Highlights
-
•
SSTs in UK shellfish harvesting waters are 18–22 °C for 8 consecutive weeks.
-
•
Increasing SSTs do not necessarily drive growth of V. parahaemolyticus in shellfish.
-
•
Whole genome sequencing identifies V. rotiferianus and V. jasicida in shellfish.
-
•
V. jascinda and V. rotiferianus show pathogenicity in insect infection model.
Keywords: Vibrio species, Vibrio jasicida, Vibrio rotiferianus, Vibrio parahaemolyticus, Shellfish, Sea-surface temperature
Abstract
Warming sea-surface temperature has led to an increase in the prevalence of Vibrio species in marine environments. This can be observed particularly in temperate regions where conditions for their growth has become more favourable. The increased prevalence of pathogenic Vibrio species has resulted in a worldwide surge of Vibriosis infections in human and aquatic animals. This study uses sea-surface temperature data around the English and Welsh coastlines to identify locations where conditions for the presence and growth of Vibrio species is favourable. Shellfish samples collected from three locations that were experiencing an increase in sea-surface temperature were found to be positive for the presence of Vibrio species. We identified important aquaculture pathogens Vibrio rotiferianus and Vibrio jasicida from these sites that have not been reported in UK waters. We also isolated human pathogenic Vibrio species including V. parahaemolyticus from these sites. This paper reports the first isolation of V. rotiferianus and V. jasicida from UK shellfish and highlights a growing diversity of Vibrio species inhabiting British waters.
1. Introduction
The aquaculture industry provides an increasing contribution to global food supplies; however, it is faced with challenges from a changing climate, globalisation, ecological constraints, and human population changes (both growth and food preference). The UK shellfish industry is worth over £350 million annually (UK Sea Fisheries Statistics, 2019), providing an important contribution to the UK aquaculture sector and economy. The farming of molluscan shellfish in nearshore ecosystems accounts for a large portion of the shellfish industry. Molluscan shellfish are filter feeders i.e., they pass water across their gills, filtering and selecting particles for consumption, including algae and bacteria. Consequently, pathogenic bacteria, including Vibrio species, can accumulate to levels where the shellfish become unsafe to eat. In the United States, 76% of seafood-associated infections, between 1973 and 2006, were due to bacteria (Iwamoto et al., 2010). In previous studies, Vibrio parahaemolyticus levels have been shown to be up to five times higher within crab tissues than in the surrounding water (Wagley et al., 2009) indicating that in some ecological habitats, Vibrio species are extremely abundant within shellfish (Parveen et al., 2008; Wagley et al., 2009). There is evidence that the increased global incidence of Vibriosis (any disease caused by a Vibrio species) is linked to climate change, with altered patterns of bacterial survival in shellfish (Baker-Austin et al., 2013; Costello et al., 2009; Froelich and Daines, 2020; Leddin and Macrae, 2020; Lindgren et al., 2012; Martinez-Urtaza et al., 2010; Takemura et al., 2014). Consequently, global increases in sea-surface temperatures in nearshore ecosystems can drive the emergence of a range of Vibrio species including pathogens of humans, marine finfish, shellfish and crustaceans in regions where this has not been previously seen (Baker-Austin et al., 2013; Froelich and Daines, 2020; Lindgren et al., 2012; Takemura et al., 2014).
Vibrio species can cause disease in shellfish in both aquaculture settings and the natural environment. These Vibrio species include V. aestuarianus, V. crassostreae and V. coralliilyticus which are responsible for the sharp decline of Pacific oyster numbers (Crassostrea gigas), resulting in serious socioeconomic losses in Europe, New Zealand and Australia (European Commission, 2015; de Lorgeril et al. 2018; Elston et al., 2008; Goudenege et al., 2015). Members of the V. harveyi clade such as V. campbelli and V. rotiferianus can also cause disease in marine fish and shrimp (European Commission, 2015; Austin and Zhang, 2006; Bruto et al., 2017; de Lorgeril et al. 2018; Elston et al., 2008; Goudenege et al., 2015; Travers et al., 2015). The poleward spread of human pathogenic Vibrio species, such as V. parahaemolyticus and V. vulnificus, is concurrent with increasing global temperature and reduced sea surface salinity, contributing to an increase in human disease burden globally (Baker-Austin et al., 2010, 2013; Martinez-Urtaza et al., 2010). Outbreaks of Vibriosis in humans have recently been reported in regions of the world previously considered to have temperate or cold climates including Peru (during its austral winter) (Fuenzalida et al., 2007), Chile (Gonzalez-Escalona et al., 2005), Alaska (McLaughlin et al., 2005), Denmark (Dalsgaard et al., 1996), Spain (Martinez-Urtaza et al., 2005, 2018) and Pacific Northwest USA (Martinez-Urtaza et al., 2013). Collectively these findings support the hypothesis that Vibrio-associated diseases are increasing and are influenced by the rise in global sea temperature.
There is a strong seasonal pattern related to temperature and abundance of Vibrio species (Kokashvili et al., 2015; Vezzulli et al., 2016). Nearly all Vibrio species can grow at temperatures between 13 and 22 °C and where salinity ranges from 5 to 25 parts per thousand (ppt) (Kaspar and Tamplin, 1993). Under these conditions, most Vibrio species will proliferate in shellfish and this coincides with elevated disease burden (Ansede-Bermejo et al., 2010; Joseph et al., 1982; McLaughlin et al., 2005; Mead et al., 1999). In the UK, Vibrio species are detected in domestically grown shellfish from May – October, with peak levels occurring at the end of August and the start of September (Ford et al., 2020a; Powell et al., 2013; Wagley et al., 2009, 2008). In this study, sea-surface temperature data were analysed to identify the locations where Vibrio species were most likely to be found around the English and Welsh coastlines. Shellfish samples were then sourced from these areas, and from a nearby region of cooler waters. The prevalence of Vibrio species in shellfish from Chichester Harbour, Osea Island, Whitstable Bay and Lyme Bay in the summer of 2018 was determined using culture-based methods. Several strains isolated from these sites were genome sequenced in order to ascertain their taxonomy and investigate their genomics.
2. Results
2.1. Sea-surface temperature at different sites in England and Wales
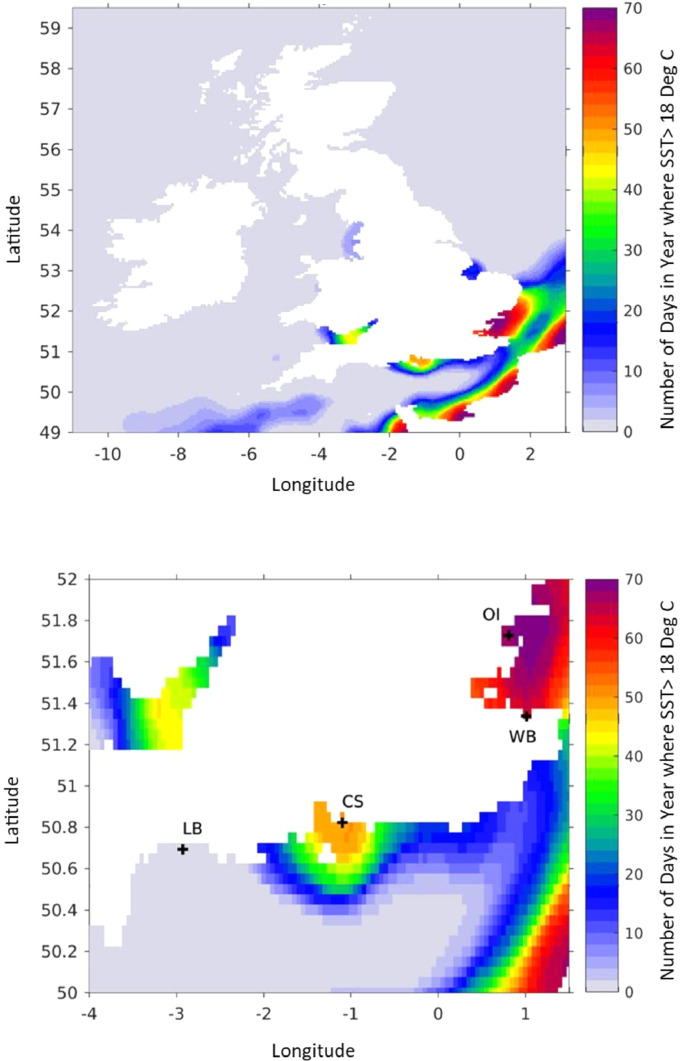
Sea-surface temperature data from the global Operational Sea-Surface Temperature and Sea Ice Analysis (OSTIA) system (Donlon et al., 2012) was used to study the sea-surface temperature around England and Wales, indicated in Figs. 1 and 2. Using data from 2015 to 2017, a map of the average number of days per year when the sea-surface temperature is warmer than 18 °C was produced (Fig. 1a). Several regions in England and Wales where commercial shellfisheries exist were analysed further (Figs. 1 and 2). The time series sea-surface temperature for these locations are in Fig. 2 and a map of location of these sites is shown in Supplementary Data 1. We identified Chichester Harbour, Whitstable Bay and Osea Island as areas were sea-surface temperatures were favourable for Vibrio Species presence. Lyme Bay was identified as areas where sea-surface temperature were lower and unfavourable for growth of Vibrio species. Fig. 2 shows the time series of daily sea-surface temperature at these four locations between 2015 and 2018 as well as several other sites in Britain and Wales where commercial shellfisheries are based.
Fig. 1.
Average number of days per year when the sea-surface temperature is above 18 deg C. Fig. 1a is a focus on Britain and Ireland, while Fig. 1b is focus on southern England with the locations of Lyme Bay (LB), Chichester/Solent (CS), Osea Island (OI) and Whitstable Bay (WB) marked with crosses. Both figures are calculated as a mean over the 3 years 2015–2017 using OSTIA analyses.
