Highlights
-
•
Collagen cleavage in tumors is primarily mediated by FAP+ cancer-associated fibroblasts.
-
•
Collagen fibers are cleaved in an MMP-dependent manner.
-
•
Released collagen fragments are internalized by M2-like tumor-associated macrophages and cancer-associated fibroblasts.
-
•
The mannose receptor is central in collagen internalization by tumor-associated macrophages.
Keywords: Extracellular matrix remodeling, Tumor microenvironment, Collagen degradation, Collagen endocytosis, Tumor-associated macrophages, Cancer-associated fibroblasts, Matrix metalloproteinases
Abbreviations: ECM, extracellular matrix; MMP, matrix metalloproteinase; FAP, fibroblast activation prot; MR, mannose receptor; uPARAP, urokinase plasminogen activator receptor-associated protein; TME, tumor microenvironment; NK, natural killer cell; IL, interleukin; TNF, tumor necrosis factor; CPM, counts per minute; ELISA, enzyme-linked immunosorbent assay; CAF, cancer-associated fibroblast; TAM, tumor-associated macrophage; α-SMA, α-smooth muscle actin; FSP-1, fibroblast-specific protein 1; PDGFR, platelet-derived growth factor receptor; ATCC, American Type Culture Collection; FMO, fluorescence minus one; CRC, colorectal cancer; LC, lung cancer; OvC, ovarian cancer
Abstract
Increased remodeling of the extracellular matrix in malignant tumors has been shown to correlate with tumor aggressiveness and a poor prognosis. This remodeling involves degradation of the original extracellular matrix (ECM) and deposition of a new tumor-supporting ECM. The main constituent of the ECM is collagen and collagen turnover mainly occurs in a sequential manner, where initial proteolytic cleavage of the insoluble fibers is followed by cellular internalization of large well-defined collagen fragments for lysosomal degradation. However, despite extensive research in the field, a lack of consensus on which cell types within the tumor microenvironment express the involved proteases still exists. Furthermore, the relative contribution of different cell types to collagen internalization is not well-established. Here, we developed quantitative ex vivo collagen degradation assays and show that the proteases responsible for the initial collagen cleavage in two murine syngeneic tumor models are matrix metalloproteinases produced by cancer-associated fibroblasts and that collagen degradation fragments are endocytosed primarily by tumor-associated macrophages and cancer-associated fibroblasts from the tumor stroma. Using tumors from mannose receptor-deficient mice, we show that this receptor is essential for collagen-internalization by tumor-associated macrophages. Together, these findings identify the cell types responsible for the entire collagen degradation pathway, from initial cleavage to endocytosis of fragments for intracellular degradation.
Introduction
Increased turnover of interstitial collagen is a key characteristic of invasive tumor growth and leads to fibrosis and the destruction of the healthy tissue [1], [2], [3]. This process includes degradation of the original extracellular matrix (ECM), generating space for the expanding tumor, as well as causing a release of matrix-embedded growth factors and bioactive fragments, further potentiating growth [4], [5]. This ECM degradation is accompanied by the deposition of a tumor-specific ECM with altered composition and increased stiffness, creating a tumor-supportive environment [6], [7], [8], [9], [10], [11]. Many collagens, including the most abundant of them, collagen type I, form large insoluble fibers. These are resistant to degradation by most proteases, except a few which mainly belong to the matrix metalloproteinase (MMP) family. However, other proteases including serine proteases (e.g. fibroblast activation protein (FAP)) and cysteine proteases (e.g. cathepsin B) have also been shown to possess some collagenolytic activity [12], [13], [14], [15], [16], [17]. Most MMPs are produced as inactive zymogens, which require activation by proteolytic removal of their prodomain, a process undertaken by other MMPs or serine proteases [18], [19]. The degradation of collagen is initiated by proteolytic cleavage generating large well-defined fragments [20]. These fragments can be further degraded by gelatinases in the extracellular space [21] or in a process involving intracellular degradation [22]. In this latter degradation pathway, the precleaved collagen fragments are internalized by receptor-mediated endocytosis and directed to the lysosomes for degradation. The internalization can be mediated by two receptors of the mannose receptor family; the mannose receptor (MR, also known as CD206) governing internalization by macrophages, and the urokinase plasminogen activator receptor-associated protein (uPARAP, also known as Endo180, CD280) mainly governing internalization by fibroblasts [23], [24], [25]. Both MR and uPARAP have been shown to be important in fibrosis during tumor growth, as their depletion leads to increased accumulation of collagen in Lewis lung carcinomas and PymT-induced ductal mammary adenocarcinomas, respectively [26], [27]. In addition, loss of uPARAP resulted in reduced tumor burden, exemplifying the importance of collagen turnover in tumor progression.
The tumor microenvironment (TME) consists of not only cancer cells but also of a wide variety of non-malignant cells. These include endothelial cells, cancer-associated fibroblasts (CAFs) and immune cells such as T cells, tumor-associated macrophages (TAMs), and dendritic cells. The most abundant non-malignant cell types of the TME are usually CAFs and TAMs [28], [29], [30]. Macrophages have traditionally been classified as either M1-like or M2-like. M1-like macrophages are pro-inflammatory and are involved in killing of pathogens and in host anti-tumor response. M2-like macrophages are anti-inflammatory cells involved in immune regulation and wound healing and are known to promote tumor growth.
Despite the fact that collagen degradation in tumors for decades has been known to be initiated by a proteolytic cleavage, the cellular sources of the involved proteases remain unclear [17], [31], [32]. Recently, intravital microscopy was used to show that the subsequent step of collagen degradation, i.e. internalization of collagen fragments, is primarily mediated by M2-like macrophages in an MR-dependent manner [26], [33]. Furthermore, MR-dependent collagen uptake by macrophage subsets in the dermis has been demonstrated using flow cytometry analysis [34]. However, these studies did not identify the cells responsible for the initial cleavage. Here, we used an ex vivo, quantitative collagen degradation assay combined with tumor single-cell suspensions to demonstrate that a native-like collagen matrix is cleaved primarily by MMPs originating from CAFs. In addition, using a variation of the assay, we were able to quantify the relative importance of two known collagen-internalizing cells, CAFs and TAMs, for the subsequent degradation of collagen fragments.
Results
Collagen cleavage by tumors is mediated by FAP+ CAFs in an MMP-dependent manner
The TME consists of many different cell types that are likely to not tolerate ex vivo culturing equally well. To investigate if all cell types originally present in the tumors were still present after several days of ex vivo culture, single-cell suspensions of murine subcutaneous Lewis lung tumors (LL/2) and orthotopic mammary EO771.LMB tumors were seeded in tissue-culture plates. The cellular composition of the tumors was determined using flow cytometry analyses (Fig. S1, gating strategies in Fig. S2A). The LL/2 and EO771.LMB cells were transduced to express GFP. In both models, the percentage of leukocytes present in the single-cell suspensions decreased with time, leading to an increase in the ratio of cancer cells to non-cancer cells after several days of culture (Fig. S1). In the EO771.LMB model, but not LL/2, CAFs also appeared to be negatively affected by ex vivo culture. These data suggest that for the purposes of this study, culture time should be kept at a minimum not exceeding two days.
