Abstract
Background
Currently, SARS‐CoV‐2 RNA detection using real‐time reverse‐transcription PCR (rRT‐PCR) is the standard diagnostic test for COVID‐19 infection. Various rRT‐PCR assays are currently used worldwide, targeting different genes of the SARS‐CoV‐2. Here, we compared the analytical sensitivity and clinical performance (sensitivity and specificity) of Allplex SARS‐CoV‐2/FluA/FluB/RSV assay (Seegene), Standard M nCoV real‐time detection kit (SD Biosensor), and U‐TOP COVID‐19 detection kit (Seasun Biomaterials) for SARS‐CoV‐2 detection.
Methods
Two hundred and forty‐nine nasopharyngeal swab samples were evaluated to compare the clinical performance of the rRT‐PCR assays. For the analytical performance evaluation, two RNA controls with known viral loads—SARS‐CoV‐2 RNA control and SARS‐COV‐2 B.1.351 RNA control—were used to investigate the potential impact of SARS‐CoV‐2 variants, particularly the B.1.351 lineage.
Results
Limits of detection ranged from 650 to 1300 copies/ml for rRT‐PCR assays, and the mean differences in cycle threshold (C t ) values of the two RNA controls were within 1.0 for each target in the rRT‐PCR assays (0.05–0.73), without any prominent C t value shift or dropouts in the SARS‐COV‐2 B.1.351 RNA control. Using the consensus criterion as the reference standard, 89 samples were positive, whereas 160 were negative. The overall clinical performance of rRT‐PCR assays was comparable (sensitivity 98.88%–100%; specificity 99.38%–100%), whereas the sensitivities of each target gene were more variable.
Conclusions
The three rRT‐PCR assays showed comparable analytical sensitivity and clinical performance. The analytical and clinical sensitivities of each target gene were influenced more by the primer and probe design than the target gene itself.
Keywords: COVID‐19, PCR, performance evaluation, SARS‐CoV‐2
For the analytical performance evaluation, two RNA controls with known viral loads—SARS‐CoV‐2 RNA control and SARS‐COV‐2 B.1.351 RNA control—were used to investigate the potential impact of SARS‐CoV‐2 variants, particularly the B.1.351 lineage. Limits of detection ranged from 650 to 1300 copies/ml for rRT‐PCR assays and the mean differences in cycle threshold (C t ) values of the two RNA controls were within 1.0 for each target in the rRT‐PCR assays (0.05–0.73), without any prominent C t value shift or dropouts in the SARS‐CoV‐2 B.1.351 RNA control. The overall clinical performance of rRT‐PCR assays was comparable (sensitivity 98.88%–100%; specificity 99.38%–100%), whereas the sensitivities of each target gene were more variable. Both analytical and clinical performances of each target gene appear to be influenced more by the primer and probe design rather than the target gene. Clinical performance results were in line with the analytical sensitivity results. The S and ORF1ab genes were the most sensitive target genes using the Allplex SARS‐CoV‐2/FluA/FluB/RSV assay and U‐TOP COVID‐19 detection kit. In the case of the Standard M nCoV real‐time detection kit, although the difference in analytical sensitivity was not evident, RdRp showed higher sensitivity than the E gene.
