Data availability) was used to pre-process GWAS summary statistics and primary quality control. This primary control step was used to uniformly transform GWAS summary statistics because of a lack of a standardized format. In short, our custom pipeline was built to automate effect/other allele alignment with 1000 Genomes phase 318 release with a maximum frequency distance of 25%, maximum minor allele frequency (MAF) of 45% for ambivalent variants and lifting genomic positions to hg19 where needed. In preparation for GWAS alignment, dbSNP153 (GCF1405) was referenced to translate MEGASTROKE rs-ids to genetic positions and to augment AF, ASD, NICM, and TAGC (GWAS abbreviations are listed in Supplemental Table 1) with allele frequency information. Standard errors for HF and NICM were recovered as follows:
Where beta is the effect size, P is the P-value of association which is quantile normalized using the qnorm-function in R. SE is the standard error calculated by dividing the beta by Z. Effect sizes were calculated from odds ratios as beta = log(OR) for BIP, insomnia, IBD, and MDD before entering our custom pipeline.
The final selection was grouped into one of three categories: atherosclerotic disease and other cardiovascular disease (12), risk factors (14), and other (20). Data were further processed with MAGMA19 (version 1.07) for genome-wide analysis, annotation and characterization of significant hits via the FUMA20 web platform (version 1.3.6, accessed June 2020, https://fuma.ctglab.nl/). We applied genome-wide significance (P < 5 × 10−8) and linkage disequilibrium (r2 > 0.05) filtering for clumping of independent loci within 1000 kb of the lead variant, and included only variants with MAF >1%. For SVS, logOnset, EvrSmk and SVD no SNP associations below the genome-wide significance threshold we found. Therefore lead variant discovery was performed with a relaxed threshold of 5 × 10−6.
Selecting cell population-specific genes
Differentially expressed genes (DEGs) were selected via the Wilcoxon sum rank test. A gene was considered a DEG if (i) 10% of cells within a cluster expressed the gene with log2 fold change of at least 0.25; (ii) the gene passed the Bonferroni adjusted significance threshold of P < 0.05 for this test. Differentially expressed genes were determined via a ‘one cluster vs. one cluster’ and ‘one cluster vs. remaining clusters’5 (Supplementary material online, Figure S2B). This method results in genes specific for a cluster compared to every other single cluster and genes specific for a cluster compared to the whole single-cell reference set.
Gene score calculation
Fully processed summary statistics and cell population-specific DEG were used for calculating gene scores. The background gene set (n = 20 112 genes) was defined as all the genes that are reported and annotated by MAGMA, regardless of their respective P-value or Z score with duplicates excluded. We calculated the random gene score per cell-population by overlapping the genes in this background set with the cell-population-specific DEG. For every anaysis, the background genes are the same per GWAS. Z-score threshold for the P-values from MAGMA was set at 3 and we included a maximum of 150 candidate genes meeting this threshold (Supplementary material online, Figure S1B). If there were more than 150 genes meeting the threshold, we only used the first 150. Calculations were made as follows: (i) per cell population, determine the overlap of the candidate genes of a GWAS with the cell population-specific DEG. (ii) Score each of these genes according to average cell population expression: assign one point for expression between 1 and 2, and two points for expression >2 and 0 points for expression <1. Only genes that are cell population-specific receive a score. Assign the points to the genes from the random candidate gene list in the same way (Supplementary material online, Figure S2Cii). (iii) Compare the scores from the candidate genes with the scores of the random genes via one-tailed Wilcoxon Rank sum test in R and adjust the results per GWAS for multiple testing using the Benjamini–Hochberg method. Adjusted P-values smaller than 0.1 (10% false discovery rate) were considered significant (Supplementary material online, Figure S2Ciii). Enrichment was defined as the fold change of the mean of the gene scores divided over the mean of the gene scores in the random set. All scripts are available on GitHub (see Data availability).
Results
Single-cell RNA-seq of atherosclerotic plaques reveals 18 cell populations
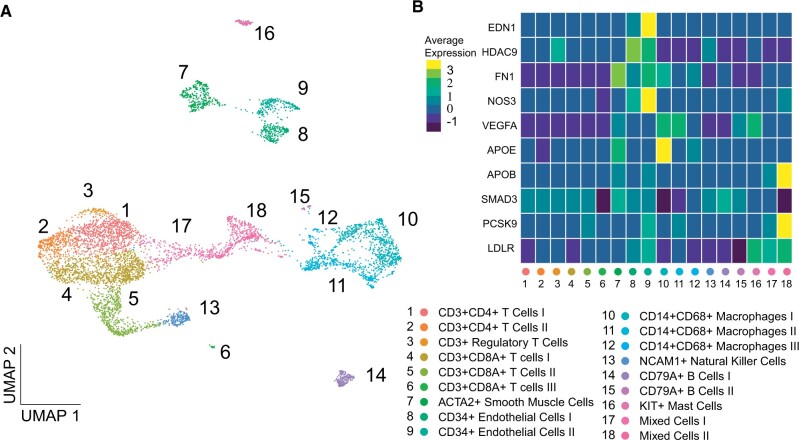
We have expanded our previously published14 scRNA-seq datasets of human atherosclerotic plaque cells in a cohort which now includes 38 patients and identified 18 cell populations amongst 6191 cells. These expanded datasets allowed us to map the genetic loci not only to main cell types (e.g. SMCs) but also to more specific cell subtypes (e.g. synthetic, contractile or transitioning SMCs)—increasing resolution compared to the previous dataset. By combining known and recently published sets of markers we identified clusters of lymphocytes, macrophages, mast cells, ECs, and SMCs (Figure 1A). This data is available for interactive exploration via the online tool Plaqview21 accessible on www.plaqview.com. Notably, the expression of selected, widely studied GWAS targets for CAD (EDN1, HDAC9, NOS3, VEGFA, FN1, APOE, APOB, SMAD3, PCSK9, and LDLR) clearly demonstrates cell-specific patterns (Figure 1B). This underscores the need for a systematic analysis using large-scale GWAS summary statistics and motivated us to devise an intuitive two-stage approach.
Figure 1.
Finding suitable therapeutic targets from genome-wide association studies with single-cell RNA sequencing. (A) UMAP visualization of 6191 cells divided into 18 cell populations derived from 38 human carotid artery plaques. (B) Heatmap of frequently studied coronary artery disease genome-wide association studies hits: EDN1, HDAC9, NOS3, VEGFA, FN1, APOE, APOB, SMAD3, PCSK9, and LDLR in atherosclerotic tissue shown in cells depicted in cell populations shown in A.
Gene-based analyses of atherosclerotic diseases and cell population-specific genes
First, we aimed to identify trait-associated genes using GWAS summary statistics by aggregating per-variant statistics and to calculate an empirically derived P-value for a given gene. Thus, gene-trait associations would be agnostically derived and solely based on genetic signals. To do this, we collected and partitioned GWAS summary statistics for 46 traits across 3 categories: atherosclerotic disease and other cardiovascular disease (12), risk factors (14), and other (20) (Supplementary material online, Table S1 and Figure S1A). From these GWASs, 40 were performed on population with European ancestry, and the remaining 6 had mixed or other ancestries. Next, to identify genes associated with individual traits we performed gene-based testing using MAGMA19 per individual GWAS. Genes mapped by gene-based testing are referred to as candidate genes (Supplementary material online, Figure S2A and Data).
Second, we aimed for an unbiased selection of genes that would best represent each cell population in a given dataset (Supplementary material online, Figure S2B). Per tissue, these genes were selected through differential gene expression and include genes that are overexpressed in specific cell populations compared to all other cells (one vs. all) or compared to any individual cell cluster (one vs. one).14 This approach eliminates ubiquitously expressed genes and genes with low expression without the need of a predefined list (Supplementary material online, Figure S4) and identifies a similar number of cell-specific genes in each population. Cell population-specific genes will be further referred to as differentially expressed genes (DEGs). For cell populations in the atherosclerotic plaque, the average number of DEG was 461, with the lowest 307 for B Cells II and highest 678 DEGs for T Cells III.