To determine the importance of different proteases in the initial cleavage of insoluble collagen fibers, a quantitative collagen degradation assay was used [35]. In this assay, a single-cell suspension of LL/2 tumors was cultured for two days on a reconstituted collagen type I matrix in which radiolabeled collagen was incorporated. Collagen fragments released from the matrix following proteolytic cleavage could then be measured as radioactivity in the culture supernatant. The addition of protease inhibitors targeting MMPs (GM6001), serine proteases (aprotinin), cysteine proteases (E-64d) or both serine- and cysteine proteases (leupeptin) showed a clear dependence on MMPs for collagen cleavage with no involvement of other protease families (Fig. 1A). The TME is known to constitute a highly inflammatory setting [36]. Therefore, to mimic such a condition, collagen degradation was also assessed in the presence of the pro-inflammatory cytokines TNF-α and IL-1β. These cytokines are widely used for stimulation of collagenolytic activity in similar assay systems [23], [35], [37], [38]. Indeed, the addition of TNF-α and IL-1β led to increased collagen degradation and an MMP-dependent degradation was again observed as evidenced by the GM6001-mediated reduction in radioactivity release (Fig. 1B). Interestingly, under these conditions aprotinin led to a small but significant reduction in collagen degradation showing that serine proteases also contribute to this process. Although this could reflect a direct collagen degradation by serine proteases, it is also possible that aprotinin inhibits the activation of other collagen degrading proteases. Indeed, serine proteases have been suggested to be involved in activation of several MMPs [39].
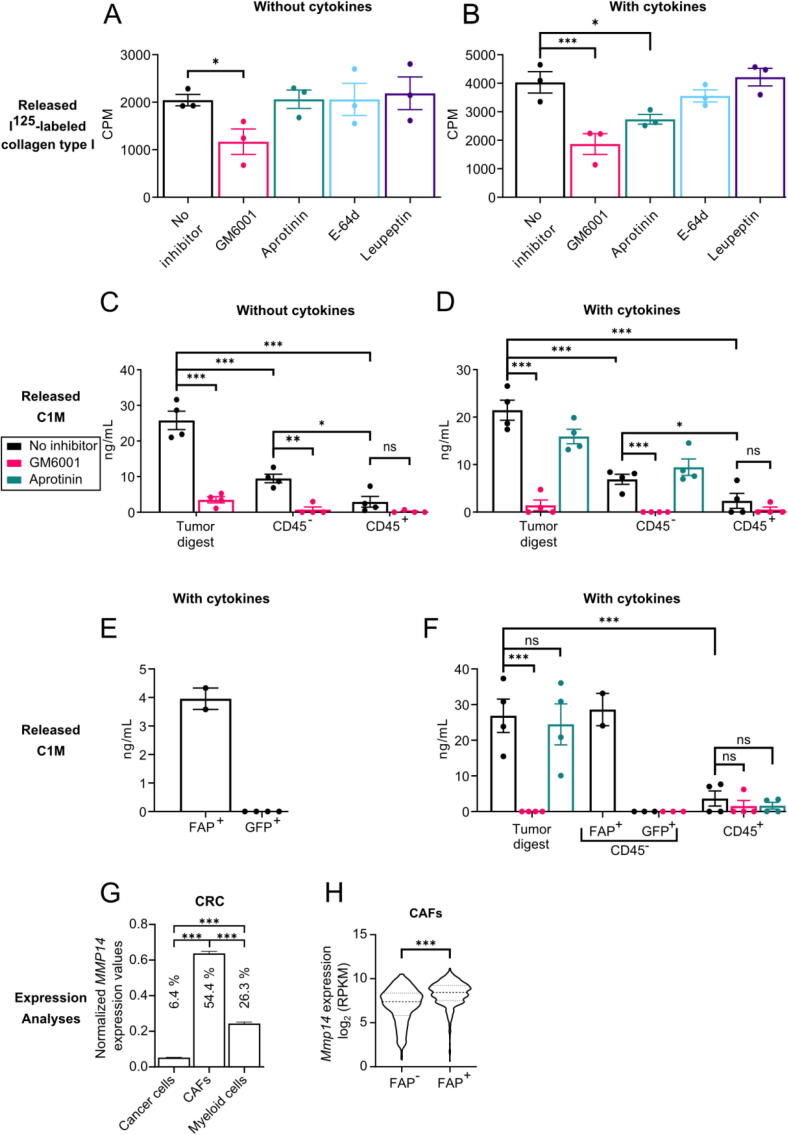
Fig. 1.
Collagen degradation by tumors is MMP-dependent and primarily mediated by FAP+ CAFs. (A-B) Degradation of collagen matrices and release of incorporated radiolabeled collagen by single-cell suspensions of LL/2 cultured for two days without (A) or with (B) the addition of 10 nM TNF-α and 1 nM IL-1β to the culture medium. GM6001 (20 µM), aprotinin (10 µM), E-64d (20 µM) or leupeptin (10 µM) was added to the culture medium where indicated. CPM = counts per minute. n = 3. Error bars = SEM. *p ≤ 0.05, ***p ≤ 0.001; one-way ANOVA with post hoc Fisher’s LSD test. (C-F) Degradation of collagen matrices and release of MMP-specific collagen type I fragments containing the neo-epitope C1M by single-cell suspensions of whole tumors (tumor digest) or sorted CD45−, CD45+, FAP+ or GFP+ cells of LL/2 (C-D), LL/2-GFP (E) or EO771.LMB-GFP (F) cultured for two days without (C) or with (D-F) the addition of 10 nM TNF-α and 1 nM IL-1β to the culture medium. GM6001 (20 µM) or aprotinin (10 µM) were added to the culture medium where indicated. C1M levels were determined using ELISA. n = 4, except for GFP+ in 1F (n = 3) and FAP+ (n = 2). Error bars = SEM. *p ≤ 0.05, ***p ≤ 0.001; one-way ANOVA with post hoc Fisher’s LSD test. (G) Expression of MMP14 in cancer cells, CAFs, and myeloid cells from colorectal cancer (CRC) from a publicly available scRNAseq data set. Error bars = SEM. *p ≤ 0.05, ***p ≤ 0.001; one-way ANOVA with post hoc Tukey’s multiple comparisons test. Percentages of analyzed cells where the transcript was detected are depicted for each cell population. (H) Violin plot showing Mmp14 expression in FAP-negative (FAP−) and FAP-positive (FAP+) CAFs from a publicly available scRNAsq data set. ***p ≤ 0.001; two-tailed Student’s t-test.
The dependence on MMPs for collagen degradation prompted us to use an innovative and highly sensitive enzyme-linked immunosorbent assay (ELISA) developed to detect a neo-epitope, C1M, generated by a site-specific cleavage of collagen type I by members of the MMP-family [40]. For this assay, single-cell suspensions of enzymatically digested whole tumors or cell suspensions enriched for leukocytes (CD45+) or non-leukocytes (CD45−) were seeded on reconstituted collagen type I matrices. After two days of culture, supernatants were collected and analyzed using the ELISA. Importantly, the addition of GM6001 to the culture medium also in this setup resulted in a significant reduction in collagen type I fragments released by the whole tumor digest (Fig. 1C). Furthermore, the CD45− cells degraded significantly more collagen type I than the CD45+ cells, and this degradation was also inhibited by GM6001 (Fig. 1C). Addition of cytokines to mimic the inflamed TME again showed an MMP-dependent collagen degradation by the whole tumor digest and the CD45− fraction (Fig. 1D). In both settings, the small collagenolytic activity of CD45+ cells also seemed to be inhibited by GM6001 although this reduction was not statistically significant (Fig. 1C and D). This might be due to the very low degradation ability of the leukocytes making it difficult to detect any effects of GM6001. We also studied the impact of aprotinin on the generation of the neo-epitope but did not observe a significant effect although a trend towards reduced collagen degradation by the whole tumor digest was observed (Fig. 1D). To determine which CD45− cells were responsible for the collagen cleavage, we performed a separate experiment using LL/2 cells transduced with GFP to generate tumors, allowing us to distinguish cancer cells from non-leukocytic stromal cells. GFP+ cancer cells and FAP+ CAFs were isolated by fluorescence-activated cell sorting (FACS) and their ability to cleave collagen was measured. Due to low abundance of the FAP+ CAFs we were only able to isolate enough of these cells for two replicates. However, it was clear that the CAFs were capable of cleaving collagen while this was not the case for the cancer cells (Fig. 1E).