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) is caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). 1 Prompt and accurate detection of SARS‐CoV‐2 is crucial to prevent its transmission and administer the appropriate treatment. SARS‐CoV‐2 RNA detection using real‐time reverse‐transcription PCR (rRT‐PCR) is the standard diagnostic test for current infections of COVID‐19. 2 Various rRT‐PCR assays are currently used worldwide, and the assays target different genes of the SARS‐CoV‐2, such as the envelope (E), ORF1ab/RNA‐dependent RNA polymerase (RdRp), nucleocapsid (N), and spike (S) genes. 3 , 4 , 5 rRT‐PCR assays targeting more than one position of the SARS‐CoV‐2 genome are recommended because of potential genetic variation in SARS‐CoV‐2, which may result in false‐negative results. 2
Since the emergence of SARS‐CoV‐2, sets of mutations have been noted, and these genetic mutations can affect the performance of the rRT‐PCR assay. Mutation analysis of the SARS‐CoV‐2 genome collected worldwide showed that mutations occurred in all SARS‐CoV‐2 diagnostic targets. 6 , 7 Moreover, novel strains with two amino acid (H69 and V70) deletion within the S gene, which includes the alpha variant of SARS‐CoV‐2 B.1.1.7 lineage, have been characterized by the S gene dropout in SARS‐CoV‐2 rRT‐PCR assays targeting the S gene. 8 , 9 Several novel mutations in either the E or N gene have been shown to affect SARS‐CoV‐2 rRT‐PCR results. 10 , 11 The beta variant of SARS‐CoV‐2 B.1.351 lineage, initially detected in South Africa, 12 is rapidly taking over the wild‐type SARS‐CoV‐2 globally, with increased transmissibility. 12 , 13 , 14 The impact of the beta variant of SARS‐CoV‐2 B.1.351 lineage on the performance of SARS‐CoV‐2 rRT‐PCR assays has not been fully elucidated. Because the primer and probe sequence information of commercial SARS‐CoV‐2 rRT‐PCR assays are not available, performance evaluation using various clinical samples is important.
The target gene of SARS‐CoV‐2 rRT‐PCR varies widely. The guidelines regarding target genes for SARS‐CoV‐2 detection differ worldwide. With the emergence of SARS‐CoV‐2, the World Health Organization recommended protocols targeting the E gene for screening and the RdRp gene for confirmation testing. 15 The US Centers for Disease Control and Prevention designed a 2019‐nCoV rRT‐PCR diagnostic panel targeting different regions of the N gene. 16 Among the target genes of the developed SARS‐CoV‐2 rRT‐PCR assays, the N gene is the most frequently selected target gene apart from ORF1ab, whereas the S gene is the least frequently selected target gene. 5
Recently, the Allplex SARS‐CoV‐2/FluA/FluB/RSV assay (Seegene), targeting the S, RdRp, and N genes for SARS‐CoV‐2 detection, have been developed. The previously introduced SARS‐CoV‐2 rRT‐PCR assay by Seegene, namely, Allplex 2019‐nCoV assay, which was approved for emergency use with authorization from the US Food and Drug Administration, has been designed to include the E gene instead of the S gene as the target gene for SARS‐CoV‐2 detection. Here, we report the analytical sensitivity and clinical performance (sensitivity and specificity) of the Allplex SARS‐CoV‐2/FluA/FluB/RSV assay in comparison with two commercially available kits: the Standard M nCoV real‐time detection kit (SD Biosensor, Osong, Korea) and U‐TOP COVID‐19 detection kit (Seasun Biomaterials). We also examined the potential impact of the SARS‐CoV‐2 variant of concern, particularly the B.1.351 lineage, on the analytical and clinical performance.
2. MATERIALS AND METHODS
2.1. Clinical samples and viral RNA
This study was performed using residual nasopharyngeal swab (NPS) specimens collected from patients who visited Korea University Guro Hospital between December 2020 and February 2021. The residual specimens were stored at −70°C until analysis and used for the performance evaluation of the three commercial kits. This study was approved by the Institutional Review Board of the Korea University Guro Hospital (2021GR0086).
For analytical sensitivity evaluation, viral RNA controls with known RNA loads, Amplirun SARS‐CoV‐2 RNA control (Vircell) derived from a Spanish clinical isolate (GISAID accession ID: EPI_ISL_429256) and Amplirun SARS‐CoV‐2 B.1.351 RNA control (Vircell) derived from a Spanish clinical isolate (GISAID accession ID: EPI_ISL_848199) were used. The two SARS‐CoV‐2 RNA controls were diluted to obtain eight concentrations as follows: 13, 130, 650, 1300, 13,000, 130,000, 1,300,000, and 13,000,000 copies/ml to analyze the linearity and amplification efficiency of each target of the rRT‐PCR assay. The preliminary limit of detection (LoD) was determined with three replicates over five concentrations ranging from 13 to 13,000 copies/ml. At the concentration level spanning the possible LoD, each concentration was replicated five times.
