Abstract
Genome-wide association studies (GWAS) have identified genetic variants associated with brain morphology and substance use behaviors (SUB). However, the genetic overlap between brain structure and SUB has not been well characterized. We leveraged GWAS summary data of 71 brain imaging measures and alcohol, tobacco, and cannabis use to investigate their genetic overlap using linkage disequilibrium score regression. We used genomic structural equation modeling to model a “common SUB genetic factor” and investigated its genetic overlap with brain structure. Furthermore, we estimated SUB polygenic risk scores (PRS) and examined whether they predicted brain imaging traits using the Adolescent Behavior and Cognitive Development (ABCD) study. We identified 8 significant negative genetic correlations, including between (1) alcoholic drinks per week and average cortical thickness, and (2) intracranial volume with age of smoking initiation. We observed 5 positive genetic correlations, including those between (1) insula surface area and lifetime cannabis use, and (2) the common SUB genetic factor and pericalcarine surface area. SUB PRS were associated with brain structure variation in ABCD. Our findings highlight a shared genetic etiology between cortical brain morphology and SUB and suggest that genetic variants associated with SUB may be causally related to brain structure differences.
Keywords: alcohol use, cannabis use, genetics, neuroimaging, smoking behavior
Introduction
Heavy use of alcohol, tobacco, or cannabis is associated with serious negative consequences, including increased risk for unemployment (Kendler et al. 2017), psychiatric comorbidity (Hasin et al. 2017; Mammen et al. 2018), substance use disorders (Danielsson et al. 2012), morbidity and mortality (Whitfield et al. 2018). Over the last few decades, noninvasive brain imaging techniques have significantly bolstered our understanding of brain structure and function and their relationship with substance use. In parallel, lower genotyping costs, advancements in statistical genetics methods, and the availability of larger samples have enabled the investigation of genetic influences on both brain structure (Hibar et al. 2017; Satizabal et al. 2019; Grasby et al. 2020) and substance use (Pasman et al. 2018; Liu et al. 2016). Despite significant developments in neuroimaging and molecular genetics technologies, few studies have examined how the genetic architectures of alcohol, tobacco, and cannabis use are associated with neuroanatomical measures.
A substantial body of work has linked brain structural variation to substance use and abuse at the phenotypic level among adults. For instance, cannabis use has been associated with reduced cortical thickness and surface area of the right entorhinal cortex (Paul and Bhattacharyya 2018), and an analysis of subcortical surface morphology identified localized differences in surface area and radial distance of the hippocampus, thalamus, putamen, and amygdala, among persons with alcohol dependence (Chye et al. 2019). The same study found surface area differences in the bilateral hippocampus, right nucleus accumbens, thalamus, and putamen among individuals with alcohol dependence (Chye et al. 2019). In another study, Gillespie et al. (2018) found an association between smaller thalamus volume and nicotine use in middle-aged males and no significant associations between subcortical volumes and cannabis use. Another study reported that higher levels of alcohol use were associated with thinner medial and dorsolateral frontal and parieto-occipital cortical regions, in addition to a larger left ventricle volume (Lange et al. 2017). In 2 of the largest meta-analyses on substance use disorders and neurological structures to date, individuals with alcohol use disorder had lower cortical thicknesses in several brain regions (e.g., insula, precuneus) (Mackey et al. 2016) and smaller cortical volumes of the thalamus, putamen, hippocampus, amygdala, and accumbens (Navarri et al. 2020). Taken together, although these studies vary widely in terms of their image acquisition methods and selection of brain regions, they highlight that alcohol, tobacco, and cannabis use are related to differences in brain morphology. However, it is unclear the extent to which these relationships are due to shared genetic factors.
The heritable nature of brain structure has been well-documented (Kremen et al. 2010; Kremen et al. 2013; Renteria et al. 2014; Swagerman et al. 2014), with genetic influences explaining close to 70% of the variance in global, subcortical, and ventricular volumes; and 45% of the variance in frontal, parietal, occipital, and temporal lobe thickness (Kremen et al. 2010). The genetic variation observed in brain structure and function is driven mainly by polygenic influences (i.e., multiple genetic loci of small individual effect sizes) (Elliott et al. 2018; van der Lee et al. 2019; Biton et al. 2020). Several studies to date have attempted to identify (1) common genetic variants underpinning brain structure, and (2) genetic variants associated with psychiatric conditions and their relationship with variability in brain morphology. In a recent genome-wide association meta-analysis, common genetic variants accounted for 34% and 26% of the variance in total cortical surface area and average cortical thickness, respectively (Grasby et al. 2020). Also, previous studies found either no or small negative genetic correlations between psychiatric phenotypes (e.g., schizophrenia, bipolar disorder) and intracranial volume (Ohi et al. 2020). To date, only one study has investigated genetic correlations between substance use phenotypes and brain structural properties and observed a very small genetic correlation between cigarette smoking frequency and cortical surface area (Grasby et al. 2020).
In addition to the limited work that has examined genetic associations between substance use and brain structure, there is a dearth of work that has investigated whether the genetics of substance use could be used to predict neuroimaging traits. Understanding whether genetic risk for substance use behaviors predicts neuroanatomical structures may elucidate potential mechanisms that contribute to substance use engagement. One study investigated whether substance use polygenic risk scores (PRS), defined as one’s aggregate genetic risk for a given phenotype (Maher 2015), was related to differences in cortical volumes; this study found that greater PRS for more frequent smoking was tied to smaller cortical volumes of the right orbitofrontal cortex in a sample of adolescents (Li et al. 2020).
The primary goal of the present study was to investigate potential genetic overlap between substance use and brain morphology by examining genetic correlations between these phenotypes. A significant genetic correlation between 2 traits is typically a sign of pleiotropy (Cho et al. 2020), of which there are 2 types, horizontal and vertical pleiotropy. There are several ways to distinguish between them, but it is an active area of research. Here, we propose PRS can be used in differentially exposed samples. For example, if the PRS derived from one trait is associated with another trait (and vice versa) regardless of whether or not the sample has been exposed to the trait for which the PRS was calculated, this might indicate horizontal pleiotropy. Horizontal pleiotropy refers to the existence of shared genetic factors that affect both traits independently (Cho et al. 2020). Vertical pleiotropy, on the other hand, occurs when there is a causal relationship between the 2 traits, which causes them to be genetically correlated as the genetic effects for the causal trait will be proportionally affecting the other trait (Geiler-Samerotte et al. 2020). Vertical pleiotropy may cause the PRS of an exposure (or causal trait) to predict the outcome trait only in a sample in which there is variance with relation to the exposure such that some proportion of the sample has been exposed for the causal changes in the outcome to occur.
With the goal of examining shared genetic factors underpinning both SUB and neuroimaging traits, we leverage the availability of recent genome-wide association studies (GWAS) summary statistics from large meta-analyses conducted by the Enhancing Neuro-Imaging Genetics through Meta-Analysis (ENIGMA) consortium (Hibar et al. 2017; Thompson et al. 2017; Thompson et al. 2020), the GWAS & Sequencing Consortium of Alcohol and Nicotine use (GSCAN) (Liu et al. 2016), and the International Cannabis Consortium (Spechler et al. 2019) to estimate pairwise genetic correlations between brain morphology measures (regional cortical surface area and thickness for 34 regions of interest and intracranial volume, total cortical surface area, and mean thickness); and tobacco use measurements (age of smoking initiation, ever being a regular smoker, smoking heaviness, and continuation vs. cessation), alcohol use (number of drinks per week), and lifetime cannabis use. To our knowledge, this is the first study to date to explore these genetic relationships. Furthermore, consistent with the common liability hypothesis (Kendler et al. 2007), genetic variants implicated in multiple substance use behaviors may be associated with specific or global neuroanatomic differences, an empirical question that remains unanswered. To address this gap, we used genomic structural equation modeling (gSEM) (Grotzinger et al. 2019) to model a common genetic factor across substance use phenotypes (i.e., tobacco, alcohol, and cannabis use) and evaluated its genetic correlation with cortical morphology measures.
