Summary
Background
Celiac Disease (CD) is a multifactorial autoimmune enteropathy (with a prevalence of approximately 1% worldwide) that exhibits a wide spectrum of clinical, serological and histological manifestations. For the diagnosis of paediatric CD, the gold standard is the combination of serological tests (with high TGA-IgA values greater than 10 times the upper limit of normal) and duodenal biopsy (with a positive TGA-IgA but low titer). Therefore, a diagnostic test that totally excludes an invasive approach has not been discovered so far and the discovery of novel biological markers would represent an undoubted advantage for the diagnosis of CD and prognostic evaluation. MicroRNAs (miRNAs), small non-coding RNAs (18–22 nucleotides) that regulate gene expression at post-transcriptional level and play important roles in many biological processes, represent a novel class of potential disease biomarkers. Their presence in biological fluids (i.e., serum, plasma, saliva, urine) provides the opportunity to employ circulating miRNAs as novel non-invasive biomarkers.
Methods
In our prospective observational study, we examined the expression of circulating miRNAs in a cohort of CD patients (both at diagnosis and on gluten-free diet, respectively referred as CD and GFD) compared to healthy controls. By small RNA-Seq we discovered a set of circulating miRNAs that were further validated by qPCR with specific assays.
Findings
We found that out of the 13 miRNAs able to discriminate the three groups (i.e., CD, GFD and controls), three of them, namely miR-192-5p, miR-215-5p and miR-125b-5p (alone or in combination), were able to discriminate these three groups with high accuracy and specificity.
Interpretation
Our conclusions emphasize that these circulating miRNAs can be employed not only for the diagnosis of CD patients with a low TGA-IgA titer but also to monitor the adherence to a gluten-free diet by CD patients. In conclusion, we suggest the use of the circulating miRNAs identified in this work as a novel diagnostic and follow-up tool for paediatric CD.
Funding
This work was supported by Fondazione Celiachia Onlus (FC) Grant n° 018/FC/2013 and by Italian Ministry of Health (Ricerca Corrente).
Research in context.
Evidence before this study
When we started this study several years ago, no previous reports in the literature were available about circulating miRNAs that characterized the pediatric population of celiac disease patients (neo-diagnosed or under a gluten-free diet). Only one study about the characterization of intestinal miRNAs (from biopsy) convinced us to undertake a study aimed at finding novel potential biomarkers of this intestinal disease. Moreover, no studies on biomarkers that can be exploited to monitor the adherence to a gluten-free diet were previously reported.
Added value of this study
The results of our study allowed us to obtain novel serological biomarkers to predict CD, and identify celiac disease also in serologically negative patients (according to ESPHGAN guidelines). Moreover, no serological biomarkers are available so far to monitor the adherence to a gluten-free diet. Last but not least, since in this study we recruited a pediatric population, the results are even more significant. Indeed, we see the possibility to suggest these circulating miRNAs as novel biomarkers without the need to perform invasive endoscopies and intestinal biopsies to confirm the disease.
Implications of all the available evidence
The implications of our study are wide both in the clinical and diagnostic fields: it could help to shorten the time-to-diagnosis, avoid biopsies to patients, and introduce in the market novel diagnostic kits. From a sanitary and economic point of view, the use of circulating miRNAs could be of primarily importance to reduce the costs of different analysis (serological and genetics), perform more diagnostic exams and to be more precise in the diagnosis of pediatric CD. From a research laboratory perspective, these findings will pave the way to further validation studies with large cohorts, the correlation of circulating miRNAs with biologically-relevant molecular and biological processes related to CD onset and progression, and with the presence of other regulatory miRNAs in different organs.
Alt-text: Unlabelled box
Introduction
Celiac Disease (CD) is an autoimmune enteropathy distributed worldwide with a prevalence of approximately 1%.11 Celiac disease is multifactorial and exhibits a wide spectrum of clinical, serological and histological manifestations. The environmental factor responsible of its onset and maintenance is gluten, a complex mixture of protein contained in grains like wheat, rye and barley. The predisposition to disease depends by HLA association with about 90–95% of CD subjects expressing the HLA-DQ2 and, the remaining 5–10% the HLA-DQ8.24 Individuals with CD develop highly specific autoantibodies as a consequence of the immunological response to gluten, including anti-tissue transglutaminase IgA (TGA-IgA), antiendomysium IgA (EMA-IgA), and anti-deamidated forms of gliadin peptides IgG (DGP-IgG), which is a determination especially requested for children younger than 2 years.1,10 The final result is a small-intestine mucosal injury characterized by villous atrophy, crypt hyperplasia and infiltration of a consistent number of lymphocytes and plasma cells in the lamina propria.15
Actually, the gold standard for the diagnosis of paediatric CD is a combination of serological tests and duodenal biopsy. In 2012, the European Society for Paediatric Gastroenterology Hepatology and Nutrition (ESPGHAN) guidelines suggested the possibility to avoid the biopsy only in adolescents and children who present HLA-DQ2/DQ8 haplotype, positivity to EMA-IgA, and levels of TGA-IgA ten-fold upper the limit of normal (ULN).13 In 2020, the new guidelines confirmed the safe no-biopsy approach for CD diagnosis in children with high TGA-IgA values greater than 10 times the upper limit of normal, and positive EMA-IgA in a second serum assessment.12 Thus, a diagnostic test that totally excludes an invasive approach has not been discovered so far and the discovery of novel biological markers would represent an undoubted advantage for the diagnosis of CD and prognostic evaluation. A novel class of potential biomarkers of disease is represented by microRNAs (miRNAs), small non-coding RNAs (18–22 nucleotides) that regulate gene expression at post-transcriptional level and play important roles in many biological processes such as development, proliferation and apoptosis.17 MiRNAs are highly stable and tissue-specific and their expression profile may constitute a specific signature of disease.21 MiRNAs also may be secreted outside cells (circulating miRNAs) in association with RNA-binding proteins, including high-density lipoproteins (HDL) and Argonaute2 (AGO2) and circulate through the bloodstream.5 Circulating miRNAs are highly stable, resistant to RNases digestion, extreme pH, high temperatures, extended storage, and multiple freeze–thaw cycles. Thus, their presence in biological fluids (i.e., serum, plasma, saliva, urine) provides the opportunity to employ circulating miRNAs as new non-invasive biomarkers.
To date, a rigorous gluten-free diet (GFD) is the only available treatment that generally alleviates gastrointestinal symptoms and reduces celiac disease-associated risks of complications.9 For a long time, clinicians discussed about the sensitivity, specificity and accuracy of serological tests employed to monitor the response to GFD, without reaching a conclusive consensus on a particular test. Although the antibodies titration can be used to monitor the adherence to the treatment, this method may have many false positives; in addition, repeated intestinal biopsies to assess the compliance are not feasible and practical.25,6 To this aim, circulating miRNAs can represent a valid alternative and can be employed effectively to monitor the response to GFD.
Our objectives were to investigate the potential of circulating miRNAs as biomarkers of CD and adherence to gluten-free diet, and to investigate if they can be employed as an alternative approach to intestinal biopsies, especially in doubtful cases where biopsy is commonly requested. Therefore, in our prospective observational study we recruited a pediatric cohort consisting in children with active CD, CD on GFD and healthy donors as controls (CTRL). We first evaluated the expression pattern of miRNAs circulating in the serum of a group of children with active CD, CD on GFD and healthy donors (discovery set) through Next Generation Sequencing (NGS).3 Then, the differentially regulated circulating miRNAs were further validated through quantitative real-time polymerase chain reaction (qPCR)4 in an independent cohort (validation set) of more than twenty individuals per group. These miRNAs were uniquely expressed in CD and not in similar autoimmune intestinal diseases (i.e., Inflammatory Bowel Diseases, IBDs)
In conclusion, our study assessed and confirmed the potential utility of circulating miRNAs as a novel tool for the diagnosis and follow-up of CD patients.