2.2. RNA extraction and SARS‐CoV‐2 rRT‐PCR
RNA extraction from clinical samples was performed using the nucleic acid extraction platform Microlab STARlet (Hamilton). Allplex SARS‐CoV‐2/FluA/FluB/RSV assay, Standard M nCoV real‐time detection kit, and U‐TOP COVID‐19 detection kit were used in this study. PCRs for all three rRT‐PCR assays were performed using the CFX96 system (Bio‐Rad).
The Allplex SARS‐CoV‐2/FluA/FluB/RSV assay has three targets for SARS‐CoV‐2, namely, the S, RdRp, and N genes. PCR was performed in a total volume of 20 µl (10 µl of extracted RNA and 10 µl of master mix). A cycle threshold (C t ) value equal to or below 38 was interpreted as positive for each target gene. The standard M nCoV real‐time detection kit detects two targets—E and RdRp—and the reaction volume was 30 µl (10 µl of extracted RNA and 20 µl of master mix). A C t value equal to or below 36 was interpreted as positive for each target gene. For the U‐TOP COVID‐19 detection kit, four genes, namely, ORF1ab, N, S, and E, were targeted. PCR was performed in a total volume of 30 µl (10 µl of extracted RNA and 20 µl of master mix), and a C t value equal to or below 38 was interpreted as positive for each target gene.
2.3. Data analysis
The clinical performance of the three rRT‐PCRs was evaluated using reference standards. The reference standard was defined using the consensus criterion, which was obtained from two out of three rRT‐PCR assays. 17 , 18 The results were considered discordant when one of the rRT‐PCR assays did not agree with the other two assays. The difference in C t values between the targets was analyzed using Student's t test. Linearity was assessed using linear regression analysis, and the amplification efficiency was evaluated as previously described. 4
2.4. Statistical analysis
The LoD was determined using a positive‐rate analysis and defined as the lowest concentration at which all replicates showed positive results (100% detection rate). Statistical significance was set at p < 0.05. All statistical analyses and visualizations were performed using the R software (version 3.4.3).
3. RESULTS
3.1. Analytical comparisons of SARS‐CoV‐2 rRT‐PCR assays
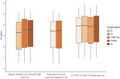
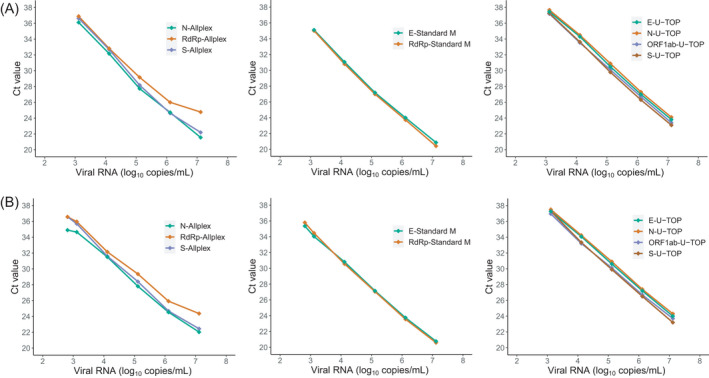
Analytical comparison of Allplex SARS‐CoV‐2/FluA/FluB/RSV kit, Standard M nCoV real‐time detection kit, and U‐TOP COVID‐19 detection kit for linearity, amplification efficiency, and LoD were determined using two RNA controls with known viral loads, SARS‐CoV‐2 RNA control and SARS‐CoV‐2 B.1.351 RNA control (Figure 1). For both RNA controls, a high degree of linearity was observed for each target of Standard M nCoV real‐time detection kit and U‐TOP COVID‐19 detection kit and the S gene of Allplex SARS‐CoV‐2/FluA/FluB/RSV kit (R 2 ≥ 0.98). A lower degree of linearity was observed in the Allplex SARS‐CoV‐2/FluA/FluB/RSV kit N gene when using SARS‐CoV‐2 B.1.351 RNA control than using the SARS‐CoV‐2 RNA control (R 2 = 0.99 versus R 2 = 0.96). However, the mean differences in C t values of the two RNA controls were all within 1.0 for each target in the rRT‐PCR assays (0.05–0.73), without any prominent C t value shift or dropouts in SARS‐CoV‐2 B.1.351 RNA control. Amplification efficiencies for all targets of rRT‐PCR assays were between 80% and 120% for both RNA controls, which was consistent with the criteria for efficient multiplex rRT‐PCR (Figure 2). 19
FIGURE 1.