Lastly, we probed significant genetic associations between substance use behaviors and neuroimaging traits by deriving substance use PRS and examined their associations with brain morphology in the Adolescent Behavior and Cognitive Development (ABCD) study, a sample of drug-naïve children aged 9–10. This work has the potential to determine whether SUB PRS predicts variation in brain structure prior to substance use engagement, thus elucidating neurobiological antecedents that may confer risk for using substances.
Methods
Datasets
Brain Imaging Measures
GWAS summary statistics for 71 neuroimaging measures were obtained through direct application to the ENIGMA Consortium (Thompson et al. 2017; Thompson et al. 2020). We specifically used summary statistics from the principal meta-analyses conducted in European ancestry individuals for 68 bilateral cortical measures (thickness and surface area) and mean cortical thickness and total cortical surface area (Grasby et al. 2020), and a large GWAS conducted on intracranial volume (Adams et al. 2016). These measurements were based on brain magnetic resonance imaging scans and genome-wide genotype data from the largest genetic studies of brain structure (Table 1). Participants in all cohorts from these studies gave written informed consent, and sites involved obtained approval from local research ethics committees or Institutional Review Boards. In the original analysis, GWAS summary statistics from each of the 50 sites had been combined using a fixed-effect inverse variance weighted meta-analysis in METAL (Willer et al. 2010).
Table 1.
Source of GWAS summary statistics datasets for phenotypes analyzed in this study
| Study | Phenotypes | N participants | N cohorts |
|---|---|---|---|
| Hibar et al. 2017 | Intracranial volume | 33 536 | 65 |
| Grasby et al. 2020 | Mean cortical thickness Total cortical surface area Surface area and thickness for 34 ROIs |
33 992 | 50 |
| Liu et al. 2016 a | Drinks per week Ever regularly smoked Smoking heaviness Smoking cessation Age at smoking initiation |
~1.2 million | 29 |
| Pasman et al. 2018 a | Lifetime cannabis use | 184 765 | 18 |
Note: Summary statistics from the studies above only included European ancestry cohorts.
The GWAS summary results also included the 23andMe cohort.
Tobacco and Alcohol Use Measures
GWAS summary statistics for alcohol and tobacco use phenotypes were obtained from the repository of their corresponding publication which included over 1 million individuals (Liu et al. 2016). Several GWAS were conducted, including (1) ever regularly smoked; (2) age of smoking initiation; (3) smoking heaviness (i.e., packs per day); (4) smoking cessation (i.e., current smoker versus former smoker); and (5) alcohol frequency (i.e., number of drinks per week). GWAS were conducted among European ancestry individuals and included samples from both the 23andMe and GSCAN cohorts. Summary statistics for the 23andMe cohort were obtained via an application and signing of a data transfer agreement between 23andMe, Inc. and the QIMR Berghofer Medical Research Institute where the genetic analyses were conducted.
Lifetime Cannabis Use
The GWAS summary statistics for lifetime cannabis use were retrieved from a meta-analysis (N = 184 765), which included European ancestry individuals from The International Cannabis Consortium, UK Biobank, and 23andMe (Pasman et al. 2018). Summary statistics excluding the 23andMe cohort were obtained from the International Cannabis Consortium’s online repository (https://www.ru.nl/bsi/research/group-pages/substance-use-addiction-food-saf/vm-saf/genetics/international-cannabis-consortium-icc/). Summary statistics for the 23andMe cohort were obtained via application and signing of a data transfer agreement between 23andMe, Inc. and the QIMR Berghofer Medical Research Institute. Across all samples used in the GWAS, participants reported on whether they had ever used cannabis or marijuana (e.g., weed, dope, draw) in their lifetime. The 23andMe summary statistics datasets were meta-analyzed with the corresponding summary statistics from GSCAN and The International Cannabis Consortium’s datasets. We used an inverse variance weighted meta-analysis implemented in METALv2011-03-25 (Willer et al. 2010).
Statistical Analyses
Linkage Disequilibrium Score Regression
Linkage disequilibrium score (LDSC) regression (Bulik-Sullivan et al. 2015a; 2015b) was used to assess pairwise genetic correlations between substance use (i.e., tobacco, alcohol, and cannabis use) and intracranial volume, as well as global and regional cortical surface area and thickness. LDSC leverages the expected relationship between the amount of LD that a variant tags and its association with a trait to model heritability and coheritability using only the distribution of variant effect sizes. LDSC regression can then assess whether inflation in GWAS test statistics is due to polygenicity or confounding biases such as cryptic relatedness or population stratification. Likewise, bivariate LDSC regression can be used to distinguish between true genetic correlations between traits and inflation due to sample overlap. For this study, each dataset was filtered to only include markers overlapping with HapMap Project Phase 3 single nucleotide polymorphisms (SNPs) (Noverlap = 1 217 312) as these tend to be well-imputed across datasets, and alleles will match those listed in the data used to estimate the LD score. We used precomputed LD scores for European populations, as provided on the LDSC website (https://github.com/bulik/ldsc). Standard errors were estimated using a block jackknife procedure and used to calculate P values.
Genomic Structural Equation Modeling (GSEM)
To gain insights into the genetic etiology of substance use in general, we performed a common factor GWAS using (gSEM) (Grotzinger et al. 2019) implemented in R. This approach leverages the genetic variance–covariance matrix between the traits under study estimated through LDSC regression. Then, structural equation models are used to partition the covariance structure and estimate latent factors. In the current study, we desired to study the common genetic etiology of substance use phenotypes and thus specified a common factor model. The effect of a genetic variant on the common factor can be estimated by including the SNPs covariance with the traits studied in the model. Repeating this procedure for all genetic variants yields a GWAS of the common factor. Genetic correlations between this common factor GWAS and the neuroimaging traits of interest were performed using bivariate LDSC regression.
Brain Plots
As shown in Figure 1, the cortical thickness and surface area results are presented by mapping the z-score for the genetic correlation between a given trait and a brain region onto a brain triangular surface plot. These plots were generated using python v.3.5 and the modules matplotlib, numpy, plotly, pandas, and scipy. All of the z-scores in Figure 1 are shown without any filtering. Statistically significant results are displayed in the tables.
Figure 1 .
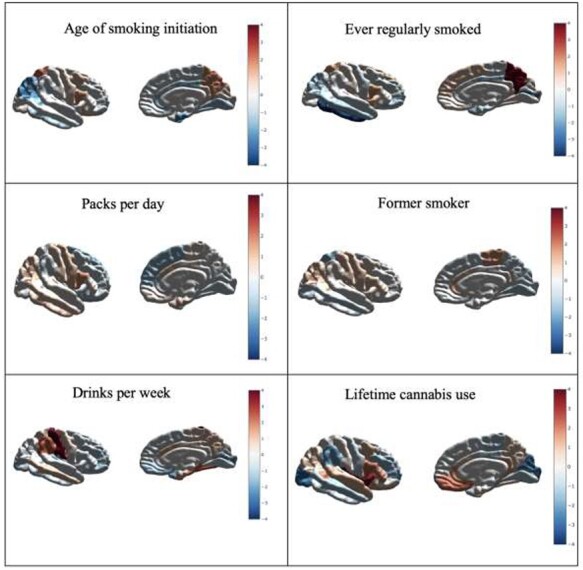
Standardized effect sizes (z-scores) reflecting the relationship between genetic risk for the substance use phenotypes and surface area. Positive effects highlighted in red denote increases in surface area and negative effects highlighted in blue denote reductions in surface area for colored regions. Results shown here correspond to all observed genetic correlations regardless of their level of statistical significance. Details for genetic correlations which surpassed multiple testing correction are shown in Table 2.
