Abstract
A newly developed PCR-based assay for the H7 variant of the Escherichia coli flagellin gene, fliC, was 100% sensitive and specific in comparison with serology and probe hybridization. It revealed broad conservation of the H7 fliC variant among phylogenetically diverse lineages of extraintestinal pathogenic E. coli (ExPEC) and superseded serotyping for certain isolates with ambiguous or non-H7 serotyping results. The H7 primers functioned well when incorporated into a multiplex PCR assay for diverse virulence-associated genes of ExPEC.
The H7 flagellar variant of Escherichia coli recently has received increasing attention because of its diagnostic importance with respect to E. coli O157:H7 (8, 9, 25, 29, 31), the major cause of hemorrhagic colitis and hemolytic-uremic syndrome in the industrialized world (6, 32, 33). Inconsistent expression of the H7 antigen and the technical difficulties associated with H antigen serotyping have led to interest in PCR-based assays for detection of the H7 allele of the E. coli flagellin gene, fliC (8, 9, 25). Three such assays have been reported. One involves amplification of the entirety of fliC using consensus primers based on the conserved 5′ and 3′ extremes of fliC, with H7 specificity obtained via a subsequent restriction digestion step (8). The other two assays utilize H7-specific primers based on unique regions within the internal variable portion of fliC that presumably encode H7-specific antigenic epitopes (9, 25). All three assays are highly sensitive and specific for detection of the H7 fliC variant among E. coli O157:H7 and O55:H7 isolates, as well as in certain nonmotile (hence H antigen-negative) shigatoxigenic E. coli isolates of serotype O157 that presumably contain, but fail to express, fliC (8, 9, 25).
However, the H7 flagellar antigen also characterizes several familiar “virulent clones” of extraintestinal pathogenic E. coli (ExPEC) (21, 27, 30). These include E. coli O1:K1:H7 and O2:K1:H7, serotypes traditionally associated with pyelonephritis and sepsis (16, 21, 27), and E. coli O18:K1:H7, a serotype traditionally associated with neonatal bacterial meningitis and neonatal sepsis but recently shown to be highly prevalent also in uncomplicated cystitis in adult women (1, 4, 12, 15, 21, 22). Although an E. coli O1:H7 isolate (strain U5-41) actually was the source of the first published H7 fliC sequence (31), all three studies of PCR-based detection of the H7 fliC variant have focused on diarrheagenic E. coli, with scant attention given to ExPEC (8, 9, 25). In these studies the only putative ExPEC isolates examined were of serotypes O1:H7 and O18:H7. No information regarding K antigens was provided, few examples of each serotype were analyzed, and the clinical sources of the isolates were not specified (8, 9, 25).
In the present study we designed and validated an H7-specific fliC PCR assay based on primers that could be incorporated into our established broad-range multiplex PCR assay for putative virulence factors (VFs) of ExPEC (11, 19). We then used the PCR assay to assess the fliC status of diverse human clinical ExPEC isolates that had not been H serotyped or that had yielded ambiguous or suspect H antigen results and that, because of clonal background or VF profile, were thought possibly to be members of an H7-positive ExPEC clonal group.
Strains and serotypes.
One hundred fifty-five independent E. coli isolates representing diverse H7-associated lineages and 35 non-H7 H antigens were obtained from various published or unpublished collections, predominantly of human extraintestinal infection isolates. Twelve were urosepsis isolates from adults (16, 19). Twelve were (unpublished) E. coli O157:H7 diarrhea isolates, provided by the Minnesota Department of Health. Twelve were (unpublished) neonatal meningitis isolates, provided by L. Spanjaard (Academic Medical Center, Amsterdam, The Netherlands). Thirteen were (unpublished) urine or fecal isolates from men with febrile urinary tract infection, provided by P. Ulleryd (University of Göteborg, Goteborg, Sweden) and F. Scheutz (Escherichia and Klebsiella Centre [World Health Organization], Copenhagen, Denmark). Twenty were diverse-source bacteremia isolates, provided by J. Maslow (Philadelphia VA Medical Center) (23). Seventeen were cystitis isolates from adult women, provided by A. Stapleton (Seattle, Wash.) (15, 17, 35) or C. Kunin (Ohio State University, Columbus) (12, 22). Seven were human urinary tract infection isolates, provided by T. Whittam (Pennsylvania State University, University Park) (37). Seven were pediatric cystitis isolates, provided by C. Johnson (Denver Children's Hospital) (14).