Mean C t values of the target genes for each rRT‐PCR assay were tested using serially diluted SARS‐CoV‐2 wild‐type RNA control (A) and SARS‐CoV‐2 B.1351 lineage RNA control (B)
FIGURE 2.
PCR efficiencies of nine target genes of three rRT‐PCR assays tested using SARS‐CoV‐2 wild‐type RNA control (A) and SARS‐CoV‐2 B.1351 lineage RNA control (B)
At the concentration level of 13–13,000 copies/ml, for the SARS‐CoV‐2 RNA control, the LoD for the target genes was estimated to be 1300 copies/ml in all three assays (Table S1). The LoD of each target gene was the lowest for the S gene when the Allplex SARS‐CoV‐2/FluA/FluB/RSV kit and ORF1ab gene of the U‐TOP COVID‐19 detection kit were used. When evaluated using the SARS‐CoV‐2 B.1.351 RNA control, the LoD of Allplex SARS‐CoV‐2/FluA/FluB/RSV kit was 650 copies/ml for detection of the target genes, and both the Standard M nCoV real‐time detection kit and U‐TOP COVID‐19 detection kit showed LoD of 1300 copies/ml. For SARS‐CoV‐2 B.1.351 RNA control, the S gene of Allplex SARS‐CoV‐2/FluA/FluB/RSV kit, E and RdRp gene of Standard M nCoV real‐time detection kit, and the ORF1ab gene were the target genes with the lowest LoD.
3.2. Clinical samples
Two hundred and forty‐nine NPS specimens collected from 215 patients were part of this study. The median age of the patients was 60 years (range, 7–89 years), and 44.19% (n = 95/215) patients were male. Among 249 samples, 42 (16.87%), 57 (22.89%), 135 (54.22%), and 15 (6.02%) samples were from inpatients, patients visiting the emergency department, the COVID‐19 screening clinic, and outpatients, respectively. All 249 NPS specimens tested negative for influenza A and B and RSV using the Allplex SARS‐CoV‐2/FluA/FluB/RSV kit.
3.3. Positivity and C t value distributions of clinical samples
All samples were analyzed using the Allplex SARS‐CoV‐2/FluA/FluB/RSV kit, Standard M nCoV real‐time detection kit, and U‐TOP COVID‐19 detection kit for comparison. The overall results of all three SARS‐CoV‐2 rRT‐PCR assays are shown in Table 1. Among the assays, the Standard M nCoV real‐time detection kit showed the highest positivity for all target genes, whereas the Allplex SARS‐CoV‐2/FluA/FluB/RSV kit showed the highest positive results for at least one target gene. For each assay, the S gene in the Allplex SARS‐CoV‐2/FluA/FluB/RSV kit, the RdRP gene in the Standard M nCoV real‐time detection kit, and the ORF1ab and E genes in the U‐TOP COVID‐19 detection kit were the most frequently detected targets with positive results.
TABLE 1.