Materials and methods
Study participants
This study was an observational prospective cohort study with a control group. A total number of 120 subjects ranging from 3 to 15 years belonging to three groups: CD (n = 40), CD on GFD for more than six months (n = 40), and CTRL (n = 40) that were consecutively recruited at the Bambino Gesù Children's Hospital of Rome at the Hepatology, Gastroenterology and Nutrition Department from January 2014 to December 2018. The inclusion criteria were: age (3–15 years), positivity to TGA-IgA and EMA-IgA, and a duodenal biopsy classified as Marsh score ≥ 3 or based on international guidelines (for CD group), and CD patients already on GFD for more than six months (for GFD group). Controls were selected among a group of children with no history/diagnosis of inflammatory or other autoimmune conditions, negative family history of CD, and negative serological antibodies for CD. The exclusion criteria were: the use of oral corticosteroids or hormones, presence of other gastrointestinal diseases or autoimmune conditions, diabetes, major psychiatric disorders and physical impairment limiting physical activity.
All patients with serious or concomitant clinical or psychiatric conditions that may affect the assessment of protocol, with diabetes, complications of any kind, other autoimmune associated conditions, or subjects treated with drugs, were not enrolled. Written informed consent was obtained from all enrolled individuals and the Ethical Committee of the Bambino Gesù Children's Hospital approved the study (669_OPBG_2013; 10/07/2013).
Serum sample collection
Blood was collected into serum separator tubes (BD Vacutainer ® SST™ II Advance). Samples were processed within 2 h from blood withdrawal. Serum was obtained after centrifugation of the tubes at 1800 g for 15 min at room temperature. To exclude haemolysis, the absorbance of serum samples was measured at λ=414 nm (Nanodrop 2000 determination) and at additional peaks at λ=541 nm and λ=576 nm. Samples with an absorbance at 414 nm greater than 0.2 (A414>0.2) were considered haemolysed and consequently discarded. After the haemolysis check we excluded one CD sample, seven GFD samples and three CTRL samples. Collected serum was stored at -80 °C until use.
RNA isolation
Total RNA was isolated by using the Plasma/Serum Circulating and Exosomal RNA Purification kit (Norgen) from 250 μl (for qPCR) or 500 μl (for small RNA sequencing) of serum depending on the particular analysis, according to manufacturer's instructions. RNA was stored at −80 °C until use.
Discovery of circulating miRNAs by RNA sequencing
Circulating miRNAs were sequenced by using a small RNA sequencing protocol. RNA samples were concentrated to approximately 5 μl using a SpeedVac instrument and all the volume was employed for small RNA library construction using TruSeq SmallRNA Sample Prep kit (Illumina, San Diego, CA), following the manufacturer's instructions. Both RNA samples and final libraries were quantified by using the Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) and quality evaluated by Agilent 2100 Bioanalyzer RNA Nano assay (Agilent technologies, Santa Clara, CA). Libraries were then processed with Illumina cBot for cluster generation on the flowcell, following the manufacturer's instructions and sequenced on single-end mode at the multiplexing level requested on HiSeq2000 (Illumina, San Diego, CA). The CASAVA 1.8.2 version of the Illumina pipeline was used to process raw data for both format conversion and de-multiplexing. Raw data have been deposited in NCBI BioProject repository with the identification code PRJNA793584 (http://www.ncbi.nlm.nih.gov/bioproject/793584).
RNA-Seq bioinformatics analysis
Quality control checks were applied to raw sequence data using package FastQC that is integrated in online data analysis platform Galaxy (https://usegalaxy.org/). Adapters were removed and low-quality bases were trimmed by the script TrimGalore. The reads were mapped against the human reference genome hg19 with Bowtie 2 (Galaxy Version 2.3.2.2) and the number of aligned reads overlapping to each transcript sequence were counted by HTSeq-count script.2 Differentially expressed miRNAs were obtained using DESeq2 package19 after applying batch effect corrections using the batch-correction methods COMBAT.
Validation of deregulated miRNAs by qPCR
Deregulated miRNAs detected by RNA sequencing were validated in the independent set by qPCR. The analyses were performed by using Serum/Plasma Focus miRNA PCR Panel I+II (Exiqon) which assessed 179 miRNAs with specific miRNA locked nucleic acid (LNA)™ PCR primers (Exiqon). In brief, RNA was reverse transcribed in 25 μl using miRCURY LNA™ universal RT cDNA Synthesis (Exiqon) following the manufacturer's instructions. A RNA spike-in control (UniSp6) was used to monitor the efficiency of the reverse transcription reaction. Then, cDNA was diluted (40X) in nuclease free water and qPCR assay was performed in 10 μl reactions by using ExiLENT SYBR® Green master mix (Exiqon), according to the protocol. The qPCR reactions were run on a QuantStudio™ 12K Flex Real-Time PCR System by using a PCR program consisting in a 95 °C denaturation step for 10 min followed by 40 cycles (95 °C for 10 s and 60 °C for 1 min). A dissociation protocol was acquired after the last qPCR cycle, to confirm the absence of unspecific amplification products.
The raw quantification cycle (Cq) values were analyzed using GenEx 6 Software (MultiD Analyses AB, Sweden) following manufacturer's instructions. qPCR data were normalized with the global mean expression of all genes20 and one-way analysis of variance (ANOVA) with correction for multiple comparisons was performed.
Statistics
All variables were expressed as mean ± standard deviation (SD), whereas qualitative data as percentage. Comparison of the mean among the groups was performed using ANOVA or non-parametric tests or χ-square (contingency tables). ANOVA has been applied for data (i.e., age) passing normality test (D'Agostino-Pearson p > 0.05). Where normality was not verified (i.e., BMI and IgA) the non-parametric Kruskal-Wallis test has been applied. Post-hoc analysis was carried out by Tukey method. Receiver operating characteristic (ROC) and area under the curve (AUC) was determined to establish sensitivity and specificity of the relevant miRNAs.26 The library Epi (R Bioconductor) was employed to generate a linear model from the miRNA expression data obtained by sequencing and qPCR, whereas the two-fold cross-validation was performed with the library pROC.23 For statistical analyses, SPSS 20.0 (SPSS, Inc., Chicago, IL, USA) was used, and a value of p < 0.05 was considered significant. Feature selection to identify relevant miRNAs across the CTRL, CD and GFD groups was performed by generalized linear models using GLMNET R package8 setting up the group as the response variable, the miRNAs as explanatory variables, elastic net mixing parameter at 1.0 and the minimum mean square error as gamma value. Best gamma value was assessed using the leave-one-out cross-validation technique. Variable importance for the glmnet model was estimated by the varImp function implemented in the CARET R package.16 For sample size (power) calculation, the software G*Power7 has been employed.
Results
Cohort's characteristics
The initial cohort consisted of 120 patients including CD (n = 40) and GFD (n = 40) patients, and age-matched CTRLs (n = 40). We performed an a priori sample size calculation with multiple groups with G*Power software (a = 0.05, b = 0.8, SD=0.5 to assess a fold change difference of at least 1.5 in the CD group compared to controls and GFD). The total sample size was calculated and resulted equal to 48. This is below the number of patients we recruited and selected for further analyses. After the haemolysis check, we excluded for further analysis one sample for CD, seven samples for GFD and three samples from CTRL. The clinical characteristics of the considered cohort of 109 subjects were reported in Table 1. EMA positivity (at diagnosis) for both CD and GFD groups was assessed only for 64 over 72 patients (n = 38 for CD and n = 26 for GFD) corresponding to a percentage of 88.8%. Although we are aware of the intrinsic variability of human samples, we estimated that the number of patients we considered in our study is adequate to have a significant representation of data. Moreover, we performed an additional statistical analysis to confirm that the subgroup of EMA positive group of patients are representative of the larger sample. We presented the results of this analysis in Table 1 together with the data already reported for the overall studied population. Therefore, we assume that missing values are missing at random.