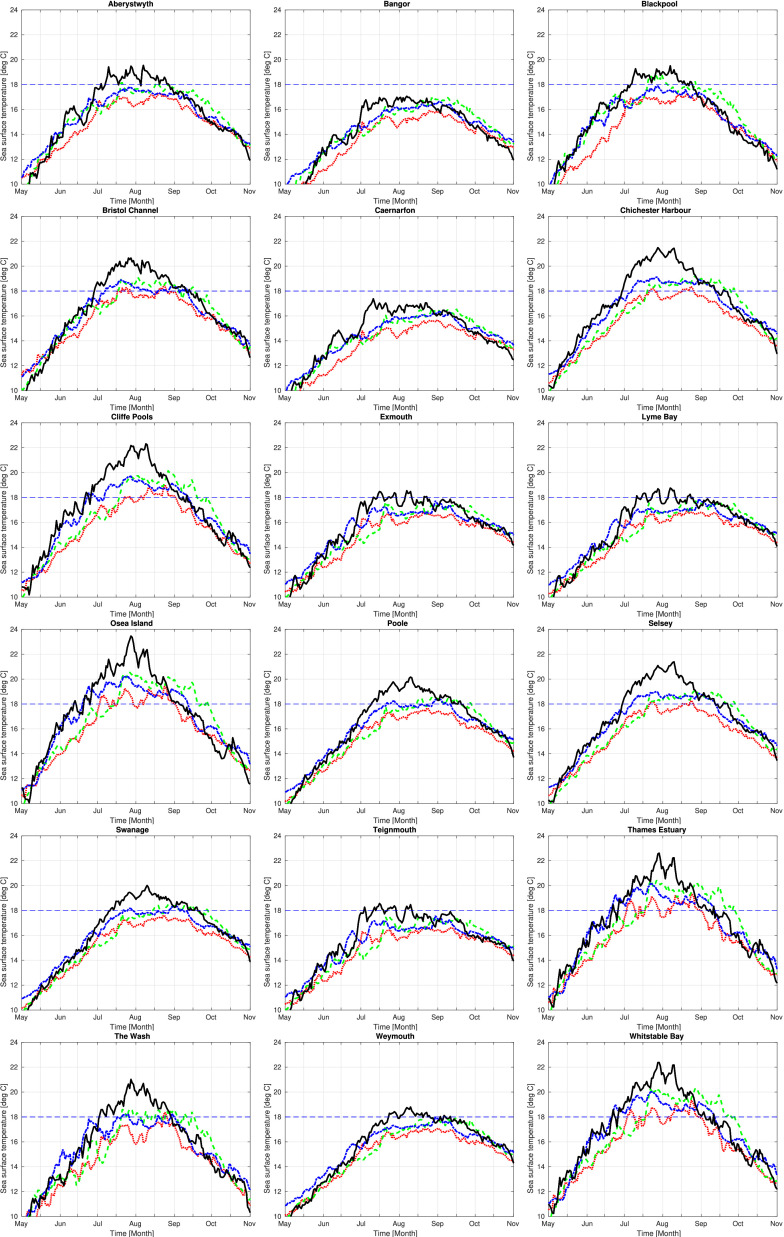
Fig. 2.
Time series of sea-surface temperature data [deg C] (at foundation depth, from OSTIA) from 2015 to 2018 in England and Wales. For 2015 (red dotted), 2016 (green dashed), 2017 (blue dash-dot) and 2018 (black solid).
The sea-surface temperature at Whitstable Bay, Osea Island and Chichester Harbour during summer 2015 (June–August) was around 18 °C but peaked around 22 °C in these areas in 2018 (Fig. 2). In Lyme Bay, the sea-surface temperature remained stable, peaking at around ∼18 °C during the summer months, during this four year period (Fig. 2). The time series data plotted for Whitstable Bay, Osea Island, Chichester Harbour, Selsey, Thames Estuary and Cliff Pools show that in 2015 the temperature occasionally exceeded 18 °C (Fig. 2). Between 2016 and 2018 the sea-surface temperature at these sites had exceeded 18 °C and remained above this temperature for eight weeks or more consecutively. Although the observed the sea-surface temperature has increased in the areas mentioned above over the four-year period, the sea-surface temperature is variable around the coast of England and Wales and there is natural variability from year to year.
2.2. Vibrio abundance peaks in July and August
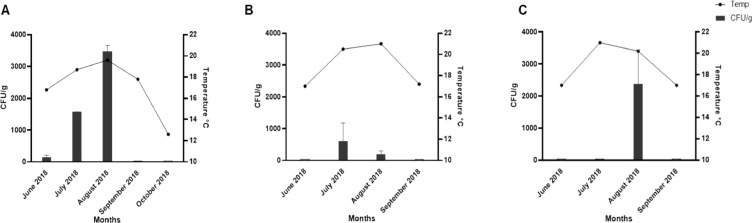
Shellfish samples from Osea Island, Whitstable Bay, Chichester Harbour and Lyme Bay were examined from June-October in 2018 and total Vibrio species were recorded from plate counts of sucrose-positive and negative colonies on thiosulphate citrate bile sucrose agar (TCBS) plates before a further enrichment step. For depurated Pacific oysters from Whitstable Bay, the total numbers of Vibrio species peaked in July to 603 CFU/g (Fig. 3B, Table 1). For depurated Pacific oysters from the Osea Island area, the total numbers of Vibrio species peaked in August at 2384 CFU/g (Fig. 3C). Non-depurated blue mussels (Mytilus edulis) from Lyme Bay, where the sea-surface temperature only briefly rose above 18 °C (Fig. 2, Table 1), were also tested and found to be negative for Vibrio species between June and September 2018 unless an enrichment step was first performed. Human pathogenic Vibrio species such as V. parahaemolyticus and V. vulnificus were not identified from samples from Osea Island, Whitstable Bay or Lyme Bay.
Fig. 3.
Total number of Vibrio species detected using direct plate enumeration from shellfish samples taken from a) Chichester Harbour b) Whitstable Bay c) Osea Island versus sea surface temperature. Samples were tested in duplicate (n = 2) and the average of both experiments is represented in datasets. Temperature data for Chichester Harbour is taken at the time of sampling at the site. Temperature taken for Whitstable Bay and Osea Island are taken from time series OSTIA sea-surface temperature values using a satellite/buoy composite.
Table 1.
Shellfish samples tested in this study.
| Sample number | Date of isolation | Sitea | Typeb | Total Vibrio species detected CFU/g | Vibrio species isolated |
|---|---|---|---|---|---|
| 01/2018 | June | B | A | < 10 | V. alginolyicus |
| 02/2018 | June | C | A | < 10 | V. alginolyicus |
| 03/2018 | June | D | B | 0 | ND |
| 04/2018 | June | A | C | 140 | V. parahaemolyticus, V. alginolyicus |
| 05/2018 | July | A | C | 1573 | Photobacterium, V. jasicida, V. rotiferianus, V. alginolyicus |
| 06/2018 | July | B | A | 603 | V. jasicida, V. alginolyicus |
| 07/2018 | July | C | A | 15 | V. jasicida, V. alginolyicus |
| 08/2018 | July | D | B | 0 | ND |
| 09/2018 | August | A | C | 3480 | V. rotiferianus, V. jasicida, V. alginolyicus |
| 10/2018 | August | B | A | 198 | Photobacterium, V. rotiferianus, V. alginolyicus |
| 11/2018 | August | C | A | 2385 | V. jasicida, V. alginolyicus |
| 12/2018 | August | D | B | 0 | ND |
| 13/2018 | September | A | C | 25 | V. rotiferianus, V. jasicida, V. alginolyicus |
| 14/2018 | September | B | A | < 10 | V. alginolyicus |
| 15/2018 | September | C | A | < 10 | V. jasicida, V. alginolyicus |
| 16/2018 | October | A | C | < 10 | V. jasicida, V. alginolyicus |
: Location of samples A – Chichester Harbour, B – Whitstable Bay, C – Osea Island, D – Lyme Bay,.
: Type of sample A – Crassostrea gigas, B – Mytilus edulis, C – Ostrea edulis
ND – Not determined.
For non-depurated European flat oysters (Ostrea edulis) samples from Chichester Harbour the total number of Vibrio species isolated in June when the temperature taken at site was an average of ∼16–17 °C, was < 140 CFU/g (Fig. 3 and Table 1). This increased to 3480 CFU/g in shellfish taken in August 2018, when the average temperature at the site was ∼20 °C (Fig. 3). A seasonal relationship was seen between increasing temperature and the total number of Vibrio species present in samples tested from Chichester Harbour for the five months of testing (r value of 0.90 using Spearman's Rho correlation) (Fig. 3a). V. parahaemolyticus was present at low levels in shellfish sample taken in June (sample EXE 04/2018 < 20 CFU/g) and representative isolates were confirmed as V. parahaemolyticus by PCR using the species marker toxR (Kim et al., 1999). The representative isolates tested were all negative for the TDH and TRH haemolysins known to be associated with virulence as determined by PCR for tdh and trh genes (Tada et al., 1992).