Detailed results of clinical samples analyzed using three rRT‐PCR assays
| Allplex SARS‐CoV‐2/FluA/FluB/RSV kit | Standard M nCoV real‐time detection kit | U‐TOP COVID‐19 detection kit | |
|---|---|---|---|
| Target genes | S, RdRp, and N genes | E and RdRp genes | ORF1ab, N, S, and E genes |
| Positive results | |||
| Any target genes | 90 | 89 | 88 |
| All target genes | 85 | 87 | 84 |
| S and RdRP genes | 1 | ||
| S gene | 3 | ||
| RdRP gene | 1 | 2 | |
| ORF1ab, N, and E genes | 1 | ||
| ORF1ab and N genes | 1 | ||
| ORF1ab and E genes | 1 | ||
| E gene | 1 | ||
| Negative results | 159 | 160 | 161 |
The positive results were defined the result of at least two of the three SARS‐CoV‐2 real‐time PCR assays.
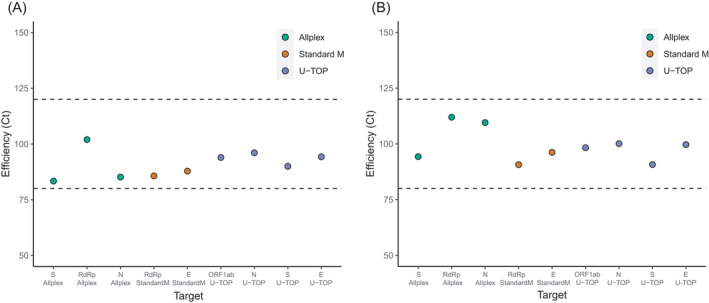
We investigated whether the difference in positivity among the targets in each assay was affected by the difference in C t values. The C t value distributions of the clinical samples are shown in Table 2 and Figure 3. When comparing the C t distribution value of each target gene for the Allplex SARS‐CoV‐2/FluA/FluB/RSV kit, the N gene showed the lowest C t value, followed by the S and RdRp genes. The difference in C t values between the targets was statistically significant (S and RdRp gene, p = 0.0011 and others all p < 0.001). In the Standard M nCoV real‐time detection kit, the E gene showed a lower C t value than that of the RdRp gene, but the difference was not statistically significant (p = 0.9709). For the U‐TOP COVID‐19 detection kit, the lowest C t value was observed in the ORF1ab gene followed by the E, N, and S genes, which was the same order as the positivity in clinical samples. The difference between the C t values of each target was significant (all p < 0.001).
TABLE 2.
C t value and clinical performance comparison of each of the target genes for real‐time reverse‐transcription PCR assays
| Genes | Allplex SARS‐CoV‐2/FluA/FluB/RSV kit | Standard M nCoV real‐time detection kit | U‐TOP COVID‐19 detection kit | ||||||
|---|---|---|---|---|---|---|---|---|---|
| S | RdRp | N | RdRp | E | ORF1ab | N | S | E | |
| C t value (mean ± SD) | 26.44 ± 6.94 | 26.52 ± 7.29 | 25.28 ± 6.72 | 25.76 ± 6.89 | 25.65 ± 6.71 | 27.62 ± 6.66 | 28.20 ± 6.49 | 28.34 ± 6.69 | 27.72 ± 6.53 |
| Sensitivity (95% CI) | 98.88 (93.90–99.97) | 97.75 (92.12–99.73) | 95.51 (88.89–98.76) | 100 (95.94–100) | 97.75 (92.12–99.73) | 97.75 (92.12–99.73) | 96.63 (90.46–99.30) | 94.38 (87.38–98.15) | 97.75 (92.12–99.73) |
| Specificity (95% CI) | 99.38 (96.57–99.98) | 100 (97.72–100) | 100 (97.72–100) | 100 (97.72–100) | 100 (97.72–100) | 100 (97.72–100) | 100 (97.72–100) | 100 (97.72–100) | 100 (97.72–100) |
The clinical performance was evaluated based on the reference standard results, which was defined by the consensus criterion.
Abbreviations: CI, confidence interval; C t , cycle threshold; SD, standard deviation
FIGURE 3.