Pyelonephritis isolates 536 (O15:K52:H31) (3), CFT073 (O6:K2:H1) (24), and J96 (O4:K-:H5) (18) were obtained, respectively, from G. Blum-Oehler (Universitat Wurzburg), H. Mobley (University of Maryland, Baltimore), and B. Minshew (formerly of University of Washington, Seattle). Urinary tract infection isolates SP57 and SP88 (O157:K-:H45) (7, 20) were obtained from H. de Ree (Duphar BV, Weesp, The Netherlands). O18:K1:H7 neonatal meningitis isolates RS218 (1), C5 (4, 5), H16 (34), and IHE3034 (28) were obtained, respectively, from K. S. Kim (Johns Hopkins University, Baltimore, Md.), R. Bortolussi (Dalhousie University, Halifax, Nova Scotia, Canada), R. Darveau (University of Washington), and T. Korhonen (University of Helsinki, Finland; via K. S. Kim). The Escherichia coli Reference collection (26) was obtained from several sources, including H. Ochman (University of Arizona, Tucson) and the American Type Culture Collection (Manassas, Va.) (11). Strains were maintained at −70°C until use.
O, K, and H antigens for these strains (as available) were as previously published or as communicated by the providers of the strains. Serotyping had been done by the E. coli Reference Center (University Park, Pa.), the International Escherichia and Klebsiella Centre (World Health Organization) (Copenhagen, Denmark), the Ontario Ministry of Health laboratory (Toronto, Ontario, Canada), The Netherlands Reference Laboratory for Bacterial Meningitis (Amsterdam, The Netherlands), the Health Canada Guelph Laboratory (Guelph, Ontario, Canada), or the Minnesota Department of Health (Minneapolis), depending on the source. Nonmotile strains were designated NM, those that were H untypeable were designated HU, and those reacting with multiple (more than three) H-specific antisera were designated HM.
For many of the strains, clonal associations had been defined previously by multilocus enzyme electrophoresis or random amplified polymorphic DNA analysis, and virulence genotypes were known, including for the K1 variant of capsule synthesis genes kpsMT II (unpublished data; 10–13, 15, 23, 37). This allowed certain strains to be classified as putative members of characteristically H7-associated clonal groups, e.g., E. coli O18:K1:H7, as previously described (12, 15).
For validation of the H7 PCR assay, positive controls included all 35 H7-seropositive isolates, whereas negative controls included all 45 strains that putatively expressed one or more specific H antigens other than the H7 antigen and that were not suspected of belonging to a characteristically H7-associated clonal group. The 75 test strains (unknowns) to which the validated H7 PCR assay was applied included eight presumably non-H7 E. coli O157 extraintestinal infection isolates plus 67 non-O157 isolates that were considered candidates for containing the H7 fliC variant based on clonal background and/or virulence genotype but for which the H antigen status had not been assessed, was ambiguous (because of the strain's reported NM, HU, or HM status), or putatively included only non-H7 H antigens.
Primer design and PCR conditions.
H7 primers were selected based on the published fliC sequence from E. coli O1:H7 strain U5-41 (GenBank accession no. L07388) (31), in comparison with other sequenced fliC variants (29, 31). Selection criteria for H7 specificity included (i) location within the internal variable region of fliC, (ii) conservation among all sequenced H7 fliC variants (29), and (iii) absence of homology with known non-H7 fliC variants. Selection criteria for compatibility with our multiplex VF PCR assay included (i) a high predicted annealing temperature (11, 19), (ii) absence of complementarity with other primers in one of the primer pools in our multiplex PCR assay, and (iii) generation of an amplicon whose size was sufficiently different from those of amplicons generated by other primers in the pool to allow visual resolution of multiple PCR products in the same lane in agarose gels.
The primers selected, H7f (5′-ACGATGCAGGCAACTTGACG-3′; nucleotides 941 to 960) and H7r (5′-GGGTTGGTCGTTGCAGAACC-3′; nucleotides 1487 to 1468), yielded a single robust ∼550-bp product from boiled-lysate template DNA, with amplification conditions as previously described for our multiplex VF PCR assay (19). Briefly, these involved 25-μl reaction mixtures containing (final concentrations or amounts) MgCl2 (4 mM), deoxynucleoside triphosphates (400 μM each), primers (0.6 μM each), Taq polymerase (1.25 U), and 2 μl of boiled-lysate template DNA. Amplification involved 25 cycles (following an initial 12-min denaturation at 95°C) of 94°C for 30 s, 63°C for 30 s, and 68°C for 3 min, followed by 72°C for 10 min, and then a hold at 4°C.