Table 1.
Patients’ data, genetics and histological characterization of CD and patients on gluten-free diet (GFD) compared to control (CTRL) individuals.
| ENTIRE COHORT | |||||
|---|---|---|---|---|---|
| Demographic characteristics | CTRL (n = 37) | CD (n = 39) | GFD (n = 33) | p-value | p-value (only EMA+ samples) |
| Gender | M = 19, F = 18 | M = 15, F = 24 | M = 13, F = 20 | 0.460 (Chi-square) | 0.595 (Chi-square) |
| Age (years) | 9.27(4.32) | 7.57(3.59) | 7.71(3.71) | 0.118 | 0.867 |
| BMI | 19.63(5.35) | 17.44(4.01) | 17.66(3.71) | 0.14 | 0.262 |
| Total serum IgA (mg/dL) | 138.8(53.1) | 116.2(58.1) | 116.9(73.6) | 0.16 | 0.400 |
| Total serum TGA-IgA at diagnosis (U/mL) | — | 2036(3585) | — | — | — |
| Total serum TGA-IgA after gluten-free diet (U/mL) | — | — | 3.7(10.4) | — | — |
| EMA-IgA positivity (n, %) (at diagnosis) | — | n = 38 (97.4%) | n = 26 (78.8%) | — | — |
| DISCOVERY COHORT | |||||
| Demographic characteristics | CTRL (n = 18) | CD (n = 22) | GFD (n = 23) | p-value | |
| Gender | M = 8, F = 10 | M = 10, F = 12 | M = 11, F = 12 | 0.975 (Chi-square) | |
| Age (years) | 9.29(3.81) | 8.01(3.19) | 6.77(3.22) | 0.074 | |
| BMI | 20.79(6.60) | 17.11(3.74) | 16.71(3.26) | 0.15 | |
| Total serum IgA (mg/dL) | 145.2(42.3) | 127.1(49.8) | 117.3(65.6) | 0.205 | |
| Total serum TGA-IgA at diagnosis (U/mL) | — | 1711(3735) | — | — | |
| Total serum TGA-IgA after gluten-free diet (U/mL) | — | — | 0.87(1.14) | — | |
| EMA-IgA positivity (n, %) (at diagnosis) | — | n = 22 (100%) | n = 17 (73.9%) | — | |
| VALIDATION COHORT | |||||
| Demographic characteristics | CTRL (n = 19) | CD (n = 17) | GFD (n = 10) | p-value | |
| Gender | M = 11, F = 8 | M = 5, F = 12 | M = 2, F = 8 | 0.081 (Chi-square) | |
| Age (years) | 9.26(4.87) | 7.01(4.08) | 10.36(3.24) | 0.118 | |
| BMI | 18.53(3.67) | 17.86(4.42) | 19.85(3.91) | 0.284 | |
| Total serum IgA (mg/dL) | 132.4(62.8) | 129.8(44.4) | 127.7(87.1) | 0.781 | |
| Total serum TGA-IgA at diagnosis (U/mL) | — | 2456(3447) | — | — | |
| Total serum TGA-IgA after gluten-free diet (U/mL) | — | — | 10.87(18.38) | — | |
| EMA-IgA positivity (n, %) (at diagnosis) | — | n = 16 (94.1%) | n = 9 (90%) | — | |
| Histology | CTRL (n = 37) | CD (n = 39) | GFD (n = 33) | ||
| Biopsy confirmed (at diagnosis) | — | n = 13 | n = 29 | ||
| Confirmed according to ESPGHAN guidelines (at diagnosis) | — | n = 26 | n = 4 | ||
| not available/not performed | n = 34 | n = 26 | n = 4 | ||
| Marsch type | |||||
| 0-2 | — | — | — | ||
| 3a | — | n = 3 | n = 4 | ||
| 3b | — | n = 8 | n = 8 | ||
| 3c | — | n = 2 | n = 17 | ||
| Genetics | |||||
| DQ2/DQ8 | n = 2 | n = 32 | n = 9 | ||
| Negative | n = 1 | n = 1 | — | ||
| n.a. | n = 34 | n = 6 | n = 24 |
Celiac disease was confirmed by intestinal biopsies in 58.3% of CD and GFD children and diagnosed according to ESPGHAN guidelines in the 41.7% of cases. Biopsies of CD and GFD children were all confirmed as Marsh 3 type (n = 7 with 3a score, n = 16 with 3b score and n = 19 with 3c score). Of note, 41 celiac patients (CD and GFD) were DQ2/DQ8 positive, one was negative (but positive for DQA1*05, DQB1*0301/04, DRB1*11, DRB1*12) and 30 were not assessed. The majority of controls were negative or not assessed (n = 35) and two were positive for DQ2 but both asymptomatic. Genetic tests for 30 CD and GFD patients were not assessed. Biopsies were not performed on CTRL individuals and on 47.2% of CD or GFD patients (CD patients according to ESPGHAN guidelines). An exploratory subgroup analyses was not possible as the number of patients we have recruited did not allow us to obtain statistically significant results. Moreover, clinical data are available only for the majority of CD and GFD patients. This exploratory analysis will be possible only after recruiting more patients. The totality of children in the GFD group were on gluten-free diet by at least six months. The cohort has been divided into two different sets, the first as the discovery set and the second as the validation set, and the clinical characteristics of the two groups have been reported in Table 1.
Sequencing of circulating microRNAs
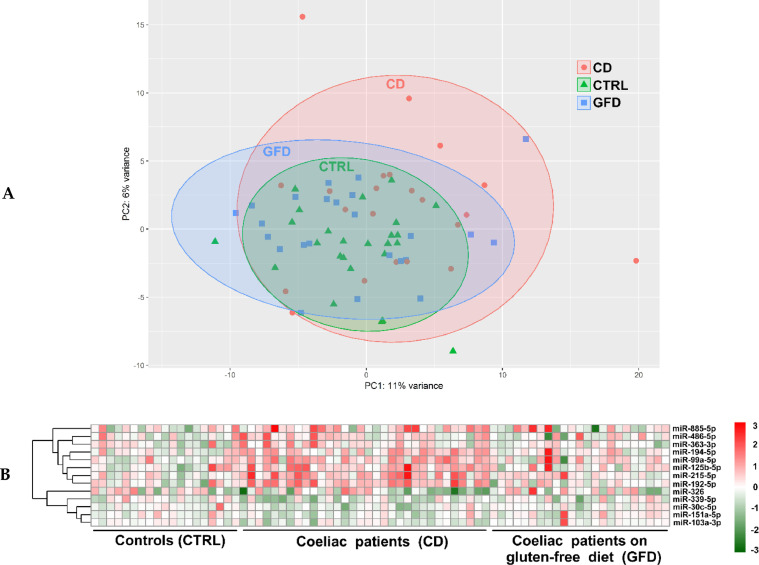
Small RNA sequencing of RNA samples from the sera of CD, GFD patients and controls afforded a large dataset of reads (more than 10M reads per sample) that have been trimmed, mapped to the human genome and intersected with the genomic positions (hg19 genome) of mature miRNAs to calculate the number of reads mapping to each feature. For each sample, the generated count files allowed us to examine the coverage of each mature miRNA (a total number of 2654 different entries) (Figure 1). Out of the 2654 mature miRNAs (miRbase rel.22.1), 1343 were not detected in all samples, and the following analyses were performed by considering only the remaining 1313 miRNAs. Next, to represent the distribution of expressed miRNAs among the three groups (CD, GFD and CTRL) we employed PCA analysis (Figure 2A). As expected, the distribution of samples in the CTRL and GFD groups are almost superimposable, whereas the CD group is wider and quite distinct from the other two groups. Interestingly, by comparing the profiles of GFD to CTRL no significant miRNAs resulted statistically significant, and this supports the results of the PCA analysis at least from a molecular point of view.