C t value of the distribution of target genes of three real‐time reverse‐transcription PCRs when analyzing the clinical samples
When the same target genes of the two different assays were compared, the mean difference in the C t values of RdRp gene between the Allplex SARS‐CoV‐2/FluA/FluB/RSV kit and the Standard M nCoV real‐time detection kit was 0.982. The mean differences in the C t values of other overlapping genes were −2.179 of E gene between the Standard M nCoV real‐time detection kit and the U‐TOP COVID‐19 detection kit, −2.282 of N gene and −2.483 of S gene between the Allplex SARS‐CoV‐2/FluA/FluB/RSV kit and the U‐TOP COVID‐19 detection kit, respectively. The differences between the C t values of same target genes were all statistically significant (p < 0.001).
3.4. Clinical samples with discordant results
Among the 249 samples, 6 (2.41%) samples showed discordant results between the rRT‐PCR assays (Table 3). With the review of medical records, six samples with discordant results were follow‐up samples from COVID‐19 patients during or after the treatment, favoring the presumptive presence of SARS‐CoV‐2 RNA. For these six COVID‐19 patient follow‐up samples, inconclusive results of rRT‐PCR assays were interpreted as positive.
TABLE 3.
Details of discordant clinical sample results
| Sample No. | Reference standard | Allplex SARS‐CoV‐2/FluA/FluB/RSV kit | Standard M nCoV real‐time detection kit | U‐TOP COVID‐19 detection kit | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Result | S (C t ) | RdRP (C t ) | N (C t ) | Result | RdRP (C t ) | E (C t ) | Result | ORF1ab (C t ) | N (C t ) | S (C t ) | E (C t ) | ||
| 1 | Positive | Target detected | ND | 37.3 | ND | Target detected | 35.6 | ND | Target not detected | ≥38.0 | ≥38.0 | ND | ND |
| 2 | Positive | Target detected | 36.0 | 37.5 | 35.7 | Target detected | 35.7 | 34.1 | Target detected | 37.0 | ≥38.0 | ≥38.0 | 37.7 |
| 3 | Positive | Target detected | 35.8 | 36.6 | ND | Target detected | 34.0 | 34.4 | Target detected | 36.8 | 37.7 | ≥38.0 | ≥38.0 |
| 4 | Positive | Target detected | 35.5 | 36.7 | 34.5 | Target detected | 35.6 | 34.0 | Target detected | 35.9 | 36.6 | ND | 35.4 |
| 5 | Positive | Target detected | 36.5 | ND | ND | Target detected | 35.1 | 33.8 | Target detected | 36.0 | 37.0 | 37.6 | 36.1 |
| 6 | Negative | Target detected | 37.4 | ND | ND | Target not detected | ND | ND | Target not detected | ≥38.0 | ≥38.0 | ND | ≥38.0 |
Abbreviations: C t , cycle threshold; ND, not detected.
3.5. Clinical performance of SARS‐CoV‐2 rRT‐PCR assays
The clinical performance of SARS‐CoV‐2 rRT‐PCR assays was evaluated for the overall results and for each target gene. As described above, the clinical performance was evaluated according to the reference standard. The consensus criterion was defined as the result of at least two of the three SARS‐CoV‐2 real‐time PCR assays. The Allplex SARS‐CoV‐2/FluA/FluB/RSV kit showed positive results in one or more targets for all six discordant samples, but one sample yielded negative results by both Standard M nCoV real‐time detection kit and U‐TOP COVID‐19 detection kit, which was later considered as true negative based on the consensus criterion. The rest of the five discordant samples were considered as true‐positive samples.
The diagnostic performances of the three assays for the overall results are shown in Table 4. Among the 249 samples, 89 were positive and 160 were negative. Both, the Allplex SARS‐CoV‐2/FluA/FluB/RSV kit and Standard M nCoV real‐time detection kit, showed the highest overall sensitivity (both 100%) for SARS‐CoV‐2 detection. Regarding specificity, the Standard M nCoV real‐time detection kit and U‐TOP COVID‐19 detection kit showed the highest overall specificity (both 100%). Considering each target gene of all three rRT‐PCR assays, the RdRp gene of Standard M nCoV real‐time detection kit showed the highest sensitivity (100%), followed by the S gene of Allplex SARS‐CoV‐2/FluA/FluB/RSV kit (98.88%). The specificities of each target gene in the three assays showed high specificities ranging from 99.38% to 100% (Table 2).