To investigate if these observations were specific for LL/2 tumors, we performed a similar experiment using orthotopic EO771.LMB breast tumors generated with GFP-transduced EO771.LMB cells. As was the case for LL/2, the tumor digest cleaved collagen in an MMP-dependent manner, as seen by the complete inhibition of cleavage by addition of GM6001 (Fig. 1F). Aprotinin did not affect the cleavage of collagen in this setup. The level of cleavage by the leukocytes was much smaller than for the complete tumor digest and this was not significantly affected by the addition of any of the two protease inhibitors although a trend of reduced collagen degradation was observed. As for LL/2, the cancer cells did not partake in collagen cleavage at all, while the CAFs cleaved collagen to a similar extent as the tumor digest, suggesting that they are the cells responsible for the collagen cleavage observed for the complete tumor digest.
Next, we used publicly available single-cell (sc) RNAseq data sets to investigate the cellular expression pattern of the collagenolytic MMPs (MMP-1, MMP-8, MMP-13, and MMP-14) in human cancers [41]. When looking into the expression of MMP14 in cancer cells, CAFs, and myeloid cells in human colorectal cancer (CRC), it was clear that CAFs were the main source of this protease (Fig. 1G). CAFs were also the main source of MMP1, MMP8, and MMP13 although with varying contributions from cancer cells and myeloid cells (Fig. S3A-C). It should also be noted that these three MMPs were expressed at much lower levels with transcripts only detected in a small fraction of analyzed cells. CAFs were also the main source of MMP14 in two other cancer types, i.e. lung cancer (LC) and ovarian cancer (OvC) (Fig. S3D-E).
We used FAP as a fibroblast marker and even though it is not a pan-fibroblast marker, we showed that FAP+ CAFs are the main source of collagenolytic MMPs. To investigate if FAP+ CAFs compared to FAP− CAFs express higher levels of collagenolytic MMPs in vivo, we used another publicly available scRNAseq data set of CAFs from murine mammary tumors [42]. By dividing CAFs into those expressing FAP and those not expressing FAP, we could compare the expression of MMPs in these subsets (Fig. 1H). Mmp14 was significantly higher expressed in FAP+ CAFs, suggesting that FAP+ CAFs are the dominant cells of the TME expressing this collagenolytic MMP. Mmp1 and Mmp8 were not expressed in this data set and MMP13 were expressed at equal levels in the FAP+ and FAP− CAFs (Fig. S3F).
Taken together, these findings show that collagen cleavage is mediated by MMPs produced by the FAP+ CAFs in the TME, with no involvement of the cancer cells. Transcriptomic analyses indicate that MMP-14 could be centrally engaged in CAF-mediated collagen degradation. Interestingly, in an inflammation-like situation, serine proteases seem to influence collagen degradation as well.
CAFs and M2-like TAMs are the main collagen-internalizing cells of the tumor microenvironment
Internalization of the proteolytically cleaved collagen fragments for complete intracellular degradation has been suggested to be mediated by MR expressed by macrophages and uPARAP expressed by fibroblasts [23], [24], [25]. However, the relative importance of the two cell types for this process has not yet been quantitatively determined. To compare the contribution of different cell types of the TME in collagen internalization, single-cell suspensions of LL/2 tumors were incubated overnight with fluorescently labeled soluble collagen type I mimicking large proteolytically released collagen fragments and analyzed by flow cytometry. For gating strategies, see supplementary Fig. S2. Consistent with previous studies, we saw that both TAMs and CAFs efficiently internalized soluble collagen (Fig. 2A, Fig. S4A) [24], [26], [43], [44].
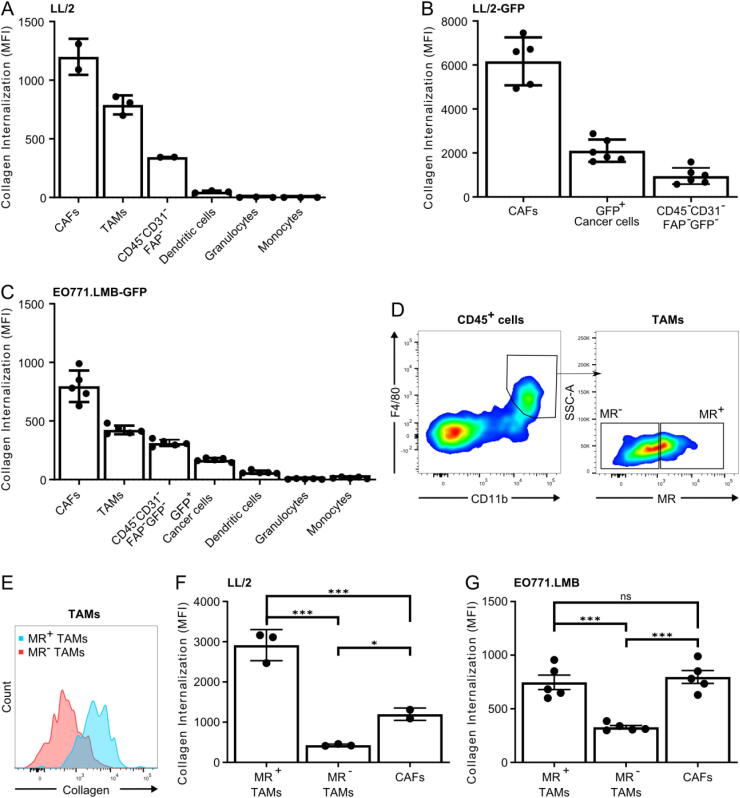
Fig. 2.
CAFs and MR+ TAMs are the main cell types in collagen internalization. (A-C) Quantification of collagen internalization by cell types of the tumor microenvironment: Single-cell suspensions of LL/2 tumors (A), LL/2-GFP tumors (B) or EO771.LMB-GFP tumors (C) were cultured overnight with soluble 10 µg/mL A647-labeled collagen type I. Flow cytometry analysis was then performed to determine the internalized fluorescence in CAFs, TAMs, dendritic cells, granulocytes, monocytes, cancer cells, CD45−CD31−FAP− cells and CD45−CD31−FAP−GFP− cells. n = 2–6. Error bars = SEM. (D) Representative density plots from flow cytometry analysis of LL/2 tumor showing how TAMs were divided into subpopulations negative and positive for MR. Gates were based on isotype control. (E) Representative overlay of histograms from flowcytometry analysis of LL/2 tumors showing the level of internalized collagen in TAMs negative and positive for MR. (F-G) Quantification of collagen internalization by MR+ macrophages, MR− macrophages and FAP+ fibroblasts from LL/2 (F) and EO771.LMB tumors (G). n = 3–5, except CAFs in 3F (n = 2). Error bars = SEM. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; one-way ANOVA with post hoc Tukey’s multiple comparisons test.
The CD45−CD31−FAP− cells, of which the majority is expected to be cancer cells, internalized only small amounts of collagen, while dendritic cells, granulocytes and monocytes did not contribute to collagen internalization (Fig. 2A, Fig. S4A). Since the CD45−CD31−FAP− population most likely also contains FAP− CAFs, we again employed GFP-expressing LL/2 cells for tumor generation to distinguish cancer cells from non-leukocytic stromal cells. Single-cell suspensions of the tumors were prepared and the collagen internalization experiment was repeated. This clearly showed that FAP+ CAFs are the main collagen internalizing non-leukocytic stromal cell type (Fig. 2B). Surprisingly, the GFP+ cancer cells internalized more collagen than the remaining CD45−CD31−GFP−FAP− fraction comprising a population most likely consisting of FAP− fibroblasts and non-cancerous epithelial cells.