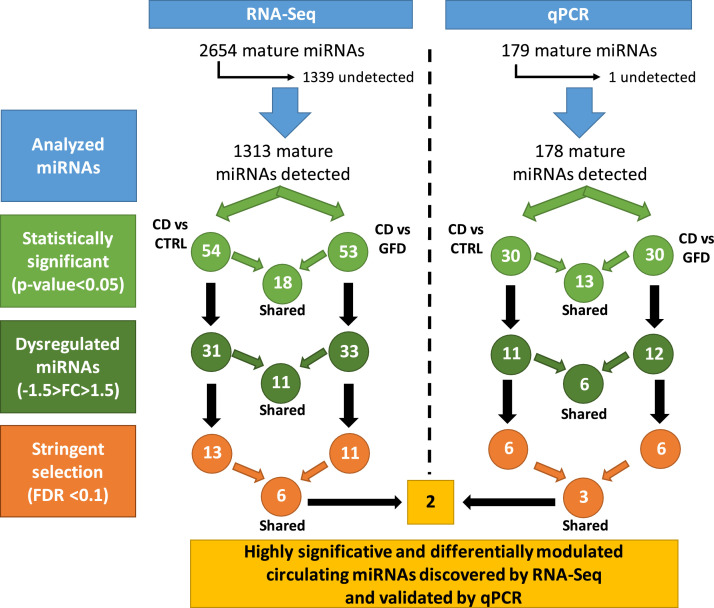
Figure 1.
Workflow of the analytical steps followed for RNA-Seq and qPCR analyses. Expression data were first filtered according to their statistical significance (p-value<0.05), then according to their expression level (-1.5>FC>1.5). Finally, a stringent selection criteria has been adopted (FDR<0.1) to restrict the number of miRNAs to validate. Only two miRNAs resulted highly significant and differentially modulated (discovered by RNA-Seq and validated by qPCR).
Figure 2.
(A) Principal component analysis (PCA) of RNA-Seq data for CTRL, CD and GFD groups. The colored regions represent the distribution of data for each category, whereas each symbol represents a patient. (B) Heatmap of the significant circulating miRNAs that are able to differentiate the three groups analyzed (i.e., CTRL, CD and GFD).
Next, we extracted the list of the statistically dysregulated miRNAs both in CD compared to CTRL and in CD compared to GFD to identify those miRNAs up- or down-regulated in the pathological condition. In Supplementary Table S1 we listed the 54 significantly (p < 0.05) dysregulated miRNAs found in CD vs CTRL, whereas in Supplementary Table S2 the 53 miRNAs dysregulated in CD compared to GFD. Only 18 miRNAs were in common between these two lists. Next, we selected those miRNAs with a fold change of at least 1.5 (-1.5>FC>1.5) in CD compared to CTRL (31 miRNAs) and in CD compared to GFD (33 miRNAs) and we calculated their sensitivity and specificity from RNA-Seq data (Table 2). Interestingly, 11 miRNAs (i.e., miR-141-3p, miR-192-5p, miR-1976, miR-215-5p, miR-224-5p, miR-24-2-5p, miR-370-3p, miR-375-3p, miR-423-5p, miR-4745-5p, miR-5683) were significantly expressed in both comparisons.
Table 2.
Statistically dysregulated (-1.5>FC>1.5) circulating miRNAs in CD compared to control (CTRL) individuals and to gluten-free diet (GFD) patients, obtained by RNA-Seq. Sensitivity and specificity have been calculated starting from RNA-Seq data. FC=Fold Change, FDR= False Discovery Rate, AUC=Area Under the Curve. Numbers in bold indicate a FDR<0.1.
| CD vs CTRL |
CD vs GFD |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| miRNA name | FC | FDR | Sensitivity | Specificity | AUC | FC | FDR | Sensitivity | Specificity | AUC |
| let-7c-5p | 1.51 | 2.631E-02 | 63.6 | 80.8 | 0.719 | |||||
| let-7f-1-3p | 1.60 | 2.816E-01 | 45.5 | 88.5 | 0.619 | |||||
| miR-100-5p | 1.52 | 6.822E-02 | 95.5 | 26.9 | 0.533 | |||||
| miR-1246 | 1.60 | 2.816E-01 | 86.4 | 57.7 | 0.698 | |||||
| miR-125a-3p | -1.55 | 3.348E-01 | 39.1 | 90.9 | 0.632 | |||||
| miR-127-5p | -1.61 | 2.816E-01 | 54.5 | 73.1 | 0.622 | |||||
| miR-134-5p | -1.62 | 7.085E-02 | 69.6 | 63.6 | 0.688 | |||||
| miR-139-3p | -1.69 | 2.005E-01 | 82.6 | 50 | 0.63 | |||||
| miR-141-3p | 2.08 | 2.985E-05 | 77.3 | 84.6 | 0.834 | 1.91 | 1.032E-03 | 73.9 | 77.3 | 0.787 |
| miR-145-5p | -1.66 | 1.433E-01 | 45.5 | 80.8 | 0.621 | |||||
| miR-150-3p | 1.55 | 2.918E-01 | 65.2 | 77.3 | 0.745 | |||||
| miR-150-5p | 1.57 | 6.096E-02 | 54.5 | 73.1 | 0.635 | |||||
| miR-185-5p | -1.56 | 7.085E-02 | 52.2 | 77.3 | 0.642 | |||||
| miR-190a-5p | -1.68 | 2.094E-01 | 65.2 | 77.3 | 0.719 | |||||
| miR-190b-5p | -1.72 | 1.837E-01 | 52.2 | 95.5 | 0.646 | |||||
| miR-192-5p | 1.68 | 9.587E-05 | 54.5 | 96.2 | 0.766 | 1.55 | 4.555E-03 | 91.3 | 54.5 | 0.709 |
| miR-1976 | -1.55 | 3.484E-01 | 72.7 | 66.4 | 0.696 | -1.72 | 1.811E-01 | 52.2 | 81.8 | 0.654 |
| miR-200a-3p | 1.74 | 1.509E-01 | 56.5 | 81.8 | 0.7 | |||||
| miR-205-5p | 1.57 | 3.134E-01 | 63.6 | 76.9 | 0.675 | |||||
| miR-215-5p | 3.55 | 1.952E-10 | 72.7 | 96.2 | 0.867 | 2.58 | 2.550E-05 | 87 | 72.7 | 0.832 |
| miR-224-5p | 1.53 | 1.586E-01 | 86.4 | 42.3 | 0.619 | 1.51 | 2.094E-01 | 52.2 | 86.4 | 0.67 |
| miR-24-2-5p | -1.51 | 5.361E-02 | 68.2 | 73.1 | 0.724 | -1.51 | 6.630E-02 | 56.5 | 90.9 | 0.747 |
| miR-3138 | 1.62 | 2.643E-01 | 30.4 | 95.5 | 0.609 | |||||
| miR-323b-3p | -1.73 | 6.822E-02 | 86.4 | 34.6 | 0.596 | |||||
| miR-335-5p | -1.52 | 6.822E-02 | 31.8 | 100 | 0.654 | |||||
| miR-337-5p | -1.63 | 2.168E-01 | 56.5 | 90.9 | 0.709 | |||||
| miR-362-5p | -1.56 | 3.146E-01 | 60.9 | 68.2 | 0.626 | |||||
| miR-3661 | 1.87 | 9.332E-02 | 95.7 | 45.5 | 0.652 | |||||
| miR-370-3p | -1.78 | 2.176E-02 | 63.6 | 73.1 | 0.652 | -1.54 | 1.585E-01 | 82.6 | 63.6 | 0.686 |
| miR-375-3p | 2.26 | 2.587E-05 | 68.2 | 92.3 | 0.809 | 1.81 | 1.159E-02 | 91.3 | 50 | 0.729 |
| miR-376c-3p | -1.58 | 3.134E-01 | 45.5 | 84.6 | 0.657 | |||||
| miR-377-5p | -1.63 | 2.643E-01 | 43.5 | 95.5 | 0.708 | |||||
| miR-423-5p | 1.67 | 6.822E-02 | 40.9 | 88.5 | 0.629 | 1.84 | 3.655E-02 | 52.2 | 86.4 | 0.694 |
| miR-424-3p | -1.77 | 7.085E-02 | 87 | 54.5 | 0.709 | |||||
| miR-431-5p | -1.51 | 3.709E-01 | 40.9 | 92.3 | 0.624 | |||||
| miR-4745-5p | 1.56 | 3.134E-01 | 22.7 | 96.2 | 0.577 | 1.53 | 3.348E-01 | 43.5 | 77.3 | 0.573 |
| miR-4775 | -1.59 | 3.134E-01 | 54.5 | 88.5 | 0.705 | |||||
| miR-487b-3p | -1.52 | 2.168E-01 | 56.5 | 81.8 | 0.698 | |||||
| miR-5010-5p | 1.54 | 3.439E-01 | 60.9 | 68.2 | 0.65 | |||||
| miR-511-5p | 1.55 | 3.484E-01 | 45.5 | 84.6 | 0.675 | |||||
| miR-539-3p | -1.57 | 3.248E-01 | 56.5 | 77.3 | 0.613 | |||||
| miR-5683 | 1.55 | 3.596E-01 | 27.3 | 100 | 0.601 | 1.56 | 3.348E-01 | 82.6 | 45.5 | 0.599 |
| miR-577 | 1.74 | 1.433E-01 | 72.7 | 69.2 | 0.703 | |||||
| miR-6735-5p | 1.81 | 1.020E-01 | 100 | 36.4 | 0.733 | |||||
| miR-6843-3p | -1.56 | 3.484E-01 | 77.3 | 61.5 | 0.67 | |||||
| miR-6847-5p | 1.88 | 9.332E-02 | 78.3 | 63.6 | 0.713 | |||||
| miR-6894-5p | 1.53 | 2.992E-01 | 31.8 | 96.2 | 0.517 | |||||