Plaque cell populations show specific enrichment of genome-wide association studies candidate genes
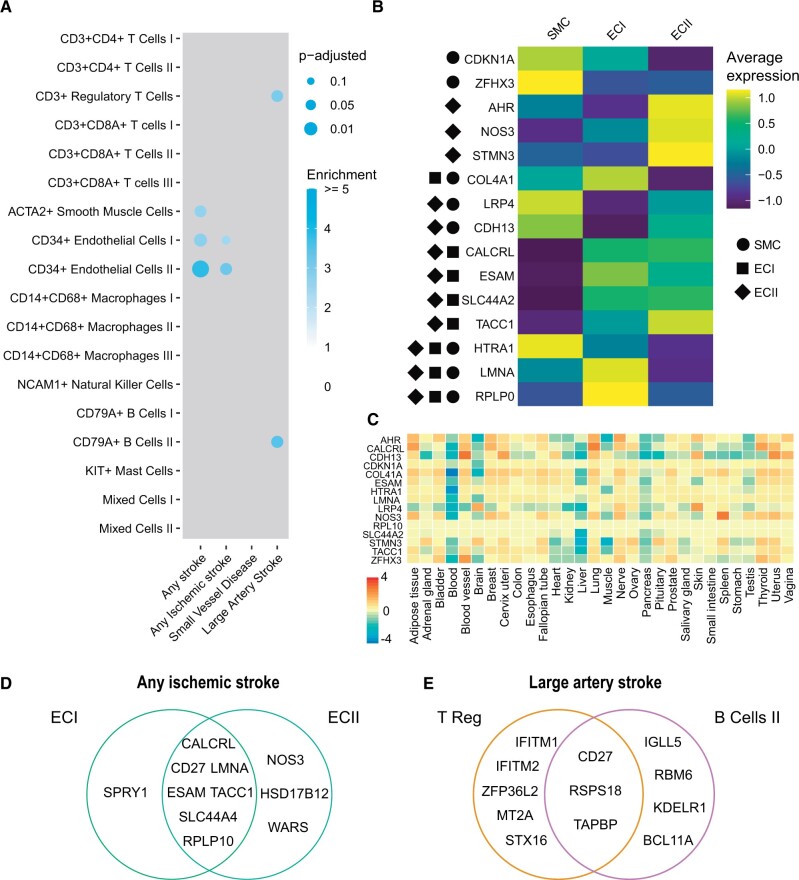
To test our hypothesis that the genetic drivers for atherosclerotic disease can be found in plaque tissue, we applied our workflow on cardiovascular candidate genes and the DEG sets from individual cell types present in the atherosclerotic plaque (Supplementary material online, Figures S3A and C, Figure S4, Table S2, and Data). The signals we observed were independent of the total number of DEGs in each cell population (Supplementary material online, Figure S3C), and did not show a bias towards GWAS sample size (Supplementary material online, Figure S1C) or trait heritability (Supplementary material online, Figure S1D). We found cell-specific enrichment for different GWAS in the atherosclerotic disease category, which was mainly observed in ECs and SMCs. Stroke candidate genes were enriched in EC populations, Regulatory T Cells, and B Cells II (Figure 3A).
Figure 3.
Genome-wide association studies related to stroke show substrate in endothelial cells. (A) Plot representing overlaps of genome-wide association studies candidate genes on stroke with genes specifically expressed in cell populations from human atherosclerotic plaque. Colour intensity indicates enrichment over random data and size indicates adjusted P-value (Benjamini–Hochberg). (B) Heatmap depicting expression of genes from any stroke from A in the respective cell populations. Only significant overlaps are plotted, symbols represent the cell populations with the significant overlaps. (C) Average of normalized expression (zero mean across samples) of genes from any stroke in from B in 30 tissues from GTEx. (D) Genes for any ischaemic stroke underlying the overlaps in ECI and ECII populations. Only significant overlaps are plotted. (E) Genes for large artery stroke underlying the overlaps in T Reg and B cells II populations. Only significant overlaps are plotted.
Smooth muscle cells are predominantly associated with coronary artery disease genes
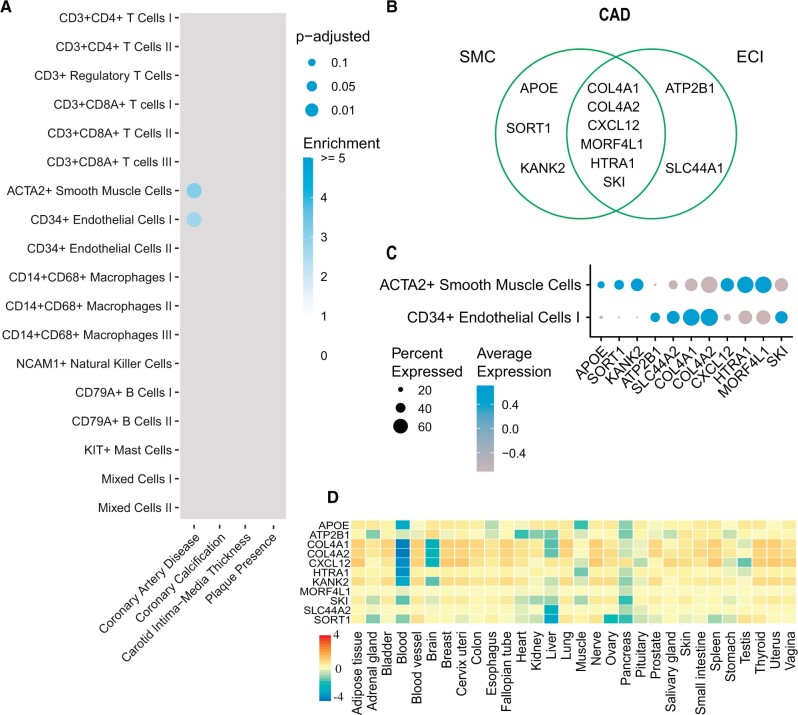
By highlighting the overlap between plaque-specific DEGs and candidate genes for CAD, we observed a significant enrichment in SMCs (P-adj. = 0.0012, 3.1 fold enrichment) and ECI (P-adj. = 0.0011, 2.7 fold enrichment) populations (Figure 2A). All P-values and fold enrichment values are listed in Supplementary material online, Table S2. This uncovered well-studied GWAS loci such as APOE, COL4A1, and COL4A2. The remainder of the overlap is driven by KANK2 and SORT1 in SMCs, SLC44A1 and ATP2B1 in ECI (Figure 2B). KANK2 belongs to the KANK protein family which plays a role in cytoskeletal function. SKI expression was present in both cell populations (Figure 2C). This proto-oncogene inhibits TGF-beta signalling.22 Interestingly, while using the single-cell transcriptomics enabled us to pinpoint specific cell (sub)populations, these potential targets exhibited ubiquitous expression in bulk transcriptomics data from GTEx tissue samples (Figure 2D). Notably, CD14+CD68+ Macrophages III showed an enrichment of 4.3 for CAD with genes APOE, SKI, and HNRNPUL1 but did not meet the significance threshold (P-adj. = 0.19).
Figure 2.
Genome-wide association studies candidate genes show cell-specific enrichment in atherosclerotic plaque cell populations. (A) Plot representing overlaps of genome-wide association studies candidate genes on cardiovascular disease with genes specifically expressed in cell populations from human atherosclerotic plaque. Colour intensity indicates enrichment over random data and size indicates adjusted P-value (Benjamini–Hochberg). (B) Genes for coronary artery disease underlying the overlaps in smooth muscle cell and ECI populations. Only significant overlaps are plotted. (C) Dotplot depicting expression of genes from B in the respective cell populations. (D) Average of normalized expression (zero mean across samples) of genes from coronary artery disease in smooth muscle cells from A in 30 tissues from GTEx.