To determine if these observations also applied to other models than the LL/2, we again used the EO771.LMB model. Specifically, GFP-transduced EO771.LMB cells were injected orthotopically and the resulting tumors were dissociated into single-cell suspensions used for assessing cell type-specific collagen internalization. Consistent with the LL/2 model, CAFs were the main collagen internalizing cells, followed by TAMs (Fig. 2C, Fig. S4B). We did observe that the involvement of cancer cells was smaller than the FAP− CAF population in this model.
We next divided the TAMs in two subpopulations, those expressing the M2-macrophage marker MR and those that do not (Fig. 2D). This analysis showed that the MR+ M2-like TAMs internalize the most collagen, while the MR− M1-like TAMs internalize significantly less collagen than both the MR+ TAMs and the CAFs (Fig. 2E and F), supporting previous findings [26], [43]. In the EO771.LMB model, the M2-like TAMs internalize collagen to the same extent as the CAFs and significantly more than the MR− TAMs (Fig. 2G).
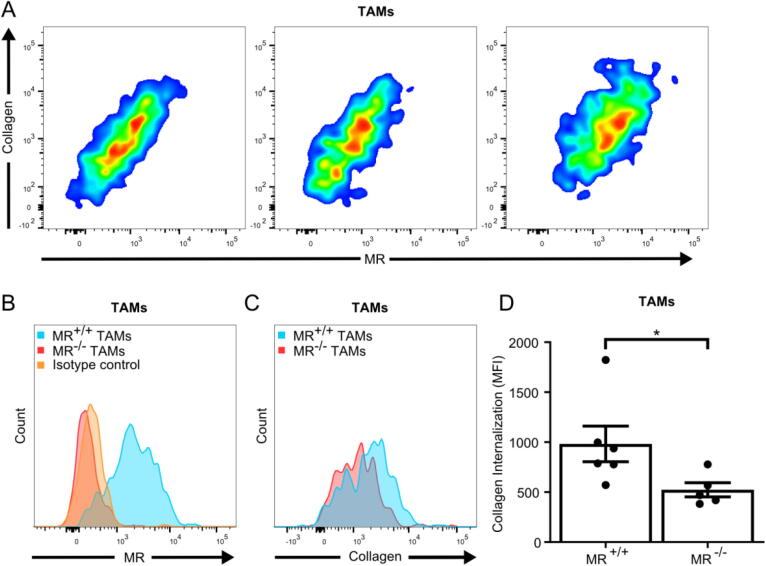
When looking into the possible dependence on MR for collagen internalization by TAMs, we found a clear positive correlation between the expression of MR on TAMs and the level of internalized collagen (Fig. 3A). A direct role of MR in collagen internalization by TAMs was therefore investigated using MR-deficient mice and wildtype littermates engrafted with LL/2 cells (Fig. 3B). Again, single-cell suspensions of the tumors were incubated overnight with fluorescently labeled soluble collagen type I before flow cytometry analysis. Loss of MR significantly diminished collagen internalization by TAMs (Fig. 3C and D). Overall, this establishes M2-like TAMs and CAFs as the main collagen-internalizing cells in the TME.
Fig. 3.
MR is essential for collagen internalization by TAMs. (A) Three independent representative examples of flow cytometry analyses of LL/2 tumor single-cell suspensions showing a correlation between MR expression and collagen internalization by TAMs. See Fig. S2 for details on gating. (B and C) Flow cytometry analysis of the expression of MR (B) or internalization of collagen (C) by TAMs in LL/2 tumor single-cell suspensions acquired from MR−/− mice or from wildtype littermates (MR+/+). (D) Quantification of internalized collagen by TAMs from tumors from MR−/− mice or from wildtype littermates. n = 5–6 Error bars = SEM. *p ≤ 0.05; Mann-Whitney test.
Discussion
Degradation of collagen, the most abundant protein of the ECM, has been extensively studied, but consensus about the mediators involved in this process has not yet been reached [32]. In this study, we used quantitative ex vivo collagen cleavage assays to investigate the initial proteolytic cleavage of collagen fibers and a flow cytometry-based internalization assay to determine the relative importance of different cell types for endocytosis of the released collagen fragments.
By using five broad-spectrum protease inhibitors to target different protease families we could show that the proteases responsible for the initial cleavage belong to the MMP family. Interestingly, in the setup with addition of pro-inflammatory cytokines we observed an effect of aprotinin, suggesting a minor contribution of serine proteases to the collagen cleavage. This could be the result of a direct cleavage of collagen fibers or an indirect effect since serine proteases are known to be able to activate latent MMPs [19], [39], [45]. In the ELISA-based cleavage assay, we only saw a weak tendency of an effect of aprotinin on collagen cleavage. Since this assay is based on detection of a neo-epitope created by specific MMP-mediated cleavage of collagen, any direct cleavage by serine proteases would not be detected.
It would be of great interest to investigate exactly which MMPs and serine proteases are responsible for the cleavage we see. However, both the MMP- and serine protease family have many members and finding potent and specific inhibitors for all will not be an easy task. Future studies dedicated to address this are needed.
Regarding the cellular source of the collagen-cleaving proteases, we saw that the CD45-negative cells, in particular FAP+ CAFs, released significantly more collagen degradation fragments compared to the leukocytes. We also showed that the cancer cells did not contribute to the cleavage at all. This was the case in both tumor models employed. However, the amount of released collagen fragments by the isolated CD45-negative cells in the LL/2 model was substantially lower than the amount released by the whole tumor digest, comprising both CD45-negative cells and leukocytes. This probably reflects the complex interactions between different cell types. Although leukocytes may not contribute directly to collagen cleavage, they may do so indirectly by production of reactive oxygen species or inflammatory cytokines, such as IL-1β and TNF-α, inducing expression of MMPs by CAFs [1], [36], [46]. In this study we observed an increase in collagen cleavage (Fig. 1A compared with 1B) in the presence of these cytokines. However, this effect was not seen in the ELISA detecting the collagen type I neo-epitope C1M (Fig. 1C compared with 1D). Since C1M is generated by a site-specific cleavage by members of the MMP-family, it is possible that the cytokines cause an increased expression of other proteases, which could lead to degradation and destruction of C1M and therefore an underestimated level of collagen type I cleavage [40].
Regarding the source of the MMPs responsible for the initial proteolytic cleavage of collagen, conflicting reports exist [18], [32]. In our previous study, CAFs and TAMs were shown to be sources of MMPs while tumor cells showed very low or no expression of these proteases [26]. The latter is in accordance with the results of this study, as we see no collagen cleavage at all by cancer cells. We also see very little collagen cleavage by the leukocytes although we previously showed expression of MMPs by TAMs. However, CAFs and TAMs had very different expression patterns of MMPs and it is possible that the MMPs expressed by TAMs are less important for collagen cleavage than the MMPs expressed by CAFs.
