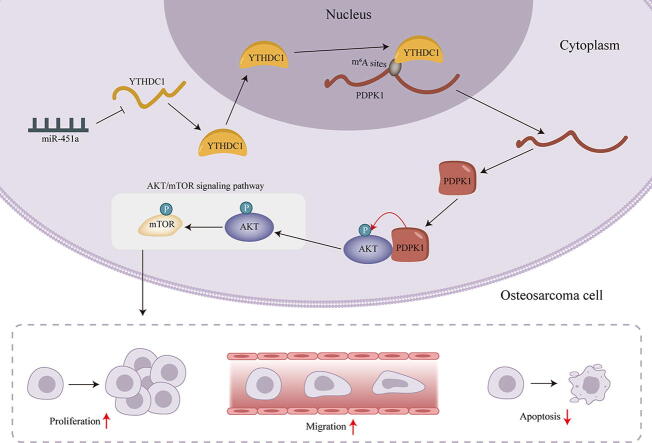
Graphical abstract
Abbreviations: mTOR, Mammalian target of rapamycin; q-PCR, Quantitative real-time polymerase chain reaction; RIP, Co-IP, Co-immunoprecipitation; miR-451a, MicroRNA-451a; PDPK1, 3-phosphoinositide dependent protein kinase 1; YTHDC1, YTH domain containing 1; GEO, Gene Expression Omnibus; Cdna, Complementary DNA; ANOVA, Analysis of variance; SD, Standard deviation
Keywords: Osteosarcoma, miR-451a, AKT/mTOR signaling pathway, m6A methylation
Highlights
-
•
It’s first proved that miR-451a can promote the malignant progression of osteosarcoma cells through AKT/mTOR pathway.
-
•
It’s first proved that YTHDC1 modifies the m6A methylation of PDPK1.
-
•
It’s first proved that YTHDC1 can promote the malignant progression of osteosarcoma cells.
Abstract
Background
Osteosarcoma is the most prevalent primary malignant bone tumor containing mesenchymal cells with poor prognosis. Being a hot spot of anti-tumor therapy researches, AKT/mammalian target of rapamycin (mTOR) signaling pathway could affect various cellular processes including transcription, protein synthesis, apoptosis, autophagy and growth.
Materials and methods
The levels of RNA and protein were detected by quantitative real-time polymerase chain reaction (q-PCR) and western blot analyses respectively. Functional assays were carried out to analyze the malignant phenotypes of osteosarcoma cells. RNA-binding protein immunoprecipitation (RIP), Co-immunoprecipitation (Co-IP), RNA pulldown, luciferase reporter and in vitro kinase assays were conducted to uncover the specific mechanism of microRNA-451a (miR-451a) in osteosarcoma cells.
Results
Functionally, miR-451a represses the malignant progression of osteosarcoma. Mechanically, miR-451a could curb the AKT/mTOR pathway via 3-phosphoinositide dependent protein kinase 1 (PDPK1)-mediated phosphorylation modification. After the certification that YTH domain containing 1 (YTHDC1) regulates the m6A phosphorylation modification of PDPK1 mRNA, we further proved that miR-451a-mediated YTHDC1 stabilizes PDPK1 mRNA via m6A-dependent regulation.
Conclusion
This study demonstrated that miR-451a regulates YTHDC1-mediated m6A methylation to activate the AKT/mTOR pathway, stimulating the malignancy of osteosarcoma.
1. Introduction
Osteosarcoma, the most common primary bone malignancy, typically afflicting children or adolescents [1]. Every year, there are approximately 1,000 new cases diagnosed with osteosarcoma in the United States according to the American Cancer Society’s statistics. Younger age, race and gender appear to increase the risk of osteosarcoma [2]. Treatment has evolved from surgical resection to systemic chemotherapy and local control surgery, which leads to an increase in long-term survival rates from <20% to 65%∼70% for patients with localized osteosarcoma [3], [4]. However, patients with metastatic disease or chemotherapy resistance exhibit only 20% survival rate, sharing a very poor prognosis [5], [6]. Thus, there is an urgent need to gain a comprehensive understanding of the molecular mechanism underlying the malignant progression of osteosarcoma.
Mammalian target of rapamycin (mTOR) is a serine/threonine kinase that involves in cellular processes including protein synthesis, growth, metabolism, aging, regeneration, autophagy, etc.; mTOR is frequently dysregulated in human cancers [7]. Remarkably, activation of mTOR enhances tumor growth and metastasis and some mTOR inhibitors have been recruited for cancer therapy while some are under assessment in clinical trials [8]. In addition, AKT (also known as protein kinase B or PKB)/mTOR signaling pathway has been suggested to regulate drug resistance to chemotherapeutic agents in cancer cells [9], [10], [11] and activation of this pathway has been reported to correlate with aggressive behaviors of tumors [12]. Moreover, an increasing number of studies have reported the anti-tumor therapies in terms of AKT/mTOR signaling pathway, including anti-osteosarcoma. For instance, XIST knockdown inhibits cell growth and autophagy in osteosarcoma by inhibiting the AKT/mTOR signaling pathway via sponging miR-375-3p [13]. Thereby, this study intended to further investigate the regulatory mechanism of AKT/mTOR signaling pathway in osteosarcoma.
MicroRNAs (miRNAs) are a type of small non-coding RNA molecules (∼22 nucleotides in length) [14], [15]. Notably, miRNA-targeted therapies have been suggested to be a promising approach to inhibit aggressive biological behaviors of osteosarcoma cells and act as regulators in multiple signaling pathways [16]. For instance, miRNA-21 silencing represses cell proliferation in osteosarcoma via targeting PTEN and regulating the TGF-β1 signaling pathway [17]. Herein, we aimed to probe into the role and mechanism of microRNA-451a (miR-451a) which is a downregulated gene in osteosarcoma according to the Gene Expression Omnibus (GEO) datasets (GSE28423). Previous studies have revealed the regulatory role of miR-451a in multiple cancers. For instance, miR-451a represses cell migration and invasion in non-small cell lung cancer [18]. In colorectal cancer, miR-451a suppresses cell proliferation and induces cell apoptosis through inhibition of BAP31 expression [19]. Particularly, miR-451a exerts its tumor suppression function in human cancers via targeting AKT/mTOR signaling pathway. For example, miR-451a is low-expressed and targets AKT/mTOR pathway in papillary thyroid carcinoma [19]. In addition, miR-451a was reported to act as a potential tumor suppressor in gastric cancer [20], [21]. But the regulatory role and mechanism of miR-451a and AKT/mTOR signaling pathway in the progression of osteosarcoma remains unknown. Hence, the current study intended to validate the hypothesis that miR-451a regulates the aggressive behaviors of osteosarcoma cells and uncover its underlying mechanism in osteosarcoma, which might provide new perspectives into osteosarcoma targeted therapy.
2. Materials and methods
2.1. Cell culture and vector construction
Human osteoblast cell (hFOB1.19) and human osteosarcoma cell lines (HOS, MG-63, U2OS and 143B) were commercially obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cell lines were incubated in RPMI 1640 medium (61870127, Gibco, USA), with the supplement of 10% FBS (10270-106, Gibco) and 100 U/ml penicillin/streptomycin. The sequences of YTH domain containing 1 (YTHDC1) and 3-phosphoinositide dependent protein kinase 1 (PDPK1) were inserted into pcDNA3.1 vector for the construction of overexpression vector, with the empty vector pcDNA3.1 as negative control (NC). Short hairpin-RNA (sh-RNA) against YTHDC1 (sh-YTHDC1) and its negative control of shRNAs (sh-NCs) were all synthesized by RiboBio (Guangzhou, China).
2.2. Quantitative real-time polymerase chain reaction (q-PCR)
The total RNAs were extracted from hFOB1.19, HOS, MG-63, U2OS and 143B by Trizol Reagent (T9108-1, Thermo Fisher Scientific), followed by reverse transcription and complementary DNA (cDNA) synthesis. Subsequently, q-PCR was implemented using ChamQ Universal SYBR qPCR Master Mix (Q711-02, Vazyme, Nanjing, China). All the procedures were performed as per the instructions of supplier. The results were calculated by the 2-ΔΔCt method. Bio-repeats were performed in triplicate.