To verify our results we used an independent scRNA-seq dataset of carotid artery plaques by Pan et al.23 (Supplementary material online, Figure S3B and D). The cell population labels broadly correspond between datasets (Supplementary material online, Figure S3E). The patterns for enrichment are similar between datasets (Supplementary material online, Figure S3A and B). We found that CAD, coronary artery calcification, and plaque presence candidate genes were enriched in the fibrochondrocyte cell population (Figure 3B), which corresponds to ACTA2+ SMCs cells in our dataset. For CAD, the enriched was driven by genes CDKN2A, COL4A1, COL4A2, APOC1, APOE, and CXCL12. Similar to the genes found in our datasets in SMCs and ECs (Supplementary material online, Table S2).
Endothelial cells are enriched for ischaemic stroke-associated genes
Atherosclerotic disease is one of the major culprits underlying stroke, with carotid plaques being the main driver of large artery stroke. We subsequently investigated overlap of GWAS-associated genes for stroke subtypes any stroke (AS) and any ischaemic stroke (IS) and found they were predominantly enriched in ECI (AS: P-adj. = 0.011, 2.7 fold enrichment; IS: P-adj = 0.077, 2.5 fold enrichment) and ECII (AS: P-adj = 1.9 × 10−5, 3.9 fold enrichment; IS: P-adj = 0.023, 3.2 fold enrichment) subtypes (Figure 3A). Overlapping genes between AS and cell populations include ESAM, LMNA, and SLC44A2 (Figure 3B). Soluble ESAM (EC adhesion molecule) levels have been correlated to myocardial infarction and heart failure.24Lmna deficient mouse models develop cardiovascular disease at an accelerated pace due to premature ageing.25 Of note, expression of these genes was not specific to vascular derived tissue of GTEx when using bulk transcriptomics data, suggesting that our findings are cell population and tissue specific (Figure 3C). No significant enrichment is found for IS in SMCs, but the cell population-specific signal in ECI and ECII is largely different from AS (Figure 3D).
Large artery stroke (LAS) is a subtype of IS. Strikingly, we see enrichment in CD3+ Regulatory T Cells and CD79A+ B Cells II (plasma cells) (P-adj = 0.026 and P-adj = 0.021, respectively) populations for LAS (Figure 3E). Suggesting the involvement of the adaptive immune system on the development of the LAS phenotype.
Transitional smooth muscle cells enriched for atherosclerotic disease candidate genes
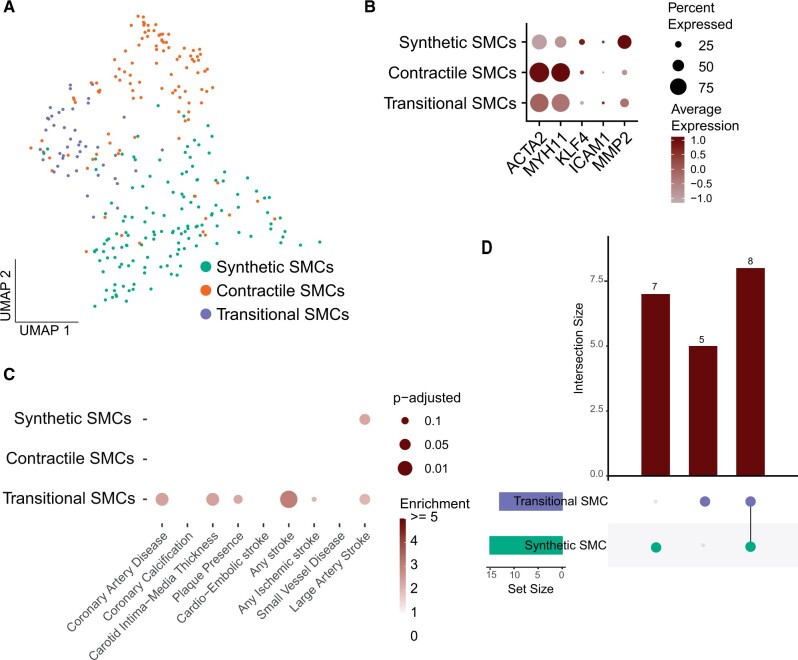
Plaque ACTA2+ SMCs can be further divided into three sub-populations with a distinct phenotype; Synthetic SMCs, contractile SMCs, and a smaller cell population consisting of SMCs with an intermediate phenotype (transitional SMCs) (Figure 4A and B, Supplementary material online, Figure S5A and Table S2). To explore if we can narrow down the signal from SMCs into a specific SMC subtype, we subjected this subset of plaque cells to our workflow (Supplementary material online, Figure S5B and C). This revealed that the majority of the signal can be found in transitional SMCs. This population is predominantly enriched for CAD (P-adj. = 0.026, 2.4 fold enrichment), carotid intima-media thickness (cIMT) (P-adj. = 0.027, 2.4 fold enrichment), and plaque presence (P-adj. = 0.081, 2.3 fold enrichment) (Figure 4C). Intima-media thickness is a commonly used tool to measure atherosclerosis.26 Signal for LAS was present in transitional and synthetic SMCs with unique hits for each cell population (Figure 4D).
Figure 4.
Analysis of three sub-populations of atherosclerotic smooth muscle cells. (A) UMAP visualisation of three sub-cell populations of smooth muscle cells in atherosclerotic plaque. (B) Dotplot depicting expression of synthetic and contractile smooth muscle cell markers in the three sub-populations. (C) Plot representing overlaps of genome-wide association studies candidate genes on cardiovascular disease and cardiometabolic traits with genes specifically expressed in the subset of smooth muscle cells from human atherosclerotic plaque. Colour intensity indicates enrichment over random data and size indicates adjusted P-value (Benjamini–Hochberg). (D) Plot depicting the number of unique and intersecting candidate genes from Large artery stroke enriched in transitional smooth muscle cells and synthetic smooth muscle cells.
To test the robustness of these results, we have repeated the analysis with the other public dataset (Pan et al.). For CAD, the overlap between transitional SMCs and corresponding ‘Fibrochondrocytes’ in Pan et al. (Supplementary material online, Figure S5D and E) showed significant enrichment and again identified a largely overlapping set of commonly studied GWAS loci COL4A1, COL4A2, APOC1, APOE, and CXCL12.
Functional assessment of smooth muscle cell-associated candidates
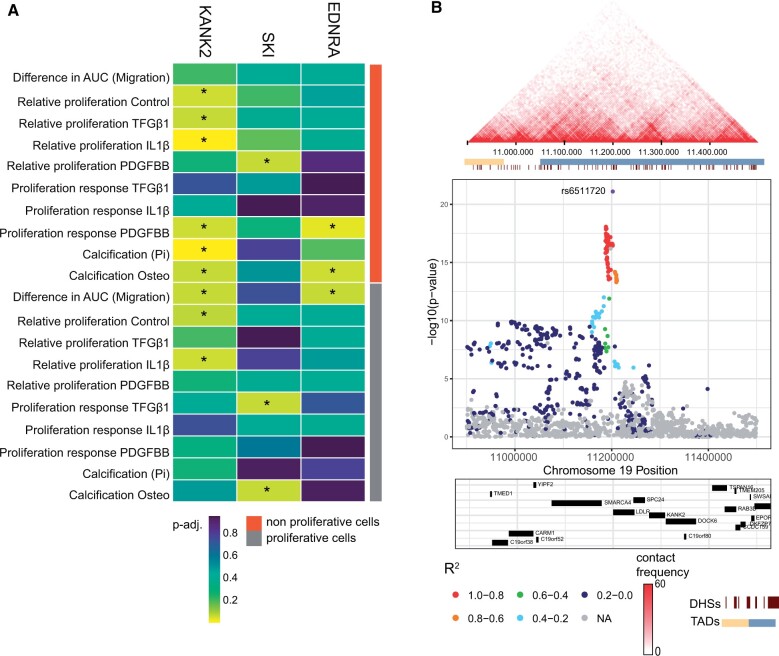
To demonstrate the potential role of newly identified gene-cell pairs in atherosclerosis-relevant process we selected three genes enriched in the SMC population of plaques. SKI, KANK2, and EDNRA are CAD candidate genes and are currently understudied in relation to atherosclerotic disease in this cell population. Making them possible interesting targets for further functional studies. To investigate the role of these genes in SMCs, we functionally characterized SMCs from the ascending aortas in 151 healthy and diverse donors for cellular migration, proliferation, and calcification.27KANK2 gene expression is correlated with proliferation under different stimuli (P-adj. < 0.1) and was negatively correlated with calcification in quiescent cells [Calcification Osteo: P-adj = 0.0058; Calcification (Pi): P-adj 0.069] (Figure 5A, Supplementary material online, Table S5). Under all stimuli, the response was positive, except upon TGFβ1 exposure. SKI expression positively correlates with calcification in proliferative cells (P-adj = 0.069) but has a negative response towards TGFβ1-induced proliferation (P-adj = 0.069, R −0.21) (Figure 5A, Supplementary material online, Table S5). EDNRA was negatively correlated with proliferation response to IL1β (P-adj. = 0.038) and calcification (P-adj = 0.062) in non-proliferative cells and showed a positive correlation in proliferative cells for migration (P-adj = 0.069) (Figure 5A, Supplementary material online, Table S5).