2.3. Biochemical characteristics of sucrose negative strains
One aim of this project was to measure the levels of the human pathogenic Vibrio species (V. parahaemolyticus or V. vulnificus) in shellfish from Whitstable Bay, Osea Island, and Chichester Harbour. Sucrose negative colonies on TCBS were further identified as presumptive V. parahaemolyticus or V vulnificus. Sucrose negative colonies were found in 10 of the 16 shellfish samples from Whitstable Bay (n = 2), Osea Island (n = 3), Lyme Bay (n = 0) and Chichester Harbour (n = 5). However, only one sample (EXE 04/2018) from Chichester Harbour, was positively identified further as V. parahaemolyticus (strain VP 04/2018). All other sucrose negative isolates were negative by PCR for V. parahaemolyticus species marker toxR (Kim et al., 1999) and tlh (Taniguchi et al., 1985) and V. vulnificus species marker vvhA (Hill et al., 1991). These sucrose negative isolates were Gram negative, oxidase positive, positive for fermentation of glucose, were not able to grow in 0% sodium chloride solution and grew well in solution containing > 2% sodium chloride. The results show that these isolates belong to the Vibrionaceae family.
The sucrose negative colonies on TCBS agar fell broadly into three groups depending on the biochemical tests applied. One group of isolates, from the Chichester Harbour and Whitstable Bay samples (EXE 05/2018, EXE 09/2018, EXE 10/2018 and EXE 13/2018) were positive for the production of indole from tryptophan and positive for the fermentation amygdalin (glycoside) (Table 2). These isolates were designated as Group A. Another group of sucrose negative colonies were designated Group B and were negative for indole production and negative for the fermentation of amygdalin. Group B isolates came from samples collected from Chichester Harbour and Osea Island. Finally, Group C included two isolates (EXE 05/2018 and EXE 10/2018) that were positively identified as Photobacterium species with a 99.8% API identification.
Table 2.
Sequenced strains used in this study.
| Strain number | Species | PCR tests |
Biochemical tests |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | ToxR | Tlh | TDH | TRH | Vvh | Oxidase | Glucose | Indole | Amygdalin | ||
| 04/2018 | V. parahaemolyticus | – | + | + | – | – | – | + | + | + | + |
| 05/2018–4 | Photobacterium Species | – | – | – | – | – | – | + | + | – | – |
| 05/2018–10 | V. jasicida | B | – | – | – | – | – | + | + | – | – |
| 05/2018–5 | V. rotiferianus | A | – | – | – | – | – | + | + | + | + |
| 07/2018–1 | V. jasicida | B | – | – | – | – | – | + | + | – | – |
| 07/2018–7 | V. jasicida | B | – | – | – | – | – | + | + | – | – |
| 09/2018–4 | V. jasicida | B | – | – | – | – | – | + | + | – | – |
| 09/2018–7 | V. rotiferianus | A | – | – | – | – | – | + | + | + | + |
| 09/2018 - 11 | V. rotiferianus | A | – | – | – | – | – | + | + | + | + |
| 10/2018–1 | V. rotiferianus | A | – | – | – | – | – | + | + | + | + |
| 10/2018–4 | V. rotiferianus | A | – | – | – | – | – | + | + | + | + |
| 16/2018–1 | V. jasicida | B | – | – | – | – | – | + | + | – | – |
| 16/2018–2 | V. jasicida | B | – | – | – | – | – | + | + | – | – |
| 16/2018–3 | V. jasicida | B | – | – | – | – | – | + | + | – | – |
2.4. Whole genome sequencing analysis identifies V. rotiferianus and V. jasicida present in shellfish samples
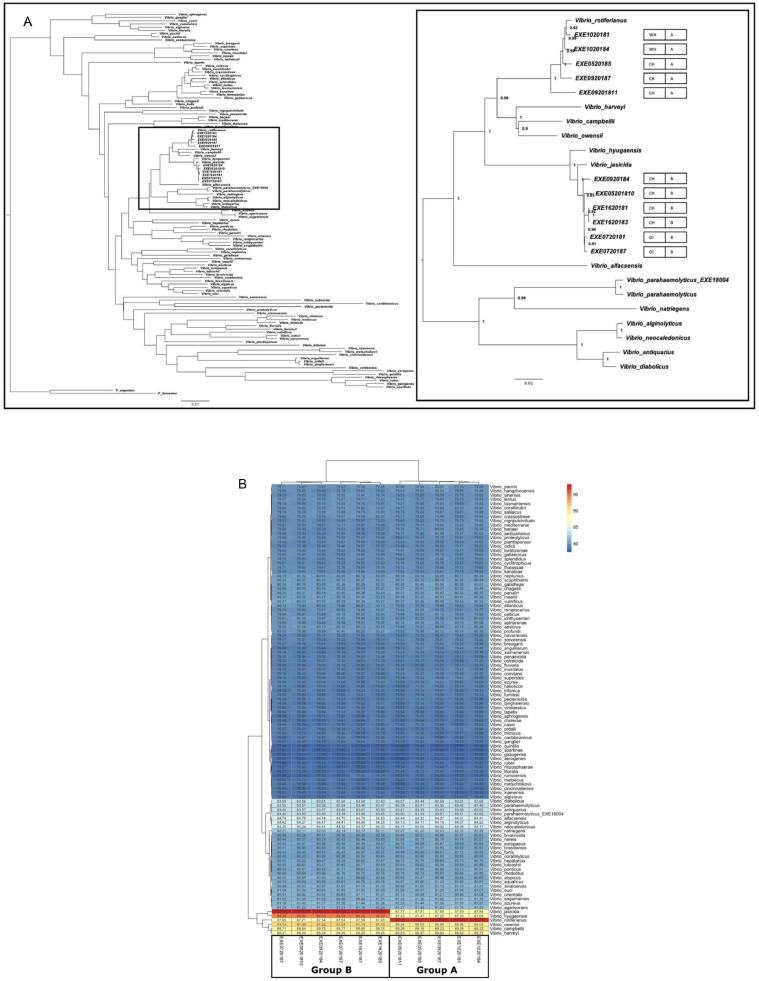
Whole genome sequencing was carried out on 14 of the isolates collected. EXE 04/2018 (thought to be V. parahaemolyticus), five representative Vibrio isolates from Group A, six representative Vibrio isolates from Group B (Table 2) and one representative Photobacterium isolate from Group C). Initial MLSA results confirmed EXE 04/2018 as V. parahaemolyticus (Witherall et al., 2019) and indicated that Group A and Group B isolates fell into the V. harveyi clade grouping with V. rotiferianus (Group A) and V. jasicida (Group B). To confirm the identify of Group A and Group B to species level, average nucleotide identity (ANI) was calculated between these strains and publicly available Vibrio strains (Supplementary Data 2) (Fig. 4a). The highest ANI was observed between the isolates of Group A and V. rotiferianus B64D1 (97%) and secondly with Group B and V. jasicida 090810c (97%), both of which are above the hypothesised species delineation threshold value of 95% (Richter and Rossello-Mora, 2009). The MLSA and ANI analyses confirm that isolates sequenced in Group A were V. rotiferianus while isolates sequenced in Group B were V. jasicida.
Fig. 4.
MLSA phylogeny of Group A and Group B strains. A - Multi Locus sequence analysis to determine the phylogenetic positions of the sequenced Vibrio isolates from Group A and Group B. Neighbour joining tree was constructed based on alignments of homologous sequences of 119 conserved ORFs. Scale bar represents 0.07 substitutions per site. B – Average nucleotide (ANI) analysis between genomes, with highest ANI observed for Group A strains as V. rotiferianus (>97%) and Group B as V. jasicida (> 97%).
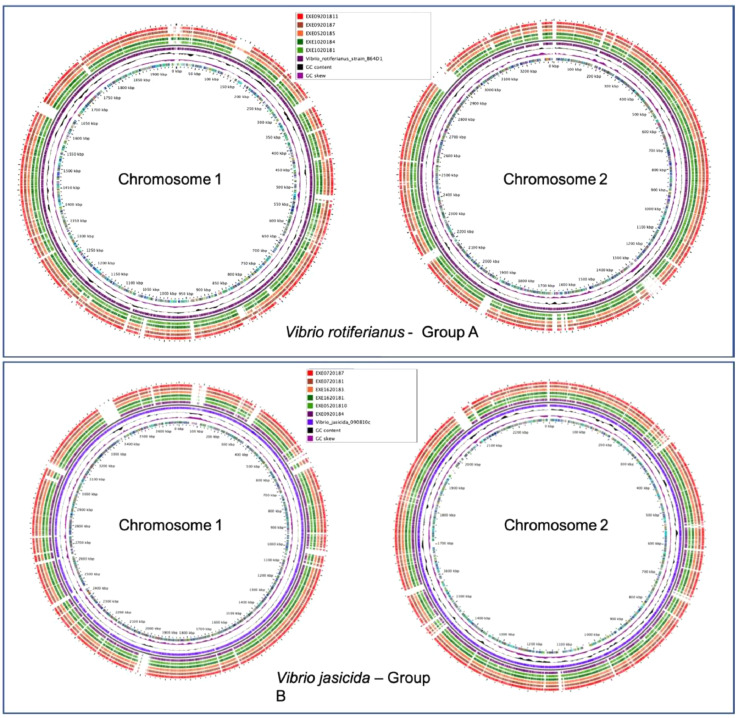
Fig. 5a shows circular genome maps comparing the assemblies of each sequenced V. rotiferianus stains in Group A aligned against that of V. rotiferianus B64D1 (GenBank accession number: CP018311.1) while Fig. 5b shows a circular genome map comparing assemblies of each sequenced V. jasicida stain in Group B aligned against that of V. jasicida 090810c (Genbank accession number GCA_002887615.1). These alignments both agree with the MLSA and ANI results. They show there is a high level of similarity between the strains from this study and the relevant reference strains used. However, these data do show that there is genomic variation within the V. rotiferianus clade and similarly within the V. jasicida clade.
Fig. 5.
Genome comparison of V. rotiferianus and V. jasicida. A – Genome comparison of 5 V. rotiferianus strains (Group A) compared to available reference V. rotiferianus B64D1 strain. B – Genome comparison of 6 V. jasicida strain (Group B) compared to available reference V. jasicida 090810c strain. The alignments are visualized using Gview (Petkau et al., 2010). The innermost ring indicates the position on the reference chromosome. Positions covered by BLASTN alignments are indicated with a solid colour; whitespace gaps represent genomic regions not covered by the BLASTN alignments.