2.3. Western blot
Total proteins were isolated from HOS and 143B cells, followed by separation on SDS-PAGE. The protein samples were then transferred onto PVDF membranes. After being blocked by skim milk, the membranes were incubated with primary antibodies and secondary antibodies (ab150077, Abcam, UK). After the washing in TBST, the blots were visualized and recorded. The antibodies used in western blot include Anti-ZO-1 (ab276131, Abcam), Anti-E-cadherin (ab40772, Abcam), Anti-N-cadherin (ab76011, Abcam), Anti-Vimentin (ab92547, Abcam), Anti-β-actin (ab8226, Abcam), Anti-AKT (ab179463, Abcam), Anti-p-AKT (ab8805, Abcam), Anti-mTOR (ab2732, Abcam), Anti-p-mTOR (ab109268, Abcam), Anti-PDPK1 (ab245451, Abcam. β-actin was used as internal control. Each experiment was repeated in triplicate.
2.4. Colony formation assay
Colony formation assay was implemented to evaluate the proliferative capacity. The transfected HOS and 143B cells were cultured in 6-well plate for 14 days. Subsequent to fixation by methanol and staining using crystal violet, colonies were visualized and counted. Each experiment was conducted in triplicate.
2.5. TUNEL assay
TUNEL assay was performed to evaluate the apoptosis of transfected HOS and 143B cells. After the fixation of paraformaldehyde, the cells undergoing apoptosis were subjected to TUNEL staining. The nucleus of cells was counterstained by DAPI. Bio-repeats were performed in triplicate.
2.6. Wound healing assay
The transfected HOS and 143B cells were inoculated into the 24-well plates. When cell confluence reached over 90%, we used a pipette tip to scratch wounds on cell layers. After PBS washing (C10010500BT, Gibco, USA), the images of wound were recorded at 0 h and 24 h respectively. Each experiment was performed in triplicate.
2.7. Transwell assays
Transwell assay was performed to assess the migratory ability of transfected HOS and 143B cells using transwell chamber without Matrigel. The medium with no serum was supplemented in the upper chamber, and the complete medium was added to the lower chamber. Afterwards, the transfected cells were plated in the upper chamber. 24 h later, we used cotton swabs to slightly abrade the non-migrated cells in the upper chamber. The migrated cells in the lower chamber were subjected to fixation and staining using methanol and crystal violet. Next, the migrated cells were counted under microscope. Each experiment was carried out three times.
2.8. Cell Counting Kit-8 (CCK-8) assay
CCK-8 kit (M4839, ABMOLE, USA) was used to evaluate the proliferative ability of transfected HOS and 143B cells in line with the supplier’s suggestions. The transfected cells were inoculated into 96-well culture plates. Next, all the transfected cells underwent 12, 24, 36, 48 and 60 h of incubation, followed by the addition of CCK-8 solution for a 4-hour incubation. The OD value was evaluated by a microplate reader (51119770DP, Thermo Fisher Scientific). Bio-repeats were performed in triplicate.
2.9. RNA-binding protein immunoprecipitation (RIP)
RIP assays was conducted using Imprint® RNA Immunoprecipitation Kit (RIP-12RXN, Sigma-Aldrich, USA). A/G magnetic beads were incubated with Anti-IgG (ab133470, Abcam) and Anti-N6-methyladenosine (Anti-m6A). Cells lysates were subjected to the incubation with the magnetic beads conjugated with Anti-IgG and Anti-m6A. After the elution, the complexes were subjected to RNA isolation and analyzed by q-PCR for the detection of PDPK1 enrichment. Each experiment was performed three times.
2.10. RNA pull-down assay
Structure buffer was added to biotinylated RNA to form secondary structure. Subsequently, biotin-labeled RNAs were subjected to heating and ice-bath for denaturation, followed by the incubation with Pierce™ Streptavidin Magnetic Beads (88816, Thermo Fisher Scientific) for 2 h at 4 °C. After HOS and 143B cells were lysed, cell lysates were used to set up three groups (Input, Bio-NC, and Bio-RNA) and incubated with bead-probe complex at 4 °C overnight. After the extraction of total RNAs or proteins from pull-down products, q-PCR or western blot was conducted to detect the enrichment. Each experiment was done in triplicate.
2.11. Luciferase reporter assay
Dual Luciferase Reporter Gene Assay Kit (RG027, Beyotime, Shanghai, China) was used to perform luciferase reporter assay. 3′untranslated region (3′UTR) of YTHDC1 was sub-cloned into pmirGLO vector for the construction of the wild type luciferase reporter vector (pmirGLO-YTHDC1 3′ UTR-Wt). Then, the binding sites of YTHDC1 3′ UTR to miR-3121-3p were mutated and mutated YTHDC1 3′ UTR was cloned into the vector to generate pmirGLO-YTHDC1 3′UTR-Mut reporter construct. The empty vector was used as negative control. After the co-transfection of the reporter vectors with miR-451a mimics or mimics NC into HOS and 143B cells, the luciferase reporter activity was determined with Renilla luciferase as internal reference. As for the detection of methylation site, luciferase reporter assay was conducted using pGL3 vector. The full sequence of PDPK1 was subcloned into pGL3 vector to construct pGL3-PDPK1 (m6A-Wt). PDPK1, with the site 1588 and 1732 mutated, were inserted into pGL3 vector to create pGL3-PDPK1 (m6A-1588 Mut) and pGL3-PDPK1 (m6A-1732 Mut). Subsequent to co-transfection with pcDNA3.1-YTHDC1 or pcDNA3.1, the reporter vectors were analyzed in terms of their luciferase activity with Renilla luciferase as internal reference. Each experiment was done three times.
2.12. Co-immunoprecipitation (Co-IP) assay
HOS and 143B cells were lysed. Then, cell lysate was incubated with A/G Agarose magnetic beads conjugated with Anti-PDPK1 and Anti-AKT. After being washed, the cell samples were subjected to western blot analysis. Bio-repeats were performed in triplicate.
2.13. In vitro kinase assay
At the condition of 30 °C, Anti-PDPK1 and Anti-AKT antibodies (8 μg) were co-incubated in the kinase buffer (20 mmol/L Tris-HCl, 50 mmol/L KCl, 10 mmol/L MgCl2 [pH 8.0], 2.5 mmol/L cold ATP and 1 μCi [γ-32P]ATP) for 30 min. The products were separated by SDS-PAGE. The phosphorylation of AKT was detected using 32p labeled probe antibody. The 32p-labeled proteins were detected by BAS-2500 Bioimaging Analyzer (Fujifilm, Tokyo, Japan). Each experiment was done three times.
2.14. Analysis of mRNA stability
HOS and 143B cells were seeded into 6-well plates before transfection. When cells grew to 70% confluency, sh-NC or sh-YTHDC1-1 was transfected into HOS and 143B cells. The transfected cells were treated with 50 mM α-amanitin for the inhibition of transcription. Afterwards, the expressions of PDPK1 and β-actin were measured at different time points (0, 6, 12, 18, and 24 h) by q-PCR. Bio-repeats were performed in triplicate.
2.15. Tumor xenograft experiments
Male BALB/c nude mice (6–8 weeks old) were purchased from Shanghai Laboratory Animal Co., Ltd. (SLAC) and were divided into four groups randomly (n = 3–5). After different transfection treatment for 48 h, HOS and 143B cells (100 μL, 2 × 106) were subcutaneously injected into each nude mouse. The tumor volume were detected every three days post one-week injection with a computational formula that 1/2 (length × width2). 28 days post injection, the mice were sacrificed. Then, the xenografts were removed and tumor weight was measured. All animal study was approved by First Hospital of Shanxi Medical University.
2.16. Statistical analysis
The experimental data were presented as mean ± SD (standard deviation) and analyzed by SPSS software. One-way ANOVA (analysis of variance), two-way ANOVA and Student’s t test were performed for the comparison of difference between groups. P value below 0.05 indicated statistically significant.