Figure 5.
Smooth muscle cell associated candidates influence migration and proliferation in vitro. (A) Correlation of KANK2, SKIi, and EDNRA expression non-proliferative and proliferative cells. (B) Plot depicting the Hi-C interactions around the main SNP associated with KANK2 (top) and regional association plot (bottom) in Hi-C data derived from IMR90 fibroblasts. AUC, area under curve; DHS, DNaseI hypersensitivity sites; Pi, inorganic phosphate; TAD, topologically associating domain.
Lipid-related risk factors are enriched in liver hepatocyte populations
Causal mechanisms for atherosclerosis have been linked to lipid metabolic processes active in the liver and blood glucose levels. This offered us the opportunity to assess the robustness of our approach across tissues and diseases. We acquired published scRNA-seq datasets of human liver cells28 and human pancreatic Langerhans islet cells29 to identify possible cell- and disease-specific targets.
Since one of the major liver functions is lipid metabolism, we expected strong signals for candidate genes involved in triglycerides levels. In line with our expectations, these GWASs indeed show the strongest signal in hepatocytes #3, #4, and #5 driven by several apolipoprotein genes (Supplementary material online, Figure S6A and C, Table S2).
Langerhans islet cells overall showed weak enrichments for all atherosclerosis-related GWASs (Supplementary material online, Figure S5B and D). Signal for CAD in endothelial and mesenchymal populations overlap with hits in comparable cell populations in plaque (Supplementary material online, Table S2). We could detect significant overlap of type 2 diabetes-associated genes with beta cells (P-adj. = 0.001) driven by INS, WFS1, and FTO amongst others (Supplementary material online, Table S2).
Altogether, we show that candidate genes for atherosclerotic disease are significantly enriched in EC and SMC populations of atherosclerotic plaque, notably in transitional SMCs. Stroke-related genes were enriched in ECs and lipid loci were found enriched in hepatocytes. Pancreatic beta cells were enriched for body mass index (BMI) candidate genes. Interestingly, only limited signals were observed in the cells of the adaptive immune system suggesting the genetic component for atherosclerosis is rooted in the structural cells of the vessel. We selected three candidate genes enriched in SMCs for functional testing in cells of ascending aortas for cellular calcification, proliferation and migration that showed that KANK2, SKI, and EDNRA expression correlated with calcification, migration, and proliferation in vascular SMCs.
Discussion
Post-GWAS analyses aiming to identify candidate genes for translation into clinical care are not straightforward. The thousands of variants associated with complex traits, including cardiovascular diseases, have made it abundantly clear that many genes contribute to disease risk and progression. For instance, GWAS for CAD have identified 163 loci linking to hundreds of candidate genes.3 Further efforts like chromatin interaction mapping linked almost 300 additional gene targets for atherosclerotic disease via distal chromatin interactions.14 This adds another layer to candidate gene prioritization. Selecting causal GWAS loci for functional testing can be deduced with vast knowledge of the trait or disease, but this remains speculative. There are numerous ways to accelerate the path to translational research. The intersection of loci found amongst different GWAS related to the same trait or disease can highlight some of the primary genes involved but, the cellular resolution is still lacking.4,30
Overlapping GWAS hits with pathways or within gene networks offers more context to study the genes. However, this is highly dependent on knowledge of the respective pathways and networks, and locating the relevant key players in the network is not indisputable.31
Colocalization of tissue-specific eQTLs10,32,33 with GWAS data provides another alternative that can be used to estimate if there are overlapping causal variants in both datasets. For example, a recent study on type 2 diabetes leveraged publicly available eQTL data and a separate pancreatic dataset to find targets for glucose- and insulin-related trait loci.10 In concordance to our data, trait-associated genes are expressed in pancreatic islet tissue. However, cell population-specific signals are lacking and eQTL signals can be confounded by variability in tissue cell composition. Currently, the first efforts are being made to study eQTLs in a single-cell space, with single-cell quantification of gene expression in disease relevant tissue. With this method, it is possible to detangle the effect of SNPs on the single-cell and patient-specific level and construct personalized gene regulatory networks.34,35
Another straightforward approach to identify the causative genes is overlapping the expression profiles of different tissues and organs to GWAS candidate genes. Utilizing bulk RNA sequencing data applicable to the GWAS helps to find targets that are directly acting in the affected tissue. This powerful approach has one major drawback: the data are composed of multiple cell populations and this signal is difficult to deconvolute into individual cell populations. It is especially limiting when looking for cells that are present in almost all tissues—like ECs or SMCs. We show here that these bulk RNA tissues approaches can be misleading due to lack of specific expression patterns for relevant GWAS loci in specific tissue, like vascular wall tissue (Figure 2E). On the other hand, the overlap with single-cell transcriptomics can detect significant overlaps in distinct cell types such as ECs.
Here, we present a possible approach to the prioritization problem. We projected GWAS loci directly into single-cell transcriptomics datasets derived from disease-relevant tissue. This allowed us to identify both known and novel gene targets together with their associated cell populations. Notably, the CAD-associated gene SKI is significantly enriched in SMC and EC populations (Figure 2A). SKI inhibits TGF-beta signalling,22 which plays a major role in atherosclerotic disease progression by controlling processes such as cell proliferation and matrix formation. Consequently, SKI increases lesion size in LDR−/− mice.36 Another identified gene—KANK2 plays a role in cytoskeletal function by reducing the cell’s ability to migrate by activating TALIN.37 This gene is specifically expressed in the SMC population of plaques. Possible impairment of cell mobility by KANK2 could lead to inefficient SMC migration towards the cap resulting in an unstable plaque phenotype. Interestingly, our functional analysis of KANK2 showed a positive association between gene expression and cell migration in quiescent and proliferative cells of SMCs from ascending aortas (R = 0.15 and 0.2, respectively) (Supplementary material online, Table S5)—suggesting more complex mechanisms. Follow-up experiments creating gene-deficient primary cell lines derived from atherosclerotic plaques to study the molecular mechanistic of those genes can provide more information of its role in atherosclerosis.38 Notably, the CAD-associated locus in the vicinity of KANK2 also contains LDLR gene (Figure 5B, bottom). Even though LDLR involvement in atherosclerosis is widely accepted, we were able to show that KANK2 is another strong candidate in this region as it also shows a strong correlation with SMC proliferation (Figure 5A). This also demonstrates that our approach can identify genes that are located more further from the lead SNP but are positioned within same broader regulatory domain (Figure 5B)—as based on Hi-C data from IMR90 fibroblast cells39 (Figure 5B, top).