TABLE 4.
Clinical performance comparison of the three rRT‐PCRs for detection of SARS‐CoV‐2
| SARS‐CoV‐2 PCR assays | Results according to reference standards | Sensitivity (95% CI) | Specificity (95% CI) | |||
|---|---|---|---|---|---|---|
| TP | FP | FN | TN | |||
| Allplex SARS‐CoV‐2/FluA/FluB/RSV kit | 89 | 1 | 0 | 159 | 100 (95.94–100) | 99.38 (96.57–99.98) |
| Standard M nCoV real‐time detection kit | 89 | 0 | 0 | 160 | 100 (95.94–100) | 100 (97.72–100) |
| U‐TOP COVID‐19 detection kit | 88 | 0 | 1 | 160 | 98.88 (93.90–99.97) | 100 (97.72–100) |
Abbreviations: CI, confidence interval; FN, false negative; FP, false positive; TN, true negative; TP, true positive.
4. DISCUSSION
The overall analytical sensitivity evaluation of all rRT‐PCR assays showed comparable performance, whereas the analytical sensitivities of each target gene were more variable. The analytical sensitivity of each target gene appears to be influenced more by the primer and probe design rather than the target gene. The importance of primer design and the optimization of primer sets and detection protocols for SARS‐CoV‐2 have been previously reported. 4 , 20 Except for the ORF1ab gene in the U‐TOP COVID‐19 detection kit, other target genes were included in each assay more than once. All rRT‐PCR assays were performed using the same RNA extraction system, and the same amount of eluate was added to the reaction (all 10 µl), and therefore, the same effective sample volume was achieved. 21 When we directly compared the analytical sensitivities of each target, the S gene in the Allplex SARS‐CoV‐2/FluA/FluB/RSV assay was approximately 10‐fold more sensitive than the S gene in the U‐TOP COVID‐19 detection kit. The RdRp and E genes in the Standard M nCoV real‐time detection kit were approximately 10‐fold more sensitive for SARS‐CoV‐2 B.1.351 detection than the RdRp gene in the Allplex SARS‐CoV‐2/FluA/FluB/RSV assay and the E gene in the U‐TOP COVID‐19 detection kit, respectively.
There are several factors known to cause false‐positive SARS‐CoV‐2 rRT‐PCR results such as contamination during sampling and processing, cross‐reaction with other viruses, and non‐specific low‐level reactions in the PCR process. 22 False‐negative results can occur due to inadequate sample collection, sample degradation, too early or too late sample collection, and mutations in primer and probe regions of rRT‐PCR. 23 In this study, the discordant results between the rRT‐PCR assays were observed in follow‐up samples from COVID‐19 patients. Persistently positive rRT‐PCR results in COVID‐19 patients' follow‐up samples do not indicate replication‐competent SARS‐CoV‐2 virus 24 , 25 , 26 ; however, viral RNA shedding has been reported in COVID‐19 patients with variable duration. 27 , 28 The positive targets detected in rRT‐PCR assays favored the presence of low number of copies of SARS‐CoV‐2 RNA in these samples. 29 However, previous history of COVID‐19 infection does not eliminate the possibility of false‐positive results; therefore, we implemented a consensus criterion as the reference standard. Interestingly, in line with the analytical sensitivity results, the S and ORF1ab genes were the most sensitive target genes using the Allplex SARS‐CoV‐2/FluA/FluB/RSV assay and U‐TOP COVID‐19 detection kit. In the case of the Standard M nCoV real‐time detection kit, although the difference in analytical sensitivity was not evident, RdRp showed higher sensitivity than the E gene.