Polygenic Risk Scoring
We estimated PRS in the ABCD study, a sample of 9–10 year olds at baseline who were recruited from various sites in the USA (Volkow et al. 2018). The ABCD sample was not included in the GWAS considered in the current study; thus, the inclusion of ABCD not only ensured sample independence, but also a lack of sample exposure to the substances studied here. Only SNPs passing quality control (minor allele frequency > 0.01, call rate > 0.9 and imputation score > 0.6) were included in the PRS analyses. To adjust for linkage disequilibrium, we used a clumping + thresholding approach. Briefly, GWAS summary statistics were clumped using PLINK1.9 using a correlation cutoff of 0.05 and a distance of 500 kilobases. Then, 8 PRS were estimated. Each of these PRS were calculated using an increasingly liberal P value threshold for variant inclusion (P < 5 × 10–8, P < 1 × 10–5, P < 0.001, P < 0.01, P < 0.05, P < 0.1, P < 0.5, P < 1). PRS were estimated by multiplying the effect size (obtained from GWAS) times the allelic dosage of the effect allele and summing across all loci for each participant. To test for the association between PRS and region of interest (ROI) morphology, we used a multiple linear regression implemented in python(v3.5) with the library statsmodels. Additional covariates included in the model were sex, age, sex×age, age2, sex×age2, and the first 10 genetic ancestry components to adjust for population stratification. Variance explained was estimated as the difference in Pearson correlation coefficient between the full model (i.e., including the PRS) and a reduced model including only the covariates.
Multiple Testing and Significance Threshold
In all analyses using LDSC regression, we applied Benjamini–Hochberg’s False Discovery Rate (FDR < 5%) to account for multiple testing (i.e., the number of cortical neuroimaging traits) within each substance use phenotype (Benjamini and Hochberg 1995). Genetic correlations that were nominally significant (i.e., P < 0.05) but did not survive multiple testing corrections are reported in the Supplementary Materials. For the analyses examining the relationship between substance use PRS and the neuroimaging traits, 8 sets of analyses (one for each PRS threshold) were conducted for each brain structure. To account for this multiple testing, we used a Bonferroni-corrected P value of 0.006 (0.05/8 tests) to evaluate statistical significance.
Results
Figure 1 shows an overview of the results as summarized by raw z-scores for all genetic correlations between substance use and brain morphology phenotypes, regardless of their level of statistical significance. Overall, we observed positive associations between alcohol use and cortical surface area. A similar pattern was observed for smoking-related phenotypes, with the exception of a negative association between smoking behaviors and surface area of the inferior temporal lobe.
Alcohol
As shown in Table 2, after correcting for multiple testing, significant positive genetic correlations were observed between alcohol use and total cortical surface area (rg = 0.12; P value = 0.023) and surface area of the postcentral gyrus’s regional area (rg = 0.17; P value = 4.3 × 10−4). A significant negative genetic correlation was observed between alcohol use and global average cortical thickness (rg = −0.10, P value = 0.047).
Table 2.
Significant genetic correlations after multiple testing correction (FDR < 5%) between brain imaging and substance use phenotypes
| Neuroimaging traits | Alcohol (drinks per week) | Ever regularly smoked | Age of smoking initiation | Lifetime cannabis use | Common substance use factor | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rg | se | P | rg | se | P | rg | se | P | rg | se | P | rg | se | P | |
| Intracranial volume | — | — | — | −0.09 | 0.03 | 0.038 | −0.21 | 0.06 | 0.027 | — | — | — | — | — | — |
| Postcentral gyrus surface rea | 0.17 | 0.04 | 5.2 × 10−4 | — | — | — | — | — | — | — | — | — | — | — | — |
| Total cortical surface area | 0.12 | 0.04 | 0.023 | — | — | — | — | — | — | — | — | — | — | — | — |
| Inferior temporal gyrus surface area | — | — | — | −0.13 | 0.03 | 1.01 × 10−3 | — | — | — | — | — | — | −0.13 | 0.03 | 1.6 × 10−3 |
| Precuneus surface area | — | — | — | 0.10 | 0.03 | 0.002 | — | — | — | — | — | — | 0.10 | 0.03 | 0.012 |
| Insula surface area | — | — | — | — | — | — | — | — | — | 0.17 | 0.05 | .009 | |||
| Pericalcarine surface area | — | — | — | — | — | — | — | — | — | — | — | — | −0.09 | 0.03 | 0.012 |
| Average cortical thickness | −0.10 | 0.04 | 0.047 | −0.09 | 0.03 | 0.020 | — | — | — | — | — | — | −0.09 | 0.03 | 0.023 |
Smoking
Significant negative genetic correlations (Table 2) were observed between ever regularly smoked and (1) intracranial volume (rg = −0.09, P value = 0.038), (2) average cortical thickness (rg = −0.09, P value = 0.02), and (3) surface area of the inferior temporal lobe (rg = −0.13, P value = 8 × 10−4). A significant negative genetic correlation was observed between age of smoking initiation and intracranial volume (rg = −0.21, P value = 0.02). Last, a significant positive genetic correlation was observed between ever regularly smoked and precuneus surface area (rg = 0.10, P value = 0.002).
Cannabis
As shown in Table 2, a significant positive genetic correlation was found between lifetime cannabis use and insula surface area (rg = 0.17, P value = 7.4 × 10−3). No other associations between cannabis use and neuroimaging traits remained significant after multiple testing correction.
Common Substance Use Genetic Factor
Negative genetic associations were observed between the common substance use genetic factor and the surface areas of the inferior temporal gyrus (rg = −0.13, P value = 0.002) and pericalcarine (rg = −0.09, P value = 0.012), and cortical thickness (rg = −0.09, P value = 0.023) (Table 2). In addition, a positive genetic correlation was observed involving the common substance use genetic factor and the precuneus surface area (rg = 0.10, P value = 0.012).
PRS Analyses
A secondary set of analyses assessing the association between substance use PRS with the neuroimaging traits were performed. Briefly, when there was evidence of a genetic association between SUB and neuroimaging traits identified in the LDSC analyses, we derived SUB PRS in the ABCD sample (see methods) and assessed whether they predicted the morphometry of the ROI. As shown in Table 3, several significant PRS associations with brain morphology were observed. Higher PRS for alcohol use predicted greater postcentral gyrus surface area and cortical surface area. Greater PRS for regularly smoking was positively associated with ICV and surface area of the inferior temporal gyrus.
Table 3.