| miR-7705 | -1.55 | 3.348E-01 | 56.5 | 77.3 | 0.68 | |||||
| miR-7976 | 1.53 | 3.484E-01 | 31.8 | 92.3 | 0.575 | |||||
| miR-873-5p | -1.52 | 3.146E-01 | 52.2 | 81.8 | 0.626 | |||||
| miR-874-3p | 1.52 | 2.096E-01 | 68.2 | 76.9 | 0.664 | |||||
| miR-887-5p | -1.51 | 3.536E-01 | 82.6 | 59.1 | 0.729 | |||||
| miR-99a-5p | 1.58 | 1.467E-02 | 77.3 | 46.2 | 0.577 | |||||
By applying the multiple comparison correction (FDR<0.1) we obtained 13 miRNAs dysregulated in CD compared to CTRL (nine upregulated and four downregulated), whereas only 11 (seven upregulated and four downregulated) when compared to GFD (Table 2). Only six miRNAs were present in both the above-mentioned lists (i.e., miR-141-3p, miR-192-5p, miR-215-5p, miR-24-2-5p, miR-375-3p and miR-423-5p) with a comparable fold change (FC) (Table 2). This finding emphasizes that these miRNAs are not only specifically modulated in celiac disease but also inversely modulated after a gluten-free diet. Therefore, these miRNA were further validated in order to find suitable biomarkers to diagnose CD or to monitor the adherence to gluten-free diet.
qPCR assessment of circulating miRNAs
We independently assessed the dysregulation of circulating miRNAs also by a high-throughput quantitative PCR (qPCR) technique by using very sensitive LNA primers.14 We employed a technology specifically designed and optimized to detect up to 179 miRNAs generally circulating in serum/plasma (Figure 1).
From qPCR data we obtained two lists of 30 significantly (p < 0.05) dysregulated circulating miRNAs in CD patients compared to CTRL (Supplementary Table S3) and 30 miRNAs when compared to GFD (Supplementary Table S4). To obtain a list of relevant miRNAs, we extracted only those miRNAs circulating in CD patients and modulated either compared to CTRL or to GFD patients. A total number of 13 miRNAs were shared between these two groups, eight miRNAs were upregulated (i.e., miR-486-5p, miR-194-5p, miR-885-5p, miR-99a-5p, miR-215-5p, miR-363-3p, miR-125b-5p, miR-192-5p) and five were donwregulated (i.e., miR-30c-5p, miR-326, miR-339-5p, miR-151a-5p and miR-103a-3p). By clustering these results, we found that these miRNAs were able to separate well the two groups of considered patients (CD and GFD) compared to controls (Figure 2B). In fact, the overexpression of some of the miRNAs are recognizable in the upper part of the central region of the dendrogram (where the CD patients clusters), whereas the downregulated miRNAs are present in the lower part of it. By contrast, in the dendrogram region where the GFD patients are clustered, a well-defined region of up- and down-regulated miRNAs was not distinguished. This is also true for controls and this fact suggests that GFD and controls have a similar expression pattern and that this pattern is different from that of CD patients.
From this initial list, we selected those miRNAs with a fold change of at least 1.5 (-1.5>FC>1.5) in CD compared to CTRL (11 miRNAs) and in CD compared to GFD (12 miRNAs) and we calculated their sensitivity and specificity from qPCR data (Table 3). A total number of 6 miRNAs were shared between these two groups, four upregulated (i.e., miR-125b-5p, miR-192-5p, miR-215-5p and miR-885-5p) and two donwregulated (i.e., miR-326 and miR-339-5p).
Table 3.
Statistically dysregulated (-1.5>FC>1.5) circulating miRNAs in CD compared to control (CTRL) individuals and to gluten-free diet (GFD) patients, obtained by qPCR. Sensitivity and specificity have been calculated starting from qPCR data. FC=Fold Change, FDR= False Discovery Rate, AUC=Area Under the Curve. Numbers in bold indicate a FDR<0.1.
| CD vs CTRL |
CD vs GFD |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| miRNA name | FC | FDR | Sensitivity | Specificity | AUC | FC | FDR | Sensitivity | Specificity | AUC |
| miR-122-5p | 1.96 | 1.990E-01 | 73.9 | 59.4 | 0.693 | |||||
| miR-125b-5p | 1.80 | 1.220E-02 | 78.1 | 78.9 | 0.803 | 1.60 | 5.893E-02 | 56.5 | 93.8 | 0.785 |
| miR-150-5p | 1.65 | 1.929E-02 | 52.2 | 90.6 | 0.757 | |||||
| miR-192-5p | 2.00 | 9.345E-04 | 62.5 | 94.7 | 0.854 | 2.04 | 4.361E-05 | 78.3 | 84.4 | 0.879 |
| miR-194-5p | 1.66 | 2.061E-02 | 71.9 | 78.9 | 0.789 | |||||
| miR-200a-3p | 1.81 | 2.752E-01 | 75 | 84.2 | 0.765 | |||||
| miR-215-5p | 1.97 | 1.727E-03 | 71.9 | 89.5 | 0.842 | 1.63 | 3.783E-02 | 87 | 71.9 | 0.783 |
| miR-223-3p | -1.57 | 6.167E-02 | 60.9 | 81.2 | 0.73 | |||||
| miR-30a-5p | 1.78 | 1.502E-01 | 69.6 | 62.5 | 0.698 | |||||
| miR-326 | -1.83 | 2.842E-01 | 75 | 57.9 | 0.656 | -1.89 | 1.844E-01 | 73.9 | 59.4 | 0.678 |
| miR-338-3p | -1.77 | 6.150E-02 | 56.5 | 84.4 | 0.73 | |||||
| miR-339-5p | -1.67 | 1.550E-01 | 56.2 | 89.5 | 0.735 | -1.51 | 2.084E-01 | 87 | 56.2 | 0.681 |
| miR-375 | 1.89 | 1.518E-01 | 82.6 | 68.8 | 0.742 | |||||
| miR-584-5p | -1.52 | 2.737E-01 | 84.4 | 42.1 | 0.63 | |||||
| miR-874-3p | 1.92 | 2.043E-01 | 78.1 | 68.4 | 0.725 | |||||
| miR-885-5p | 1.92 | 8.186E-02 | 71.9 | 73.7 | 0.737 | 1.79 | 1.659E-01 | 82.6 | 53.1 | 0.702 |
| miR-99a-5p | 1.71 | 8.089E-03 | 84.4 | 73.7 | 0.808 | |||||
By selecting the most significant (FDR<0.1) miRNAs from these lists, we found that six miRNAs were differentially expressed in CD compared to CTRL (i.e., miR-125b-5p, miR-192-5p, miR-194-5p, miR-215-5p, miR-885-5p and miR-99a-5p) and all of them were upregulated. When CD were compared to GFD we obtained six dysregulated miRNAs: four were upregulated (i.e., miR-125b-5p, miR-150-5p, miR-192-5p and miR-215-5p) and two were downregulated (i.e., miR-223-3p and miR-338-3p).