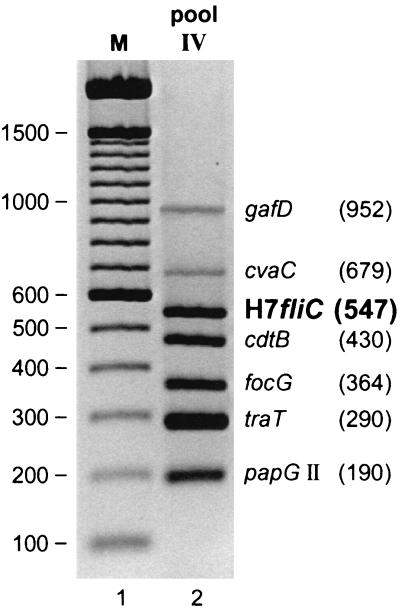
The new H7 primers were next incorporated into primer pool IV of our multiplex VF PCR assay (19), which as currently formulated includes primers specific for gafD (952 bp), cvaC (679 bp), cdtB (430 bp), focG (364 bp), traT (290 bp), and papG allele II (190 bp). With positive-control template DNA containing each of these genes plus the H7 fliC variant, modified primer pool IV (now containing eight primer pairs, i.e., two pairs for cdtB [19] plus one pair each for the other six genes) generated the predicted seven discrete amplicons, including the H7 fliC product. The seven amplicons were readily resolved by size when run together in the same lane of an agarose gel (Fig. 1).
FIG. 1.
H7-specific fliC primers as part of a multiplex VF PCR assay. VF-specific PCR products (lane 2), labeled as to gene and size (base pairs), were amplified simultaneously in a single PCR using primer pool IV, which contains eight primer pairs (two pairs for cdtB, which yield products of the same size, plus one pair each for the other six genes). The template DNA contained all seven genes of interest. Lane 1 (M),100-bp molecular weight ladder.
All assays were done in duplicate using boiled lysates that were prepared independently from separate colonies of each isolate. Discrepant results were investigated with additional determinations as needed.
Probe hybridization.
To genetically confirm the sensitivity and specificity of the new H7 primers, the fliC amplicon from an O157:H7 positive-control strain was labeled with digoxigenin by PCR as previously described (17) and used as a probe in dot blot hybridization assays, which were done under stringent conditions, as previously described (17, 19).
Restriction fragment length polymorphism (RFLP) analysis of PCR products.
To further confirm the H7 specificity of amplicons generated from the unknown strains and to assess fliC sequence diversity among all H7-positive E. coli isolates, H7 fliC PCR products were restricted with HincII and, for selected isolates, with RsaI. Buffers and conditions were as specified by the manufacturer (New England Biolabs, Beverly, Mass.). Restriction fragments were resolved by size using agarose gel electrophoresis. As a negative control, the ∼510-bp bmaE PCR product from control strain IHE11165 (19) was similarly analyzed.
Observed sensitivity and specificity of H7 PCR assay.
Among the control strains the newly designed H7-specific primers were 100% sensitive and specific for the H7 fliC variant (Table 1). They detected all 35 positive-control strains, including 22 human ExPEC isolates representing seven different O:K:H serotypes and four different clinical syndromes, 12 isolates of E. coli O157:H7 from patients with diarrhea, and an E. coli O55:H7 isolate (ECOR 37) (Table 1; Fig. 2). In contrast, they failed to amplify fliC from the 45 negative controls, which collectively expressed 35 unique non-H7 H antigens (Table 1; Fig. 2). Dot blot hybridization with a DNA probe generated from the H7 fliC amplicon yielded complete concordance with H7 fliC PCR (not shown).
TABLE 1.
H7 fliC status of 155 predominantly extraintestinal E. coli isolates
| Category (no.)a | Clonal groupb | H antigenc | Clinical sourced | No. of isolates | No. of H7 fliC-positive isolatese |
|---|---|---|---|---|---|
| H7-positive control (35) | O18:K1:H7 | H7 | b, c, m | 7 | 7 |
| O1:K1:H7 | H7 | b | 6 | 6 | |
| O2:K1:H7 | H7 | b | 4 | 4 | |
| O6:K53:H7 | H7 | b, i | 2 | 2 | |
| O15/45:K1:H7 | H7 | i | 1 | 1 | |
| O23:K1:H7 | H7 | i | 1 | 1 | |
| O75:K-:H7 | H7 | i | 1 | 1 | |
| O157:H7 | H7 | d | 12 | 12 | |
| O55:H7 | H7 | f | 1 | 1 | |
| H7-negative control (45) | Various | Variousf | b, c, f, i, p | 45 | 0 |
| H unknown (75) | O18:K1:(H7) | Not tested | c, f, m, u | 13 | 12 |
| HU | b, c, u | 7 | 7 | ||
| HM | c | 3 | 3 | ||
| NM | b, u | 3 | 3 | ||
| Non-H7g | c, f, u | 10 | 7 | ||
| O2:K1:(H7) | HU | b | 4 | 3 | |
| NM | b | 1 | 1 | ||
| Non-H7h | f, p | 2 | 0 | ||
| O1:K1:(H7) | Not tested | m | 2 | 2 | |
| HU | b | 1 | 1 | ||
| O12,O16:K1 | Variousi | c | 5 | 0 | |
| O16:K1 | Variousi | b | 4 | 0 | |
| O1:K1:H- | NM | b, f | 4 | 4 | |
| O7:K1:H- | NM | a, c, f, m, p | 6 | 0 | |
| O-:K1 | Not tested | m | 2 | 1 | |
| O157 (non-H7) | Variousj | c, b, u | 8 | 0 |
Positive controls, H7-seropositive isolates, negative controls, isolates seropositive for one or more specific non-H7 H antigens and not suspected of belonging to an H7-associated clonal group; unknown, isolates with H antigen not tested, absent, indeterminate, or other than H7 (if the strain was suspected of belonging to an H7-associated clonal group or was an extraintestinal O157 isolate).