CAFs are a highly heterogeneous group of cells and no single marker can reliably be used to detect all subtypes. Markers commonly used to detect CAFs include α-smooth muscle actin (α-SMA), fibroblast-specific protein 1 (FSP-1), platelet-derived growth factor receptor (PDGFR)–α and -β and FAP [47], [48]. Different subtypes of CAFs display varying levels of activity as well as distinct properties [48]. It is therefore likely that some subtypes express higher levels of MMPs than others and, therefore, will be more important for collagen cleavage. In this study, FAP was used as a marker for CAFs as it has been shown to be expressed by activated CAFs in the majority of human epithelial cancers [49], [50]. In addition to being a marker for activated CAFs, FAP is itself a membrane-bound serine protease with some collagenolytic activity [12], [49]. Due to this heterogeneity of CAFs, using FAP as our CAF marker will not captivate the full picture of CAF functions. However, for both cleavage and internalization of collagen we see that the FAP+ CAFs are the main non-leukocytic stromal cells. It should also be noted that FAP has been shown to also be expressed by a few other cell types other than CAFs, although CAFs are the predominant FAP-expressing cells [51].
Our initial investigations of the impact of ex vivo culturing on different cell types show that CAFs and especially leukocytes are negatively affected by the cell culture conditions, or they simply become overgrown by the cancer cells. In either case, this could potentially cause an underestimation of the importance of mainly leukocytes, but also CAFs, for internalization of collagen. Although their abundance diminished, the use of a live/dead marker in the initial gating ensures that the cells identified are viable. Still, the large differences in abundance between the different populations could affect the interpretation of their importance. To account for this, collagen internalization was depicted as MFI to reflect the average uptake per individual cell.
Using intravital microscopy it was previously shown that collagen in skin and in tumors is mainly internalized by M2-like macrophages with little contribution from fibroblasts [26], [43]. This method was also used to determine that TAMs are the main tumor-associated collagen-internalizing cell type when this process is modeled in vivo [26]. However, the visualization of collagen internalization in situ does not permit accurate quantification of collagen internalization nor does it allow for determination of the relative contribution of the involved cell types. In this study, using a flow cytometry-based internalization assay, we were able to compare the contribution of TAMs and CAFs to the total process of collagen internalization. The findings of this study show a more prominent role of CAFs in collagen internalization when compared with all TAMs (Fig. 2A and C). However, it cannot be excluded that the relatively large contribution from fibroblasts observed in our study was due to an increased activation caused by the ex vivo culture, since high levels of collagen internalization by fibroblasts have been shown before in in vitro studies [23], [24], [44]. When comparing collagen internalization by CAFs with TAMs divided into MR+ and MR− subpopulations, it was evident that the MR+ TAMs were the most efficient collagen-internalizing cell type (Fig. 2F and G). This supports the previous notion that MR+ M2-like macrophages are the predominant collagen-internalizing cell type, with a minor contribution from MR− M1-like macrophages [26]. Although MR+ macrophages internalized collagen more efficiently than MR− macrophages in our study, the internalization by the MR-deficient macrophages was not completely ablated, pointing towards the existence of an alternative internalization route. This could potentially involve uPARAP as it has been shown to be expressed on macrophages and to mediate collagen internalization by macrophages in vivo, although to a lesser extent than MR [43], [52], [53]. Furthermore, macropinocytosis has been suggested to constitute an additional pathway of collagen internalization in macrophages [54], [55].
Altogether, these findings provide insights into the complete collagen degradation pathway active in tumors. Our study establishes CAFs, as the central cell type responsible for the initial cleavage of collagen, and CAFs and M2-like TAMs as the predominant cells engaged in the internalization of the released collagen fragments for complete intracellular degradation.
Experimental procedures
Animal experiments
Animal experiments were performed in accordance with institutional guidelines and approved by the animal experiments inspectorate in Denmark. Mrc1+/− mice in a C57BL/6 background were interbred to generate Mrc1−/− mice and littermate Mrc1+/+ control mice. Genotyping was performed as previously described [43].
Tumor models
Lewis lung carcinoma (LL/2) cells (American Type Culture Collection (ATCC)) were cultured in DMEM (Thermo Scientific) with 10% fetal bovine serum (FBS) and 1% Penicillin-Streptomycin (P/S) (Gibco by Life Technologies). The EO771.LMB cell line was kindly gifted from Prof. Robin L. Anderson (Olivia Newton-John Cancer Research Institute, Heidelberg, Australia) and cultured in DMEM with 20% FBS, 1% P/S and 20 mM HEPES (Gibco). For some experiments, the cell lines were transduced to stably express GFP by lentiviral transduction.
Female C57BL/6 mice or mixed-sex Mrc1−/− mice and littermate Mrc1+/+ control mice were injected subcutaneously with 500,000 LL/2 cells in 100 μL DMEM on each flank. After 10–14 days, the mice were sacrificed, and the tumors were excised. For orthotopic injections with EO771.LMB, mice were anaesthetized using isoflurane and 500,000 cells in 50 μL PBS were injected in the mammary fat pad around the 4th nipple.
Multicolor flow cytometry
After excision, tumors were finely diced and incubated in digestion buffer (2.1 mg/mL bacterial collagenase type I (Corning) and 75 μg/mL DNase I (Worthington) in RPMI (Gibco by Life Technologies) supplemented with 1% Penicillin-Streptomycin) with rotation overnight at 4 °C for single cell isolation. The following day, the tumors were incubated at 37 °C with gentle shaking for 15 min before the suspension was pipetted up and down to mechanically dissolve any remaining clumps. The suspension was filtered twice through 70 μm cell strainers (Corning), centrifuged at 300xg for 5 min, and the supernatant was discarded. Red blood cells were lysed by resuspending the cells in 2 mL RBC lysis buffer (Sigma-Aldrich), pipetting gently up and down for 30 s and leaving it still for 2 min before diluting the RBC lysis buffer with RPMI with 10% FBS and 1% P/S. The cells were centrifuged at 300xg and resuspended in staining buffer (5 mM EDTA, 0.5% bovine serum albumin in PBS (Lonza)) before staining. Antibody panels are listed in table S1. When using non-GFP-expressing tumors, ZombieAqua was used for dead cell exclusion. When using GFP expressing tumors, the Invitrogen™ LIVE/DEAD™ Fixable Near-IR Dead Cell Stain was used for dead cell exclusion. Fluorescence minus one (FMO) controls or isotype controls were included. Stained cells were washed in staining buffer and analyzed using a FACSCanto II (BD Biosciences). Data was analyzed using FlowJo software.
CD45 enrichment
Cells were blocked for 10 min at 4 °C with murine FcR-blocking reagent (Miltenyi Biotec) before the CD45+ and CD45− fractions of tumor suspensions were isolated with anti-CD45 MicroBeads (Miltenyi Biotec) according to the manufacturer’s instructions.
Fluorescence-activated cell sorting (FACS)
After CD45 enrichment, the CD45-negative fraction was used to sort FAP+ CAFs and GFP+ cancer cells. The cell suspensions were stained with Invitrogen™ LIVE/DEAD™ Fixable Near-IR Dead Cell Stain (Fisher Scientific) and biotinylated anti-FAP/Streptavidin-BV421. After staining, the cells were resuspended in staining buffer at 107 cells/mL and sorted using a FACSMelody (BD Biosciences).
Radioidodination
Labeling of collagen with I125 was performed by a modification of a procedure previously described [56]. Briefly, 3 mg of Iodogen was dissolved in 25 mL chloroform. 100 μL was added to a glass tube and left overnight on an agitation table in a fume hood to let the solution evaporate. 20 μg of rat tail tendon collagen type I (Corning) was dissolved in 0.1 M Tris-HCl, pH 7.6. 1 mCi of I125 (Perkin Elmer) was added to the tube to a total volume of 100 μL and the labeling proceeded at room temperature with agitation for 10 min. The reaction was then stopped by addition of 900 μL 0.1 M Tris-HCl, pH 8.1, 0.01% Tween 80. Finally, the solution was desalted by gel filtration.