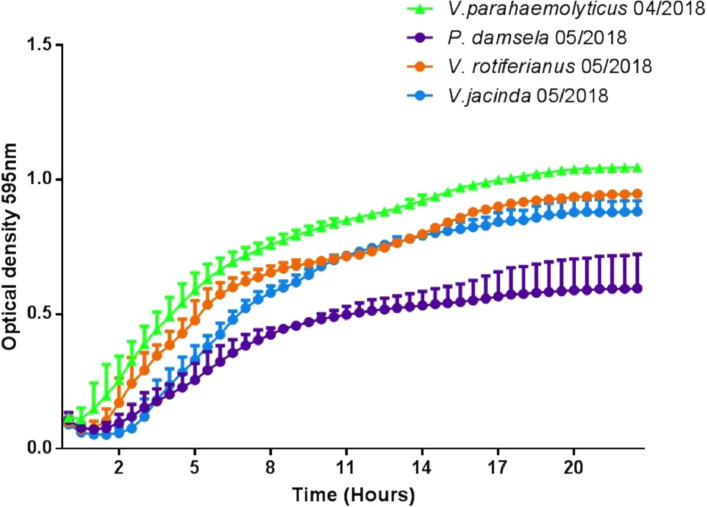
Overall, V. rotiferianus was found in four of the 16 samples tested and was isolated from two locations tested in the UK in 2018 (Chichester Harbour and Osea Island). Vibrio jasicida was found in eight out of the 16 samples tested in this study and were also isolated from two locations in the UK in 2018 (Chichester Harbour and Whitstable Bay) (Table 1). Biochemically the only way to distinguish between V. rotiferianus and V. jasicida isolates was on the production of indole from tryptophan and the fermentation amygdalin (glycoside) (V. rotiferianus positive and V. jasicida negative). The growth of V. rotiferianus and V. jasicida was tested at 30 °C in Marine Broth and compared to growth of V. parahaemolyticus strains to determine and characteristic growth phenotypes. The V. parahaemolyticus strain 05/2018 grew at similar rates to reference RIMD2210633 (Fig. 6). Compared to the Vibrio strains tested, the growth of Photobacterium damsela in Marine Broth was slower (Fig. 6).
Fig. 6.
Growth curve of representative strains isolated from native oysters (Ostrea edulis) harvested from Chichester Harbour. Strains were tested with six technical repeats and two experimental repeats. Data shown is the average of both experiments with mean error.
2.5. Pathogenic potential of V. parahaemolyticus
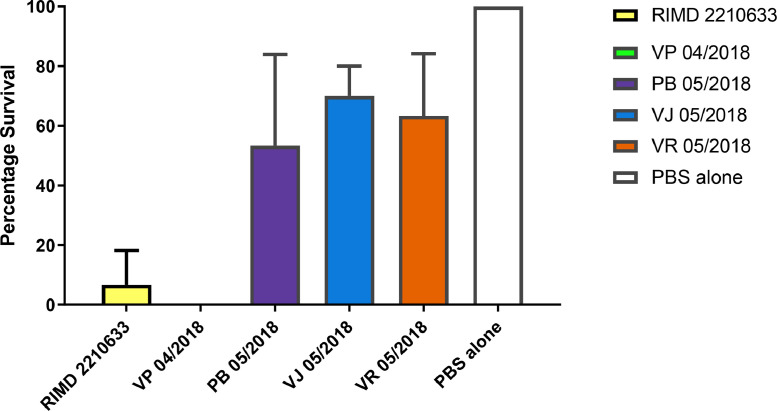
Previous work has shown that the insect infection model Galleria mellonella can be used to assess the virulence of V. parahaemolyticus strains (Wagley et al., 2017) and could distinguish clinical toxigenic strains from non-toxigenic strains from the environment. The V. parahaemolyticus strain RIMD 22,106,333, (a typical and well-referenced clinical TDH positive reference strain) was compared with the V. parahaemolyticus strain VP 05/2018 isolated from sample EXE 04/2018. Strain VP 04/2018 was negative for both TDH and TRH haemolysin genes. Both RIMD 2,210,633 and VP 04/2018 caused lethal infections at a dose of 106 CFU per larvae (Fig. 7). The LD50 for RIMD2210633 was 10,000 CFU per larvae while the LD50 dose for EXE 04/2018 was 1000 CFU per larvae when the experiment was carried out at 37 °C indicating that VP 04/2018 was more virulent than the reference strain even without the presence of known virulence genes (tdh and trh) needed for human infection.
Fig. 7.
Galleria mellonella infection with strains isolated from native oysters (Ostrea edulis) harvested from Chichester Harbour. Stains were grown at 37 °C and a dose of 106 CFU of each strain was injected into the larvae. Percentage survival was measured after 48 h. The results shown here are the means of three experiments, each using groups of 10 larvae paper strain. The error bars indicate standard deviation.
2.6. Pathogenic potential of V. rotiferianus, V. jasicida and Ph. damsela
Environmental strains Ph. damsela (PB 05/2018), V. jasicida (VJ 05/2018) and V. rotiferianus (VR 05/2018) were injected into G. mellonella at a dose of 106 CFU per larvae. The results are shown in Fig. 7. Strain Ph. damsela (PB 05/2018) showed 50% survival while V. jasicida (VJ 05/2018) and V. rotiferianus (VR 05/2018) showed 70% and 60% survival, respectively at 106 CFU per larvae. At a higher dose of 107 CFU per larvae, VJ 05/2018 and VR 05/2018 showed 40% survival after 48 h (data not shown).
3. Discussion
One of the potential consequences of climate change will be changed patterns of parasite and microbial disease in aquaculture settings. One concern is the potential for an increased incidence of Vibriosis - diseases caused by Vibrio species. Vibriosis caused by some species, such as V. parahaemolyticus, can occur in humans causing gastroenteritis originating from raw or undercooked seafood. Vibriosis is also one of the main bacterial diseases of larvae and juvenile bivalves, affecting the early development stages of shellfish growth. The presence of Vibrio species, and their growth, is dependant on the water temperature of the environment. Thus, an increase in sea surface temperatures could lead to increased bacterial growth, leading to more disease.
In this study, sea-surface temperature data were assessed in several nearshore ecosystems in English and Welsh waters that accommodate UK shellfisheries (Fig. 2 and Supplementary Data 1). In several areas, an increase in peak summer sea-surface temperature from 18 °C to 22° was recorded over this four-year period. Furthermore, for some of these shellfish harvesting areas, the sea-surface temperature remained above 18 °C for up to eight consecutive weeks. Fluctuations at shellfish sites cannot be ruled out when comparing to sea-surface temperatures of wider regions however, temperature increases for prolonged periods could alter habitats and change the distribution of bacterial species within them. Vibrio species are predominantly found in locations where the temperatures are above 18 °C, a temperature that is suitable for their growth and proliferation. Increased sea-surface temperatures in nearshore ecosystems in the UK could allow for favourable growth conditions that support the growth of Vibrio species. Research carried out in the UK has mainly focussed on the presence of human pathogenic Vibrio species, such as V. parahaemolyticus, in the UK marine environment. These have been readily detected in some nearshore ecosystems since 2001 (Baker-Austin et al., 2010; Ford et al., 2020b; Martinez-Urtaza et al., 2013; Powell et al., 2013; Wagley et al., 2009, 2008), thus it is unsurprising that V. parahaemolyticus was detected in samples from Chichester Harbour when the temperature was suitable for bacterial growth. However, why V. parahaemolyticus was not detected in later months, when sea-surface temperature continued to be suitable for growth suggests that other factors such as competition from other Vibrio species and environmental abiotic factors in the community play a vital role in V. parahaemolyticus establishment. Efforts to use temperature and salinity data alone to predict the occurrence of pathogenic Vibrio species in an ecosystem have not always been reliable indicators of Vibrio emergence (McLaughlin et al., 2005). In some disease outbreaks, Vibrio species have appeared in shellfish where waters were less than < 18 °C, indicating that simplistic environmental conditions alone are not reliable indicators of disease prediction (McLaughlin et al., 2005). Conversely, in this study, increasing temperatures in nearshore ecosystems did not necessarily drive the occurrence of V. parahaemolyticus in the shellfish. Thus, further research is required to combine key environmental, biological, and molecular factors to ascertain the drivers behind the seasonal occurrence of V. parahaemolyticus in the natural environment.
During this study, our focus was on detecting the main human pathogens such as V. parahaemolyticus and V. vulnificus. However, during shellfish testing, V. alginolyticus was also detected in nearly all shellfish samples and can be considered part of normal marine flora (Xie et al., 2005). It should be noted that V. alginolyticus can increase health risk for consumers (Baker-Austin et al., 2018; Reilly et al., 2011) and further studies need to be conducted to ascertain whether V. alginolyticus strains from the UK are a risk to consumers and recreational users of waterways where V. alginolyitcus are widely present.