Although a certain number of miRNAs (i.e., 21 by RNA-Seq and 17 by qPCR) were statistically significant (p-value<0.05) in GFD compared to CTRL (see Supplementary Table S5) after the application of the multiple comparisons correction, these miRNAs resulted no more significantly modulated. This result confirmed again that the differences between GFD and CTRL are negligible, at least for what concern the circulating miRNAs.
By qPCR analysis, we confirmed that the dysregulation of the circulating miRNAs observed by small RNA-Seq was also detectable by other techniques. All of the miRNAs statistically dysregulated (FDR<0.1) obtained by small RNA-Seq (Table 2) were also present in the panel of miRNAs that we assessed by qPCR.
Interestingly, among the most significant miRNAs (FDR<0.1) obtained by qPCR, three of them (i.e., miR-192-5p, miR-215-5p and miR-125b-5p) (Table 3) that we previously detected by RNA-Seq, were present in both the miRNAs list of CD vs CTRL and CD vs GFD patients. Therefore, by qPCR analysis, we confirmed the expression levels of these three miRNAs and this prompted us to continue the study of their predictive value for diagnostic applications.
Circulating miRNAs as biomarkers of CD
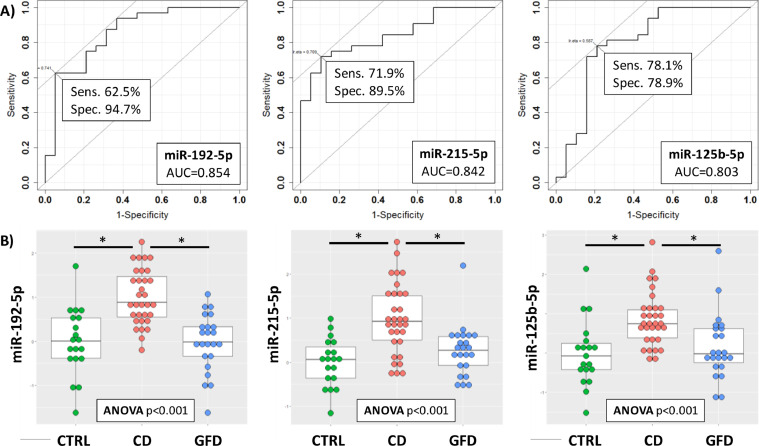
In order to evaluate the diagnostic ability of the three miRNAs distinctive of CD that we have found, we performed a Receiver Operating Characteristic (ROC) curve analysis. As illustrated in Figure 3A, all of the three miRNAs identified in this study and predictive of celiac disease, had a very good performance. In particular, miR-192-5p reached a sensitivity of 62.5% and a specificity of 94.7% (AUC=0.854), miR-215-5p a sensitivity of 71.9% and a specificity of 89.5% (AUC=0.842) and miR-125b-5p a sensitivity of 78.1% and a specificity of 78.9% (AUC=0.803).
Figure 3.
ROC curves for the discrimination of CD patients compared to CTRL (A) and DotPlots (B) of miR-192-5p, miR-215-5p and miR-125b-5p for the three groups of patients (CTRL, CD and GFD). The asterisk indicates a significant difference (p < 0.001)
The dot plots of Figure 3B illustrate the differences of expression of these three miRNAs in the three groups of patients (CTRL, CD and GFD). Parametric and non-parametric ANOVA test with Tukey post-hoc analysis has been used to assess significant differences among groups. According to previous results, we found significant differences (p < 0.001) only among the CD group compared to CTRL or to GFD, and no significant difference was observed between CTRL and GFD groups.
Then, we investigated the association between hsa-miR-125b-5p, hsa-miR-215-5p, or hsa-miR-192-5p expression and group classification (CTRL vs CD) adjusting for confounders (Age, BMI and Total serum IgA) by logistic regression. This analysis showed a significant association only for miRNA expression alone and even by adjusting for potential confounders we did not find any other significative data (Odd ratio > 1 and p-value < 0.05). The odd ratio indicates the constant effect of each covariate on the likelihood that sample is assigned to the CD group. The odds ratio (OR), the 95% confidence interval (CI) and the logistic regression p-value for each analysed miRNAs are shown in Supplementary Tables S6–S8.
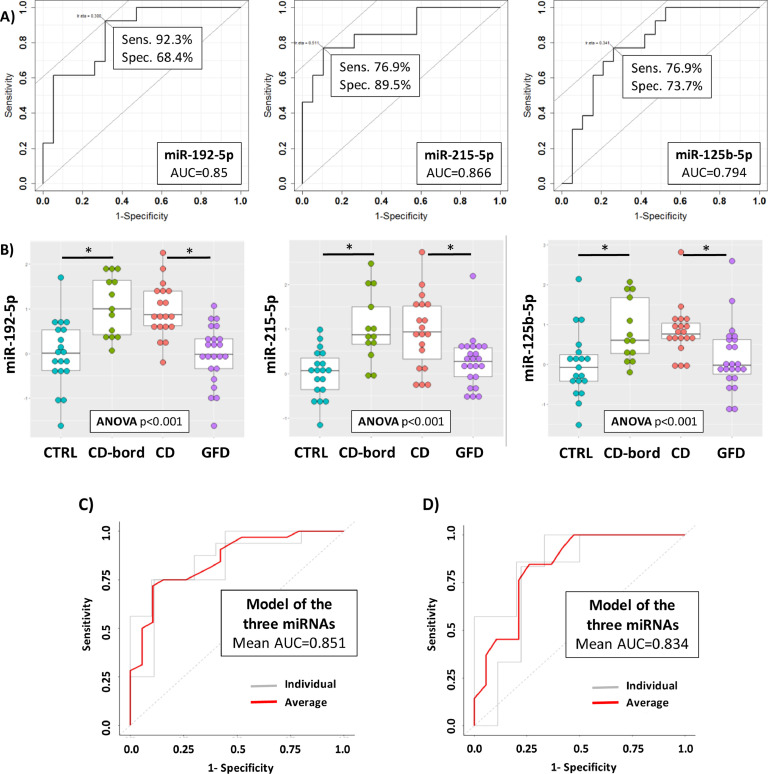
As a further investigation, we assessed if these three miRNAs are useful also to predict CD in those cases with an uncertain diagnosis (i.e., patients with levels of TGA-IgA below 10 times the limit of normal). To this aim, from the recruited cohort of CD patients we extracted only those patients (n = 13) with a value of TGA-IgA (at diagnosis) less than ten times the upper limit of normal (according to ESPHGAN guidelines) who underwent intestinal biopsy to confirm CD diagnosis. We referred these patients as ‘borderline’ patients (CD-bord). We examined the expression of the three circulating miRNAs analysed so far. As shown in Figure 4A all of the three miRNAs identified in the larger cohort of CD patients (i.e., miR-192-5p, miR-215-5p and miR-125b-5p) are also able to discriminate with good scores this groups of patients. In particular, miR-192-5p reached a sensitivity of 92.3% and a specificity of 68.4% (AUC=0.85), miR-215-5p a sensitivity of 76.9% and a specificity of 89.5% (AUC=0.866) and miR-125b-5p a sensitivity of 76.9% and a specificity of 73.7% (AUC=0.794). The dot plots of Figure 4B illustrate the differences of expression of these three miRNAs in the four groups of patients (CTRL, CD-bord, CD and GFD).