3. Results
3.1. MiR-451a suppresses the malignant progression of osteosarcoma in vitro
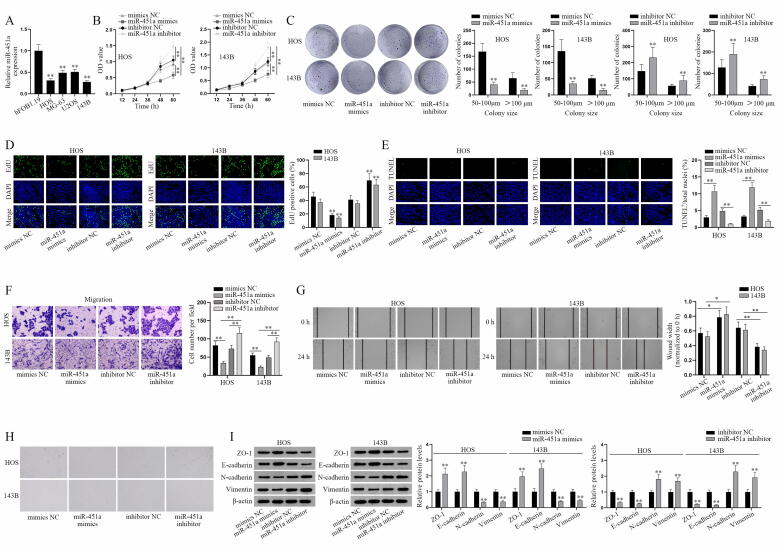
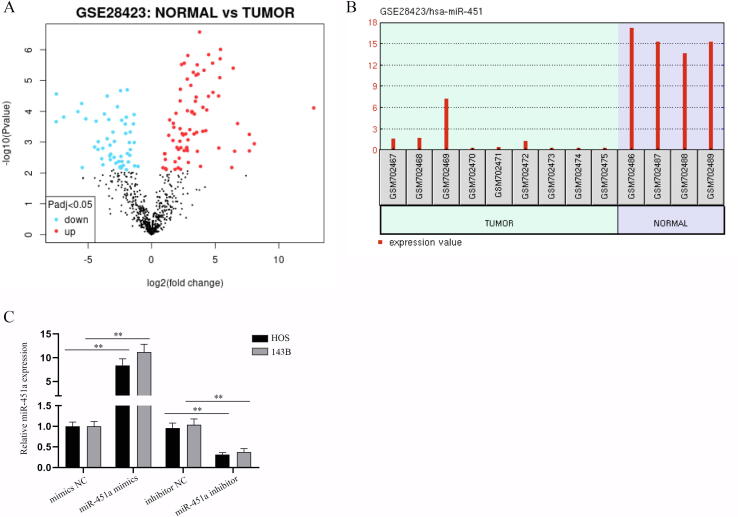
To begin with, GEO database (GSE28423) was firstly used to sift out the target miRNA as miR-451a and the volcano plot of miR-451a was shown in Fig. S1A. The expression of miR-451a in osteosarcoma and normal bone tissues proved that miR-451a was relatively low-expressed in osteosarcoma cell lines compared to that in normal bones (Fig. S1B). Also, we detected miR-451a expression in normal osteoblast hFOB1.19 and osteosarcoma cell lines (HOS, MG-63, U2OS, and 143B) via q-PCR (Fig. 1A). The results indicated that miR-451a was notably downregulated in osteosarcoma cell lines, especially in HOS and 143B cells. After the overexpression and knockdown efficiency of miR-451a being certified (Fig. S1C), we conducted a series of functional assays in HOS and 143B cells to determine whether miR-451a affects the malignant progression of osteosarcoma cells. Cell proliferation assays certified that miR-451a overexpression suppressed OD values and reduced colonies in number and colony size while miR-451a knockdown caused a totally opposite effect (Fig. 1B-C). Likewise, we found the decreased EdU positive cells induced by miR-451a overexpression but increased ones caused by miR-451a deficiency (Fig. 1D). TUNEL assays showed that upregulated miR-451a increased total apoptosis rate whereas downregulated miR-451a decreased it (Fig. 1E). Then Transwell assay indicated that miR-451a negatively modulated cell migration ability by curbing cell number in miR-451 mimics groups and increasing cell number in miR-451a inhibitor groups (Fig. 1F). Wound healing assay further verified this negative regulation of miR-451a on cell migration (Fig. 1G). Cell morphology was observed via microscopy to evaluate EMT. After transfection of miR-451a mimics, the cells changed from a spindle shape to a round or rectangular shape while miR-451a inhibitor led to the opposite effect (Fig. 1H). Further, ZO-1, E-cadherin, N-cadherin and Vimentin protein levels were detected by western blot analysis to analyze the EMT of osteosarcoma cells since ZO-1 and E-cadherin are EMT negative associated proteins, N-cadherin and Vimentin are EMT positive associated proteins. We found the enhanced ZO-1/E-cadherin protein levels and reduced N-cadherin/Vimentin protein levels in miR-451a mimics group, with the opposite result in miR-451a inhibitor group (Fig. 1I), which verified that miR-451a inhibits the EMT of osteosarcoma cells. Collectively, miR-451a could suppress osteosarcoma cell proliferation, migration, EMT and facilitate osteosarcoma cell apoptosis.
Fig. 1.
MiR-451a inhibits the malignant progression of osteosarcoma in vitro. (A) Q-PCR detection of miR-451a expression level in human osteosarcoma cell lines. (B-D) CCK-8, colony formation and EdU assays were performed to assess the effects of miR-451a on the proliferative ability of transfected osteosarcoma cells (HOS and 143B). (E) TUNEL assay was conducted to evaluate the apoptosis of transfected osteosarcoma cells. (F-G) Cell migration was determined by Transwell migration and wound healing assays. (H) Representative images of osteosarcoma cell morphology. (I) The protein levels of EMT-related markers (ZO-1, E-cadherin, N-cadherin and Vimentin) in transfected HOS and 143B cells with miR-451a mimics or miR-451a inhibitor were detected. *P < 0.05, **P < 0.01.
3.2. MiR-451a inhibits AKT/mTOR pathway via PDPK1-mediated phosphorylation modification
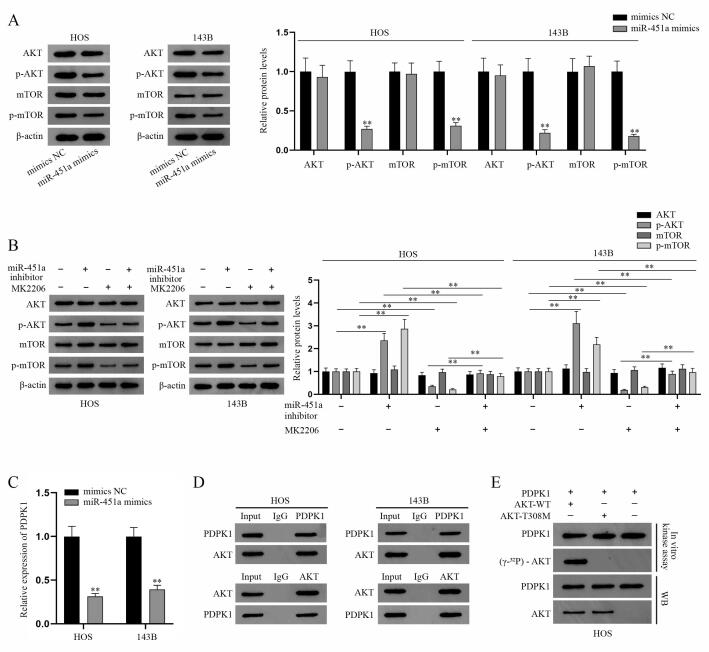
Although we have certified that miR-451a could impede the malignant progression of osteosarcoma cell, the mechanism of miR-451a needs further exploration given that miR-451a has no protein-coding ability. Previous study has reported that miR-451a plays a tumor suppressive role in gastric cancer via targeting AKT/mTOR pathway [21]. Nevertheless, the specific mechanism remains to be explored. Therefore, we conducted experiment to assess the mechanism of the regulation of miR-451a on AKT/mTOR pathway in osteosarcoma. The key proteins associated with AKT/mTOR pathway were firstly detected via western blot analysis. The results indicated that in HOS and 143B cells, the protein levels of phosphorylated AKT and mTOR (p-AKT and p-mTOR) were significantly impaired in miR-451a mimics group whereas the protein levels of AKT and mTOR were almost not changed (Fig. 2A). Further, we found that miR-451a knockdown could impede the inhibition of MK2206 (AKT inhibitor) on the protein levels of p-AKT and p-mTOR (Fig. 2B). Consequently, we concluded that miR-451a suppresses AKT/mTOR pathway. PPK1 (also known as PDPK1) was reported to play a critical role in AKT/mTOR pathway [22], [23] and facilitate the malignant progression of osteosarcoma [24]. Given that PDPK1 shares the alias PDK1 with another enzyme, we chose the “PDPK1” in this research to avoid ambiguity. We speculated that miR-451a could modulate PDPK1 to suppress AKT/mTOR pathway. To verify this assumption, PDPK1 expression was firstly measured via q-PCR and the results showed that overexpressed miR-451a curbed PDPK1 expression (Fig. 2C). The results of Co-IP assays proved the interaction between AKT and PDPK1 in osteosarcoma cells (Fig. 2D), providing evidence for PDPK1 phosphorylation of AKT. According to the previous literature [25], PDPK1 could phosphorylate AKT at Thr308 site. Then, in vitro phosphorylation assays proved that the phosphorylation site of PDPK1 interacting with AKT in osteosarcoma is Thr308 site since PDPK1 could phosphorylate the wild type AKT instead of AKT-T308 mutated protein (Fig. 2E). To sum up, PDPK1 phosphorylates the Thr308 site on AKT and miR-451a curbs AKT/mTOR pathway via PDPK1-mediated phosphorylation modification.