Currently, 32 stroke loci have been implicated by GWAS.4 From the new main stroke loci found by Malik et al., only SLC44A2 was significantly enriched in scRNA-seq populations. However, we identified other potential targets in SMCs, ECs and in B cells, and regulatory T cell populations (Figure 3A). One of the targets, ESAM, was not yet directly linked to atherosclerosis but is known to enhance vascular permeability in selective tissues40 and could therefore contribute to the influx of cholesterol and inflammatory cells in the arteries contributing to the stroke phenotype. Another target, Gs-coupled receptor calcitonin receptor-like receptor (CALCRL) is expressed in both ECI and ECII populations. Activation of this receptor leads to an anti-inflammatory response in ECs. Subsequently, CALCRL deficient mice present with more advanced atherosclerotic lesions.41 A recent study by Örd et al.42 researched the enrichment of CAD, IS, and seven other cardiometabolic GWAS signals in scATAC-seq peaks of carotid artery plaques. Similarly, they report enrichment of CAD and stroke loci in SMCs and ECs and total cholesterol loci in macrophages.
Remarkable is the absence of significant enrichment in the cells of the adaptive immune system of the plaque for CAD. Out of six T-cell populations, four exhibited the enrichment of 0.00 and CD3+CD8A+ T cells II and II showed enrichment of 1.3 and 0.37, respectively, indicating that there is no or very little overlap between CAD candidate genes and cell population-specific DEGs. This suggests that the genetic component of atherosclerotic disease is not rooted in the adaptive immune system but rather in the structural cells of the vessel and influences the integrity of the vascular wall or plaque stability.
Our workflow can be generalized to other single-cell transcriptomics datasets and complex traits. We applied it to scRNA-seq data from tissues with expected causal involvement in atherosclerotic disease: human liver cells and human pancreatic Langerhans Islet cells. Genome-wide association studies studying lipid levels were associated with hepatocyte subsets but in particular subsets Hep3, Hep4, and Hep5 (Supplementary material online, Figure S6A). It suggests that these risk factors for atherosclerosis have a stronger biological foundation in lipid metabolism in the liver rather than local lipid metabolism in the plaque. Langerhans Islet cells show less prominent signals for our selected GWAS, suggesting that the genetic mechanisms involved in atherosclerosis do not have a local component in the pancreas (Supplementary material online, Figure S6B).
Little to no signal can be observed for GWASs that have no relation to the cells composing the atherosclerotic plaque, such as height, cancers and congnition-related traits such as bipolar disorder and neuroticism (Supplementary material online, Figure S3A). Yet, these GWASs have ample candidate genes and considerable GWAS sample size—suggesting the specificity of our approach. (Supplementary material online, Figure S1B and C, Table S1). Similarly, no signal for BMI is found in plaque cell populations, intuitively confirming that the mechanisms underlying obesity cannot be accounted for in plaque tissue.43
In highly heterogeneous tissues, such as atherosclerotic plaque, multiple genes can be shared amongst the same class of cell populations (e.g. all ECs), whilst they are lacking in others. To resolve this problem and to keep such genes as cell type-specific, we implemented an approach that can unambiguously uncover cell-specific genes without the need of a predefined list of ubiquitously expressed genes. Although this method is inclusive, the balance between the sufficient number of DEGs and their specificity can be adapted depending on the scRNA-seq dataset in question.
Our approach, however, is subject to several limitations. In our workflow, a bias may be introduced by low-powered GWAS, or by threshold introduced in the selection of candidate genes. The depth and quality of the scRNA sequencing is a another limiting factor. The amount and kind of RNA that can be detected and favours the most abundant genes with highest mRNA counts per cell. As a result certain gene classes, like transcription factors or signalling molecules are more difficult to detect. At the same time, cell types like neutrophils and foam cells in atherosclerotic plaque are difficult to capture by single-cell transcriptomics and might be underrepresented or completely missed. Digestion protocols, tissue quality, and sample handling also affect the quality and/or cell composition of scRNA-seq data. Finally, our workflow does not utilize the existing information about eQTLs, non-coding DNA regulatory units and chromatin organization. Integration of these datasets has potential to increase the accuracy and number of identified target genes. However, we aimed to remain agnostic with respect to identifying candidate genes based on genetic data, and thus designed our experiment using GWAS data only. For instance, integration with Hi-C data would be biased towards a specific cell type in a specific tissue, and favour the selection of genes expression in that given tissue and thus bias the cell type enrichment analysis. Our approach moves atherosclerotic candidate genes forward based on the gene-based genetic association and the differential expression plaques.
In conclusion, we systematically projected GWAS summary statistics for 46 traits directly into single-cell transcriptomic profiles across 18 distinct cell populations derived from plaques. We devised a two-step workflow that (i) leverages large-scale human genetic data to prioritize trait-associated genes, and (ii) calculates a cell-type-specific enrichment score. We uncovered 11 potential cell population-specific targets for CAD and provided functional evidence for KANK2, SKI, and EDNRA in SMCs. Additionally, we verified the robustness and transportability of our approach by confirming the tissue- and trait-specific enrichment of circulating lipid genes in liver and glycaemic disorder genes in pancreatic tissue. The principal strength of our framework is the cell-specific resolution it offers for disease-associated genes mapped to GWAS loci, thus putting the emphasis on prioritization of candidates (instead of adding to an ever-growing candidate-list) and accelerating the path to functional testing.
Lead author biography
Lotte Slenders earned a BS in Biology and a MS in Molecular and Cellular Life Sciences from Utrecht University in the Netherlands. In 2018, she became a PhD candidate in the lab of Gerard Pasterkamp at the Utrecht University Medical Centre. During her PhD, she further developed as a data-scientist with a biology background. Her main research topic revolves around the omics data of human atherosclerotic plaques from the AtheroExpress biobank study. She is using single-cell RNAseq data to study complex genetic loci and molecular mechanisms of cell plasticity such as endoMT.
Supplementary material
Supplementary material is available at European Heart Journal Open online.
Supplementary Material
Acknowledgements
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from: FUMA GTEx Portal on 12 July 2020. The authors thank Sonya A. MacParland and Mauro J. Muraro for providing their scRNA-seq data on human liver and pancreatic islet cells at our request. The authors are thankful for the support of the ERA-CVD programme ‘druggable-MI-targets’ (grant number: 01KL1802), the EU H2020 TO_AITION (grant number: 848146), and the Leducq Fondation ‘PlaqOmics’ (grant number: 18CVD02).
Funding
This work was supported by grants from: Fondation Leducq (‘PlaqOmics’ 18CVD02 to G.P. and C.L.M.), the National Institutes of Health (R01HL148239 and R00HL125912 to C.L.M), the Netherlands CardioVascular Research Initiative of the Netherlands Heart Foundation (CVON 2011/B019 and CVON 2017-20 supporting S.W.v.d.L.), generating the best evidence-based pharmaceutical targets for atherosclerosis (GENIUS I&II supporting S.W.v.d.L.), Interuniversity Cardiology Institute of the Netherlands (ICIN, 09.001 supporting S.W.v.d.L), ERA-CVD program ‘druggable-MI-targets’ (01KL1802, supporting supporting S.W.v.d.L.), the EU H2020 TO_AITION (848146 supporting S.W.v.d.L.) and American Heart Association Transformational Project Award (19TPA34910021 supporting R.A. and M.C.).
Conflict of interest: Authors have no conflicts of interest to disclose.
Data availability
Custom scripts used for this publication are available on GitHub https://github.com/CirculatoryHealth/gwas2single and https://github.com/CirculatoryHealth/gwas2cojo. Raw summary statistics can be accessed through their original publications (Supplementary material online, Table S4). Quality controlled and/or FUMA-MAGMA processed summary statistics are available for download at Zenodo (https://zenodo.org/record/4823040). Raw human carotid artery plaque scRNA-seq data can be accessed via https://dataverse.nl/dataverse/umculab. The plaque scRNA-seq data after quality control, i.e. counts, scores, etc., used in this paper can be found at https://doi.org/10.34894/TYHGEF. Other data supplements can be found in the Supplementary material online, Data section.