Figure 4.
ROC curves for the discrimination of borderline CD patients (CD-bord) compared to CTRL (A) and DotPlots (B) of miR-192-5p, miR-215-5p and miR-125b-5p for the four groups of patients (CTRL, CD-bord, CD and GFD). The asterisk indicates a significant difference (p < 0.001). ROC curves of the model obtained by using the three miRNAs (i.e., miR-192-5p, miR-215-5p and miR-125b-5p) and the two-fold cross-validation for the diagnosis of CD patients compared to controls (C) and borderline CD patients compared to controls (D). Individual folds and average are colored in gray or red, respectively.
Finally, in order to devise a comprehensive diagnostic model that included all of the three miRNAs at the same time and potentially had the highest predictive power, we calculated the ROC curve analysis by using a linear model function to include all the three miRNAs. In order to limit data overfitting due to the limited number of patients, we performed a two-fold cross-validation and we reported the ROC curves for individual iterations (gray lines) and the average value (red line). For CD vs CTRL the average AUC was 0.851, and for CD-bord vs CTRL was 0.834. These values were in line with the previous ones, and supported the conclusions previously obtained. Results of this elaboration were reported in Figure 4C for the prediction of CD disease (i.e., CD vs. CTRL), whereas Fig. 4D reports the prediction of only the CD borderline patients (i.e., CD-bord vs. CTRL).
Circulating miRNAs as biomarkers of adherence to gluten-free diet
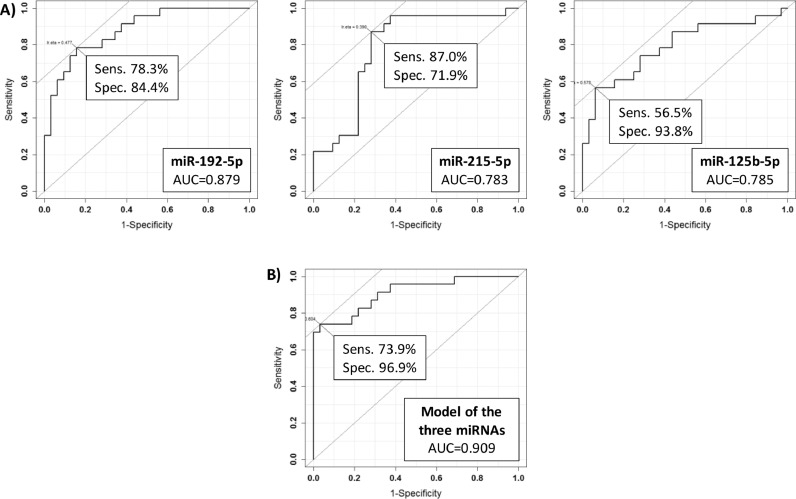
In order to evaluate the diagnostic ability of the three miRNAs discussed so far (i.e., miR-192-5p, miR-215-5p and miR-125b-5p) to discriminate CD patients on a GFD from CD patients, we performed a ROC curve analysis by considering the two categories of CD and GFD patients. As shown in Figure 5, miR-192-5p reached a sensitivity of 78.3% and a specificity of 84.4% (AUC=0.879), miR-215-5p a sensitivity of 87.0% and a specificity of 71.9% (AUC=0.783) and miR-125b-5p a sensitivity of 56.5% and a specificity of 93.8% (AUC=0.785). Next, we performed the analysis using a linear model of the three miRNAs, reaching a good predictive power (i.e., sensitivity of 73.9%, specificity of 96.9% and AUC=0.909). These results suggest that the combination of the three miRNAs identified in CD, can be successfully used also to discriminate with a good reliability CD patients undergoing a prolonged GFD. This result is relevant, as nowadays no reliable analytical tests are available to monitor the adherence to GFD.
Figure 5.
ROC curves for the discrimination of CD patients under a gluten-free diet (GFD patients) compared to CD patients (A) and ROC curve of the model obtained by using the three miRNAs (i.e., miR-192-5p, miR-215-5p and miR-125b-5p) for the discrimination of GFD patients compared to CD patients.
Finally, to determine whether differences in the expression of miRNAs could discriminate among the CTRL, CD and GFD groups, we performed an ordinal regression with CD, GFD and CTRL as outcome variables. We applied the generalized linear model (GLMNET) algorithm8 to the expression profiles in order to find the relevant miRNAs with the highest contribution to the separation among the distinct groups. Variable importance was estimated for the glmnet model by the varImp function, which was implemented in the CARET R package.16 The resulting variable importance for each miRNA and patient group was reported in Supplementary Table S9. Analysis identified 12, 12 and 10 miRNAs with importance higher than zero for CTRL, CD, and GFD groups, respectively. None of the miRNAs obtained an importance greater than zero in all three groups. Of note, hsa-miR-215-5p and hsa-miR-125-5p were the only two miRNAs with relevance greater than zero in CTRL and CD groups, whereas hsa-miR-192-5p was the miRNA that obtained the highest importance in the group of CD patients. Therefore, these results strongly suggest that miRNAs identified with this algorithm can represent biomarkers with a high discriminating power across various groups and deserving further validation on a wider cohort.
Discussion
In this prospective observational study we investigated the presence of circulating miRNAs in the sera of patients affected by celiac disease or adhering to a gluten-free diet (GFD) by employing RNA-Seq and qPCR analyses, two high throughput techniques that allowed us to identify potential biomarkers of disease and progression. These two techniques are technically complimentary but allowed obtaining similar information about the expressed circulating miRNAs (Figure 1). We identified at least three miRNAs (i.e., miR-192-5p, miR-215-5p and miR-125b-5p) that could be employed successfully not only to identify patients with biopsy-confirmed celiac disease, but also affected patients with low levels of TGA-IgA (referred as ‘borderline’ patients). These three miRNAs (reported in Table 3) are uniquely expressed in the sera of celiac disease patients and may have an active role in regulating many important biological processes implicated in CD disease as their potential gene targets belong to apoptotic processes, cell cycle phase transitions and other immune-related processes. Among the three miRNAs, miR-192-5p was already found dysregulated in duodenal biopsies of CD patients compared to controls, although the expression was downregulated and not upregulated as we found in serum. Moreover, to exclude that the regulation of these miRNAs is not a common feature of other intestinal inflammatory diseases (i.e., Inflammatory Bowel Diseases, IBDs) we compared their expression with that of a small cohort of patients (n = 14) affected by either Ulcerative Colitis or Crohn's disease already characterized by our group (data not shown). Although Ulcerative Colitis and Crohn's disease represent the two main forms of IBDs,18 two forms of multifactorial diseases in which autoimmune and immune-mediated processes are involved,22 none of the miRNAs we found specifically regulated in CD patients are expressed in these two diseases.