Fig. 2.
MiR-451a inhibits AKT/mTOR signaling pathway via PDPK1-induced phosphorylation modification. (A) Western blot was applied to measure the expression of AKT/mTOR signaling pathway associated proteins (AKT, p-AKT, mTOR, and p-mTOR) in transfected HOS and 143B cells with miR-451a mimics, taking β-actin as internal reference. (B) AKT/mTOR signaling pathway associated protein levels were evaluated by western blot analysis. (C) Q-PCR detection of PDPK1 expression level in transfected HOS and 143B cells with miR-451a mimics or mimics NC. (D) Co-IP assay was used to detect the interaction between PDPK1 and AKT proteins in osteosarcoma cells. (E) In vitro phosphorylation experiment was carried out to verify Thr308 as PDPK1-specific phosphorylation site in the sequence of AKT. **P < 0.01.
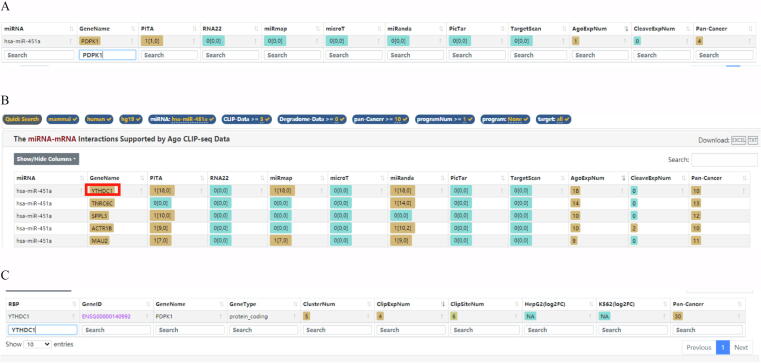
3.3. YTHDC1 modifies PDPK1 mRNA with m6A methylation
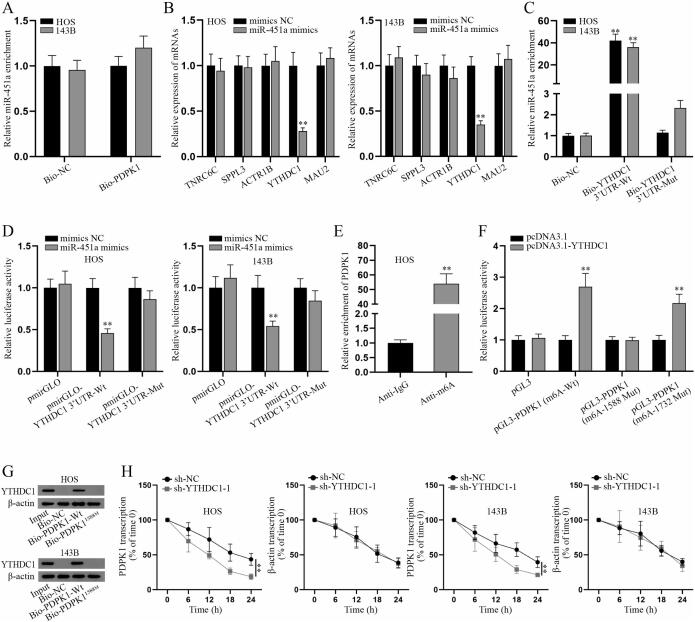
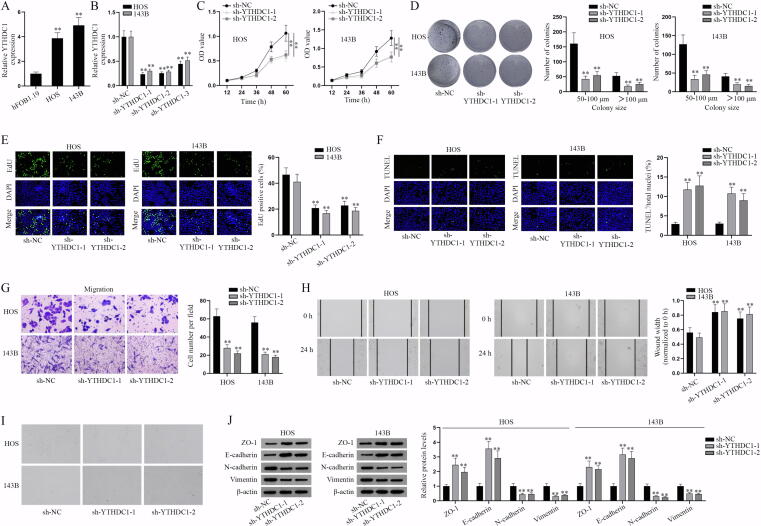
According to the bioinformatics prediction on starBase, miR-451a could bind to PDPK1, but with low predicted interactions (Fig. S2A). Hence, we focused on the specific mechanism between miR-451a and PDPK1 via a series of experiments. RNA pull-down assay detected that there was no enrichment of miR-451a in biotin-labeled PDPK1 (Bio-PDPK1) group (Fig. 3A). Subsequently, downstream mRNAs of miR-451a were screened on starBase database (CLIP-Data>=3, pan-Cancer>=10; Fig. S2B) and q-PCR was used to measure the expression of candidate mRNAs (Fig. 3B). It turned out that only YTHDC1 expression was markedly decreased after miR-451a overexpression, suggesting that YTHDC1 may be the downstream gene of miR-451a. Further, RNA pull-down assay showed that miR-451a was notably enriched in the complex pulled down by biotinylated wild type YTHDC1 3′UTR (Bio-YTHDC1 3′UTR-Wt) instead of mutant type (Bio-YTHDC1 3′UTR-Mut) versus control group (Fig. 3C). Luciferase reporter assay indicated that the luciferase activity of wild type YTHDC1 3′UTR (pmirGLO-YTHDC1 3′UTR-Wt) was impaired with overexpressed miR-451a while that of mutant YTHDC1 3′UTR (pmirGLO-YTHDC1 3′UTR-Mut) was not changed (Fig. 3D). Thus, it was concluded that miR-451a interacts with YTHDC1 3′UTR in osteosarcoma cells. Based on the prediction from starBase database, we found that RNA binding protein (RBP) YTHDC1 could bind with PDPK1 (Fig. S2C). We also found that YTHDC1 plays a role as an m6A reader in a variety of cancers [26], [27]. We then explored the effect of YTHDC1 on the malignancy of osteosarcoma. Firstly, YTHDC1 expression was detected in osteosarcoma cell lines (HOS and 143B) and normal bone cell hFOB1.19 via q-PCR (Fig. S3A). The results showed that YTDHC1 was highly expressed in osteosarcoma cell lines (HOS and 143B) versus hFOB1.19. After the verification of the knockdown efficiency of sh-YTHDC1-1/2/3 in Fig. S3B, functional assays were implemented in HOS and 143B cells with downregulated YTHDC1. CCK-8 and colony formation assay certified that YTHDC1 knockdown impeded proliferation of osteosarcoma cells (Fig. S3C-D). Similarly, EdU positive cells were reduced by YTHDC1 deficiency (Fig. S3E), further proving the inhibitory effect of YTHDC1 inhibition on cell proliferation. Inversely, TUNEL assay indicated that YTHDC1 knockdown facilitates cell apoptosis (Fig. S3F). Then the results of Transwell and wound healing assays indicated that YTHDC1 knockdown could propel cell migration (Fig. S3G-H). The ablation of YTHDC1 caused the change of cell morphology that cells changed from spindle shape to a round or rectangular shape (Fig. S3I), suggesting the suppression of YTHDC1 knockdown on EMT of HOS and 143B cells. Likewise, western blot analysis further verified this finding accompanied with the increased ZO-1, E-cadherin expressions and decreased N-cadherin and Vimentin expressions (Fig. S3J). In a word, YTHDC1 promotes the malignant progression of osteosarcoma cells.