References
- 1. Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J.. Complement factor H polymorphism in age-related macular degeneration. Science 2005;308:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, Suveges D, Vrousgou O, Whetzel PL, Amode R, Guillen JA, Riat HS, Trevanion SJ, Hall P, Junkins H, Flicek P, Burdett T, Hindorff LA, Cunningham F, Parkinson H.. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019;47:D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, Giannakopoulou O, Jiang T, Hamby SE, Di Angelantonio E, Assimes TL, Bottinger EP, Chambers JC, Clarke R, Palmer CNA, Cubbon RM, Ellinor P, Ermel R, Evangelou E, Franks PW, Grace C, Gu D, Hingorani AD, Howson JMM, Ingelsson E, Kastrati A, Kessler T, Kyriakou T, Lehtimäki T, Lu X, Lu Y, März W, McPherson R, Metspalu A, Pujades-Rodriguez M, Ruusalepp A, Schadt EE, Schmidt AF, Sweeting MJ, Zalloua PA, AlGhalayini K, Keavney BD, Kooner JS, Loos RJF, Patel RS, Rutter MK, Tomaszewski M, Tzoulaki I, Zeggini E, Erdmann J, Dedoussis G, Björkegren JLM, Schunkert H, Farrall M, Danesh J, Samani NJ, Watkins H, Deloukas P; EPIC-CVD Consortium. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet 2017;49:1385–1391. [DOI] [PubMed] [Google Scholar]
- 4. Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese A-K, van der Laan SW, Gretarsdottir S, Anderson CD, Chong M, Adams HHH, Ago T, Almgren P, Amouyel P, Ay H, Bartz TM, Benavente OR, Bevan S, Boncoraglio GB, Brown RD, Butterworth AS, Carrera C, Carty CL, Chasman DI, Chen W-M, Cole JW, Correa A, Cotlarciuc I, Cruchaga C, Danesh J, de Bakker PIW, DeStefano AL, den Hoed M, Duan Q, Engelter ST, Falcone GJ, Gottesman RF, Grewal RP, Gudnason V, Gustafsson S, Haessler J, Harris TB, Hassan A, Havulinna AS, Heckbert SR, Holliday EG, Howard G, Hsu F-C, Hyacinth HI, Ikram MA, Ingelsson E, Irvin MR, Jian X, Jiménez-Conde J, Johnson JA, Jukema JW, Kanai M, Keene KL, Kissela BM, Kleindorfer DO, Kooperberg C, Kubo M, Lange LA, Langefeld CD, Langenberg C, Launer LJ, Lee J-M, Lemmens R, Leys D, Lewis CM, Lin W-Y, Lindgren AG, Lorentzen E, Magnusson PK, Maguire J, Manichaikul A, McArdle PF, Meschia JF, Mitchell BD, Mosley TH, Nalls MA, Ninomiya T, O'Donnell MJ, Psaty BM, Pulit SL, Rannikmäe K, Reiner AP, Rexrode KM, Rice K, Rich SS, Ridker PM, Rost NS, Rothwell PM, Rotter JI, Rundek T, Sacco RL, Sakaue S, Sale MM, Salomaa V, Sapkota BR, Schmidt R, Schmidt CO, Schminke U, Sharma P, Slowik A, Sudlow CLM, Tanislav C, Tatlisumak T, Taylor KD, Thijs VNS, Thorleifsson G, Thorsteinsdottir U, Tiedt S, Trompet S, Tzourio C, van Duijn CM, Walters M, Wareham NJ, Wassertheil-Smoller S, Wilson JG, Wiggins KL, Yang Q, Yusuf S, Bis JC, Pastinen T, Ruusalepp A, Schadt EE, Koplev S, Björkegren JLM, Codoni V, Civelek M, Smith NL, Trégouët DA, Christophersen IE, Roselli C, Lubitz SA, Ellinor PT, Tai ES, Kooner JS, Kato N, He J, van der Harst P, Elliott P, Chambers JC, Takeuchi F, Johnson AD, Sanghera DK, Melander O, Jern C, Strbian D, Fernandez-Cadenas I, Longstreth WT, Rolfs A, Hata J, Woo D, Rosand J, Pare G, Hopewell JC, Saleheen D, Stefansson K, Worrall BB, Kittner SJ, Seshadri S, Fornage M, Markus HS, Howson JMM, Kamatani Y, Debette S, Dichgans M; MEGASTROKE Consortium. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 2018;50:524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Setten J, Isgum I, Smolonska J, Ripke S, de Jong PA, Oudkerk M, de Koning H, Lammers J-WJ, Zanen P, Groen HJM, Boezen HM, Postma DS, Wijmenga C, Viergever MA, Mali WPTM, de Bakker PIW.. Genome-wide association study of coronary and aortic calcification implicates risk loci for coronary artery disease and myocardial infarction. Atherosclerosis 2013;228:400–405. [DOI] [PubMed] [Google Scholar]
- 6. Franceschini N, Giambartolomei C, de Vries PS, Finan C, Bis JC, Huntley RP, Lovering RC, Tajuddin SM, Winkler TW, Graff M, Kavousi M, Dale C, Smith AV, Hofer E, van Leeuwen EM, Nolte IM, Lu L, Scholz M, Sargurupremraj M, Pitkänen N, Franzén O, Joshi PK, Noordam R, Marioni RE, Hwang S-J, Musani SK, Schminke U, Palmas W, Isaacs A, Correa A, Zonderman AB, Hofman A, Teumer A, Cox AJ, Uitterlinden AG, Wong A, Smit AJ, Newman AB, Britton A, Ruusalepp A, Sennblad B, Hedblad B, Pasaniuc B, Penninx BW, Langefeld CD, Wassel CL, Tzourio C, Fava C, Baldassarre D, O'Leary DH, Teupser D, Kuh D, Tremoli E, Mannarino E, Grossi E, Boerwinkle E, Schadt EE, Ingelsson E, Veglia F, Rivadeneira F, Beutner F, Chauhan G, Heiss G, Snieder H, Campbell H, Völzke H, Markus HS, Deary IJ, Jukema JW, de Graaf J, Price J, Pott J, Hopewell JC, Liang J, Thiery J, Engmann J, Gertow K, Rice K, Taylor KD, Dhana K, Kiemeney LALM, Lind L, Raffield LM, Launer LJ, Holdt LM, Dörr M, Dichgans M, Traylor M, Sitzer M, Kumari M, Kivimaki M, Nalls MA, Melander O, Raitakari O, Franco OH, Rueda-Ochoa OL, Roussos P, Whincup PH, Amouyel P, Giral P, Anugu P, Wong Q, Malik R, Rauramaa R, Burkhardt R, Hardy R, Schmidt R, de Mutsert R, Morris RW, Strawbridge RJ, Wannamethee SG, Hägg S, Shah S, McLachlan S, Trompet S, Seshadri S, Kurl S, Heckbert SR, Ring S, Harris TB, Lehtimäki T, Galesloot TE, Shah T, de Faire U, Plagnol V, Rosamond WD, Post W, Zhu X, Zhang X, Guo X, Saba Y, Dehghan A, Seldenrijk A, Morrison AC, Hamsten A, Psaty BM, van Duijn CM, Lawlor DA, Mook-Kanamori DO, Bowden DW, Schmidt H, Wilson JF, Wilson JG, Rotter JI, Wardlaw JM, Deanfield J, Halcox J, Lyytikäinen L-P, Loeffler M, Evans MK, Debette S, Humphries SE, Völker U, Gudnason V, Hingorani AD, Björkegren JLM, Casas JP, O'Donnell CJ; MEGASTROKE Consortium. GWAS and colocalization analyses implicate carotid intima-media thickness and carotid plaque loci in cardiovascular outcomes. Nat Commun 2018;9:5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khera AV, Kathiresan S.. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet 2017;18:331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kamb A, Harper S, Stefansson K.. Human genetics as a foundation for innovative drug development. Nat Biotechnol 2013;31:975–978. [DOI] [PubMed] [Google Scholar]
- 9. Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, Cardon LR, Whittaker JC, Sanseau P.. The support of human genetic evidence for approved drug indications. Nat Genet 2015;47:856–860. [DOI] [PubMed] [Google Scholar]
- 10. Hormozdiari F, van de Bunt M, Segrè AV, Li X, Joo JWJ, Bilow M, Sul JH, Sankararaman S, Pasaniuc B, Eskin E.. Colocalization of GWAS and eQTL signals detects target genes. Am J Hum Genet 2016;99:1245–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mancuso N, Shi H, Goddard P, Kichaev G, Gusev A, Pasaniuc B.. Integrating gene expression with summary association statistics to identify genes associated with 30 complex traits. Am J Hum Genet 2017;100:473–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen G, Ning B, Shi T.. Single-cell RNA-Seq technologies and related computational data analysis. Front Genet 2019;10:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verhoeven BAN, Velema E, Schoneveld AH, de Vries JPPM, de Bruin P, Seldenrijk CA, de Kleijn DPV, Busser E, van der Graaf Y, Moll F, Pasterkamp G.. Athero-express: differential atherosclerotic plaque expression of mRNA and protein in relation to cardiovascular events and patient characteristics. Rationale and design. Eur J Epidemiol 2004;19:1127–1133. [DOI] [PubMed] [Google Scholar]
- 14. Depuydt MAC, Prange KHM, Slenders L, Örd T, Elbersen D, Boltjes A, de Jager SCA, Asselbergs FW, de Borst GJ, Aavik E, Lönnberg T, Lutgens E, Glass CK, den Ruijter HM, Kaikkonen MU, Bot I, Slütter B, van der Laan SW, Yla-Herttuala S, Mokry M, Kuiper J, de Winther MPJ, Pasterkamp G.. Microanatomy of the human atherosclerotic plaque by single-cell transcriptomics. Circ Res 2020;127:1437–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. R Core Team. R: A language and environment for statistical computing. 2021. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
- 16. Stuart T, Satija R.. Integrative single-cell analysis. Nat Rev Genet 2019;20:257–272. [DOI] [PubMed] [Google Scholar]
- 17. van Rossum G. Python tutorial, Technical Report CS-R9526. 1995.