The use of a linear model containing information of the expression levels of circulating miRNAs within the various groups, can further improve the sensitivity of the method, and allowed to obtain a sensitivity of 68.8% and a specificity of 94.7 (AUC=0.867) for the diagnosis of CD patients. For the diagnosis of CD in borderline patients, the model was able to reach a sensitivity of 84.6% and a specificity of 84.2 (AUC=0.883). However, we emphasize that although the latter results are relevant, a wider population is needed in order to validate further these results and suggest these miRNAs for the diagnosis of CD in borderline patients.
Moreover, we confirmed that GFD might restore the dysregulated levels of some miRNAs and that, at the end, their expression is not significantly different from unaffected individuals. Therefore, this change parallels the restoring of a normal intestinal phenotype during a GFD regimen. The model of the three miRNAs can identify patients under GFD with a sensitivity of 73.9% and a specificity of 96.9% (AUC=0.909), thus representing an innovative way to monitor patient's adherence to diet. Nowadays, no other methods can allow to obtain this kind of information objectively, nor other analytical techniques or serological biomarkers are available yet to follow the adherence to a gluten-free diet for CD patients. Therefore, we strongly support the use of circulating miRNAs as a supplementary tool for the diagnosis of celiac disease without recurring to intestinal biopsy, a procedure that, especially for children, may result quite invasive and not very tolerated. Moreover, the application of a generalized linear model (Glmnet) confirmed that the miRNAs identified in this study, have the potentiality to be employed to discriminate the considered groups of patients, confirming the validity of the experimental techniques that we employed.
Anyway, our solid and consistent results should be further validated by recruiting a larger cohort of patients, especially those presenting TGA-IgA levels less than ten times the upper limit of normal, or GFD patients observed in a longitudinal study.
In conclusion, we have identified three valuable novel non-invasive biomarkers that alone or in combination may be employed successfully in the clinical practice for the diagnosis and follow-up of pediatric CD patients.
Authors’ contributions
C.F. designed the experiments, performed all the practical work, verified the underlying data and drafted the manuscript; A.B. performed part of the experimental work, verified the underlying data and collaborated with P.U. and D.C. to perform bioinformatics and statistical analyses; P.U. designed and performed many of the bioinformatics analyses, A.A. revised the analyses, drafted and edited the manuscript text, D.C. performed statistical analyses and collaborated with P.U. and A.M., M.A. and M.C. cooperated to clinical recruitment, A.P. collaborated to the bioinformatics analyses, Al.Mo. was responsible of clinical data entry, informed consent, ethical questions, M.S. and O.V. revised the manuscript, C.C., E.L., S.G. and F.F. recruited the patients and revised the manuscript, A.M. coordinated the wet-lab and bioinformatics experiments, coordinated the multicenter recruitment of patients and checked the clinical data, drafted and revised the manuscript. All authors read and approved the final version of the manuscript.
Declaration of interests
All of the authors declare no conflict of interest.
Acknowledgments
This work was supported by Fondazione Celiachia Onlus (FC) Grant n° 018/FC/2013 and by Italian Ministry of Health (Ricerca Corrente). The authors at Bambino Gesù Children's Hospital are particularly indebted to Prof. Valerio Nobili, head of the Hepatology and Gastroenterology Department for his help and suggestions. Dr. Nobili passed away prematurely few years ago and the authors would like to dedicate this manuscript to his memory.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103851.
Appendix. Supplementary materials
References
- 1.Amarri S., Alvisi P., De Giorgio R., et al. Antibodies to deamidated gliadin peptides: an accurate predictor of coeliac disease in infancy. J Clin Immunol. 2013;33:1027–1030. doi: 10.1007/s10875-013-9888-z. [DOI] [PubMed] [Google Scholar]
- 2.Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgos K.L., Javaherian A., Bomprezzi R., et al. Identification of extracellular miRNA in human cerebrospinal fluid by next-generation sequencing. RNA. 2013;19:712–722. doi: 10.1261/rna.036863.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C., Ridzon D.A., Broomer A.J., et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X., Liang H., Zhang J., Zen K., Zhang C.Y. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Fasano A., Catassi C. Current approaches to diagnosis and treatment of celiac disease: an evolving spectrum. Gastroenterology. 2001;120:636–651. doi: 10.1053/gast.2001.22123. [DOI] [PubMed] [Google Scholar]
- 7.Faul F., Erdfelder E., Buchner A., Lang A.G. Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav Res Methods. 2009;41:1149–1160. doi: 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
- 8.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 9.Green P.H., Lebwohl B., Greywoode R. Celiac disease. J Allergy Clin Immunol. 2015;135:1099–1106. doi: 10.1016/j.jaci.2015.01.044. quiz 1107. [DOI] [PubMed] [Google Scholar]
- 10.Guandalini S., Assiri A. Celiac disease: a review. JAMA Pediatr. 2014;168:272–278. doi: 10.1001/jamapediatrics.2013.3858. [DOI] [PubMed] [Google Scholar]
- 11.Gujral N., Freeman H.J., Thomson A.B. Celiac disease: prevalence, diagnosis, pathogenesis and treatment. World J Gastroenterol. 2012;18:6036–6059. doi: 10.3748/wjg.v18.i42.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Husby S., Koletzko S., Korponay-Szabo I., et al. European Society Paediatric Gastroenterology, Hepatology and nutrition guidelines for diagnosing coeliac disease 2020. J Pediatr Gastroenterol Nutr. 2020;70:141–156. doi: 10.1097/MPG.0000000000002497. [DOI] [PubMed] [Google Scholar]
- 13.Husby S., Koletzko S., Korponay-Szabo I.R., et al. European Society for Pediatric Gastroenterology, Hepatology, and nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr. 2012;54:136–160. doi: 10.1097/MPG.0b013e31821a23d0. [DOI] [PubMed] [Google Scholar]
- 14.Jacobsen N., Andreasen D., Mouritzen P. Profiling microRNAs by real-time PCR. Methods Mol Biol. 2011;732:39–54. doi: 10.1007/978-1-61779-083-6_4. [DOI] [PubMed] [Google Scholar]
- 15.Kagnoff M.F. Celiac disease: pathogenesis of a model immunogenetic disease. J Clin Invest. 2007;117:41–49. doi: 10.1172/JCI30253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28:1–26. [Google Scholar]
- 17.Li M., Marin-Muller C., Bharadwaj U., Chow K.H., Yao Q., Chen C. MicroRNAs: control and loss of control in human physiology and disease. World J Surg. 2009;33:667–684. doi: 10.1007/s00268-008-9836-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loddo I., Romano C. Inflammatory bowel disease: genetics, epigenetics, and pathogenesis. Front Immunol. 2015;6:551. doi: 10.3389/fimmu.2015.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. 550-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mestdagh P., Van Vlierberghe P., De Weer A., et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10 doi: 10.1186/gb-2009-10-6-r64. R64-2009-10-6-r64. Epub 2009 Jun 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell P.S., Parkin R.K., Kroh E.M., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pascual V., Dieli-Crimi R., Lopez-Palacios N., Bodas A., Medrano L.M., Nunez C. Inflammatory bowel disease and celiac disease: overlaps and differences. World J Gastroenterol. 2014;20:4846–4856. doi: 10.3748/wjg.v20.i17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robin X., Turck N., Hainard A., et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12 doi: 10.1186/1471-2105-12-77. 77-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuppan D., Junker Y., Barisani D. Celiac disease: from pathogenesis to novel therapies. Gastroenterology. 2009;137:1912–1933. doi: 10.1053/j.gastro.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Sugai E., Nachman F., Vaquez H., et al. Dynamics of celiac disease-specific serology after initiation of a gluten-free diet and use in the assessment of compliance with treatment. Dig Liver Dis. 2010;42:352–358. doi: 10.1016/j.dld.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 26.Swets J.A. Measuring the accuracy of diagnostic systems. Science. 1988;240:1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.