This study shows that members of the V. harveyi clade are also present in nearshore ecosystems in the UK and that Vibrio species may play a role in both human and shellfish associated diseases. The core of the V. harveyi clade is made up of V. harveyi itself and closely related species including V. alginolyticus, V. azureus, V. mytili, V. natriegens, V. campbellii, V. parahaemolyticus, V. owensii, V. jascinda, V. sagamiensis and V. rotiferianus. The members of V. harveyi clade share a high degree of genetic and phenotypic similarity, which can often lead to the misidentification of species within this clade (Gomez-Gil et al., 2004; Lin et al., 2010; Sawabe et al., 2007). The analysis of shellfish samples from the UK in the summer of 2018 identified the presence of V. parahaemolyticus, V. jasicida, and V. rotiferianus in three shellfish sites around the UK. To the best of our knowledge, this study is the first to identify and report V. rotiferianus and V. jasicida presence in shellfish harvesting areas in England. Previous testing of shellfish (prior to 2008) for the presence of Vibrio species in UK sites did not detect the presence of V. rotiferianus and V. jasicida in UK shellfisheries (Wagley et al., 2008). V. rotiferianus was first described by Gomez-Gil and co-workers in 2003, isolated from the rotifer Brachionus plicatilis (Gomez-Gil et al., 2003). This species was described as growing yellow sucrose positive colonies on TCBS agar and was positive for the production of indole. In this study, V. rotiferianus strains isolated were positive for indole production but grew as sucrose negative (green) colonies on TCBS agar indicating that these strains are biochemically different from those first described in the literature. V. jasicida, first described in 2012, has been recovered from marine invertebrates and vertebrates, including Atlantic salmon and flounder (Yoshizawa et al., 2012). In this study, we examined the pathogenic potential of the V. jasicida and V. rotiferianus in an insect infection model using G. mellonella larvae. This model has been shown to be a useful for studying human pathogenic Vibrio species as well as aquaculture Vibrio and non-Vibrio species (Djainal et al., 2020; McMillan et al., 2015). The data here indicate that the strains of V. jasicida and V. rotiferianus tested are able to cause infection and mortality in G. mellonella, and this model could be used in the future to further investigate the virulence of these aquaculture pathogens. Members of the V. harveyii clade are a major concern for sustainability of the aquaculture industry (King et al., 2019; Vezzulli et al., 2010; Wendling et al., 2014), thus, it is vital to understand the epidemiological significance of these new emerging Vibrio species before any associated disease burden becomes a problem in aquaculture settings.
Using a ‘One Health’ aquaculture approach is needed to understand the decline in native oysters at Chichester Harbour.
Chichester Harbour is home to the native oysters (Ostrea edulis) and once supported a substantial fishery, but stocks of native oysters have declined significantly over the last 20 years (Helmer et al., 2019). Globally, 85% of all oyster beds and reefs have been lost, and across Europe and England up to 95% of native oyster reefs have been extirpated (Airoldi and Beck, 2007; Beck et al., 2011). This trend has been reflected in the Solent, once home to one of Europe's largest native oyster fisheries, with seabed densities of 32m2, supporting 450 commercial vessels in the 1980′s (Key and Davidson, 1981). Native oyster stocks have since declined, suffering a population crash in 2007 from which they have not recovered, and remain severely depleted in many harbours, including Chichester Harbour. A long-term study found numbers of O. edulis within Chichester Harbour had decreased by 96%, between 1998 and 2017 (Helmer et al., 2019). Sussex Inshore Fisheries and Conservation Authority (IFCA) has been conducting annual stock assessments (catch per unit effort (CPUE) estimates) for the oysters in Chichester Harbour since 2013. In the main fishery area (Emsworth Channel), the CPUE decreased from 50 kg/h/1 min dredge width in 2013 to 22 kg/h/1 min in 2017. In 2018, the CPUE dropped to 3.9 kg/h/1 min and the fishery was closed. CPUE remained low in 2019 (1.4 kg/h/1 min) and the fishery has remained closed. The continued low CPUE levels indicates a lack of continued oyster recruitment in Chichester Harbour and efforts to restore the populations of native oysters have been unsuccessful in recent years.
Several reports of the implications of Vibrio species acting as a potential disease causing agent involved in oyster mortality have been published (King et al., 2019; Waechter et al., 2002; Wendling et al., 2014) but in the majority of records no clear aetiological agent can be identified (Garnier et al., 2007). In this study, the presence of Vibrio species such as V. jasicida, and V. rotiferianus belonging to the V. harveyi clade is reported and this may play a role in oyster disease in this area. Further contributions to disease in oysters may be attributed to abiotic factors such as temperature, nutrient concentrations, and salinity as well as host oyster genetic backgrounds that make them sensitive to disease (de Lorgeril et al. 2018; Lang et al., 2010; Malham et al., 2009; Samain et al., 2007). The low levels of oyster recruitment in Chichester Harbour may also be due to the level of harvesting exceeding the reproductive capacity of the species as well as the presence of the invasive slipper limpet (Crepidula fornicate) that can outcompete O. edulis larvae leading to a dominance of C. fornicata in the Solent (Preston et al., 2020). Leakage of partially treated sewage into Chichester Harbour between 2010 and 2015 may also have contributed to the low levels of oyster recruitment in the harbour, which needs to be further investigated (Environment Agency v Southern Water Services Ltd (2021) All ER (D) 62 (Aug)). Furthermore, oysters are typically populated with multiple bacterial and viral species. In vivo infection studies to find the causative agents of oyster mortality do not consider the complexity in the natural environment or how microorganisms interact with each other to cause host disease.
Bivalve larvae can naturally exhibit a high mortality during their transition to a juvenile state. Whether the Vibrio strains isolated in this study are responsible for bivalve larvae death in Chichester Harbour requires more investigation. Metagenomic time-series studies targeting the microbiome of oysters and the surrounding environments coupled with a survey of environmental abiotic factors and host susceptibility such as those conducted by King and co-workers to study the mortality events in the Pacific oyster in Australia (King et al., 2019) are needed in this area to identify causes of oyster mortality events. To understand why native oysters populations have declined in Chichester Harbour, it is thus important to use a ‘One Health’ approach (Stentiford et al., 2020). This approach examines all hazards that may contribute to water quality and the health of the oysters if aquaculture is going to thrive again in Chichester Harbour. These hazards may include chemicals such as naturally occurring biotoxins and partially treated sewage leaks entering the harbour, as well as the presence of increasing numbers and species of animal and human pathogens.
4. Conclusion
This study assessed the presence of culturable bacterial species in shellfish from multiple sites around England and found them to be populated with multiple Vibrio and non-Vibrio species. A number of factors could explain the increasing amount of Vibrio species now present in temperate regions including the UK marine and estuarine environments. Firstly, favourable growth conditions for Vibrio species such as increasing sea-surface temperatures and heavy rainfall events during the summer, (which can bring the water salinity down to optimum), are currently present in some areas around the UK (Baker-Austin et al., 2018, 2013). Secondly, globalisation and the importing of more seafood from regions that have a higher incidence of Vibrio species in the environment may favour the introduction of a variety of species into our waters. Connected to this is the increase in shipping and the release of ship ballast water that has been shown to be a transport vector for Vibrio species (Drake et al., 2007; Mimura et al., 2005; Rivera et al., 2012; Ruiz et al., 2000). If ballast water is contaminated with Vibrio species and is discharged into harbours and coastal waterways then this could lead to the introduction and establishment of new pathogenic and non-pathogenic Vibrio species into the environment. Finally, the improvement of detection methods of Vibrio species from water and shellfish in the UK has improved vastly over the past 15 years and in the advent of cheaper and faster genome sequencing, scientists are able to sequence strains that we previously may have overlooked. Consequentially, the conditions that favour the growth and establishment of Vibrio species as a seasonal human and aquaculture pathogen in temperate regions, is a public health concern and should be monitored in the environment.
5. Methods and materials
5.1. Shellfish collection and bacterial isolation
Bacterial strains used in this study are shown in Table 1. V. parahaemolyticus strains were initially cultured aerobically onto selective media Thiosulphate Citrate Bile Sucrose (TCBS) agar (Oxoid) at 37 °C for 24 h. For enumeration of colony counts, routine sub culturing and growth, Marine Agar (Conda Labs, Spain) was used and incubated at 30 °C for 24 h.
For shellfish testing, non-depurated samples were sought initially from all sites but could only be obtained from Chichester Harbour and Lyme Bay while commercially depurated shellfish were obtained from Osea Island and Whitstable Bay. In the five months of June to October 2018, a total of five shellfish samples were tested for the presence of Vibrio species from Chichester Harbour. Shellfish were supplied as part of ongoing research programmes and were received confidentially directly from harvesting areas. Samples comprised of non-depurated O. edulis and were held between 4 and 8 °C during transit and, on arrival in the laboratory, were placed in the fridge until analysis. Further, commercially available shellfish samples that had been harvested from Osea Island and Whitstable Bay and non-depurated M. edulis from Lyme Bay were also obtained and tested for the presence of Vibrio species. Samples were tested in duplicate and the average of both experiments was used to calculate the total Vibrio species count. All raw shellfish samples were analysed within 24 h of collection and according to ISO 21,872–1 with minor modifications. A total of 25 g of shellfish meat (taken from a minimum of eight animals) was disrupted using a stomacher before the addition of 225 ml of alkaline salt peptone water (ASPW; Oxoid Ltd., Basingstoke, Hampshire, UK). Sequential log dilutions of stomached shellfish meat were carried out in APSW to a maximum of 10−1 and 100 µl was spread onto the surface of TCBS plates to determine direct enumeration of Vibrio species. Total Vibrio species were recorded from plate counts of sucrose-positive and negative colonies on thiosulphate citrate bile sucrose agar (TCBS) plates before a further enrichment step. All samples were then incubated at 41 °C for 6 h for an enrichment step, after which a 5 µl loopful was taken from directly below the surface of the broth and streaked onto TCBS plates. All TCBS plates were incubated at 37 °C for 24 h. Typical sucrose negative (green) colonies were subcultured onto marine agar (Conda labs, Spain) and incubated at 30 °C for 24 h. Presumptive colonies were identified as V. parahaemolyticus if they met the following criteria: positive for oxidase, negative for Voges Proskauer and Ortho-nitrophenyl-β-d-galactopyranoside, no growth in 0% NaCl and no acid from sucrose. Further identification by API 20E strips (BioMerieux) was also carried out on sucrose negative colonies. All biochemically identified V. parahaemolyticus strains were further analysed by PCR amplification using the species target toxR (Kim et al., 1999). All sucrose negative colonies that were also negative for V. parahaemolyticus PCR tests were checked for V. vulnificus identification using PCR amplification using the species target vvh (Hill et al., 1991).