Clonal groups all represent E. coli phylogenetic group B2 except O157:H7 and O55:H7 (ungrouped lineages); O1:K1:H-, O7:K1:H-, and O157:(non-H7) (phylogenetic group D); and O-:K1 (non-group B2, non-O7:K1:H-).
HU, H untypeable; HM, H multiple; NM, nonmotile; non-H7, specific H antigen(s) other than H7.
Clinical source codes: a, asymptomatic bacteriuria; b, bacteremia; c, cystitis; d, diarrhea; f, feces (healthy host); i, invasive (febrile) male urinary tract infection; m, neonatal meningitis; p, pyelonephritis; u, urinary tract infection (syndrome not otherwise specified).
Probe hybridization and PCR yielded concordant results.
Non-H7 antigens (n = 35) included 1 to 6, 8 to 12, 14 to 18, 21, 23, 25, 27 to 32, 38, 40, 41, 43 to 45, 48, 49, 53, and 56.
H antigens 1, 15, 23, 24, and 32. All but the three O18:H1 strains, from ET 4 of Whittam et al. (37), were H7 fliC positive.
HU (three isolates), NM (four isolates), and H48 (two isolates).
H not tested (one isolate), HU (one isolate), HM (two isolates), H45 (three isolates), and H32 (one isolate).
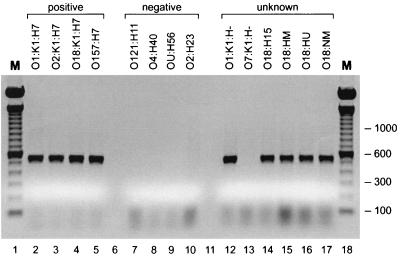
FIG. 2.
PCR detection of the H7 fliC variant in phylogenetically diverse E. coli lineages. Thirty-five H7-positive control isolates from various H7-associated clonal groups were uniformly positive for the 547-bp H7 fliC PCR product (lanes 2 to 5), whereas 45 negative-control strains representing 35 other H types were uniformly negative (lanes 7 to 10). Among 75 unknown strains (lanes 12 to 17), all representatives of the O1:K1:H- clonal group were H7 fliC positive (lane 12), whereas all O7:K1:H- strains were H7 fliC negative (lane 13). Most strains of uncertain H status that were suspected of being members of the O18:K1:H7 clonal group were H7 fliC positive, whether they had been typed as having another H antigen (lane 14), as having multiple H antigens (lane 15), or as being H untypeable (lane 16) or were nonmotile (lane 17). Similar results were obtained for unknown strains putatively representing the O1:K1:H7 or O2:K1:H7 clonal groups (not shown). M, 100-bp molecular weight ladder (lanes 1 and 18).
Determination of H7 fliC analysis of unknown ExPEC isolates.
We next used the H7 PCR assay to evaluate the fliC status of 75 additional E. coli strains, predominantly human clinical extraintestinal isolates. Eight of these strains were E. coli O157 extraintestinal infection isolates that were expected to be H7 fliC negative since, unlike E. coli O157:H7, they were from phylogenetic group D (not shown), contained ExPEC-associated VFs (not shown), had caused extraintestinal infections (Table 1), and in some instances had been typed as positive for H antigens other than H7 (Table 1) and/or as negative for Shiga toxin production (not shown). All eight proved to be H7 fliC negative by PCR, which was confirmed by probe hybridization (Table 1).
On the basis of other characteristics (e.g., O:K serotype, multilocus enzyme electrophoresis- or PCR-derived phylotype, and virulence genotype), the remaining 67 unknown strains were considered likely to be members of potentially H7-positive clonal groups of ExPEC (Table 1). For these strains the H antigen had not been determined, was ambiguous (i.e., NM, HM, or HU), or was putatively not H7. Forty-six of these 67 strains were from the O1:K1, O2:K1, or O18:K1 clonal group of E. coli phylogenetic group B2 (Table 1). Thirty-nine (85%) of the 46 strains proved to be H7 fliC positive by both PCR and probe hybridization, whereas the remaining 7 were concordantly H7 fliC negative (Table 1; Fig. 2). The 39 H7 fliC-positive isolates included 12 of 13 that previously had not been tested for the H antigen, 19 of 20 that had been H typed as NM, HM, or HU, and 7 of 12 that had been assigned one or more specific H types other than H7. Among the last 12 strains, the 5 isolates previously serotyped as H1 (representatives of ET 4, described by Whittam et al. [37]; putatively O18:K1) or H4 (ECOR 61 and 62; putatively O2:K1 [2, 11]) were the only ones found to be H7 fliC negative. In contrast, those that had been serotyped as H15, H23, H24, or H32 were H7 fliC positive (Fig. 2).