- 18. 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR.. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leeuw CA, de Mooij JM, Heskes T, Posthuma D.. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015;11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Watanabe K, Taskesen E, van Bochoven A, Posthuma D.. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma WF, Hodonsky CJ, Turner AW, Wong D, Song Y, Mosquera JV, Ligay AV, Slenders L, Gancayco C, Pan H, Barrientos NB, Alencar GF, Owsiany K, Owens GK, Reilly MP, Li M, Pasterkamp G, Mokry M, van der Laan SW, Khomtchouk BB , Miller CL.. Enhanced single-cell RNA-seq workflow reveals coronary artery disease cellular cross-talk and candidate drug targets. Atherosclerosis 2022;doi: 10.1016/j.atherosclerosis.2021.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suzuki H, Yagi K, Kondo M, Kato M, Miyazono K, Miyazawa K.. c-Ski inhibits the TGF-β signaling pathway through stabilization of inactive Smad complexes on Smad-binding elements. Oncogene 2004;23:5068–5076. [DOI] [PubMed] [Google Scholar]
- 23. Pan H, Xue C, Auerbach BJ, Fan J, Bashore AC, Cui J, Yang DY, Trignano SB, Liu W, Shi J, Ihuegbu CO, Bush EC, Worley J, Vlahos L, Laise P, Solomon RA, Connolly ES, Califano A, Sims PA, Zhang H, Li M, Reilly MP.. Single-cell genomics reveals a novel cell state during smooth muscle cell phenotypic switching and potential therapeutic targets for atherosclerosis in mouse and human. Circulation 2020;142:2060–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park M, Kulkarni A, Beatty A, Ganz P, Regan M, Vittinghoff E, Whooley M.. Soluble endothelial cell selective adhesion molecule and cardiovascular outcomes in patients with stable coronary disease: a report from the Heart and Soul Study. Atherosclerosis 2015;243:546–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hamczyk MR, Andrés V.. Vascular smooth muscle cell loss underpins the accelerated atherosclerosis in Hutchinson-Gilford progeria syndrome. Nucleus 2019;10:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Leary DH, Polak JF.. Intima-media thickness: a tool for atherosclerosis imaging and event prediction. Am J Cardiol 2002;90:18L–21L. [DOI] [PubMed] [Google Scholar]
- 27. Aherrahrou R, Guo L, Nagraj VP, Aguhob A, Hinkle J, Chen L, Yuhl Soh J, Lue D, Alencar GF, Boltjes A, van der Laan SW, Farber E, Fuller D, Anane-Wae R, Akingbesote N, Manichaikul AW, Ma L, Kaikkonen MU, Björkegren JLM, Önengüt-Gümüşcü S, Pasterkamp G, Miller CL, Owens GK, Finn A, Navab M, Fogelman AM, Berliner JA, Civelek M.. Genetic regulation of atherosclerosis-relevant phenotypes in human vascular smooth muscle cells. Circ Res 2020;127:1552–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, Gupta G, Cheng ML, Liu LY, Camat D, Chung SW, Seliga RK, Shao Z, Lee E, Ogawa S, Ogawa M, Wilson MD, Fish JE, Selzner M, Ghanekar A, Grant D, Greig P, Sapisochin G, Selzner N, Winegarden N, Oyedele Adeyi O, Keller G, Bader GD, McGilvray ID. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 2018;9:4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Muraro MJ, Dharmadhikari G, Grün D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, de Koning EJP, van Oudenaarden A.. A single-cell transcriptome atlas of the human pancreas. Cell Syst 2016;3:385–394.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nguyen T-M, Shafi A, Nguyen T, Draghici S.. Identifying significantly impacted pathways: a comprehensive review and assessment. Genome Biol 2019;20:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shadrina AS, Shashkova TI, Torgasheva AA, Sharapov SZ, Klarić L, Pakhomov ED, Alexeev DG, Wilson JF, Tsepilov YA, Joshi PK, Aulchenko YS.. Prioritization of causal genes for coronary artery disease based on cumulative evidence from experimental and in silico studies. Sci Rep 2020;10:10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ongen H, Brown AA, Delaneau O, Panousis NI, Nica AC, Dermitzakis ET; GTEx Consortium. Estimating the causal tissues for complex traits and diseases. Nat Genet 2017;49:1676–1683. [DOI] [PubMed] [Google Scholar]
- 34. Petretto E. Single cell expression quantitative trait loci and complex traits. Genome Med 2013;5:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van der Wijst MGP, de Vries DH, Groot HE, Trynka G, Hon CC, Bonder MJ, Stegle O, Nawijn MC, Idaghdour Y, van der Harst P, Ye CJ, Powell J, Theis FJ, Mahfouz A, Heinig M, Franke L.. The single-cell eQTLGen consortium. eLife 2020;9:e52155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Potì F, Ceglarek U, Burkhardt R, Simoni M, Nofer J-R.. SKI-II—a sphingosine kinase 1 inhibitor—exacerbates atherosclerosis in low-density lipoprotein receptor-deficient (LDL-R−/−) mice on high cholesterol diet. Atherosclerosis 2015;240:212–215. [DOI] [PubMed] [Google Scholar]
- 37. Sun Z, Tseng H-Y, Tan S, Senger F, Kurzawa L, Dedden D, Mizuno N, Wasik AA, Thery M, Dunn AR, Fässler R.. Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation. Nat Cell Biol 2016;18:941–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buono MF, Slenders L, Wesseling M, Hartman RJG, Monaco C, den Ruijter HM, Pasterkamp G, Mokry M.. The changing landscape of the vulnerable plaque: a call for fine-tuning of preclinical models. Vascul Pharmacol 2021;141:106924. [DOI] [PubMed] [Google Scholar]
- 39. Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014;159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duong CN, Nottebaum AF, Butz S, Volkery S, Zeuschner D, Stehling M, Vestweber D.. Interference with ESAM (Endothelial Cell-Selective Adhesion Molecule) plus vascular endothelial-cadherin causes immediate lethality and lung-specific blood coagulation. Arterioscler Thromb Vasc Biol 2020;40:378–393. [DOI] [PubMed] [Google Scholar]