5.2. Genome sequencing, assembly and annotation
Thirteen unidentified strains and one V. parahaemolyticus strain (Table 1) were genome sequenced. Bacterial cells were harvested and genomic DNA was extracted using Wizard Genomic DNA Purification Kit (Promega). Library preparation was carried out by Exeter DNA sequencing service. In brief, DNA was concentrated using GeneRead kit (Lot No. 145,025,210) and end repair and adenylation of fragments was carried using NEXTFLEX® Rapid DNAseq kit (#5144–02) according to manufactures instructions. Purification and concentration of PCR amplified library was carried out according to GeneRead kit instructions. Genome wide sequence data (150 bp, paired end) was generated using an Illumina NovaSeq. Raw sequencing reads were screened for contamination and quality filtered using Trimmomatic (Bolger et al., 2014). Sequencing metrics were generated using FastQC (Wingett and Andrews, 2018). This filtered dataset was then assembled de-novo using SPADES v3.15.1 (Bankevich et al., 2012). De-novo assemblies were assessed for completeness using BUSCO (Seppey et al., 2019) (Supplementary Data 3) and annotated using PROKKA (Seemann, 2014). Assembly metrics were generated using the QUAST (Gurevich et al., 2013).
5.3. Multi Locus sequence analysis (MLSA)
Multi Locus sequence analysis (MLSA) was conducted on all sequenced strains and a panel of reference strains downloaded from the NCBI genomes database (Feb 2021). The MLSA was conducted using six conserved housekeeping genes (efp, glnA, gyrB, metG, purM, pntA). The sequences of these genes were extracted from the genome annotations for each strain and concatenated. The concatenated MLSA sequences were aligned using MUSCLE (Edgar, 2004) and phylogenetic analysis was conducted using MEGA 10 (Kumar et al., 2016). Maximum likelihood trees were generated using the general time reversible model and 100 bootstrap replicates were performed. Photobacterium species were initially used as the outgroup to root the tree.
5.4. Evolutionary analysis by maximum likelihood method
The evolutionary history was inferred by using the Maximum Likelihood method and General Time Reversible model (Nei and Kumar, 2000). The tree with the highest log likelihood (−291,299.34) is shown in Fig. 4a. Initial tree(s) for the heuristic search were obtained automatically by applying neighbour-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences amongst sites (five categories (+G, parameter = 0.7879)). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 45.80% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 119 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions with less than 95% site coverage were eliminated, i.e., fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position (partial deletion option). There was a total of 8965 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018).
5.5. Average nucleotide identity (ANI)
Average nucleotide identity was calculated using fastANI (Jain et al., 2018) between the sequenced isolates and a reference panel of Vibrio genomes downloaded from the NCBI genomes database (February 2021).
5.6. Determining sea surface temperatures for UK coastal waters
The sea-surface temperature data is a daily value, valid at foundation depth. This is the depth, several meters down, where there are no variations on the sub-daily timescale and hence can be treated as a daily-mean value. The data is from the Operational Sea Surface Temperatures and Sea Ice Analysis (OSTIA), which is used to provide the sea-surface temperature in Met-Office global weather forecasts. The sea temperature dataset is produced by combining observations from satellite and buoys and filling gaps between these observations in a physically consistent manner. The time-series plotted in Fig. 2 were extracted for the locations nearest to the sample collections sites (Supplementary data 1). However, since the sea-surface temperature data are available on a 0.05 × 0.05 latitude-longitude grid, corresponding to roughly a 5 km x 5 km spatial average, the nearest location may be a couple of kilometres away from the sampling site. Additionally, they are more likely to be representative of the offshore sea-surface temperature than the temperature within a sheltered harbour. Despite these caveats, the OSTIA data can be used as a guide of where sea-surface temperatures are likely to be suitable for the presence of Vibrio species. During 2018, the sea-surface temperature plots were updated every few weeks to track the warming of the seawaters as summer approached and to help inform when to acquire the shellfish samples.
5.7. Infection of Galleria mellonella larvae
Galleria mellonella larvae were purchased from TruLarv™ (Biosystems Technology, Exeter, Devon, UK). Larvae weighing between 0.2 and 0.35 g were chosen for experiments. For each experiment a total of ten larvae were used per strain to be tested. The larvae were infected by micro-injection (Hamilton Ltd) into the right foremost proleg with 106 CFU per larvae of V. parahaemolyticus in 10 µl volume which had been grown in Marine Broth at 37 °C and washed twice in PBS. Bacterial cell counts were carried out by plating serial dilutions of the inoculum onto Marine agar. For control purposes, ten larvae were inoculated with PBS and a further ten were left inoculated. The larvae were incubated at 37 °C and survival was recorded for all strains after 24 and 48 h. Larvae were scored as dead when they ceased moving or failed to respond when gently manipulated with a pipette tip. Observation findings were also recorded if larvae changed colour from their normal pale cream coloration to brown or black indicative of melanisation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was funded by a research grant from the Biotechnology and Biological Sciences Research Council BBSRC (BB/N016513/1) to SW, RWT and CSB. We would like to thank Chichester and Havant Council for their advise and guidance and acquisition of shellfish samples. We would also like to thank John Holmyard and Dr Ross Brown for providing us with samples from Lyme Bay.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2021.117942.
Appendix. Supplementary materials
References
- Airoldi L., Beck M.W. Loss, status and trends for coastal marine habitats of Europe. Oceanogr. Mar. Biol. 2007;45(45):345–405. Vol. [Google Scholar]
- Ansede-Bermejo J., Gavilan R.G., Trinanes J., Espejo R.T., Martinez-Urtaza J. Origins and colonization history of pandemic Vibrio parahaemolyticus in South America. Mol. Ecol. 2010;19(18):3924–3937. doi: 10.1111/j.1365-294X.2010.04782.x. [DOI] [PubMed] [Google Scholar]
- Austin B., Zhang X.H. Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett. Appl. Microbiol. 2006;43(2):119–124. doi: 10.1111/j.1472-765X.2006.01989.x. [DOI] [PubMed] [Google Scholar]
- Baker-Austin C., Oliver J.D., Alam M., Ali A., Waldor M.K., Qadri F., Martinez-Urtaza J. Vibrio spp. infections. Nat. Rev. Dis. Primers. 2018;4(1):8. doi: 10.1038/s41572-018-0005-8. [DOI] [PubMed] [Google Scholar]
- Baker-Austin C., Stockley L., Rangdale R., Martinez-Urtaza J. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: a European perspective. Environ. Microbiol. Rep. 2010;2(1):7–18. doi: 10.1111/j.1758-2229.2009.00096.x. [DOI] [PubMed] [Google Scholar]
- Baker-Austin C., Trinanes J.A., Taylor N.G.H., Hartnell R., Siitonen A., Martinez-Urtaza J. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Clim. Chang. 2013;3(1):73–77. [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M.W., Brumbaugh R.D., Airoldi L., Carranza A., Coen L.D., Crawford C., Defeo O., Edgar G.J., Hancock B., Kay M.C., Lenihan H.S., Luckenbach M.W., Toropova C.L., Zhang G.F., Guo X.M. Oyster reefs at risk and recommendations for conservation, restoration, and management. Bioscience. 2011;61(2):107–116. [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruto M., James A., Petton B., Labreuche Y., Chenivesse S., Alunno-Bruscia M., Polz M.F., Le Roux F. Vibrio crassostreae, a benign oyster colonizer turned into a pathogen after plasmid acquisition. ISME J. 2017;11(4):1043–1052. doi: 10.1038/ismej.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello A., Abbas M., Allen A., Ball S., Bell S., Bellamy R., Friel S., Groce N., Johnson A., Kett M., Lee M., Levy C., Maslin M., McCoy D., McGuire B., Montgomery H., Napier D., Pagel C., Patel J., de Oliveira J.A., Redclift N., Rees H., Rogger D., Scott J., Stephenson J., Twigg J., Wolff J., Patterson C. Managing the health effects of climate change: lancet and university college London institute for global health commission. Lancet. 2009;373(9676):1693–1733. doi: 10.1016/S0140-6736(09)60935-1. [DOI] [PubMed] [Google Scholar]
- Dalsgaard A., Frimodt-Moller N., Bruun B., Hoi L., Larsen J.L. Clinical manifestations and molecular epidemiology of Vibrio vulnificus infections in Denmark. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 1996;15(3):227–232. doi: 10.1007/BF01591359. [DOI] [PubMed] [Google Scholar]