Nine other unknown strains represented the O12,O16:K1 and O16:K1 clonal groups of phylogenetic group B2, which are genetically distinct from E. coli O1:K1, O2:K1, and O18:K1 (unpublished data). Although these serotypes are not associated with the H7 antigen, many members are nonmotile, leaving open the possibility of a silent copy of the H7 fliC variant. However, all nine strains proved to be H7 fliC negative by both PCR and probe hybridization (Table 1).
We next evaluated the H7 fliC status of eight nonmotile or “H not tested” E. coli K1 isolates from phylogenetic groups other than group B2 that on the basis of their K1 status we considered candidates for containing a silent copy of the H7 fliC variant (Table 1). All four of the O1:K1:H- isolates tested (including ECOR 35 and 36; phylogenetic group D [10]) were H7 fliC positive, whereas all six O7:K1:H- isolates tested (including ECOR 38 to 41; also phylogenetic group D [10]) were H7 fliC negative (Table 1; Fig. 2). Of the two O-:K1:(H not tested) neonatal meningitis isolates from The Netherlands, which according to PCR-based phylotyping were non-group B2 and non-O7:K1, one was H7 fliC positive (Table 1). For all eight of these strains, H7 PCR and probe hybridization yielded concordant results.
Conservation of H7 fliC in diverse lineages.
To assess H7 fliC sequence diversity, the H7 fliC amplicons from all H7 PCR-positive isolates were subjected to RFLP analysis with HincII. All amplicons yielded a uniform two-band RFLP pattern (estimated fragment sizes, 392 and 155 bp; data not shown). Selected H7 fliC amplicons also were restricted with RsaI, which in previous studies has demonstrated diversity among H7 fliC variants from H7-positive E. coli isolates of diverse O serogroups (8, 9). Representatives of each of the H7-associated clonal groups in the present study yielded a uniform three-band RsaI RFLP pattern (estimated fragment sizes, 285, 196, and 74 bp: data not shown). Taken together, these findings demonstrate broad conservation of H7 fliC sequence across the species within the region flanked by the new H7-specific primers.
Implications of findings.
In the present study we developed and validated a novel PCR-based assay for the E. coli H7 fliC variant and then used the assay to assess the fliC status of diverse (predominantly extraintestinal) wild-type E. coli isolates. The assay was simple to use and interpret. Among validation set strains it was 100% sensitive and specific in comparison with both serology and probe hybridization. It revealed broad conservation of fliC among phylogenetically diverse lineages of E. coli, including certain familiar virulent clones of ExPEC, and provided novel evidence of a serologically unapparent copy of the H7 fliC variant in members of the (group D-derived) O1:K1:H- clonal group. It superseded serotyping by identifying as H7 positive certain strains that yielded ambiguous H antigen results or in which only H antigens other than H7 had previously been identified. The primers also were successfully incorporated into an established multiplex PCR assay for virulence genes of ExPEC.
Our assay represents the fourth reported PCR-based assay capable of detecting the H7 fliC variant of E. coli (8, 9, 25). Like two previously described assays (9, 25), for H7 specificity the new assay relies on detection of unique sequences within the internal variable portion of fliC. Its novelty lies in the use of primers that are compatible with our multiplex PCR assay for extended virulence genotyping of ExPEC (19). This permits the addition of the (extraintestinal virulence-associated) H7 fliC variant to the already-broad array of virulence-associated genes or their alleles (n = 34) that our VF assay can simultaneously detect (11, 19).
The H7 PCR assay was fully sensitive and specific, compared with serological detection of the H antigen (control strains) or probe detection of the H7 fliC variant (total population). This confirms the technical adequacy of the assay for diagnostic and molecular-epidemiology purposes. At a more fundamental level, the assay's precise specificity demonstrates that, in addition to the selected primer-binding sites, the intervening fliC region, which was used as an H7-specific probe, is indeed specific for the H7 allele of fliC. Conversely, the assay's high degree of sensitivity, as demonstrated by the detection of the H7 fliC variant among all H7 sero- or probe-positive strains, irrespective of clonal background, provides additional evidence that the H7 fliC variant is broadly conserved across the species. Supporting this conclusion were the uniform HincII and RsaI RFLP profiles obtained for H7 fliC amplicons irrespective of clonal origin.