Fluorescence-labeling of collagen
For the internalization assay, collagen was labeled by a modification of a procedure previously described [57]. Briefly, rat tail tendon collagen type I (Corning) (1 mg/mL final concentration) was polymerized by gently mixing with 13 mM HCl before addition of a neutralizing buffer to reach a final pH of 7.4. The mix was incubated at 37 °C for 1½-2 h, washed once with sterile PBS before addition of PBS to a total volume of 6 mL and then 0.8 mL 1 M sodium bicarbonate. A vial of Alexa Fluor 647 (A647) fluorescent dye (Life Technologies) dissolved in 0.5 mL PBS was immediately added and incubated with gentle shaking for two hours at room temperature. Unbound dye was removed by regular washing with PBS for 2–3 days. The labeled collagen was washed once with water to remove PBS and then washed very briefly in 13 mM HCl. The collagen was left overnight stirring at 4 °C in 13 mM HCl. The following day any small leftover fibrils were removed by centrifugation at 20000xg for 60 min at 4 °C.
Collagen degradation assays
For some degradation assays, 125I-labeled collagen was incorporated into trypsin-resistant collagen matrices [23], [35]. Briefly, rat tail tendon collagen type I (500 μg/mL final concentration) was acidified and then brought to neutral pH by addition of appropriate volumes of 13 mM HCl followed by a neutralizing buffer. A trace amount of 125I-labeled collagen was immediately added to obtain a final radioactivity of 500,000 CPM/mL before each well of a 24-well tissue culture plate was coated with 350 μL of this solution. The collagen was incubated for 2–4 h at 37 °C to allow the collagen to polymerize before being left to dry in a flow hood for two days. Before use, the collagen matrices were washed three times in sterile water and left 24 h at room temperature in PBS. Tumor single-cell suspensions were added, using 0.5 × 106 cells/well in 500 µL RPMI with 10% FBS and 1% P/S in 24-well plates. Where indicated, 10 nM TNF-α (PeproTech), 1 nM IL-1β (PeproTech), 20 µM GM6001 (Calbiochem), 10 µM aprotinin (Sigma-Aldrich), 20 µM E-64d (Sigma-Aldrich) or 10 µM leupeptin (Sigma-Aldrich) were added to the culture medium. After two days of culture, collagen degradation was measured as the release of radioactivity into the supernatant using a Wizard2 gamma counter (Perkin Elmer).
Alternatively, collagen matrices were prepared as described above, without incorporation of 125I-labeled collagen and without 24 h of incubation with PBS [37], [38]. Complete tumor single-cell suspensions, the CD45+ fraction, CD45− fractions, sorted FAP+ CAFs or GFP+ cancer cells were added, using 500,000 cells/well in 350 µL RPMI with 10% FBS and 1% P/S in 24-well plates. Due to the larger size of these cells, 300,000 cells/well of sorted CAFs and cancer cells were used. Where indicated, 10 nM TNF-α, 1 nM IL-1β, 20 µM GM6001 or 10 µM aprotinin were added to the culture medium. After two days of culture, the culture supernatants were collected and an MMP-generated collagen type I degradation product (C1M) released to the culture medium was measured using a competitive ELISA developed by Nordic Bioscience [40], [58]. Briefly, 96-well pre-coated streptavidin plates were coated with biotinylated peptides specific for the C1M peptide and incubated for 30 min at 20 °C. Standard peptide or supernatant were added followed by addition of peroxidase-conjugated monoclonal antibodies and incubated for 20 h at 4 °C. Next, tetramethylbenzinidine (Kem-En-Tec Diagnostics, Denmark) was added and incubated for 15 min at 20 °C followed by adding 1% sulfuric acid to stop the reaction. The absorbance was measured at 450 nm with 650 nm as reference and analyzed using the Softmax Pro v. 6.3 software.
Ex vivo collagen internalization assay
For quantification of ex vivo collagen internalization by cells of the TME, LL/2 or EO771.LMB tumors were dissociated to single-cell suspensions and directly cultured overnight with A647-labeled soluble collagen and analyzed by flow cytometry, as described above.
Statistical analyses
All statistical tests were performed using GraphPad Prism 8. Unpaired two-tailed Student’s t-test or Mann-Whitney test were used where appropriate for comparison of two groups. One-way ANOVA was used for comparison of three groups or more with post hoc Fisher’s LSD test or Tukey’s multiple comparisons test.
CRediT authorship contribution statement
Marie-Louise Thorseth: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft, Visualization, Writing – review & editing. Marco Carretta: Formal analysis, Investigation, Visualization, Writing – review & editing. Christina Jensen: Methodology, Investigation, Writing – review & editing. Kasper Mølgaard: Methodology, Writing – review & editing. Henrik J. Jürgensen: Resources, Methodology, Investigation, Writing – review & editing. Lars H. Engelholm: Resources, Methodology, Writing – review & editing. Niels Behrendt: Resources, Methodology, Writing – review & editing. Nicholas Willumsen: Methodology, Resources, Writing – review & editing. Daniel H. Madsen: Conceptualization, Methodology, Formal analysis, Resources, Writing – original draft, Supervision, Project administration, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: CJ and NW are employed by Nordic Bioscience A/S, which is a company involved in discovery and development of biochemical biomarkers.
Acknowledgements
We thank Signe Ingvarsen for critically reviewing the manuscript. This work was supported by the Danish Cancer Society (DHM and MLT grant no. R174-A11581-17-S52; DHM and MC grant no. R231-A14035; NB grant no. R146-A9326-16-S2, R231-A13820; HJJ grant no. R90-A5989), The Lundbeck Foundation (DHM grant no. R307-2018-3326) The Danish Medical Research Council (NB grant no. DFF-4004-00340), The Novo Nordisk Foundation (NB), Krista and Viggo Petersen’s Foundation (NB), The Research Foundation of the Danish Capital Region (NB), The Danish Research Foundation (NW and CJ), Independent Research Fund Denmark (HJJ grant no. 4092-00387B).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mbplus.2022.100101.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Kessenbrock K., Plaks V., Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.P. Lu, K. Takai, V.M. Weaver, Z. Werb, Extracellular Matrix Degradation and Remodeling in Development and Disease, Cold Spring Harb. Perspect. Biol. 3 (2011) a005058–a005058. 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed]
- 3.Iozzo R.V., Gubbiotti M.A. Extracellular matrix: The driving force of mammalian diseases. Matrix Biol. 2018;71–72:1–9. doi: 10.1016/j.matbio.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hynes R.O. The Extracellular Matrix: Not Just Pretty Fibrils. Science. 2009;326(5957):1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricard-Blum S., Vallet S.D. Fragments generated upon extracellular matrix remodeling: Biological regulators and potential drugs. Matrix Biol. 2019;75–76:170–189. doi: 10.1016/j.matbio.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Lu P., Weaver V.M., Werb Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.P. Schedin, P.J. Keely, Mammary gland ECM remodeling, stiffness, and mechanosignaling in normal development and tumor progression, Cold Spring Harb. Perspect. Biol. 3 (2011) 1–22. 10.1101/cshperspect.a003228. [DOI] [PMC free article] [PubMed]