Substance use PRS associations with brain imaging traits in the ABCD study
| PRS phenotype and threshold | Neuroimaging trait | beta | se | P | rsq |
|---|---|---|---|---|---|
| Age of smoking initiation P < 5 × 10–8 | ICV | 234858.981 | 140317.946 | 0.0942175 | 0.00027157 |
| Age of smoking initiation P < 5 × 10–5 | ICV | 47480.4985 | 21525.4542 | 0.02742835 | 0.00047153 |
| Age of smoking initiation P < 0.001 | ICV | 362.899012 | 4812.83657 | 0.93989656 | 5.51E-07 |
| Age of smoking initiation P < 0.01 | ICV | 913.671368 | 2128.03885 | 0.66768118 | 1.79E-05 |
| Age of smoking initiation P < 0.05 | ICV | 0.84520238 | 1343.31729 | 0.99949799 | 3.84E-11 |
| Age of smoking initiation P < 0.10 | ICV | 254.083657 | 1133.04516 | 0.82256987 | 4.88E-06 |
| Age of smoking initiation P < 0.50 | ICV | 148.33005 | 873.235382 | 0.86512261 | 2.80E-06 |
| Age of smoking initiation P < 1 | ICV | 258.738116 | 861.626692 | 0.7639638 | 8.74E-06 |
| Lifetime cannabis use P < 5 × 10–8 | Insula SA | 37.4306302 | 26.6738751 | 0.1605766 | 0.00020745 |
| Lifetime cannabis use P < 5 × 10–5 | Insula SA | 5.65217963 | 12.7767595 | 0.65822678 | 2.06E-05 |
| Lifetime cannabis use P < 0.001 | Insula SA | 1.01071199 | 3.12938501 | 0.74672316 | 1.10E-05 |
| Lifetime cannabis use P < 0.01 | Insula SA | 1.06632932 | 1.43571811 | 0.45767678 | 5.81E-05 |
| Lifetime cannabis use P < 0.05 | Insula SA | −0.3878843 | 0.82967746 | 0.64014661 | 2.30E-05 |
| Lifetime cannabis use P < 0.10 | Insula SA | 0.14259272 | 0.71478884 | 0.84188525 | 4.19E-06 |
| Lifetime cannabis use P < 0.50 | Insula SA | 0.11972915 | 0.55560989 | 0.82938989 | 4.89E-06 |
| Lifetime cannabis use P < 1 | Insula SA | 0.17710138 | 0.54598012 | 0.74566418 | 1.11E-05 |
| Alcohol (drinks per week) P < 5 × 10–8 | Postcentral gyrus SA | 314.0491 | 241.34794 | 0.19321851 | 0.00018698 |
| Alcohol (drinks per week) P < 5 × 10–5 | Postcentral gyrus SA | 242.191884 | 157.208433 | 0.12346028 | 0.00026207 |
| Alcohol (drinks per week) P < 0.001 | Postcentral gyrus SA | 122.137472 | 76.1193971 | 0.10863333 | 0.00028428 |
| Alcohol (drinks per week) P < 0.01 | Postcentral gyrus SA | 94.2982005 | 46.2256129 | 0.04138856 | 0.0004594 |
| Alcohol (drinks per week) P < 0.05 | Postcentral gyrus SA | 90.5536151 | 32.4186689 | 0.00523085 | 0.00086092 |
| Alcohol (drinks per week) P < 0.10 | Postcentral gyrus SA | 83.2904156 | 28.6206291 | 0.00362279 | 0.00093441 |
| Alcohol (drinks per week) P < 0.50 | Postcentral gyrus SA | 68.464345 | 24.0737467 | 0.00446757 | 0.00089242 |
| Alcohol (drinks per week) P < 1 | Postcentral gyrus SA | 69.2765934 | 23.7780433 | 0.00358458 | 0.00093653 |
| Alcohol (drinks per week) P < 5 × 10–8 | Cortical SA | 9124.44994 | 7610.78436 | 0.23060957 | 0.00014092 |
| Alcohol (drinks per week) P < 5 × 10–5 | Cortical SA | 7455.72118 | 4957.00023 | 0.13260169 | 0.00022178 |
| Alcohol (drinks per week) P < 0.001 | Cortical SA | 3792.39809 | 2400.82514 | 0.11423406 | 0.00024461 |
| Alcohol (drinks per week) P < 0.01 | Cortical SA | 4060.76871 | 1457.95714 | 0.0053617 | 0.00075999 |
| Alcohol (drinks per week) P < 0.05 | Cortical SA | 3152.539 | 1022.41403 | 0.0020535 | 0.00093121 |
| Alcohol (drinks per week) P < 0.10 | Cortical SA | 2884.8708 | 902.673758 | 0.00139946 | 0.00100031 |
| Alcohol (drinks per week) P < 0.50 | Cortical SA | 2060.42849 | 759.34238 | 0.00667361 | 0.00072134 |
| Alcohol (drinks per week) P < 1 | Cortical SA | 2053.69802 | 750.018751 | 0.00619183 | 0.00073455 |
| Alcohol (drinks per week) P < 5 × 10–8 | Cortical thickness | 0.04544648 | 0.05265596 | 0.3881184 | 9.23E-05 |
| Alcohol (drinks per week) P < 5 × 10–5 | Cortical thickness | 0.05690662 | 0.0342854 | 0.09699681 | 0.0003413 |
| Alcohol (drinks per week) P < 0.001 | Cortical thickness | −0.0013141 | 0.01660991 | 0.93694271 | 7.76E-07 |
| Alcohol (drinks per week) P < 0.01 | Cortical thickness | −0.0050523 | 0.01009038 | 0.61659566 | 3.11E-05 |
| Alcohol (drinks per week) P < 0.05 | Cortical thickness | −0.0043889 | 0.00707866 | 0.53526241 | 4.76E-05 |
| Alcohol (drinks per week) P < 0.10 | Cortical thickness | −0.0049285 | 0.00624998 | 0.43038693 | 7.71E-05 |
| Alcohol (drinks per week) P < 0.50 | Cortical thickness | −0.003584 | 0.00525614 | 0.49533922 | 5.76E-05 |
| Alcohol (drinks per week) P < 1 | Cortical thickness | −0.0045353 | 0.0051915 | 0.38236757 | 9.46E-05 |
| Ever regularly smoked P < 5 × 10–8 | ICV | −1978.042 | 5072.99229 | 0.69660919 | 1.47E-05 |
| Ever regularly smoked P < 5 × 10–5 | ICV | −3458.9854 | 3759.51187 | 0.35756877 | 8.21E-05 |
| Ever regularly smoked P < 0.001 | ICV | −4331.9386 | 2443.59965 | 0.07630616 | 0.00030464 |
| Ever regularly smoked P < 0.01 | ICV | −4509.9739 | 1860.97659 | 0.01539681 | 0.0005691 |
| Ever regularly smoked P < 0.05 | ICV | −3515.4547 | 1528.28333 | 0.02145969 | 0.00051276 |
| Ever regularly smoked P < 0.10 | ICV | −4043.4712 | 1416.89768 | 0.0043321 | 0.00078891 |
| Ever regularly smoked P < 0.50 | ICV | −3683.6723 | 1277.07865 | 0.00393175 | 0.00080596 |
| Ever regularly smoked P < 1 | ICV | −3629.1407 | 1270.23747 | 0.00428726 | 0.00079074 |
| Ever regularly smoked P < 5 × 10–8 | Inferior temporal gyrus SA | −8.4774256 | 16.8143476 | 0.61415056 | 2.76E-05 |
| Ever regularly smoked P < 5 × 10–5 | Inferior temporal gyrus SA | −11.001419 | 12.4625704 | 0.37739505 | 8.45E-05 |
| Ever regularly smoked P < 0.001 | Inferior temporal gyrus SA | −17.476902 | 8.09826778 | 0.03095036 | 0.00050474 |
| Ever regularly smoked P < 0.01 | Inferior temporal gyrus SA | −17.940266 | 6.16611144 | 0.00363043 | 0.00091696 |
| Ever regularly smoked P < 0.05 | Inferior temporal gyrus SA | −7.9418555 | 5.06553519 | 0.11696357 | 0.00026647 |
| Ever regularly smoked P < 0.10 | Inferior temporal gyrus SA | −7.802245 | 4.69788283 | 0.09679399 | 0.000299 |
| Ever regularly smoked P < 0.50 | Inferior temporal gyrus SA | −6.3497341 | 4.23451398 | 0.13377975 | 0.00024376 |
| Ever Regularly Smoked P < 1 | Inferior temporal gyrus SA | −6.3576148 | 4.21178636 | 0.13121644 | 0.00024701 |
| Ever regularly smoked P < 5 × 10–8 | Precuneus SA | −6.668509 | 18.5119273 | 0.71868667 | 1.39E-05 |
| Ever regularly smoked P < 5 × 10–5 | Precuneus SA | −8.3536585 | 13.7222474 | 0.54269637 | 3.96E-05 |
| Ever regularly smoked P < 0.001 | Precuneus SA | −10.0019 | 8.91705899 | 0.26204213 | 0.00013444 |
| Ever regularly smoked P < 0.01 | Precuneus SA | −10.59728 | 6.79150769 | 0.11871272 | 0.00026014 |
| Ever regularly smoked P < 0.05 | Precuneus SA | −3.1817718 | 5.5778535 | 0.56840299 | 3.48E-05 |
| Ever regularly smoked P < 0.10 | Precuneus SA | −2.7015228 | 5.17231723 | 0.60147349 | 2.92E-05 |
| Ever regularly smoked P < 0.50 | Precuneus SA | −2.1866766 | 4.66179669 | 0.63903775 | 2.35E-05 |
| Ever regularly smoked P < 1 | Precuneus SA | −2.1720255 | 4.63682002 | 0.6394903 | 2.35E-05 |
| Ever regularly smoked P < 5 × 10–8 | Cortical thickness | −0.0026998 | 0.00424612 | 0.52491442 | 5.01E-05 |
| Ever regularly smoked P < 5 × 10–5 | Cortical thickness | −0.000504 | 0.00314707 | 0.87277414 | 3.18E-06 |
| Ever regularly smoked P < 0.001 | Cortical thickness | 0.0001948 | 0.00204637 | 0.92416299 | 1.12E-06 |
| Ever regularly smoked P < 0.01 | Cortical thickness | −0.0009318 | 0.00155876 | 0.55002245 | 4.43E-05 |
| Ever regularly smoked P < 0.05 | Cortical thickness | −0.0024819 | 0.00127979 | 0.0525028 | 0.00046586 |