Cross-lineage conservation of the H7 fliC variant has been suggested previously by the high level of nucleotide identity between H7 fliC variants from E. coli O157:H7 and O55:H7, E. coli O1:H7, and E. coli O128:H7 (9, 29, 31) and the ability of H7-specific primers to generate fliC amplicons from H7-positive strains of diverse O types (9). However, the only published direct evidence of conservation of fliC across phylogenetic boundaries involved a single O128:H7 diarrhea isolate (DEC 13a), in comparison with multiple E. coli O157:H7 diarrhea isolates (29, 36). On the contrary, the undefined phylogenetic origin of the single O1:H7 strain for which the fliC sequence is available (31) and the previously noted diversity of RsaI fliC RFLP patterns among various H7-positive strains (8, 9) have left uncertainty as to the degree of sequence conservation among fliC variants from different E. coli lineages. The broad phylogenetic conservation of the H7 fliC variant demonstrated here strongly supports horizontal transfer of fliC, rather than convergent evolution, as the explanation for the occurrence of the H7 antigen in distantly related lineages of E. coli (29). Our data confirm that this applies to ExPEC as well as to diarrheagenic E. coli pathotypes (29).
The present study provides the first assessment of the ability of a PCR or probe hybridization assay to detect the H7 fliC variant among well-defined clonal groups of ExPEC. The ExPEC lineages we found to contain the H7 fliC variant include several from E. coli phylogenetic group B2, i.e., E. coli O1:K1:H7, O2:K1:H7, O18:K1:H7, O6:K53:H7, O15/45:K1:H7, O23:K1:H7, and O75:K-:H7, and one from phylogenetic group D, i.e., E. coli O1:K1:H-. In contrast, members of two other group D-derived ExPEC clonal groups, i.e., O7:K1:H- and O157:(non-H7), were consistently H7 fliC negative.
The new PCR assay detected the H7 fliC variant in most strains that for various reasons were predicted to possibly be H7 but which had not been H typed, that had yielded ambiguous H serotypes (i.e., NM, HU, or HM), or that ostensibly expressed only non-H7 H antigens. Detection of the H7 fliC variant in nonmotile strains has been described previously for E. coli O157:H- (8, 9, 25, 29). However, such analysis has been reported for only eight nondiarrheagenic E. coli isolates, none of which presumably represented ExPEC (8, 25). Likewise, detection of the H7 fliC variant in H-untypeable strains has been assessed for E. coli O157 (five isolates) (21) and certain other non-ExPEC serogroups (three isolates) (17), but not for ExPEC-associated serogroups (8, 25). Thus, our detection of the H7 fliC variant among candidate O18:K1:H7 or O2:K1:H7 isolates that were nonmotile (5 of 5 isolates) or that had been serotyped as HU (10 of 11 isolates) is novel. We likewise confirmed as H7 fliC positive all 3 suspected O18:K1:H7 isolates that carried the ambiguous designation HM and 7 of the 10 O18:K1 isolates that putatively expressed only non-H7 flagellar antigens. Although the presence of the H7 fliC variant is not necessarily inconsistent with a non-H7 phenotype, the most plausible explanation for these discrepancies is false serotype results. Taken together, our findings suggest that PCR-based H7 genotyping is actually more accurate than H serotyping for identifying H7-positive strains.
In summary, we devised and validated a novel PCR-based assay for the E. coli H7 fliC variant that proved to be 100% sensitive and specific in comparison with both serology and probe hybridization. The assay revealed broad conservation of the H7 fliC variant among phylogenetically diverse lineages of E. coli, including certain familiar virulent clones of ExPEC, and provided novel evidence that the H7 fliC variant is characteristically present in E. coli O1:K1:H-. It superseded serotyping by confirming as H7 positive certain strains with ambiguous H antigen results or for which only non-H7 H antigens had previously been identified. The H7 primers were successfully incorporated into an established multiplex PCR assay for virulence genes of ExPEC and functioned well in combination with seven other primer pairs. The new H7 fliC PCR assay should make H7 “molecular serotyping,” with or without detection of additional ExPEC virulence factors, available to any laboratory equipped for diagnostic PCR.
Acknowledgments
This work was supported by Office of Research and Development, Medical Research Service, Department of Veterans Affairs, National Institutes of Health, grant DK-47504, and National Research Initiative (NRI) Competitive Grants Program/United States Department of Agriculture grant 00-35212-9408 (J.R.J.).
We thank the colleagues who generously provided strains and serotype data. Dave Prentiss and Ann Emery (VA Medical Center, Minneapolis), respectively, prepared the figures and helped with manuscript preparation.