- 8.Levental K.R., Yu H., Kass L., Lakins J.N., Egeblad M., Erler J.T., Fong S.F.T., Csiszar K., Giaccia A., Weninger W., Yamauchi M., Gasser D.L., Weaver V.M. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139(5):891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D.E. Kuczek, A.M.H. Larsen, M.-L. Thorseth, M. Carretta, A. Kalvisa, M.S. Siersbæk, A.M.C. Simões, A. Roslind, L.H. Engelholm, E. Noessner, M. Donia, I.M. Svane, P. thor Straten, L. Grøntved, D.H. Madsen, Collagen density regulates the activity of tumor-infiltrating T cells, J. Immunother. Cancer. 7 (2019) 68. 10.1186/s40425-019-0556-6. [DOI] [PMC free article] [PubMed]
- 10.Jürgensen H.J., van Putten S., Nørregaard K.S., Bugge T.H., Engelholm L.H., Behrendt N., Madsen D.H. Cellular uptake of collagens and implications for immune cell regulation in disease. Cell. Mol. Life Sci. 2020 doi: 10.1007/s00018-020-03481-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsen A.M.H., Kuczek D.E., Kalvisa A., Siersbæk M.S., Thorseth M.-L., Johansen A.Z., Carretta M., Grøntved L., Vang O., Madsen D.H. Collagen density modulates the immunosuppressive functions of macrophages. J. Immunol. 2020;205(5):1461–1472. doi: 10.4049/jimmunol.1900789. [DOI] [PubMed] [Google Scholar]
- 12.Mazur A., Holthoff E., Vadali S., Kelly T., Post S.R., Engler A.J. Cleavage of type i collagen by fibroblast activation protein-α enhances class a scavenger receptor mediated macrophage adhesion. PLoS One. 2016;11(3):e0150287. doi: 10.1371/journal.pone.0150287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brömme D., Okamoto K., Wang B.B., Biroc S. Human cathepsin O2, a matrix protein-degrading cysteine protease expressed in osteoclasts family with a proline in the position adjacent to the. Biochemistry. 1996;271:2126–2132. doi: 10.1074/jbc.271.4.2126. [DOI] [PubMed] [Google Scholar]
- 14.Burleigh M.C., Barrett A.J., Lazarus G.S., Cathepsin B. A lysosomal enzyme that degrades native collagen. Biochem. J. 1974;137:387–398. doi: 10.1042/bj1370387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirschke H., Kembhavi A.A., Bohley P., Barrett A.J. Action of rat liver cathepsin L on collagen and other substrates. Biochem. J. 1982;201:367–372. doi: 10.1042/bj2010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan M.-H., Zhu Q., Li H.-H., Ra H.-J., Majumdar S., Gulick D.L., Jerome J.A., Madsen D.H., Christofidou-Solomidou M., Speicher D.W., Bachovchin W.W., Feghali-Bostwick C., Puré E. Fibroblast activation protein (FAP) accelerates collagen degradation and clearance from lungs in mice. J. Biol. Chem. 2016;291(15):8070–8089. doi: 10.1074/jbc.M115.701433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sloane B.F., Honn K.V. Cysteine proteinases and metastasis. Cancer Metastasis Rev. 1984;3(3):249–263. doi: 10.1007/BF00048388. [DOI] [PubMed] [Google Scholar]
- 18.Egeblad M., Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer. 2002;2(3):161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 19.Sternlicht M.D., Werb Z. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 2001;17(1):463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song F. Matrix metalloproteinase dependent and independent collagen degradation. Front. Biosci. 2006;11:3100. doi: 10.2741/2036. [DOI] [PubMed] [Google Scholar]
- 21.G. Murphy, J.J. Reynolds, U. Bretz, M. Baggiolini, Partial purification of collagenase and gelatinase from human polymorphonuclear leucocytes. Analysis of their actions on soluble and insoluble collagens, Biochem. J. 203 (1982) 209–221. 10.1042/bj2030209. [DOI] [PMC free article] [PubMed]
- 22.D.E. Kuczek, M.L. Hübbe, D.H. Madsen, Internalization of collagen: an important matrix turnover pathway in cancer, in: Extracell. Matrix Tumor Biol., 2017: pp. 17–38. 10.1007/978-3-319-60907-2_2.
- 23.Madsen D.H., Engelholm L.H., Ingvarsen S., Hillig T., Wagenaar-Miller R.A., Kjøller L., Gårdsvoll H., Høyer-Hansen G., Holmbeck K., Bugge T.H., Behrendt N. Extracellular collagenases and the endocytic receptor, urokinase plasminogen activator receptor-associated protein/endo180, cooperate in fibroblast-mediated collagen degradation. J. Biol. Chem. 2007;282(37):27037–27045. doi: 10.1074/jbc.M701088200. [DOI] [PubMed] [Google Scholar]
- 24.Madsen D.H., Ingvarsen S., Jürgensen H.J., Melander M.C., Kjøller L., Moyer A., Honoré C., Madsen C.A., Garred P., Burgdorf S., Bugge T.H., Behrendt N., Engelholm L.H. The non-phagocytic route of collagen uptake: A distinct degradation pathway. J. Biol. Chem. 2011;286:26996–27010. doi: 10.1074/jbc.M110.208033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behrendt N., Jensen O.N., Engelholm L.H., Mørtz E., Mann M., Danø K. A urokinase receptor-associated protein with specific collagen binding properties. J. Biol. Chem. 2000;275(3):1993–2002. doi: 10.1074/jbc.275.3.1993. [DOI] [PubMed] [Google Scholar]
- 26.Madsen D.H., Jürgensen H.J., Siersbæk M.S., Kuczek D.E., Grey Cloud L., Liu S., Behrendt N., Grøntved L., Weigert R., Bugge T.H. Tumor-associated macrophages derived from circulating inflammatory monocytes degrade collagen through cellular uptake. Cell Rep. 2017;21(13):3662–3671. doi: 10.1016/j.celrep.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curino A.C., Engelholm L.H., Yamada S.S., Holmbeck K., Lund L.R., Molinolo A.A., Behrendt N., Nielsen B.S., Bugge T.H. Intracellular collagen degradation mediated by uPARAP/Endo180 is a major pathway of extracellular matrix turnover during malignancy. J. Cell Biol. 2005;169:977–985. doi: 10.1083/jcb.200411153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao Y., Keller E.T., Garfield D.H., Shen K., Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013;32(1-2):303–315. doi: 10.1007/s10555-012-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gajewski T.F., Schreiber H., Fu Y.-X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013;14(10):1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noy R., Pollard J.W. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liotta L.A., Thorgeirsson U.P., Garbisa S. Role of collagenases in tumor cell invasion. Cancer Metastasis Rev. 1982;1(4):277–288. doi: 10.1007/BF00124213. [DOI] [PubMed] [Google Scholar]
- 32.Madsen D.H., Bugge T.H. The source of matrix-degrading enzymes in human cancer: Problems of research reproducibility and possible solutions. J. Cell Biol. 2015;209:195–198. doi: 10.1083/jcb.201501034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madsen D.H., Bugge T.H. Imaging collagen degradation in vivo highlights a key role for M2-polarized macrophages in extracellular matrix degradation. Oncoimmunology. 2013;2 doi: 10.4161/onci.27127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jürgensen H.J., Silva L.M., Krigslund O., van Putten S., Madsen D.H., Behrendt N., Engelholm L.H., Bugge T.H. CCL2/MCP-1 signaling drives extracellular matrix turnover by diverse macrophage subsets. Matrix Biol. Plus. 2019;1:1–20. doi: 10.1016/j.mbplus.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingvarsen S., Porse A., Erpicum C., Maertens L., Jürgensen H.J., Madsen D.H., Melander M.C., Gårdsvoll H., Høyer-Hansen G., Noel A., Holmbeck K., Engelholm L.H., Behrendt N. Targeting a single function of the multifunctional matrix metalloprotease MT1-MMP Impact on Lymphangiogenesis. J. Biol. Chem. 2013;288(15):10195–10204. doi: 10.1074/jbc.M112.447169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coussens L.M., Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Havemose-Poulsen A., Holmstrup P., Stoltze K., Birkedal-Hansen H., Birfcedal-Hansen H. Dissolution of type I collagen fibrils by gingival fibroblasts isolated from patients of various periodontitis categories. J. Periodontal Res. 1998;33:280–291. doi: 10.1111/j.1600-0765.1998.tb02201.x. [DOI] [PubMed] [Google Scholar]