Fig. 3.
YTHDC1 modifies the m6A methylation of PDPK1. (A) RNA pulldown followed by q-PCR was performed to determine the binding between miR-451a and PDPK1 in HOS and 143B cells. (B) The levels of candidate mRNAs in transfected HOS and 143B cells with miR-451a mimics or NC mimics were measured. (C) RNA pulldown assay was carried out to validate the binding between miR-451a and YTHDC1 3′UTR. (D) Luciferase reporter assay was conducted to further verify that miR-451a binds to YTHDC1 3′UTR. (E) RIP followed by q-PCR was performed to determine whether m6A modification occurs in the sequence of PDPK1. (F) Luciferase reporter assay was used to determine the YTHDC1-specific methylation site in the sequence of PDPK1. (G) RNA pulldown assay was done to further verify m6A-1588 site as the YTHDC1-specific methylation site in the sequence of PDPK1. (H) Remaining levels of PDPK1 and β-actin mRNA were detected via q-PCR in the transfected HOS and 143B cells after 50 mM α-amanitin treatment, with 18S rRNA as internal reference. **P < 0.01.
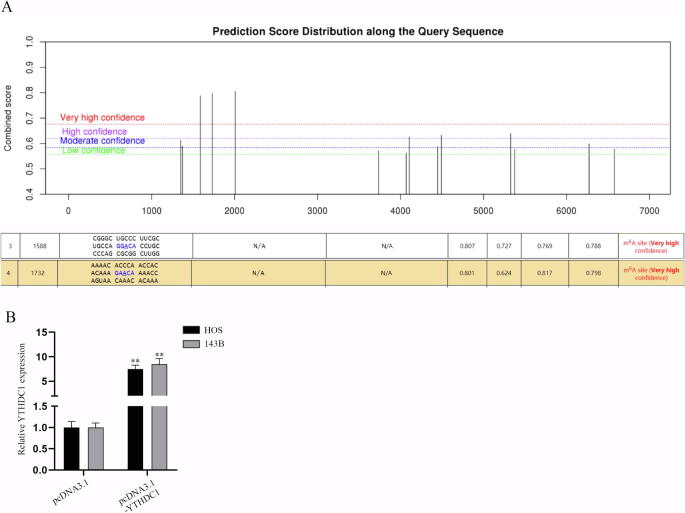
According to the information shown in Fig. S4A, PDPK1 has multiple m6A methylation sites. Then, m6A-1588 and m6A-1732 sites were chosen in the subsequent studies. We then speculated that YTHDC1 influences PDPK1 expression through regulation of PDPK1 m6A methylation. RIP assays certified that PDPK1 was notably enriched in anti-m6A group, suggesting that PDPK1 mRNA could undergo m6A methylation (Fig. 3E). The overexpression efficiency of pcDNA3.1-YTHDC1 was verified by q-PCR analysis (Fig. S4B). Luciferase reporter assay was performed to determine the YTHDC1 specific methylation site in the sequence of PDPK1. The results showed that the luciferase activity of pGL3-PDPK1 (m6A-Wt) or pGL3-PDPK1 (m6A-1732 Mut) was markedly strengthened in pcDNA3.1-YTHDC1 group versus control groups while that of pGL3-PDPK1 (m6A-1588 Mut) had no marked change, revealing that YTHDC1 mediates the methylation of PDPK1 at site 1588 (Fig. 3F). RNA pull-down assay further proved this finding with the verification that YTHDC1 was pulled down by wild type PDPK1 while there was no abundance of YTHDC1 in PDPK11588M group (Fig. 3G). In HOS and 143B cells treated with α-amanitin, the stability of PDPK1 and β-actin mRNA was assessed by q-PCR analysis with YTHDC1 knockdown (Fig. 3H). The results verified that YTHDC1 could stabilize PDPK1 mRNA. To sum up, YTHDC1 regulates PDPK1 mRNA m6A methylation at site 1588.
3.4. MiR-451a inhibits AKT/mTOR signaling pathway via YTHDC1-mediated PDPK1
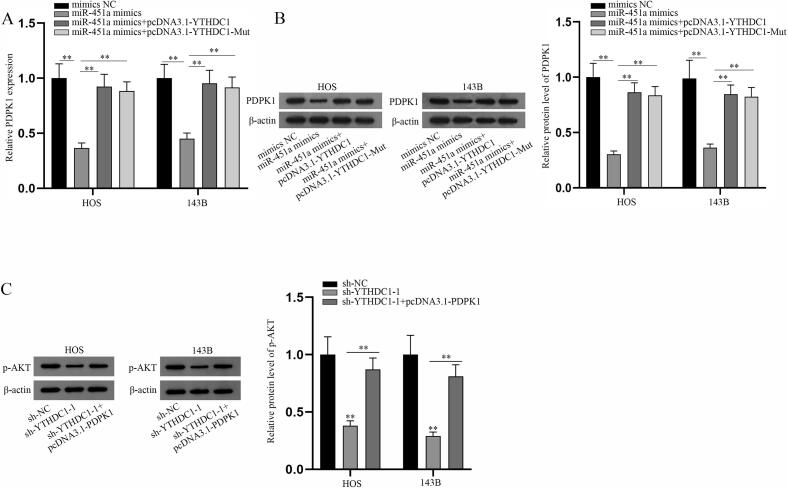
PDPK1 RNA and protein levels were detected via q-PCR and western blot analyses under different transfections (Fig. 4A-B). The results showed that overexpressed YTHDC1 could reverse the inhibition of miR-451a overexpression on RNA and protein levels of PDPK1 and YTHDC1 overexpression (mutated one also included) gave a higher level of PDPK1, indicating that miR-451a modulates PDPK1 level via YTHDC1. Then, protein levels of p-AKT after the indicated transfections were also measured by western blot (Fig. 4C). The results certified that PDPK1 overexpression could partly counteract the suppression of YTHDC1 knockdown on p-AKT level, revealing that YTHDC1 regulates p-AKT level by PDPK1. In a word, miR-451a represses AKT/mTOR signaling pathway via YTHDC1-mediated PDPK1.
Fig. 4.
MiR-451a inhibits AKT/mTOR signaling pathway via YTHDC1-mediated PDPK1. (A-B) Rescue experiments were performed in HOS and 143B cells transfected with mimics NC, miR-451a mimics, miR-451a mimics + pcDNA3.1-YTHDC1, or miR-451a mimics + pcDNA3.1-YTHDC1-Mut. Q-PCR and western blot analyses of PDPK1 RNA and protein levels in the transfected HOS and 143B cells respectively. (C) Rescue experiments were performed in the indicated transfected HOS and 143B cells. Western blot analysis of p-AKT expression in the transfected HOS and 143B cells was shown. **P < 0.01.