REFERENCES
- 1.Achtman M, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver R P. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun. 1983;39:315–335. doi: 10.1128/iai.39.1.315-335.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amor K, Heinrisch D E, Frirdich E, Ziebell K, Johnson R P, Whitfield C. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect Immun. 2000;68:1116–1124. doi: 10.1128/iai.68.3.1116-1124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonacorsi S P P, Clermont O, Tinsley C, Le Gall I, Beaudoin J-C, Elion J, Nassif X, Bingen E. Identification of regions of the Escherichia coli chromosome specific for neonatal meningitis-associated strains. Infect Immun. 2000;68:2096–2101. doi: 10.1128/iai.68.4.2096-2101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bortolussi R, Ferrieri P, Wannamaker L W. Dynamics of Escherichia coli infection and meningitis in infant rats. Infect Immun. 1978;22:480–485. doi: 10.1128/iai.22.2.480-485.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyce T G, Swerdlow D L, Griffin P M. Escherichia coli O157:H7 and the hemolytic-uremic syndrome. N Engl J Med. 1995;333:364–368. doi: 10.1056/NEJM199508103330608. [DOI] [PubMed] [Google Scholar]
- 7.de Ree J M, Schwillens P, van den Bosch J F. Monoclonal antibodies for serotyping the P fimbriae of uropathogenic Escherichia coli. J Clin Microbiol. 1986;24:121–125. doi: 10.1128/jcm.24.1.121-125.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fields P I, Blom K, Hughes H J, Helsel L O, Feng P, Swaminathan B. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J Clin Microbiol. 1997;35:1066–1070. doi: 10.1128/jcm.35.5.1066-1070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gannon V P J, D'Souza S, Graham T, King R K, Rahn K, Read S. Use of the flagellar H7 gene as a target in multiplex PCR assays and improved specificity in identification of enterohemorrhagic Escherichia coli strains. J Clin Microbiol. 1997;35:656–662. doi: 10.1128/jcm.35.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herzer P J, Inouye S, Inouye M, Whittam T S. Phylogenetic distribution of branched RNS-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson J R, Delavari P, Kuskowski M, Stell A L. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis. 2001;183:78–88. doi: 10.1086/317656. [DOI] [PubMed] [Google Scholar]
- 12.Johnson J R, Delavari P, O'Bryan T. Escherichia coli O18:K1:H7 isolates from acute cystitis and neonatal meningitis exhibit common phylogenetic origins and virulence factor profiles. J Infect Dis. 2001;183:425–434. doi: 10.1086/318086. [DOI] [PubMed] [Google Scholar]
- 13.Johnson J R, Delavari P, Stell A L, Whittam T S, Carlino U, Russo T A. Molecular comparison of extraintestinal Escherichia coli isolates from the same electrophoretic lineages from humans and domestic animals. J Infect Dis. 2001;183:154–159. doi: 10.1086/317662. [DOI] [PubMed] [Google Scholar]
- 14.Johnson J R, Johnson C E, Maslow J N. Clinical and bacteriologic correlates of the papG alleles among Escherichia coli strains from children with acute cystitis. Pediatr Infect Dis J. 1999;18:446–451. doi: 10.1097/00006454-199905000-00009. [DOI] [PubMed] [Google Scholar]
- 15.Johnson J R, O'Bryan T T, Delavari P, Kuskowski M, Stapleton A, Carlino U, Russo T. Clonal relationships and extended virulence genotypes among Escherichia coli isolates from women with first episode or recurrent cystitis. J Infect Dis. 2001;183:1508–1517. doi: 10.1086/320198. [DOI] [PubMed] [Google Scholar]
- 16.Johnson J R, Orskov I, Orskov F, Goullet P, Picard B, Moseley S L, Roberts P L, Stamm W E. O, K, and H antigens predict virulence factors, carboxylesterase B pattern, antimicrobial resistance, and host compromise among Escherichia coli strains causing urosepsis. J Infect Dis. 1994;169:119–126. doi: 10.1093/infdis/169.1.119. [DOI] [PubMed] [Google Scholar]
- 17.Johnson J R, Russo T A, Brown J J, Stapleton A. papG alleles of Escherichia coli strains causing first episode or recurrent acute cystitis in adult women. J Infect Dis. 1998;177:97–101. doi: 10.1086/513824. [DOI] [PubMed] [Google Scholar]
- 18.Johnson J R, Russo T A, Scheutz F, Brown J J, Zhang L, Palin K, Rode C, Bloch C, Marrs C F, Foxman B. Discovery of disseminated J96-like strains of uropathogenic Escherichia coli O4:H5 containing genes for both PapGJ96 (“class I”) and PrsGJ96 (“class III”) Gal(α1-4)Gal-binding adhesins. J Infect Dis. 1997;175:983–988. doi: 10.1086/514006. [DOI] [PubMed] [Google Scholar]