| Ever regularly smoked P < 0.10 | Cortical thickness | −0.0021716 | 0.00118668 | 0.06729459 | 0.00041484 |
| Ever regularly smoked P < 0.50 | Cortical thickness | −0.0018017 | 0.00106948 | 0.09208759 | 0.00035161 |
| Ever regularly smoked P < 1 | Cortical thickness | −0.0018479 | 0.00106367 | 0.08236926 | 0.00037392 |
| Common SUB factor P < 5 × 10–8 | Inferior temporal gyrus SA | −285.23808 | 271.72387 | 0.29387263 | 0.00011948 |
| Common SUB factor P < 5 × 10–5 | Inferior temporal gyrus SA | −245.08906 | 168.354548 | 0.14549085 | 0.00022976 |
| Common SUB factor P < 0.001 | Inferior temporal gyrus SA | −106.53498 | 101.973457 | 0.2961792 | 0.00011834 |
| Common SUB factor P < 0.01 | Inferior temporal gyrus SA | 10.6785604 | 73.5891492 | 0.88462738 | 2.28E-06 |
| Common SUB factor P < 0.05 | Inferior temporal gyrus SA | −18.147942 | 58.4265476 | 0.75610453 | 1.05E-05 |
| Common SUB factor P < 0.10 | Inferior temporal gyrus SA | −30.149333 | 53.3313944 | 0.5718721 | 3.47E-05 |
| Common SUB factor P < 0.50 | Inferior temporal gyrus SA | −41.969645 | 47.3527678 | 0.37547328 | 8.52E-05 |
| Common SUB factor P < 1 | Inferior temporal gyrus SA | −32.905934 | 47.1182847 | 0.48496759 | 5.29E-05 |
| Common SUB factor P < 5 × 10–8 | Precuneus SA | −380.97908 | 299.149805 | 0.20286484 | 0.00017331 |
| Common SUB factor P < 5 × 10–5 | Precuneus SA | −133.06232 | 185.367588 | 0.47288403 | 5.51E-05 |
| Common SUB factor P < 0.001 | Precuneus SA | 20.2493511 | 112.278233 | 0.85688271 | 3.48E-06 |
| Common SUB factor P < 0.01 | Precuneus SA | 135.433412 | 80.9894219 | 0.09451816 | 0.00029876 |
| Common SUB factor P < 0.05 | Precuneus SA | 144.306263 | 64.2938244 | 0.02482955 | 0.00053806 |
| Common SUB factor P < 0.10 | Precuneus SA | 134.479338 | 58.6958016 | 0.02198295 | 0.00056064 |
| Common SUB factor P < 0.50 | Precuneus SA | 90.548278 | 52.1227025 | 0.08238913 | 0.00032242 |
| Common SUB factor P < 1 | Precuneus SA | 92.6382002 | 51.8617832 | 0.07409755 | 0.00034087 |
| Common SUB factor P < 5 × 10–8 | Pericalcarine SA | −106.33581 | 136.11675 | 0.43470263 | 7.47E-05 |
| Common SUB factor P < 5 × 10–5 | Pericalcarine SA | −83.22776 | 84.3292338 | 0.32370361 | 0.00011924 |
| Common SUB factor P < 0.001 | Pericalcarine SA | −72.053948 | 51.0860628 | 0.15844919 | 0.0002435 |
| Common SUB factor P < 0.01 | Pericalcarine SA | −28.6623 | 36.8530616 | 0.43674282 | 7.41E-05 |
| Common SUB factor P < 0.05 | Pericalcarine SA | −46.868987 | 29.2560626 | 0.10919047 | 0.00031412 |
| Common SUB factor P < 0.10 | Pericalcarine SA | −49.474937 | 26.7060453 | 0.06398133 | 0.00042002 |
| Common SUB factor P < 0.50 | Pericalcarine SA | −57.746032 | 23.7081432 | 0.01488518 | 0.00072581 |
| Common SUB factor P < 1 | Pericalcarine SA | −56.845659 | 23.5901382 | 0.01598784 | 0.00071042 |
| Common SUB factor P < 5 × 10–8 | Cortical thickness | −0.108783 | 0.06862599 | 0.11297138 | 0.00031131 |
| Common SUB factor P < 5 × 10–5 | Cortical thickness | −0.0286126 | 0.04253268 | 0.50114585 | 5.61E-05 |
| Common SUB factor P < 0.001 | Cortical thickness | −0.0035863 | 0.02575566 | 0.88926085 | 2.40E-06 |
| Common SUB factor P < 0.01 | Cortical thickness | −0.0247286 | 0.01858003 | 0.18325307 | 0.00021948 |
| Common SUB factor P < 0.05 | Cortical thickness | −0.022517 | 0.0147516 | 0.12694879 | 0.00028866 |
| Common SUB factor P < 0.10 | Cortical thickness | −0.0228674 | 0.01346731 | 0.08954865 | 0.00035719 |
| Common SUB factor P < 0.50 | Cortical thickness | −0.021622 | 0.01195656 | 0.07058626 | 0.00040511 |
| Common SUB factor P < 1 | Cortical thickness | −0.0221886 | 0.01189683 | 0.06220768 | 0.00043091 |
Note: ICV = intracranial volume; SUB = substance use behavior; SA = surface area.
There was no evidence of statistically significant associations between (1) age of initiation PRS and ICV, (2) lifetime cannabis use PRS and insula surface area; (3) alcohol use PRS and cortical thickness, (4) ever regularly smoked PRS with inferior temporal gyrus surface area, precuneus surface area, and cortical thickness, and (5) common substance use genetic factor with surface area of the inferior temporal gyrus, precuneus surface area, pericalcarine surface area, and cortical thickness.
Discussion
Although alcohol, tobacco, and cannabis use have previously been linked to brain structural differences (Lange et al. 2017; Paul and Bhattacharyya 2018; Chye et al. 2019), there is a dearth of work that has examined whether the genetic architecture of substance use overlaps with that of brain structure. The present study examined whether genetic liability for alcohol, tobacco, or cannabis use is associated with cortical brain morphology and probed significant genetic associations by creating SUB PRS and examining their association with brain morphology in a substance use naïve sample. Such work has the potential to shed light onto the genetic and neurobiological precursors of substance use behaviors.
In terms of the relationship between alcohol use and brain morphology, we found a genetic correlation between alcoholic drinks per week and a thinner average cortical thickness. These findings are consistent with previous work indicating that individuals with alcohol dependence display decreased cortical thickness compared to nonalcohol dependent individuals (Fortier et al. 2011). There is some evidence that reduced cortical thickness is associated with characteristics such as poorer executive function, which may exacerbate the risk of drinking more frequently (Burzynska et al. 2012). We also found a genetic correlation between drinks per week and larger cortical surface areas and surface areas of the postcentral gyrus, in line with previous work showing that alcohol abuse is associated with structural differences in the bilateral postcentral gyrus (Jang et al. 2007). Future work should investigate the precise mechanisms that account for the association between alcohol consumption and these brain phenotypes.
Regarding the relationship between smoking phenotypes and neuroanatomical traits, intracranial volume was negatively associated with both smoking initiation (i.e., ever being a regular smoker) and age at smoking initiation. Ever being a regular smoker was (a) positively genetically correlated with the precuneus surface area, and (b) negatively genetically correlated with average cortical thickness and surface area of the inferior temporal gyrus. Previous observational studies have identified a relationship between phenotypic smoking and structural variation in the inferior temporal cortex (a region implicated in object and face recognition) (Conway 2018) and the precuneus (a region involved in motor imagery, directing attention, and processing abstract mental images) (Ogiso et al. 2000). For instance, an increased number of years of smoking has been associated with smaller cortical volumes in the left middle temporal gyrus and right inferior temporal gyrus (Thiel and Fink 2007) and cortical perfusion levels in the left precuneus (Durazzo et al. 2015). These findings suggest that the genetic association between brain morphology and tobacco smoking depends mainly on the age at which the behavior started or having a lifetime history of smoking regularly, rather than on its frequency or being a former smoker.