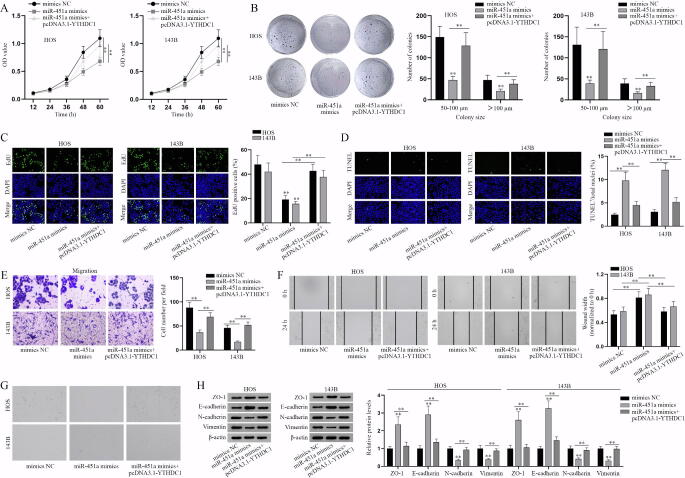
3.5. MiR-451a impairs the progression of osteosarcoma cells via YTHDC1/PDPK1 axis
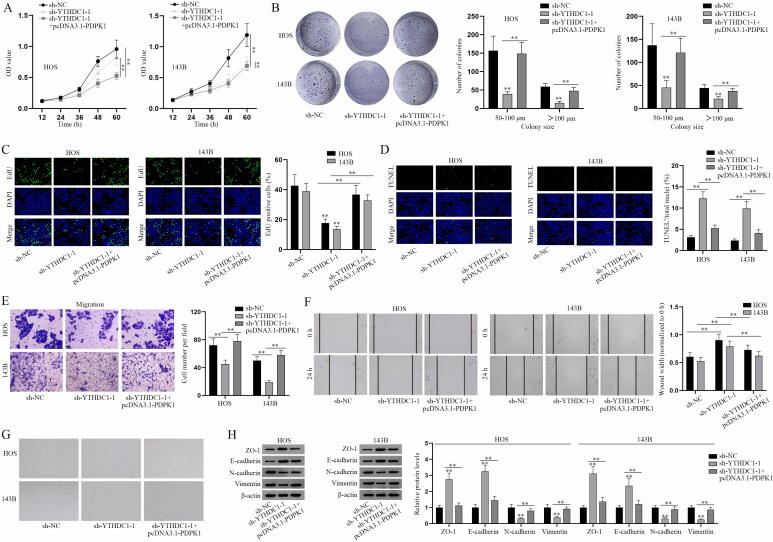
Next, we designed rescue experiments to determine whether miR-451a/YTHDC1/PDPK1 axis affects the progression of osteosarcoma cells. A series of functional assays were performed to evaluate the proliferation, apoptosis, migration and EMT of osteosarcoma cells transfected with the indicated plasmids. As the results of CCK-8, colony formation and EdU assays showed, overexpressing PDPK1 altered the inhibitory effect of YTHDC1 deficiency on osteosarcoma cell proliferation (Fig. 5A-C). Meanwhile, overexpressing YTHDC1 counteracted the repressive effect of miR-451a mimics on cell proliferation (Fig. S5A-C). TUNEL assays showed that PDPK1 overexpression altered the promoting effect of YTHDC1 silencing on cell apoptosis, and YTHDC1 upregulation counteracted the facilitating role of miR-451a mimics on cell apoptosis in osteosarcoma (Fig. 5D, Fig. S5D). Transwell migration and wound healing assays showed that PDPK1 overexpression countervailed the suppressive effect of inhibited YTHDC1 on the migratory ability of osteosarcoma cells (Fig. 5E-F). Ihe upregulation of YTHDC1 altered the suppressive effect of miR-451a mimics on cell migration (Fig. S5E-F). Through the observation of osteosarcoma cell morphology and western blot analysis of EMT-related markers, it was indicated that miR-451a represses EMT in osteosarcoma via regulating YTHDC1-mediated PDPK1 (Fig. 5G-H, Fig. S5G-H). In conclusion, miR-451a inhibits the progression of osteosarcoma cells via YTHDC1/PDPK1 axis.
Fig. 5.
YTHDC1 regulates the progression of osteosarcoma via PDPK1. (A-C) The proliferative ability of osteosarcoma cells transfected with sh-NC, sh-YTHDC1-1 or sh-YTHDC1-1 + pcDNA3.1-PDPK1 was evaluated by proliferation assays. (D) The apoptosis of transfected osteosarcoma cells in different groups was determined by TUNEL assay. (E-F) The migratory ability of differently transfected osteosarcoma cells was assessed by Transwell migration and wound healing assays. (G) The morphology of transfected osteosarcoma cells was observed via microscopy. (H) Western blot analysis of EMT markers in the transfected osteosarcoma cells. **P < 0.01.
3.6. MiR-451a hampers the tumorgenesis of osteosarcoma in vivo
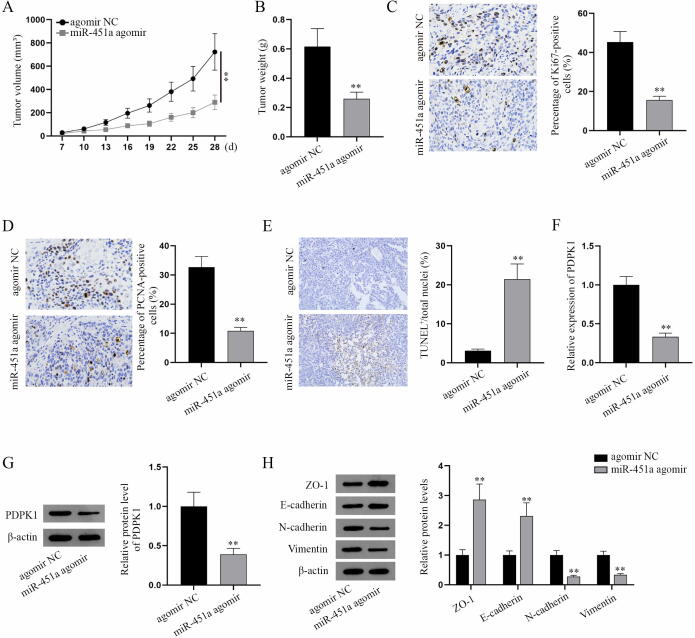
Besides in vitro assays, we performed in vivo experiments to verify this mechanism. BALB/c nude mice (male, 6 ∼ 8-week-old) were subcutaneously injected with HOS and 143B cells transfected with agomir NC or miR-451a agomir. Tumor volume and tumor weight were overtly reduced by miR-451a agomir (Fig. 6A-B). IHC analysis of cell proliferation markers (Ki67 and PCNA) in xenografts showed that in mice model, miR-451a agomir significantly hampers tumor growth of HOS and 143B tumor xenografts accompanied by decreased Ki67-positive cells and PCNA-positive cells (Fig. 6C-D). TUNEL assay indicated that miR-451a agomir induces apoptosis in osteosarcoma xenografts in mice (Fig. 6E). The above results indicated that miR-451a inhibits tumor growth but triggers apoptosis. According to q-PCR and western blot analyses, PDPK1 expression in xenografts was decreased in miR-451a agomir groups (Fig. 6F-G), which indicated that miR-451a downregulates PDPK1 expression in osteosarcoma xenografts in mice. Additionally, ZO-1 and E-cadherin expressions were increased while N-cadherin and Vimentin expressions were reduced in miR-451a agomir group, which indicated that miR-451a suppresses EMT in osteosarcoma xenografts in mice (Fig. 6H). Collectively, miR-451a impedes the tumorgenesis of osteosarcoma in vivo.
Fig. 6.
MiR-451a hampers the tumorgenesis of osteosarcoma in vivo. (A) The volume of tumor xenografts in mice was detected every three days after seven days after treatment (i.e. injection with HOS or 143B cells expressing agomir NC or miR-451a agomir). (B) The weight of tumors excised from the mice sacrificed on the 28th day for xenografts. The weight of tumor xenografts was measured. (C-D) IHC analysis of cell proliferation markers (Ki67 and PCNA) in tumor xenografts in mice was shown. (E) TUNEL assay was performed to evaluate the apoptosis of paraffin-embedded tissues. (F-G) QPCR and western blot were applied to detect PDPK1 expression in mice tumor xenografts. (H) Western blot analysis of EMT markers in mice tumor xenografts. **P < 0.01.
4. Discussion
Previous studies have reported the crucial roles of miRNAs in the diagnosis, treatment, and prognosis of osteosarcoma [28]. Herein, we validated the markedly downregulated expression of miR-451a in osteosarcoma cell lines via q-PCR analysis, which is consistent with the expression profile of miR-451a in osteosarcoma tissues as identified in GEO datasets (GSE28423) and the finding in the previous study [29]. Compared to the former studies on the suppressive role of miR-451a in osteosarcoma cells from the aspects of cell growth and invasion [29], our study also verified its inhibitory effect on osteosarcoma cell proliferation through CCK-8, colony formation, and EdU assays. Additionally, our study further validated that miR-451a induces cell apoptosis and hampers cell migration, EMT in osteosarcoma through TUNEL, transwell migration, wound healing assays and observation of tumor cell morphology as well as western blot analysis of EMT-associated markers (ZO-1, E-cadherin, N-cadherin, and Vimentin).
Subsequent to the investigation into its biological role, we also explored the regulatory mechanism of miR-451a in osteosarcoma. Unlike the exploration of downstream targets in the previous study [29], we dug into the regulation mechanism of miR-451a on the oncogenic AKT/mTOR signaling pathway in osteosarcoma. Through western blot analysis of AKT/mTOR pathway related proteins, we found that miR-451a inhibits the AKT/mTOR signaling pathway via PDPK1 which was previously reported as a potential diagnostic biomarker for osteosarcoma [30] and a tumor-propeller in osteosarcoma [24]. Further, we uncovered that miR-451a suppresses the AKT/mTOR signaling pathway via PDPK1-induced phosphorylation of AKT at Thr308 site through q-PCR analysis, Co-IP assay and in vitro phosphorylation experiment.