- de Lorgeril J., Lucasson A., Petton B., Toulza E., Montagnani C., Clerissi C., Vidal-Dupiol J., Chaparro C., Galinier R., Escoubas J.M., Haffner P., Degremont L., Charriere G.M., Lafont M., Delort A., Vergnes A., Chiarello M., Faury N., Rubio T., Leroy M.A., Perignon A., Regler D., Morga B., Alunno-Bruscia M., Boudry P., Le Roux F., Destoumieux-Garzomicronn D., Gueguen Y., Mitta G. Immune-suppression by OsHV-1 viral infection causes fatal bacteraemia in Pacific oysters. Nat. Commun. 2018;9(1):4215. doi: 10.1038/s41467-018-06659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djainal W.A.S., Shahin K., Metselaar M., Adams A., Desbois A.P. Larva of greater wax moth Galleria mellonella is a suitable alternative host for the fish pathogen Francisella noatunensis subsp. orientalis. BMC Microbiol. 2020;20(1):8. doi: 10.1186/s12866-020-1695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlon C.J., Martin M., Stark J., Roberts-Jones J., Fiedler E., W W. The operational sea surface temperature and sea ice analysis (OSTIA) system. Remote Sens. Environ. 2012;116:140–158. [Google Scholar]
- Drake L.A., Doblin M.A., Dobbs F.C. Potential microbial bioinvasions via ships' ballast water, sediment, and biofilm. Mar. Pollut. Bull. 2007;55(7–9):333–341. doi: 10.1016/j.marpolbul.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston R.A., Hasegawa H., Humphrey K.L., Polyak I.K., Hase C.C. Re-emergence of Vibrio tubiashii in bivalve shellfish aquaculture: severity, environmental drivers, geographic extent and management. Dis. Aquat. Organ. 2008;82(2):119–134. doi: 10.3354/dao01982. [DOI] [PubMed] [Google Scholar]
- European Commission Oyster mortality EFSA panel on animal health and welfare (AHAW) EFSA J. 2015;13(6) [Google Scholar]
- Ford C.L., Powell A., Lau D.Y.L., Turner A.D., Dhanji-Rapkova M., Martinez-Urtaza J., Baker-Austin C. Isolation and characterisation of potentially pathogenic Vibrio species in a temperate, higher latitude hotspot. Environ. Microbiol. Rep. 2020;12(4):424–434. doi: 10.1111/1758-2229.12858. [DOI] [PubMed] [Google Scholar]
- Ford C.L., Powell A., Lau D.Y.L., Turner A.D., Dhanji-Rapkova M., Martinez-Urtaza J., Baker-Austin C. Isolation and characterisation of potentially pathogenic Vibrio species in a temperate, higher latitude hotspot. Environ. Microbiol. Rep. 2020;12(4):424–434. doi: 10.1111/1758-2229.12858. [DOI] [PubMed] [Google Scholar]
- Froelich B.A., Daines D.A. In hot water: effects of climate change on Vibrio-human interactions. Environ. Microbiol. 2020;22(10):4101–4111. doi: 10.1111/1462-2920.14967. [DOI] [PubMed] [Google Scholar]
- Fuenzalida L., Armijo L., Zabala B., Hernandez C., Rioseco M.L., Riquelme C., Espejo R.T. Vibrio parahaemolyticus strains isolated during investigation of the summer 2006 seafood related diarrhea outbreaks in two regions of Chile. Int. J. Food Microbiol. 2007;117(3):270–275. doi: 10.1016/j.ijfoodmicro.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Garnier M., Labreuche Y., Garcia C., Robert M., Nicolas J.L. Evidence for the involvement of pathogenic bacteria in summer mortalities of the Pacific oyster Crassostrea gigas. Microb. Ecol. 2007;53(2):187–196. doi: 10.1007/s00248-006-9061-9. [DOI] [PubMed] [Google Scholar]
- Gomez-Gil B., Soto-Rodriguez S., Garcia-Gasca A., Roque A., Vazquez-Juarez R., Thompson F.L., Swings J. Molecular identification of Vibrio harveyi-related isolates associated with diseased aquatic organisms. Microbiology. 2004;150(Pt 6):1769–1777. doi: 10.1099/mic.0.26797-0. [DOI] [PubMed] [Google Scholar]
- Gomez-Gil B., Thompson F.L., Thompson C.C., Swings J. Vibrio rotiferianus sp. nov., isolated from cultures of the rotifer Brachionus plicatilis. Int. J. Syst. Evolut. Microbiol. 2003;53(Pt 1):239–243. doi: 10.1099/ijs.0.02430-0. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Escalona N., Cachicas V., Acevedo C., Rioseco M.L., Vergara J.A., Cabello F., Romero J., Espejo R.T. Vibrio parahaemolyticus diarrhea, Chile, 1998 and 2004. Emerg. Infect. Dis. 2005;11(1):129–131. doi: 10.3201/eid1101.040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudenege D., Travers M.A., Lemire A., Petton B., Haffner P., Labreuche Y., Tourbiez D., Mangenot S., Calteau A., Mazel D., Nicolas J.L., Jacq A., Le roux F. A single regulatory gene is sufficient to alter Vibrio aestuarianus pathogenicity in oysters. Environ. Microbiol. 2015;17(11):4189–4199. doi: 10.1111/1462-2920.12699. [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmer L., Farrell P., Hendy I., Harding S., Robertson M., Preston J. Active management is required to turn the tide for depleted Ostrea edulis stocks from the effects of overfishing, disease and invasive species. PeerJ. 2019;7:e6431. doi: 10.7717/peerj.6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W.E., Keasler S.P., Trucksess M.W., Feng P., Kaysner C.A., Lampel K.A. Polymerase chain reaction identification of Vibrio vulnificus in artificially contaminated oysters. Appl. Environ. Microbiol. 1991;57(3):707–711. doi: 10.1128/aem.57.3.707-711.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M., Ayers T., Mahon B.E., Swerdlow D.L. Epidemiology of seafood-associated infections in the United States. Clin. Microbiol. Rev. 2010;23(2):399–411. doi: 10.1128/CMR.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C., Rodriguez R.L., Phillippy A.M., Konstantinidis K.T., Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018;9(1):5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S.W., Colwell R.R., Kaper J.B. Vibrio parahaemolyticus and related halophilic Vibrios. Crit. Rev. Microbiol. 1982;10(1):77–124. doi: 10.3109/10408418209113506. [DOI] [PubMed] [Google Scholar]
- Kaspar C.W., Tamplin M.L. Effects of temperature and salinity on the survival of Vibrio vulnificus in seawater and shellfish. Appl. Environ. Microbiol. 1993;59(8):2425–2429. doi: 10.1128/aem.59.8.2425-2429.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key, D., Davidson, P.E., 1981. A Review of Development of the Solent Oyster Fishery, 1971–80. Laboratory Leaflet, No. 52. Lowestoft. 40 pp.
- Kim Y.B., Okuda J., Matsumoto C., Takahashi N., Hashimoto S., Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 1999;37(4):1173–1177. doi: 10.1128/jcm.37.4.1173-1177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King W.L., Jenkins C., Go J., Siboni N., Seymour J.R., Labbate M. Characterisation of the pacific oyster microbiome during a summer mortality event. Microb. Ecol. 2019;77(2):502–512. doi: 10.1007/s00248-018-1226-9. [DOI] [PubMed] [Google Scholar]
- Kokashvili T., Whitehouse C.A., Tskhvediani A., Grim C.J., Elbakidze T., Mitaishvili N., Janelidze N., Jaiani E., Haley B.J., Lashkhi N., Huq A., Colwell R.R., Tediashvili M. Occurrence and diversity of clinically important Vibrio species in the aquatic environment of Georgia. Front. Public Health. 2015;3:232. doi: 10.3389/fpubh.2015.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evolut. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evolut. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R.P., Langdon C.J., Taris N.G., Camara M.D. Use of laboratory assays to predict subsequent growth and survival of Pacific oyster (Crassostrea gigas) families planted in coastal waters. Aquaculture. 2010;306(1–4):68–79. [Google Scholar]
- Leddin D., Macrae F. Climate change: implications for gastrointestinal health and disease. J. Clin. Gastroenterol. 2020;54(5):393–397. doi: 10.1097/MCG.0000000000001336. [DOI] [PubMed] [Google Scholar]
- Lin B., Wang Z., Malanoski A.P., O'Grady E.A., Wimpee C.F., Vuddhakul V., Alves N., Thompson F.L., Gomez-Gil B., Vora G.J. Comparative genomic analyses identify the Vibrio harveyi genome sequenced strains BAA-1116 and HY01 as Vibrio campbellii. Environ. Microbiol. Rep. 2010;2(1):81–89. doi: 10.1111/j.1758-2229.2009.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren E., Andersson Y., Suk J.E., Sudre B., Semenza J.C. Public health. Monitoring EU emerging infectious disease risk due to climate change. Science. 2012;336(6080):418–419. doi: 10.1126/science.1215735. [DOI] [PubMed] [Google Scholar]
- Malham S.K., Cotter E., O'Keeffe S., Lynch S., CUlloty S.C., King J.W., Latchford J.W., Beaumont A.R. Summer mortality of the Pacific oyster, Crassostrea gigas, in the Irish Sea: the influence of temperature and nutrients on health and survival. Aquaculture. 2009;287(1–2):128–138. [Google Scholar]
- Martinez-Urtaza J., Baker-Austin C., Jones J.L., Newton A.E., Gonzalez-Aviles G.D., DePaola A. Spread of pacific northwest Vibrio parahaemolyticus strain. N. Engl. J. Med. 2013;369(16):1573–1574. doi: 10.1056/NEJMc1305535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Urtaza J., Bowers J.C., Trinanes J., DePaola A. Climate anomalies and the increasing risk of Vibrio parahaemolyticus and Vibrio vulnificus illnesses. Food Res. Int. 2010;43(7):1780–1790. [Google Scholar]