Lifetime cannabis use was negatively genetically correlated with insula surface area. Individuals who have greater genetic liability for using cannabis may use cannabis more frequently, which has been associated with cortical differences (Chye et al. 2020; De Niz et al. 2020), such as variation in the insula. For example, work by Chye et al. (2019, 2020) indicated that adolescents who used cannabis showed reduced thickness in the bilateral insula, a brain region that has been closely linked to addictive behaviors, including craving and drug-seeking, interoceptive processing, response to reward, and impulsive decision making (Battistella et al. 2014; Naqvi et al. 2014). Other work has shown that individuals who initiate cannabis use had a smaller insula surface area in the right hemisphere (Infante et al. 2018).
The common substance use genetic factor was positively genetically correlated with precuneus surface area and was negatively genetically associated with average cortical thickness and surface areas of the pericalcarine and the inferior temporal gyrus. These findings are in line with the genetic correlations observed between neuroimaging traits and alcohol, tobacco and cannabis use in isolation. However, the correlation with pericalcarine surface area was unique to the common substance use genetic factor. The pericalcarine cortex, which is involved in visual and spatial processing of information (Holmes et al. 2016), has previously been linked to impulsivity and sensation-seeking (Holmes et al. 2016; Kubera et al. 2018) and substance use (Jacobus et al. 2014). A study reported that individuals who use more than one substance (i.e., alcohol and tobacco) had thicker left pericalcarine cortices relative to controls. However, this association did not remain significant upon controlling for alcohol use (Jacobus et al. 2014).
Several substance use PRS were associated with variation in brain structure in the ABCD sample. For example, higher alcohol use PRS were associated with larger postcentral gyrus surface area and cortical surface area, whereas higher genetic propensity to be a regular smoker was positively associated with ICV and inferior temporal gyrus surface area. The fact that these PRS were associated with structural brain morphology differences in a sample of children prior to substance use engagement suggests that these are either shared genetic factors (horizontal pleiotropy) (Solovieff et al. 2013) or potentially causal associations whereby brain morphology differences predispose certain individuals to engage in substance use. For genetic correlations discovered only though LDSC but not observed through PRS, a potential explanation is that substance use behaviors affect the regional brain morphology. As such, these effects would only be observed in samples where there is variance in relation to substance use behaviors, such as the adult discovery samples for substance use and neuroimaging GWAS, but not for the ABCD sample which includes children naïve to substance use.
Limitations regarding the interpretation of the genetic correlations presented here must be acknowledged. These include the potential influence of pleiotropic effects. It is established that genetic variants associated with a given trait can also be related to other attributes as well (Biton et al. 2020). In our case, a potential mediator is that of cognitive ability and educational attainment. Cognitive ability has been robustly linked to increased total surface area and ICV (Cox et al. 2018; Nave et al. 2019; Mitchell et al. 2020), but has also been negatively associated with substance abuse phenotypes (Gustavson et al. 2017; Schepis et al. 2018; Beverly et al. 2019). Recent studies have shown that a genetic predisposition for higher cognitive ability or educational attainment is linked to variation in regional cortical morphology, particularly in the frontal and temporal lobes (Lett et al. 2020; Mitchell et al. 2020). It is thus plausible that the associations we observe may, in part, be mediated by educational attainment—especially as many of the identified regions for both sets of traits are involved in cognitive processing, impulse control, and decision making. For example, one study found that the surface area and thickness of the prefrontal, insula, and medial temporal cortices were significant mediators of the relationship between PRS for intelligence and general cognitive performance in 2 independent cohorts (Lett et al. 2020). Although we cannot exclude the possibility that some of our observations are influenced by cognitive ability, several of our observations are in contrast to what would be expected if the relationship was entirely driven by cognitive ability. For example, we observed a positive genetic correlation between alcohol use and total cortical surface area. Future studies would benefit from examining the relationship among cortical morphology, substance use, and cognitive ability together.
In addition, longitudinal data on substance use or neuroimaging traits were not available; thus, we were unable to investigate the potential effect of prolonged exposure to the substances investigated here. For instance, in analyses where significant genetic correlations were revealed between substance use and neuroimaging traits, it is possible that the associations observed may be due to changes in neural structures as a result of acute or chronic substance use. Indeed, modifications in glial cell number, reductions in the neuronal size and volume of the neuropile, as well as epigenetic alterations may be possible mechanisms through which substance use contributes to changes in brain structure and function (Cecil et al. 2015; Kroenke and Bayly 2018), something the current study was not able to examine. Future longitudinal studies are needed to examine the direction of effects between substance use and brain structure, as well as investigate brain-based pathways and ROI that may influence the gene-substance use relationship.
Nevertheless, this study is one of few studies to examine genetic correlations between tobacco, alcohol, and cannabis use with neuroimaging traits, elucidating the relationship between the genetic architecture of substance use and brain structure. Moreover, to our knowledge, this is one of the only studies to examine whether substance use PRS could be used to predict variation in brain organization in children, elucidating potential neural mechanisms that predispose individuals to engage in substance use. With the increasing emphasis on precision medicine and personalized health initiatives, our work is a first step in helping to elucidate the complex relationship between genes and brain morphology in relation to substance use; however, at this time, clinical or prevention applications are limited as much remains unknown about the biological and molecular pathways through which substance use influences neural structure and vice versa. In terms of next steps, future research should consider examining genetics, brain structure, substance use and related behaviors, and environments over time to help determine the causal relationships among these variables. Such work has the potential to contribute to a better understanding the mechanisms involved in the pathogenesis of substance use, enable the detection of individuals at heightened risk for substance use problems, and aid in more precise diagnosis and treatments.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. To apply and access 23andMe summary statistics, please visit research.23andme.com/dataset-access/ for more information.
Supplementary Material
Contributor Information
Jill A Rabinowitz, Department of Mental Health, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD 21205, USA.
Adrian I Campos, Department of Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, Queensland 4006, Australia; School of Biomedical Sciences, Faculty of Medicine, The University of Queensland, Brisbane, Queensland 4072, Australia.
Jue-Sheng Ong, Department of Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, Queensland 4006, Australia.
Luis M García-Marín, Department of Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, Queensland 4006, Australia; School of Biomedical Sciences, Faculty of Medicine, The University of Queensland, Brisbane, Queensland 4072, Australia.
Sarael Alcauter, Instituto de Neurobiología, Universidad Nacional Autónoma de México, Querétaro 76230, México.
Brittany L Mitchell, Department of Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, Queensland 4006, Australia; School of Biomedical Science, Faculty of Health, Queensland University of Technology (QUT), Brisbane, Queensland 4059, Australia.
Katrina L Grasby, Department of Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, Queensland 4006, Australia.
Gabriel Cuéllar-Partida, The University of Queensland Diamantina Institute, The University of Queensland, Brisbane, Queensland 4102, Australia.
Nathan A Gillespie, Virginia Institute for Psychiatric and Behavior Genetics, Department of Psychiatry, Virginia Commonwealth University, Richmond, VA 23284, USA.
Andrew S Huhn, Department of Psychiatry and Behavioral Sciences, School of Medicine, Baltimore, MD 21205, USA.
Nicholas G Martin, Department of Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, Queensland 4006, Australia.
Paul M Thompson, Imaging Genetics Center, Mark & Mary Stevens Institute for Neuroimaging & Informatics, Keck School of Medicine, University of Southern California, Los Angeles, CA 90007, USA.
Sarah E Medland, Department of Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, Queensland 4006, Australia.
Brion S Maher, Department of Mental Health, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD 21205, USA.
Miguel E Rentería, Department of Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, Queensland 4006, Australia; School of Biomedical Sciences, Faculty of Medicine, The University of Queensland, Brisbane, Queensland 4072, Australia; School of Biomedical Science, Faculty of Health, Queensland University of Technology (QUT), Brisbane, Queensland 4059, Australia.
Funding
The Australian National Health & Medical Research Council and the Australian Research Council through an NHMRC-ARC Dementia Research Development Fellowship (GNT1002821 to M.E.R.). The ABCD Study® is supported by the National Institutes of Health and additional federal partners under award numbers U01DA041048, U01DA050989, U01DA051016, U01DA041022, U01DA051018, U01DA051037, U01DA050987, U01DA041174, U01DA041106, U01DA041117, U01DA041028, U01DA041134, U01DA050988, U01DA051039, U01DA041156, U01DA041025, U01DA041120, U01DA051038, U01DA041148, U01DA041093, U01DA041089, U24DA041123, U24DA041147.