- 19.Johnson J R, Stell A L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 20.Johnson J R, Stell A L, Scheutz F, O'Bryan T T, Russo T A, Carlino U B, Fasching C C, Kavle J, van Dijk L, Gaastra W. Analysis of F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex PCR-based assay. Infect Immun. 2000;68:1587–1599. doi: 10.1128/iai.68.3.1587-1599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korhonen T K, Valtonen M V, Parkkinen J, Vaisanen-Rhen V, Fine J, Orskov F, Orskov I, Svenson S B, Makela P H. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect Immun. 1985;48:486–491. doi: 10.1128/iai.48.2.486-491.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunin C M, Hua T H, Krishnan C, Van Arsdale White L, Hacker J. Isolation of a nicotinamide-requiring clone of Escherichia coli O18:K1:H7 from women with acute cystitis: resemblance to strains found in neonatal meningitis. Clin Infect Dis. 1993;16:412–416. doi: 10.1093/clind/16.3.412. [DOI] [PubMed] [Google Scholar]
- 23.Maslow J N, Whittam T S, Gilks C F, Wilson R A, Mulligan M E, Adams K S, Arbeit R D. Clonal relationships among bloodstream isolates of Escherichia coli. Infect Immun. 1995;63:2409–2417. doi: 10.1128/iai.63.7.2409-2417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mobley H L T, Jarvis K G, Elwood J P, Whittle D I, Lockatell C V, Russell R G, Johnson D E, Donnenberg M S, Warren J W. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of αGal(1-4)-βGal binding in virulence of a wild-type strain. Mol Microbiol. 1993;10:143–155. doi: 10.1111/j.1365-2958.1993.tb00911.x. [DOI] [PubMed] [Google Scholar]
- 25.Nagano I, Kunishima M, Itoh Y, Wu Z, Takahashi Y. Detection of verotoxin-producing Escherichia coli O157:H7 by multiplex polymerase chain reaction. Microbiol Immunol. 1998;42:371–376. doi: 10.1111/j.1348-0421.1998.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 26.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orskov I, Orskov F, Birch-Andersen A, Kanamori M, Svanborg Eden C. O, K, H and fimbrial antigens in Escherichia coli serotypes associated with pyelonephritis and cystitis. Scand J Infect Dis. 1982;33:18–25. [PubMed] [Google Scholar]
- 28.Pouttu R, Puustinen T, Virkola R, Hacker J, Klemm P, Korhonen T K. Amino acid residue Ala-62 in the FimH fimbrial adhesins is critical for the adhesiveness of meningitis-associated Escherichia coli to collagens. Mol Microbiol. 1999;31:1747–1757. doi: 10.1046/j.1365-2958.1999.01311.x. [DOI] [PubMed] [Google Scholar]
- 29.Reid S D, Selander R K, Whittam T S. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J Bacteriol. 1999;181:153–160. doi: 10.1128/jb.181.1.153-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo T A, Johnson J R. A proposal for an inclusive designation for extraintestinal pathogenic Escherichia coli: ExPEC. J Infect Dis. 2000;181:1753–1754. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 31.Schoenhals G, Whitfield C. Comparative analysis of flagellin sequences from Escherichia coli strains possessing serologically distinct flagellar filaments with a shared complex surface pattern. J Bacteriol. 1993;175:5395–5402. doi: 10.1128/jb.175.17.5395-5402.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slutsker L, Altekruse S F, Swerdlow S F. Foodborne diseases. Emerging pathogens and trends. Infect Dis Clin N Am. 1998;12:199–216. doi: 10.1016/s0891-5520(05)70418-9. [DOI] [PubMed] [Google Scholar]
- 33.Slutsker L, Ries A A, Greene K D, Wells J G, Hutwagner L, Griffin P M. Escherichia coli O157:H7 diarrhea in the United States: clinical and epidemiologic features. Ann Intern Med. 1997;126:505–513. doi: 10.7326/0003-4819-126-7-199704010-00002. [DOI] [PubMed] [Google Scholar]
- 34.Somerville J E, Casiano L, Darveau R P. Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect Immun. 1999;67:6583–6590. doi: 10.1128/iai.67.12.6583-6590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stapleton A, Moseley S, Stamm W E. Urovirulence determinants in Escherichia coli isolates causing first-episode and recurrent cystitis in women. J Infect Dis. 1991;163:773–779. doi: 10.1093/infdis/163.4.773. [DOI] [PubMed] [Google Scholar]
- 36.Wang G, Whittam T S, Berg C M, Berg D E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993;21:5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whittam T S, Wolfe M L, Wilson R A. Genetic relationships among Escherichia coli isolates causing urinary tract infections in humans and animals. Epidemiol Infect. 1989;102:37–46. doi: 10.1017/s0950268800029666. [DOI] [PMC free article] [PubMed] [Google Scholar]