Moreover, we probed into the regulatory mechanism of miR-451a on PDPK1 in osteosarcoma. PDPK1 has been reported to be implicated in malignancy of cervical cancer [31], papillary thyroid cancer [32], prostate cancer [33], hepatocellular carcinoma [34]. Herein, bioinformatics prediction showed that miR-451a could bind to PDPK1, but with low predicted interactions. Given that, we turned to investigate the indirect regulation approach of miR-451a on PDPK1 in osteosarcoma. In this study, we firstly verified YTHDC1 as the downstream gene of miR-451a in osteosarcoma through a combination of bioinformatics prediction, q-PCR analysis, and RNA pulldown as well as luciferase reporter assays. Also, we validated the promoting role of YTHDC1 in osteosarcoma.
Moreover, m6A modifications and their regulators have been associated with the development of multiple human cancers [35], [36]. For instance, reduction of RNA m6A methylation activates Wnt/PI3K-Akt signaling pathway to facilitate malignant phenotypes of gastric cancer cells [37]. Some studies focused on nuclear m6A reader YTHDC1 and m6A modification. For example, PDPK1 was predicted to bind to RBP YTHDC1 which could regulate human cancer via acting as m6A reader [26]. In this study, several putative m6A methylation sites in the sequence of PDPK1 were discovered owing to the bioinformatics prediction. Subsequently, we validated that YTHDC1 upregulates PDPK1 expression via m6A methylation modification and verified m6A-1588 site as the YTHDC1-specific methylation site in the sequence of PDPK1 through RIP, luciferase reporter and RNA pulldown assays. Plus, we found that YTHDC1 could stabilize PDPK1 mRNA. Further, we performed rescue experiments followed by q-PCR and western blot analyses and found that miR-451a-mediated YTHDC1 stabilizes PDPK1 mRNA via the m6A-dependent regulation. In addition to in vitro assays, we conducted in vivo experiments and found that miR-451a represses the tumor growth and induces apoptosis. The mRNA and protein levels of PDPK1 and EMT markers were also validated to be inhibited by miR-451a in osteosarcoma in vivo.
5. Conclusions
The current study manifested the suppressive effect of miR-451a in osteosarcoma and demonstrated that miR-451a regulates the aggressive behaviors of osteosarcoma cells via modulating YTHDC1-mediated m6A methylation and PDPK1-mediated phosphorylation modifications to activate the AKT/mTOR signaling pathway, which might provide new perspectives into osteosarcoma treatment.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Dong Cao: Data curation. Shanshan Ge: . Mengchun Li: .
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We appreciate all supports.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbo.2022.100412.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
Expression profiles of miR-451a and overexpression/knockdown efficiency of miR-451a mimics/inhibitor. (A) Volcano plot of differently expressed miRNAs in osteosarcoma (tumor) and normal samples on the GEO datasets (GSE28423). Red dots presented upregulated miRNAs in tumor samples relative to normal samples while blue ones displayed downregulated miRNAs. (B) The expression profile of miR-451 (named miR-451a in this study) in osteosarcoma cell lines (Tumor) compared with normal bones (Normal). (C) The overexpression efficiency of miR-451a mimics and knockdown efficiency of miR-451a inhibitor in HOS and 143B cells were verified via q-PCR. **P < 0.01.
Supplementary Fig. 2.
Bioinformatics prediction. (A) The starBase database predicted that miR-451a combines with PDPK1. (B) The target mRNAs of miR-451a were obtained through starBase prediction (CLIP-Data>=3; pan-Cancer>=10). (C) YTHDC1 was predicted as one of RNA binding proteins (RBPs) for PDPK1 on the starBase.
Supplementary Fig. 3.
The loss-of-function effects of YTHDC1 on the malignant phenotypes of osteosarcoma cells. (A) The expression level of YTHDC1 in human normal hFOB1.19 osteoblasts and osteosarcoma cell lines (HOS and 143B). (B) The knockdown efficiency of sh-YTHDC1-1/2/3 was validated via q-PCR. (C-E) CCK8, colony formation and EdU assays were performed to assess the effects of silenced YTHDC1 on the proliferative ability of transfected osteosarcoma cells (HOS and 143B). (F) TUNEL assay was conducted to evaluate the apoptosis of transfected osteosarcoma cells with sh-YTHDC1-1/2. (G-H) The migratory capacity of transfected osteosarcoma cells with sh-YTHDC1-1/2 was determined by Transwell migration and wound healing assays. (I) Osteosarcoma cell morphology was captured with optical microscope after transfection with sh-YTHDC1-1/2. (J) Western blot was applied to measure the protein levels of EMT-related markers in transfected HOS and 143B cells with sh-YTHDC1-1/2. **P < 0.01.
Supplementary Fig. 3J.
Supplementary Fig. 4.
YTHDC1 propels the malignant progression of osteosarcoma. (A) Bioinformatics site SRAMP predicts m6A methylation sites in the sequence of PDPK1. (B) The upregulation efficiency of pcDNA3.1-YTHDC1 was validated via q-PCR. **P < 0.01.
Supplementary Fig. 5.
MiR-451a inhibits the malignant progression of osteosarcoma via YTHDC1. (A-C) CCK-8, colony formation and EdU assays were performed to analyze the proliferation of HOS and 143B transfected cells with mimics NC, miR-451a mimics or miR-451a mimics+pcDNA3.1-YTHDC1. (D) The apoptosis of the indicated HOS and 143B cells was detected. (E-F) The migratory ability of the indicated HOS and 143B cells was analyzed by Transwell and wound healing assays. (G) The osteosarcoma cell morphology in different groups was observed via microscopy. (H) Western blot analysis of EMT markers in the indicated HOS and 143B cells. **P < 0.01.
Supplementary Fig. 5H.