- Martinez-Urtaza J., Simental L., Velasco D., DePaola A., Ishibashi M., Nakaguchi Y., Nishibuchi M., Carrera-Flores D., Rey-Alvarez C., Pousa A. Pandemic Vibrio parahaemolyticus O3:K6. Eur. Emerg. Infect. Dis. 2005;11(8):1319–1320. doi: 10.3201/eid1108.050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Urtaza J., Trinanes J., Abanto M., Lozano-Leon A., Llovo-Taboada J., Garcia-Campello M., Pousa A., Powell A., Baker-Austin C., Gonzalez-Escalona N. Epidemic dynamics of Vibrio parahaemolyticus illness in a hotspot of disease emergence, Galicia, Spain. Emerg. Infect. Dis. 2018;24(5):852–859. doi: 10.3201/eid2405.171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J.B., DePaola A., Bopp C.A., Martinek K.A., Napolilli N.P., Allison C.G., Murray S.L., Thompson E.C., Bird M.M., Middaugh J.P. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N. Engl. J. Med. 2005;353(14):1463–1470. doi: 10.1056/NEJMoa051594. [DOI] [PubMed] [Google Scholar]
- McMillan S., Verner-Jeffreys D., Weeks J., Austin B., Desbois A.P. Larva of the greater wax moth, Galleria mellonella, is a suitable alternative host for studying virulence of fish pathogenic Vibrio anguillarum. BMC Microbiol. 2015;15:127. doi: 10.1186/s12866-015-0466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead P.S., Slutsker L., Dietz V., McCaig L.F., Bresee J.S., Shapiro C., Griffin P.M., Tauxe R.V. Food-related illness and death in the United States. Emerg. Infect. Dis. 1999;5(5):607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura H., Katakura R., Ishida H. Changes of microbial populations in a ship's ballast water and sediments on a voyage from Japan to Qatar. Mar. Pollut. Bull. 2005;50(7):751–757. doi: 10.1016/j.marpolbul.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Nei M., Kumar S. Oxford University Press; New York: 2000. Molecular Evolution and Phylogenetics. [Google Scholar]
- Parveen S., Hettiarachchi K.A., Bowers J.C., Jones J.L., Tamplin M.L., McKay R., Beatty W., Brohawn K., Dasilva L.V., Depaola A. Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake bay oysters and waters. Int. J. Food Microbiol. 2008;128(2):354–361. doi: 10.1016/j.ijfoodmicro.2008.09.019. [DOI] [PubMed] [Google Scholar]
- Petkau A., Stuart-Edwards M., Stothard P., Van Domselaar G. Interactive microbial genome visualisation with GView. Bioinformatics. 2010;26(24):3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell A., Baker-Austin C., Wagley S., Bayley A., Hartnell R. Isolation of pandemic Vibrio parahaemolyticus from UK water and shellfish produce. Microb. Ecol. 2013;65(4):924–927. doi: 10.1007/s00248-013-0201-8. [DOI] [PubMed] [Google Scholar]
- Preston J., Fabra M., Helmer L., Johnson E., Harris-Scott E., Hendy I.W. Interactions of larval dynamics and substrate preference have ecological significance for benthic biodiversity and Ostrea edulisLinnaeus, 1758 in the presence of Crepidula fornicata. Aquat. Conserv. Mar. Freshw. Ecosyst. 2020;30(11):2133–2149. [Google Scholar]
- Reilly G.D., Reilly C.A., Smith E.G., Baker-Austin C. Vibrio alginolyticus-associated wound infection acquired in British waters Guernsey. Eurosurveillance. 2011;16(42) doi: 10.2807/ese.16.42.19994-en. July 2011. [DOI] [PubMed] [Google Scholar]
- Richter M., Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera I.N., Souza K.M., Souza C.P., Lopes R.M. Free-living and plankton-associated Vibrios: assessment in ballast water, harbor areas, and coastal ecosystems in Brazil. Front. Microbiol. 2012;3:443. doi: 10.3389/fmicb.2012.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz G.M., Rawlings T.K., Dobbs F.C., Drake L.A., Mullady T., Huq A., Colwell R.R. Global spread of microorganisms by ships. Nature. 2000;408(6808):49–50. doi: 10.1038/35040695. [DOI] [PubMed] [Google Scholar]
- Samain J.F., Degremont L., Soletchnik P., Haure J., Bedier E., Ropert M., Moal J., Huvet A., Bacca H., Van Wormhoudt A., Delaporte M., Costil K., Pouvreau S., Lambert C., Boulo V., Soudant P., Nicolas J.L., Le Roux F., Renault T., Gagnaire B., Geret F., Boutet I., Burgeot T., Boudry P. Genetically based resistance to summer mortality in the Pacific oyster (Crassostrea gigas) and its relationship with physiological, immunological characteristics and infection processes. Aquaculture. 2007;268(1–4):227–243. [Google Scholar]
- Sawabe T., Kita-Tsukamoto K., Thompson F.L. Inferring the evolutionary history of Vibrios by means of multi Locus sequence analysis. J. Bacteriol. 2007;189(21):7932–7936. doi: 10.1128/JB.00693-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Seppey M., Manni M., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- Stentiford G.D., Bateman I.J., Hinchliffe S.J., Bass D., Hartnell R., Santos E.M., Devlin M.J., Feist S.W., Taylor N.G.H., Verner-Jeffreys D.W., van Aerle R., Peeler E.J., Higman W.A., Smith L., Baines R., Behringer D.C., Katsiadaki I., Froehlich H.E., Tyler C.R. Sustainable aquaculture through the one health lens. Nat. Food. 2020;1(8):468–474. doi: 10.1038/s43016-020-0127-5. [DOI] [PubMed] [Google Scholar]
- Tada J., Ohashi T., Nishimura N., Shirasaki Y., Ozaki H., Fukushima S., Takano J., Nishibuchi M., Takeda Y. Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol. Cell. Probes. 1992;6(6):477–487. doi: 10.1016/0890-8508(92)90044-x. [DOI] [PubMed] [Google Scholar]
- Takemura A.F., Chien D.M., Polz M.F. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 2014;5:38. doi: 10.3389/fmicb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H., Ohta H., Ogawa M., Mizuguchi Y. Cloning and expression in Escherichia coli of Vibrio parahaemolyticus thermostable direct hemolysin and thermolabile hemolysin genes. J. Bacteriol. 1985;162(2):510–515. doi: 10.1128/jb.162.2.510-515.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers M.A., Boettcher Miller K., Roque A., Friedman C.S. Bacterial diseases in marine bivalves. J. Invertebr. Pathol. 2015;131:11–31. doi: 10.1016/j.jip.2015.07.010. [DOI] [PubMed] [Google Scholar]
- UK Sea Fisheries Statistics, 2019. https://www.gov.uk/government/statistics/uk-sea-fisheries-annual-statistics-report-2019.
- Vezzulli L., Grande C., Reid P.C., Helaouet P., Edwards M., Hofle M.G., Brettar I., Colwell R.R., Pruzzo C. Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc. Natl. Acad. Sci. U. S. A. 2016;113(34):E5062–E5071. doi: 10.1073/pnas.1609157113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli L., Previati M., Pruzzo C., Marchese A., Bourne D.G., Cerrano C., VibrioSea C. Vibrio infections triggering mass mortality events in a warming Mediterranean sea. Environ. Microbiol. 2010;12(7):2007–2019. doi: 10.1111/j.1462-2920.2010.02209.x. [DOI] [PubMed] [Google Scholar]
- Waechter M., Le Roux F., Nicolas J.L., Marissal E., Berthe F. Characterisation of pathogenic bacteria of the cupped oyster Crassostrea gigas. C. R. Biol. 2002;325(3):231–238. doi: 10.1016/s1631-0691(02)01428-2. [DOI] [PubMed] [Google Scholar]
- Wagley S., Borne R., Harrison J., Baker-Austin C., Ottaviani D., Leoni F., Vuddhakul V., Titball R.W. Galleria mellonella as an infection model to investigate virulence of Vibrio parahaemolyticus. Virulence. 2017;9(1):197–207. doi: 10.1080/21505594.2017.1384895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagley S., Koofhethile K., Rangdale R. Prevalence and potential pathogenicity of Vibrio parahaemolyticus in Chinese mitten crabs (Eriocheir sinensis) harvested from the river Thames estuary, England. J. Food Prot. 2009;72(1):60–66. doi: 10.4315/0362-028x-72.1.60. [DOI] [PubMed] [Google Scholar]
- Wagley S., Koofhethile K., Wing J.B., Rangdale R. Comparison of V. parahaemolyticus isolated from seafoods and cases of gastrointestinal disease in the UK. Int. J. Environ. Health Res. 2008;18(4):283–293. doi: 10.1080/09603120801911064. [DOI] [PubMed] [Google Scholar]
- Wendling C.C., Batista F.M., Wegner K.M. Persistence, seasonal dynamics and pathogenic potential of Vibrio communities from Pacific oyster hemolymph. PLoS One. 2014;9(4):e94256. doi: 10.1371/journal.pone.0094256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingett S.W., Andrews S. FastQ Screen: a tool for multi-genome mapping and quality control. F1000Res. 2018;7:1338. doi: 10.12688/f1000research.15931.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherall L., Wagley S., Butler C., Tyler C.R., Temperton B. Genome sequences of four Vibrio parahaemolyticus strains isolated from the English channel and the river Thames. Microbiol. Resour. Announc. 2019;8(24) doi: 10.1128/MRA.00392-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z.Y., Hu C.Q., Chen C., Zhang L.P., Ren C.H. Investigation of seven Vibrio virulence genes amongst Vibrio alginolyticus and Vibrio parahaemolyticus strains from the coastal mariculture systems in Guangdong. Chin. Lett. Appl. Microbiol. 2005;41(2):202–207. doi: 10.1111/j.1472-765X.2005.01688.x. [DOI] [PubMed] [Google Scholar]
- Yoshizawa S., Tsuruya Y., Fukui Y., Sawabe T., Yokota A., Kogure K., Higgins M., Carson J., Thompson F.L. Vibrio jasicida sp. nov., a member of the Harveyi clade, isolated from marine animals (packhorse lobster, abalone and Atlantic salmon) Int. J. Syst. Evol. Microbiol. 2012;62(Pt 8):1864–1870. doi: 10.1099/ijs.0.025916-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.