- 41. Nakayama A, Albarrán-Juárez J, Liang G, Roquid KA, Iring A, Tonack S, Chen M, Müller OJ, Weinstein LS, Offermanns S.. Disturbed flow–induced Gs-mediated signaling protects against endothelial inflammation and atherosclerosis. JCI Insight 2020;5(23):e140485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Örd T, Õunap K, Stolze LK, Aherrahrou R, Nurminen V, Toropainen A, Selvarajan I, Lönnberg T, Aavik E, Ylä-Herttuala S, Civelek M, Romanoski CE, Kaikkonen MU.. Single-cell epigenomics and functional fine-mapping of atherosclerosis GWAS loci. Circ Res 2021;129:240–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Mägi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Zhao JH, Zhao W, Chen J, Fehrmann R, Hedman ÅK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Leach IM, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stančáková A, Strawbridge RJ, Sung YJ, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Ärnlöv J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Blüher M, Böhringer S, Bonnycastle LL, Böttcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Chen Y-DI, Clarke R, Daw EW, de Craen AJM, Delgado G, Dimitriou M, Doney ASF, Eklund N, Estrada K, Eury E, Folkersen L, Fraser RM, Garcia ME, Geller F, Giedraitis V, Gigante B, Go AS, Golay A, Goodall AH, Gordon SD, Gorski M, Grabe H-J, Grallert H, Grammer TB, Gräßler J, Grönberg H, Groves CJ, Gusto G, Haessler J, Hall P, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heard-Costa NL, Helmer Q, Hengstenberg C, Holmen O, Hottenga J-J, James AL, Jeff JM, Johansson Å, Jolley J, Juliusdottir T, Kinnunen L, Koenig W, Koskenvuo M, Kratzer W, Laitinen J, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindström J, Lo KS, Lobbens S, Lorbeer R, Lu Y, Mach F, Magnusson PKE, Mahajan A, McArdle WL, McLachlan S, Menni C, Merger S, Mihailov E, Milani L, Moayyeri A, Monda KL, Morken MA, Mulas A, Müller G, Müller-Nurasyid M, Musk AW, Nagaraja R, Nöthen MM, Nolte IM, Pilz S, Rayner NW, Renstrom F, Rettig R, Ried JS, Ripke S, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Scott WR, Seufferlein T, Shi J, Smith AV, Smolonska J, Stanton AV, Steinthorsdottir V, Stirrups K, Stringham HM, Sundström J, Swertz MA, Swift AJ, Syvänen A-C, Tan S-T, Tayo BO, Thorand B, Thorleifsson G, Tyrer JP, Uh H-W, Vandenput L, Verhulst FC, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Warren HR, Waterworth D, Weedon MN, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wright AF, Zhang QLifeLines Cohort Study Brennan EP, Choi M, Dastani Z, Drong AW, Eriksson P, Franco-Cereceda A, Gådin JR, Gharavi AG, Goddard ME, Handsaker RE, Huang J, Karpe F, Kathiresan S, Keildson S, Kiryluk K, Kubo M, Lee J-Y, Liang L, Lifton RP, Ma B, McCarroll SA, McKnight AJ, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Okada Y, Perry JRB, Dorajoo R, Reinmaa E, Salem RM, Sandholm N, Scott RA, Stolk L, Takahashi A, Tanaka T, van 't Hooft FM, Vinkhuyzen AAE, Westra H-J, Zheng W, Zondervan KTADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium Heath AC, Arveiler D, Bakker SJL, Beilby J, Bergman RN, Blangero J, Bovet P, Campbell H, Caulfield MJ, Cesana G, Chakravarti A, Chasman DI, Chines PS, Collins FS, Crawford DC, Cupples LA, Cusi D, Danesh J, de Faire U, den Ruijter HM, Dominiczak AF, Erbel R, Erdmann J, Eriksson JG, Farrall M, Felix SB, Ferrannini E, Ferrières J, Ford I, Forouhi NG, Forrester T, Franco OH, Gansevoort RT, Gejman PV, Gieger C, Gottesman O, Gudnason V, Gyllensten U, Hall AS, Harris TB, Hattersley AT, Hicks AA, Hindorff LA, Hingorani AD, Hofman A, Homuth G, Hovingh GK, Humphries SE, Hunt SC, Hyppönen E, Illig T, Jacobs KB, Jarvelin M-R, Jöckel K-H, Johansen B, Jousilahti P, Jukema JW, Jula AM, Kaprio J, Kastelein JJP, Keinanen-Kiukaanniemi SM, Kiemeney LA, Knekt P, Kooner JS, Kooperberg C, Kovacs P, Kraja AT, Kumari M, Kuusisto J, Lakka TA, Langenberg C, Le Marchand L, Lehtimäki T, Lyssenko V, Männistö S, Marette A, Matise TC, McKenzie CA, McKnight B, Moll FL, Morris AD, Morris AP, Murray JC, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Madden PAF, Pasterkamp G, Peden JF, Peters A, Postma DS, Pramstaller PP, Price JF, Qi L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ridker PM, Rioux JD, Ritchie MD, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schunkert H, Schwarz PEH, Sever P, Shuldiner AR, Sinisalo J, Stolk RP, Strauch K, Tönjes A, Trégouët D-A, Tremblay A, Tremoli E, Virtamo J, Vohl M-C, Völker U, Waeber G, Willemsen G, Witteman JC, Zillikens MC, Adair LS, Amouyel P, Asselbergs FW, Assimes TL, Bochud M, Boehm BO, Boerwinkle E, Bornstein SR, Bottinger EP, Bouchard C, Cauchi S, Chambers JC, Chanock SJ, Cooper RS, de Bakker PIW, Dedoussis G, Ferrucci L, Franks PW, Froguel P, Groop LC, Haiman CA, Hamsten A, Hui J, Hunter DJ, Hveem K, Kaplan RC, Kivimaki M, Kuh D, Laakso M, Liu Y, Martin NG, März W, Melbye M, Metspalu A, Moebus S, Munroe PB, Njølstad I, Oostra BA, Palmer CNA, Pedersen NL, Perola M, Pérusse L, Peters U, Power C, Quertermous T, Rauramaa R, Rivadeneira F, Saaristo TE, Saleheen D, Sattar N, Schadt EE, Schlessinger D, Slagboom PE, Snieder H, Spector TD, Thorsteinsdottir U, Stumvoll M, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Walker M, Wallaschofski H, Wareham NJ, Watkins H, Weir DR, Wichmann H-E, Wilson JF, Zanen P, Borecki IB, Deloukas P, Fox CS, Heid IM, O'Connell JR, Strachan DP, Stefansson K, van Duijn CM, Abecasis GR, Franke L, Frayling TM, McCarthy MI, Visscher PM, Scherag A, Willer CJ, Boehnke M, Mohlke KL, Lindgren CM, Beckmann JS, Barroso I, North KE, Ingelsson E, Hirschhorn JN, Loos RJF, Speliotes EK.. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Custom scripts used for this publication are available on GitHub https://github.com/CirculatoryHealth/gwas2single and https://github.com/CirculatoryHealth/gwas2cojo. Raw summary statistics can be accessed through their original publications (Supplementary material online, Table S4). Quality controlled and/or FUMA-MAGMA processed summary statistics are available for download at Zenodo (https://zenodo.org/record/4823040). Raw human carotid artery plaque scRNA-seq data can be accessed via https://dataverse.nl/dataverse/umculab. The plaque scRNA-seq data after quality control, i.e. counts, scores, etc., used in this paper can be found at https://doi.org/10.34894/TYHGEF. Other data supplements can be found in the Supplementary material online, Data section.