References
- 1.Ferguson J.L., Turner S.P. Bone cancer: diagnosis and treatment principles. Am. Fam. Phys. 2018;98(4):205–213. [PubMed] [Google Scholar]
- 2.Sadykova L.R., Ntekim A.I., Muyangwa-Semenova M., Rutland C.S., Jeyapalan J.N., Blatt N., Rizvanov A.A. Epidemiology and risk factors of osteosarcoma. Cancer Invest. 2020;38(5):259–269. doi: 10.1080/07357907.2020.1768401. [DOI] [PubMed] [Google Scholar]
- 3.Isakoff M.S., Bielack S.S., Meltzer P., Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33(27):3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson M.E. Update on survival in osteosarcoma. Orthoped Clin N Am. 2016;47(1):283–292. doi: 10.1016/j.ocl.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 5.Meazza C., Scanagatta P. Metastatic osteosarcoma: a challenging multidisciplinary treatment. Expert Rev. Anticancer Ther. 2016;16(5):543–556. doi: 10.1586/14737140.2016.1168697. [DOI] [PubMed] [Google Scholar]
- 6.Lilienthal I., Herold N. Targeting molecular mechanisms underlying treatment efficacy and resistance in osteosarcoma: A review of current and future strategies. Int. J. Mol. Sci. 2020;21(18):6885. doi: 10.3390/ijms21186885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murugan A.K. mTOR: Role in cancer, metastasis and drug resistance. Semin. Cancer Biol. 2019;59:92–111. doi: 10.1016/j.semcancer.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Hua H., Kong Q., Zhang H., Wang J., Luo T., Jiang Y. Targeting mTOR for cancer therapy. J. Hematol. Oncol. 2019;12(1):71. doi: 10.1186/s13045-019-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kagawa S., Takano S., Yoshitomi H., Kimura F., Satoh M., Shimizu H., Yoshidome H., Ohtsuka M., Kato A., Furukawa K., Matsushita K., Nomura F., Miyazaki M. Akt/mTOR signaling pathway is crucial for gemcitabine resistance induced by Annexin II in pancreatic cancer cells. J. Surg. Res. 2012;178(2):758–767. doi: 10.1016/j.jss.2012.05.065. [DOI] [PubMed] [Google Scholar]
- 10.Teng X., Fan X.F., Li Q., Liu S., Wu D.Y., Wang S.Y., Shi Y., Dong M. XPC inhibition rescues cisplatin resistance via the Akt/mTOR signaling pathway in A549/DDP lung adenocarcinoma cells. Oncol. Rep. 2019;41(3):1875–1882. doi: 10.3892/or.2019.6959. [DOI] [PubMed] [Google Scholar]
- 11.Fu R., Tong J.-S. miR-126 reduces trastuzumab resistance by targeting PIK3R2 and regulating AKT/mTOR pathway in breast cancer cells. J. Cell Mol. Med. 2020;24(13):7600–7608. doi: 10.1111/jcmm.15396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pal S.K., Quinn D.I. Differentiating mTOR inhibitors in renal cell carcinoma. Cancer Treat. Rev. 2013;39(7):709–719. doi: 10.1016/j.ctrv.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun X., Wei B., Peng Z.H., Fu Q.L., Wang C.J., Zheng J.C., Sun J.C. Knockdown of lncRNA XIST suppresses osteosarcoma progression by inactivating AKT/mTOR signaling pathway by sponging miR-375-3p. Int. J. Clin. Exp. Path. 2019;12(5):1507–1517. [PMC free article] [PubMed] [Google Scholar]
- 14.Romano G., Veneziano D., Acunzo M., Croce C.M. Small non-coding RNA and cancer. Carcinogenesis. 2017;38(5):485–491. doi: 10.1093/carcin/bgx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Correia de Sousa M., Gjorgjieva M., Dolicka D., Sobolewski C., Foti M. Deciphering miRNAs' Action through miRNA Editing. Int. J. Mol. Sci. 2019;20(24):6249. doi: 10.3390/ijms20246249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otoukesh B., Abbasi M., Gorgani H.O., Farahini H., Moghtadaei M., Boddouhi B., Kaghazian P., Hosseinzadeh S., Alaee A. MicroRNAs signatures, bioinformatics analysis of miRNAs, miRNA mimics and antagonists, and miRNA therapeutics in osteosarcoma. Cancer Cell Int. 2020;20:254. doi: 10.1186/s12935-020-01342-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu X., Li L., Lu Y., Yu X., Chen H., Yin Q., Zhang Y. miRNA-21 inhibition inhibits osteosarcoma cell proliferation by targeting PTEN and regulating the TGF-β1 signaling pathway. Oncol. Lett. 2018;16(4):4337–4342. doi: 10.3892/ol.2018.9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Y.Y., Cui J.Y., Yuan J., Wang X. MiR-451a suppressed cell migration and invasion in non-small cell lung cancer through targeting ATF2. Eur. Rev. Med. Pharmacol. Sci. 2018;22(17):5554–5561. doi: 10.26355/eurrev_201809_15818. [DOI] [PubMed] [Google Scholar]
- 19.K. Xu, B. Han, Y. Bai, X.Y. Ma, Z.N. Ji, Y. Xiong, S.K. Miao, Y.Y. Zhang, L.M. Zhou, MiR-451a suppressing BAP31 can inhibit proliferation and increase apoptosis through inducing ER stress in colorectal cancer, Cell Death Dis. 10(3) (2019) 152. [DOI] [PMC free article] [PubMed]
- 20.Riquelme I., Tapia O., Leal P., Sandoval A., Varga M.G., Letelier P., Buchegger K., Bizama C., Espinoza J.A., Peek R.M., Araya J.C., Roa J.C. miR-101-2, miR-125b-2 and miR-451a act as potential tumor suppressors in gastric cancer through regulation of the PI3K/AKT/mTOR pathway. Cell. Oncol. (Dordrecht) 2016;39(1):23–33. doi: 10.1007/s13402-015-0247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Streleckiene G., Inciuraite R., Juzenas S., Salteniene V., Steponaitiene R., Gyvyte U., Kiudelis G., Leja M., Ruzgys P., Satkauskas S., Kupcinskiene E., Franke S., Thon C., Link A., Kupcinskas J., Skieceviciene J. miR-20b and miR-451a are involved in gastric carcinogenesis through the PI3K/AKT/mTOR signaling pathway: data from gastric cancer patients, cell lines and ins-gas mouse model. Int. J. Mol. Sci. 2020;21(3) doi: 10.3390/ijms21030877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segerström L., Baryawno N., Sveinbjörnsson B., Wickström M., Elfman L., Kogner P., Johnsen J.I. Effects of small molecule inhibitors of PI3K/Akt/mTOR signaling on neuroblastoma growth in vitro and in vivo. Int. J. Cancer. 2011;129(12):2958–2965. doi: 10.1002/ijc.26268. [DOI] [PubMed] [Google Scholar]
- 23.Liu R., Chen Z., Yi X., Huang F., Hu G., Liu D., Li X.i., Zhou H., Liu Z. 9za plays cytotoxic and proapoptotic roles and induces cytoprotective autophagy through the PDK1/Akt/mTOR axis in non-small-cell lung cancer. J. Cell. Physiol. 2019;234(11):20728–20741. doi: 10.1002/jcp.28679. [DOI] [PubMed] [Google Scholar]
- 24.Li Z., Shen J., Chan M.T., Wu W.K. MicroRNA-379 suppresses osteosarcoma progression by targeting PDK1. J. Cell Mol. Med. 2017;21(2):315–323. doi: 10.1111/jcmm.12966. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Sarbassov D.D., Guertin D.A., Ali S.M., Sabatini D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science (New York N.Y.) 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 26.I.A. Roundtree, G.Z. Luo, Z. Zhang, X. Wang, T. Zhou, Y. Cui, J. Sha, X. Huang, L. Guerrero, P. Xie, E. He, B. Shen, C. He, YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs, eLife 6 (2017). [DOI] [PMC free article] [PubMed]
- 27.Chen H., Li Y., Li L.i., Zhu J., Yang Z., Zhang J., Li S., Xin Y., Xia H., He J. YTHDC1 gene polymorphisms and hepatoblastoma susceptibility in Chinese children: A seven-center case-control study. J. Gene Med. 2020;22(11):e3249. doi: 10.1002/jgm.3249. [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Liu S., Shi J., Li J., Wang S., Liu H., Zhao S., Duan K., Pan X., Yi Z. The role of miRNA in the diagnosis, prognosis, and treatment of osteosarcoma. Cancer Biother. Radiopharm. 2019;34(10):605–613. doi: 10.1089/cbr.2019.2939. [DOI] [PubMed] [Google Scholar]
- 29.X. Ma, D. Li, Y. Gao, C. Liu, miR-451a inhibits the growth and invasion of osteosarcoma via targeting TRIM66, Technol. Cancer Res. Treat. 18 (2019) 1533033819870209. [DOI] [PMC free article] [PubMed] [Retracted]
- 30.Zhang Y., Yang F. Analyzing the disease module associated with osteosarcoma via a network- and pathway-based approach. Exp. Ther. Med. 2018;16(3):2584–2592. doi: 10.3892/etm.2018.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu T., Zhang Q., Gao L. LncRNA CAR10 upregulates PDPK1 to promote cervical cancer development by sponging miR-125b-5p. Biomed. Res. Int. 2020;2020:4351671. doi: 10.1155/2020/4351671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Qi M. miR-718 is involved in malignancy of papillary thyroid cancer through repression of PDPK1. Pathol. Res. Pract. 2018;214(11):1787–1793. doi: 10.1016/j.prp.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 33.Nalairndran G., Hassan Abdul Razack A., Mai C.-W., Fei‐Lei Chung F., Chan K.-K., Hii L.-W., Lim W.-M., Chung I., Leong C.-O. Phosphoinositide-dependent Kinase-1 (PDPK1) regulates serum/glucocorticoid-regulated Kinase 3 (SGK3) for prostate cancer cell survival. J. Cell Mol. Med. 2020;24(20):12188–12198. doi: 10.1111/jcmm.15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang Y., Chen J., Feng W., Qiao C., Han W., Nie Y., Wu K., Fan D., Xia L. Interleukin 1β-mediated HOXC10 overexpression promotes hepatocellular carcinoma metastasis by upregulating PDPK1 and VASP. Theranostics. 2020;10(8):3833–3848. doi: 10.7150/thno.41712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z.X., Li L.M., Sun H.L., Liu S.M. Link between m6A modification and cancers. Front. Bioeng. Biotechnol. 2018;6:89. doi: 10.3389/fbioe.2018.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He L., Li H., Wu A., Peng Y., Shu G., Yin G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer. 2019;18(1):176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C., Zhang M., Ge S., Huang W., Lin X., Gao J., Gong J., Shen L. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019;8(10):4766–4781. doi: 10.1002/cam4.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]