Notes
We thank the research participants and employees of 23andMe for their contribution to this study.
Data used in the preparation of this article were obtained from the Adolescent Brain Cognitive DevelopmentSM (ABCD) Study (https://abcdstudy.org), held in the NIMH Data Archive ((NDA). This is a multisite, longitudinal study designed to recruit more than 10 000 children age 9–10 and follow them over 10 years into early adulthood.
A full list of supporters is available at https://abcdstudy.org/federal-partners.html. A listing of participating sites and a complete listing of the study investigators can be found at https://abcdstudy.org/consortium_members/. ABCD consortium investigators designed and implemented the study and/or provided data but did not necessarily participate in the analysis or writing of this report. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or ABCD consortium investigators.
The ABCD data repository grows and changes over time. The ABCD data used in this report came from DOI: 10.15154/1522627.
References
- Adams HH, Hibar DP, Chouraki V, Stein JL, Nyquist PA, Rentería ME, Trompet S, Arias-Vasquez A, Seshadri S, Desrivières S, et al. 2016. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat Neurosci. 19:1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistella G, Fornari E, Annoni JM, Chtioui H, Dao K, Fabritius M, Favrat B, Mall JF, Maeder P, Giroud C. 2014. Long-term effects of cannabis on brain structure. Neuropsychopharmacology. 39:2041–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini U, Hochberg Y. 1995. Controlling the first discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Series B. 57:289–300. [Google Scholar]
- Beverly HK, Castro Y, Opara I. 2019. Age of first marijuana use and its impact on education attainment and employment status. J Drug Issues. 49:228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton A, Traut N, Poline JB, Aribisala BS, Bastin ME, Bulow R, Cox SR, Deary IJ, Fukunaga M, Grabe HJ, et al. 2020. Polygenic architecture of human neuroanatomical diversity. Cereb Cortex. 30:2307–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neal BM, Schizophrenia Working Group of the Psychiatric Genomics Consortium . 2015a. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, Perry JR, Patterson N, Robinson EB, et al. 2015b. An atlas of genetic correlations across human diseases and traits. Nat Genet. 47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzynska AZ, Nagel IE, Preuschhof C, Gluth S, Backman L, Li SC, Lindenberger U, Heekeren HR. 2012. Cortical thickness is linked to executive functioning in adulthood and aging. Hum Brain Mapp. 33:1607–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil CAM, Walton E, Viding E. 2015. DNA methylation, substance use and addiction: a systematic review of recent animal and human research from a developmental perspective. Curr Addict Rep. 2:331–346. [Google Scholar]
- Cho Y, Haycock PC, Sanderson E, Gaunt TR, Zheng J, Morris AP, Davey Smith G, Hemani G. 2020. Exploiting horizontal pleiotropy to search for causal pathways within a Mendelian randomization framework. Nat Commun. 11:1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chye Y, Kirkham R, Lorenzetti V, McTavish E, Solowij N, Yucel M. 2020. Cannabis, cannabinoids, and brain morphology: a review of the evidence. Biol Psychiatry Cogn Neurosci Neuroimaging. 6:627–635. [DOI] [PubMed] [Google Scholar]
- Chye Y, Gutman BA, Mackey S, Ching CRK, Batalla A, Blaine S, Brooks S, Caparelli EC, Cousijn J, Dagher A, et al. 2019. Subcortical surface morphometry in substance dependence: an ENIGMA addiction working group study. Addict Biol. 25:e12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway BR. 2018. The organization and operation of inferior temporal cortex. Annu Rev Vis Sci. 4:381–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox SR, Bastin ME, Ritchie SJ, Dickie DA, Liewald DC, Maniega SM, Redmond P, Royle NA, Pattie A, Hernández MV. 2018. Brain cortical characteristics of lifetime cognitive ageing. Brain Struct Funct. 223:509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson AK, Wennberg P, Hibell B, Romelsjo A. 2012. Alcohol use, heavy episodic drinking and subsequent problems among adolescents in 23 European countries: does the prevention paradox apply? Addiction. 107:71–80. [DOI] [PubMed] [Google Scholar]
- De Niz M, Kehrer J, Brancucci NMB, Moalli F, Reynaud EG, Stein JV, Frischknecht F. 2020. 3D imaging of undissected optically cleared Anopheles stephensi mosquitoes and midguts infected with plasmodium parasites. PLoS One. 15:e0238134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durazzo TC, Meyerhoff DJ, Murray DE. 2015. Comparison of regional brain perfusion levels in chronically smoking and non-smoking adults. Int J Environ Res Public Health. 12:8198–8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott LT, Sharp K, Alfaro-Almagro F, Shi S, Miller KL, Douaud G, Marchini J, Smith SM. 2018. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 562:210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier CB, Leritz EC, Salat DH, Venne JR, Maksimovskiy AL, Williams V, Milberg WP, McGlinchey RE. 2011. Reduced cortical thickness in abstinent alcoholics and association with alcoholic behavior. Alcohol Clin Exp Res. 35:2193–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Li S, Lazaris C, Taylor A, Ziv N, Ramjeawan C, Paaby AB, Siegal ML. 2020. Extent and context dependence of pleiotropy revealed by high-throughput single-cell phenotyping. PLoS Biol. 18:e3000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie NA, Neale MC, Bates TC, Eyler LT, Fennema-Notestine C, Vassileva J, Lyons MJ, Prom-Wormley EC, McMahon KL, Thompson PM, et al. 2018. Testing associations between cannabis use and subcortical volumes in two large population-based samples. Addiction. 113(9):1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasby KL, Jahanshad N, Painter JN, Colodro-Conde L, Bralten J, Hibar DP, Lind PA, Pizzagalli F, Ching CRK, McMahon MAB, et al. 2020. The genetic architecture of the human cerebral cortex. Science. 367:eaay6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotzinger AD, Rhemtulla M, de Vlaming R, Ritchie SJ, Mallard TT, Hill WD, Ip HF, Marioni RE, McIntosh AM, Deary IJ, et al. 2019. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 3:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavson DE, Stallings MC, Corley RP, Miyake A, Hewitt JK, Friedman NP. 2017. Executive functions and substance use: relations in late adolescence and early adulthood. J Abnorm Psychol. 126:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin DS, Sarvet AL, Cerda M, Keyes KM, Stohl M, Galea S, Wall MM. 2017. US adult illicit cannabis use, cannabis use disorder, and medical marijuana laws: 1991-1992 to 2012-2013. JAMA Psychiat. 74:579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Adams HHH, Jahanshad N, Chauhan G, Stein JL, Hofer E, Renteria ME, Bis JC, Arias-Vasquez A, Ikram MK, et al. 2017. Novel genetic loci associated with hippocampal volume. Nat Commun. 8:13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AJ, Hollinshead MO, Roffman JL, Smoller JW, Buckner RL. 2016. Individual differences in cognitive control circuit anatomy link sensation seeking, impulsivity, and substance use. J Neurosci. 36:4038–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante M, Courtney KE, Castro N, Squeglia LM, Jacobus J. 2018. Adolescent brain surface area pre- and post-cannabis and alcohol initiation. J Stud Alcohol Drugs. 79:835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobus J, Squeglia LM, Sorg SF, Nguyen-Louie TT, Taper SF. 2014. Cortical thickness and neurocognition in adolescent marijuana and alcohol users following 28 days of monitored abstinence. J Stud Alcohol Drugs. 75:729–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang DP, Namkoong K, Kim JJ, Park S, Kim IY, Kim SI, Kim YB, Cho ZH, Lee E. 2007. The relationship between brain morphometry and neuropsychological performance in alcohol dependence. Neurosci Lett. 428:21–26. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Myers J, Prescott CA. 2007. Specificity of genetic and environmental risk factors for symptoms of cannabis, cocaine, alcohol, caffeine, and nicotine dependence. Arch Gen Psychiatry. 