- 38.Holmbeck K., Bianco P., Caterina J., Yamada S., Kromer M., Kuznetsov S.A., Mankani M., Gehron Robey P., Poole A.R., Pidoux I., Ward J.M., Birkedal-Hansen H. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99(1):81–92. doi: 10.1016/S0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- 39.Matrisian L.M. The matrix-degrading metalloproteinases. BioEssays. 1992;14(7):455–463. doi: 10.1002/bies.950140705. [DOI] [PubMed] [Google Scholar]
- 40.Leeming D.J., He Y., Veidal S.S., Nguyen Q.H.T., Larsen D.V., Koizumi M., Segovia-Silvestre T., Zhang C., Zheng Q., Sun S., Cao Y., Barkholt V., Hägglund P., Bay-Jensen A.C., Qvist P., Karsdal M.A. A novel marker for assessment of liver matrix remodeling: An enzyme-linked immunosorbent assay (ELISA) detecting a MMP generated type I collagen neo-epitope (C1M) Biomarkers. 2011;16:616–628. doi: 10.3109/1354750X.2011.620628. [DOI] [PubMed] [Google Scholar]
- 41.Qian J., Olbrecht S., Boeckx B., Vos H., Laoui D., Etlioglu E., Wauters E., Pomella V., Verbandt S., Busschaert P., Bassez A., Franken A., Bempt M.V., Xiong J., Weynand B., van Herck Y., Antoranz A., Bosisio F.M., Thienpont B., Floris G., Vergote I., Smeets A., Tejpar S., Lambrechts D. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020;30(9):745–762. doi: 10.1038/s41422-020-0355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartoschek M., Oskolkov N., Bocci M., Lövrot J., Larsson C., Sommarin M., Madsen C.D., Lindgren D., Pekar G., Karlsson G., Ringnér M., Bergh J., Björklund Å., Pietras K. Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-07582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madsen D.H., Leonard D., Masedunskas A., Moyer A., Jürgensen H.J., Peters D.E., Amornphimoltham P., Selvaraj A., Yamada S.S., Brenner D.A., Burgdorf S., Engelholm L.H., Behrendt N., Holmbeck K., Weigert R., Bugge T.H. M2-like macrophages are responsible for collagen degradation through a mannose receptor-mediated pathway. J. Cell Biol. 2013;202:951–966. doi: 10.1083/jcb.201301081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engelholm L.H., List K., Netzel-Arnett S., Cukierman E., Mitola D.J., Aaronson H., Kjøller L., Larsen J.K., Yamada K.M., Strickland D.K., Holmbeck K., Danø K., Birkedal-Hansen H., Behrendt N., Bugge T.H. uPARAP/Endo180 is essential for cellular uptake of collagen and promotes fibroblast collagen adhesion. J. Cell Biol. 2003;160:1009–1015. doi: 10.1083/jcb.200211091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Netzel-Arnett S., Mitola D.J., Yamada S.S., Chrysovergis K., Holmbeck K., Birkedal-Hansen H., Bugge T.H. Collagen dissolution by keratinocytes requires cell surface plasminogen activation and matrix metalloproteinase activity. J. Biol. Chem. 2002;277(47):45154–45161. doi: 10.1074/jbc.M206354200. [DOI] [PubMed] [Google Scholar]
- 46.Siwik D.A., Chang D.-L.-F., Colucci W.S. Interleukin-1beta and tumor necrosis factor-alpha decrease collagen synthesis and increase matrix metalloproteinase activity in cardiac fibroblasts in vitro. Circ. Res. 2000;86:1259–1265. doi: 10.1161/01.RES.86.12.1259. [DOI] [PubMed] [Google Scholar]
- 47.Öhlund D., Elyada E., Tuveson D. Fibroblast heterogeneity in the cancer wound. J. Exp. Med. 2014;211:1503–1523. doi: 10.1084/jem.20140692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costa A., Kieffer Y., Scholer-Dahirel A., Pelon F., Bourachot B., Cardon M., Sirven P., Magagna I., Fuhrmann L., Bernard C., Bonneau C., Kondratova M., Kuperstein I., Zinovyev A., Givel A.-M., Parrini M.-C., Soumelis V., Vincent-Salomon A., Mechta-Grigoriou F. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33(3):463–479.e10. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 49.Scanlan M.J., Raj B.K.M., Calvo B., Garin-Chesa P., Sanz-Moncasi M.P., Healey J.H., Old L.J., Rettig W.J. Molecular cloning of fibroblast activation protein α, a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5657–5661. doi: 10.1073/pnas.91.12.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santos A.M., Kissil J.L., Puré E., Santos A.M., Jung J., Aziz N., Kissil J.L., Puré E. Targeting fibroblast activation protein inhibits tumor stromagenesis and growth in mice. J. Clin. Invest. 2009;119:3613–3625. doi: 10.1172/JCI38988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puré E., Blomberg R. Pro-tumorigenic roles of fibroblast activation protein in cancer: back to the basics. 2018;37:4343–4357. doi: 10.1038/s41388-018-0275-3.Pro-tumorigenic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheikh H., Yarwood H., Ashworth A., Isacke C.M. Endo180, an endocytic recycling glycoprotein related to the macrophage mannose receptor is expressed on fibroblasts, endothelial cells and macrophages and functions as a lectin receptor. J. Cell Sci. 2000;113(6):1021–1032. doi: 10.1242/jcs.113.6.1021. [DOI] [PubMed] [Google Scholar]
- 53.Nielsen B.S., Rank F., Engelholm L.H., Holm A., Danø K., Behrendt N. Urokinase receptor-associated protein (uPARAP) is expressed in connection with malignant as well as benign lesions of the human breast and occurs in specific populations of stromal cells. Int. J. Cancer. 2002;98:656–664. doi: 10.1002/ijc.10227. [DOI] [PubMed] [Google Scholar]
- 54.Sprangers S., Everts V. Molecular pathways of cell-mediated degradation of fibrillar collagen. Matrix Biol. 2019;75–76:190–200. doi: 10.1016/j.matbio.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 55.von Delwig A., Hilkens C.M.U., Altmann D.M., Holmdahl R., Isaacs J.D., Harding C.V., Robertson H., McKie N., Robinson J.H. Inhibition of macropinocytosis blocks antigen presentation of type II collagen in vitro and in vivo in HLA-DR1 transgenic mice. Arthritis Res. Ther. 2006;8:1–11. doi: 10.1186/ar1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nielsen L.S., Kellerman G.M., Behrendt N., Picone R., Dano K., Blasi F. A 55000–60000 M(r) receptor protein for urokinase-type plasminogen activator. Identification in human tumor cell lines and partial purification. J. Biol. Chem. 1988;263:2358–2363. [PubMed] [Google Scholar]
- 57.H. Birkedal-Hansen, S. Yamada, J. Windsor, A.H. Poulsen, G. Lyons, W. Stetler-Stevenson, B. Birkedal-Hansen, Matrix Metalloproteinases, Curr. Protoc. Cell Biol. 17 (2003) 10.8.1-10.8.23. 10.1002/0471143030.cb1008s17. [DOI] [PubMed]
- 58.Jensen C., Madsen D.H., Hansen M., Schmidt H., Svane I.M., Karsdal M.A., Willumsen N. Non-invasive biomarkers derived from the extracellular matrix associate with response to immune checkpoint blockade (anti-CTLA-4) in metastatic melanoma patients. J. Immunother. Cancer. 2018;6:1–10. doi: 10.1186/s40425-018-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.