64:1313–1320. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Ohlsson H, Karriker-Jaffe KJ, Sundquist J, Sundquist K. 2017. Social and economic consequences of alcohol use disorder: a longitudinal cohort and co-relative analysis. Psychol Med. 47:925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremen WS, Fennema-Notestine C, Eyler LT, Panizzon MS, Chen CH, Franz CE, Lyons MJ, Thompson WK, Dale AM. 2013. Genetics of brain structure: contributions from the Vietnam era twin study of aging. Am J Med Genet B Neuropsychiatr Genet. 162B:751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremen WS, Prom-Wormley E, Panizzon MS, Eyler LT, Fischl B, Neale MC, Franz CE, Lyons MJ, Pacheco J, Perry ME, et al. 2010. Genetic and environmental influences on the size of specific brain regions in midlife: the VETSA MRI study. NeuroImage. 49:1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke CD, Bayly PV. 2018. How forces fold the cerebral cortex. J Neurosci. 38:767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubera KM, Schmitgen MM, Maier-Hein KH, Thomann PA, Hirjak D, Wolf RC. 2018. Differential contributions of cortical thickness and surface area to trait impulsivity in healthy young adults. Behav Brain Res. 350:65–71. [DOI] [PubMed] [Google Scholar]
- Lange EH, Nerland S, Jorgensen KN, Morch-Johnsen L, Nesvag R, Hartberg CB, Haukvik UK, Osnes K, Melle I, Andreassen OA, et al. 2017. Alcohol use is associated with thinner cerebral cortex and larger ventricles in schizophrenia, bipolar disorder and healthy controls. Psychol Med. 47:655–668. [DOI] [PubMed] [Google Scholar]
- Lett TA, Vogel BO, Ripke S, Wackerhagen C, Erk S, Awasthi S, Trubetskoy V, Brandl EJ, Mohnke S, Veer IM. 2020. Cortical surfaces mediate the relationship between polygenic scores for intelligence and general intelligence. Cereb Cortex. 30:2708–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu B, Banaschewski T, Bokde ALW, Quinlan EB, Desrivieres S, Flor H, Frouin V, Garavan H, et al. 2020. Orbitofrontal cortex volume links polygenic risk for smoking with tobacco use in healthy adolescents. Psychol Med. 1–8. [DOI] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, et al. 2016. Genetic imaging consortium for addiction medicine: from neuroimaging to genes. Prog Brain Res. 224:203–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey S, Kan KJ, Chaarani B, Alia-Klein N, Batalla A, Brooks S, Cousijn J, Dagher A, de Ruiter M, Desrivieres S, et al. 2016. Genetic imaging consortium for addiction medicine: From neuroimaging to genes. Prog Brain Res. 224:203–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher BS. 2015. Polygenic scores in epidemiology: risk prediction, etiology, and clinical utility. Curr Epidemiol Rep. 2:239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammen G, Rueda S, Roerecke M, Bonato S, Lev-Ran S, Rehm J. 2018. Association of cannabis with long-term clinical symptoms in anxiety and mood disorders: a systematic review of prospective studies. J Clin Psychiatry. 79:e1-e12. [DOI] [PubMed] [Google Scholar]
- Mitchell BL, Cuéllar-Partida G, Grasby KL, Campos AI, Strike LT, Hwang L-D, Okbay A, Thompson PM, Medland SE, Martin NG, et al. 2020. Educational attainment polygenic scores are associated with cortical total surface area and regions important for language and memory. NeuroImage. 212:116691. [DOI] [PubMed] [Google Scholar]
- Naqvi NH, Gaznick N, Tranel D, Bechara A. 2014. The insula: a critical neural substrate for craving and drug seeking under conflict and risk. Ann N Y Acad Sci. 1316:53–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarri X, Afzali MH, Lavoie J, Sinha R, Stein DJ, Momenan R, Veltman DJ, Korucuoglu O, Sjoerds Z, van Holst RJ, et al. 2020. How do substance use disorders compare to other psychiatric conditions on structural brain abnormalities? A cross-disorder meta-analytic comparison using the ENIGMA consortium findings. Hum Brain Mapp. 10.1002/hbm.25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave G, Jung WH, Karlsson Linnér R, Kable JW, Koellinger PD. 2019. Are bigger brains smarter? Evidence from a large-scale preregistered study. Psychol Sci. 30:43–54. [DOI] [PubMed] [Google Scholar]
- Ogiso T, Kobayashi K, Sugishita M. 2000. The precuneus in motor imagery. Neuroreport. 11:1345–1349. [DOI] [PubMed] [Google Scholar]
- Ohi K, Shimada T, Kataoka Y, Yasuyama T, Kawasaki Y, Shioiri T, Thompson PM. 2020. Genetic correlations between subcortical brain volumes and psychiatric disorders. Br J Psychiatry. 216:280–283. [DOI] [PubMed] [Google Scholar]
- Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL, Abdellaoui A, Nivard MG, Baselmans BML, Ong JS, et al. 2018. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci. 21:1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Bhattacharyya S. 2018. Does thinner right entorhinal cortex underlie genetic liability to cannabis use? Psychol Med. 48:2766–2775. [DOI] [PubMed] [Google Scholar]
- Renteria ME, Hansell NK, Strike LT, McMahon KL, de Zubicaray GI, Hickie IB, Thompson PM, Martin NG, Medland SE, Wright MJ. 2014. Genetic architecture of subcortical brain regions: common and region-specific genetic contributions. Genes Brain Behav. 13:821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satizabal CL, Adams HHH, Hibar DP, White CC, Knol MJ, Stein JL, Scholz M, Sargurupremraj M, Jahanshad N, Roshchupkin GV, et al. 2019. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat Genet. 51:1624–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepis TS, Teter CJ, McCabe SE. 2018. Prescription drug use, misuse and related substance use disorder symptoms vary by educational status and attainment in US adolescents and young adults. Drug Alcohol Depend. 189:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. 2013. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 14:483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spechler PA, Allgaier N, Chaarani B, Whelan R, Watts R, Orr C, Albaugh MD, D'Alberto N, Higgins ST, Hudson KE, et al. 2019. The initiation of cannabis use in adolescence is predicted by sex-specific psychosocial and neurobiological features. Eur J Neurosci. 50:2346–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swagerman SC, Brouwer RM, de Geus EJ, Hulshoff Pol HE, Boomsma DI. 2014. Development and heritability of subcortical brain volumes at ages 9 and 12. Genes Brain Behav. 13:733–742. [DOI] [PubMed] [Google Scholar]
- Thiel CM, Fink GR. 2007. Visual and auditory alertness: modality-specific and supramodal neural mechanisms and their modulation by nicotine. J Neurophysiol. 97:2758–2768. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Andreassen OA, Arias-Vasquez A, Bearden CE, Boedhoe PS, Brouwer RM, Buckner RL, Buitelaar JK, Bulayeva KB, Cannon DM, et al. 2017. ENIGMA and the individual: Predicting factors that affect the brain in 35 countries worldwide. Neuroimage. 145:389–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Andreassen OA, Arias-Vasquez A, Bearden CE, Boedhoe PS, Brouwer RM, Buckner RL, Buitelaar JK, Bulayeva KB, Cannon DM, et al. 2020. ENIGMA and global neuroscience: a decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl Psychiatry. 10:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee SJ, Knol MJ, Chauhan G, Satizabal CL, Smith AV, Hofer E, Bis JC, Hibar DP, Hilal S, van den Akker EB, et al. 2019. A genome-wide association study identifies genetic loci associated with specific lobar brain volumes. Commun Biol. 2:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Koob GF, Croyle RT, Bianchi DW, Gordon JA, Koroshetz WJ, Perez-Stable EJ, Riley WT, Bloch MH, Conway K, et al. 2018. The conception of the ABCD study: from substance use to a broad NIH collaboration. Dev Cogn Neurosci. 32:4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB, Heath AC, Madden PAF, Landers JG, Martin NG. 2018. Effects of high alcohol intake, alcohol-related symptoms and smoking on mortality. Addiction. 113:158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. 2010. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. To apply and access 23andMe summary statistics, please visit research.23andme.com/dataset-access/ for more information.