The analytical comparison using SARS‐CoV‐2 RNA and SARS‐CoV‐2 B.1.351 RNA controls showed a low degree of linearity in the RdRp gene of the Allplex SARS‐CoV‐2/FluA/FluB/RSV assay. However, the difference in linearity did not result in analytical sensitivity. Our data showed that the presence of other mutations in B.1.351 did not have an impact on the analytical sensitivity and that the C t value shift was minimal. Wollschläger et al. have reported a D3L mutation in B.1.1.7‐positive samples causing an N gene dropout or C t value shift in the Allplex SARS‐CoV‐2/FluA/FluB/RSV assay but not in the Allplex 2019‐nCoV assay. 30 As expected, N gene dropout or C t value shift was not observed in the B.1.351 lineage, which does not carry the D3L mutation.
This study had some limitations. The impact of the B.1.351 lineage on the clinical performance of rRT‐PCR could not be evaluated. We initially aimed to evaluate the clinical performance of B.1.351‐positive samples. Therefore, the clinical performance was evaluated using patient samples collected during December 2020 and February 2021 when COVID‐19 infection by the SARS‐CoV‐2 B.1.351 lineages were reported in Korea. Variant screening using variant‐specific PCR (SARS‐CoV‐2 Variants I and II assay, Seegene) revealed that all 89 SARS‐CoV‐2 positive samples were negative for the targeted variants, suggesting that B.1.351 was not present in our clinical samples (data not shown). Because of the low prevalence of SARS‐CoV‐2 B.1.351 lineage in our clinical samples, only the effect of the SARS‐CoV‐2 B.1.351 lineage on analytical sensitivities was evaluated using RNA controls.
In summary, comparative evaluation of rRT‐PCR assays, including the recently developed Allplex SARS‐CoV‐2/FluA/FluB/RSV assay, showed that all three rRT‐PCR assays showed comparable overall analytical and clinical performances. For each target of the rRT‐PCR assay, the analytical and clinical sensitivity of each target gene appeared to be influenced more by the primer and probe design than the target gene itself. Additionally, the B.1.351 lineage did not have an impact on the analytical sensitivity, and the C t value shift was minimal. However, further studies using B.1.351‐positive clinical samples are warranted to confirm its impact on clinical performance.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Tab S1
ACKNOWLEDGEMENT
None.
Kim HN, Yoon S‐Y, Lim CS, Yoon J. Comparison of three molecular diagnostic assays for SARS‐CoV‐2 detection: Evaluation of analytical sensitivity and clinical performance. J Clin Lab Anal. 2022;36:e24242. doi: 10.1002/jcla.24242
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. World Health Organization , Coronavirus disease (COVID‐19) outbreak, 2021. https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019. Accessed September 09, 2021.
- 2. World Health Organization . Diagnostic Testing for SARS‐CoV‐2: Interim Guidance. WHO. 2020;2020. [Google Scholar]
- 3. Hong KH, Lee SW, Kim TS, et al. Guidelines for laboratory diagnosis of coronavirus disease 2019 (COVID‐19) in Korea. Ann Lab Med. 2019;40(2020):351‐360. doi: 10.3343/alm.2020.40.5.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vogels CBF, Brito AF, Wyllie AL, et al. Analytical sensitivity and efficiency comparisons of SARS‐CoV‐2 RT‐qPCR primer‐probe sets. Nat Microbiol. 2020;5:1299‐1305. doi: 10.1038/s41564-020-0761-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. FIND, the global alliance for diagnostics, 2021. https://www.finddx.org/test‐directory/. Accessed September 09, 2021.
- 6. Wang R, Hozumi Y, Yin C, Wei GW. Mutations on COVID‐19 diagnostic targets. Genomics. 2020;112:5204‐5213. doi: 10.1016/j.ygeno.2020.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nalla AK, Casto AM, Huang MW, et al. Comparative performance of SARS‐CoV‐2 detection assays using seven different primer‐probe sets and one assay kit. J Clin Microbiol. 2020;58:e00557‐20. doi: 10.1128/JCM.00557-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kidd M, Richter A, Best A, et al. S‐variant SARS‐CoV‐2 lineage B1.1.7 is associated with significantly higher viral load in samples tested by TaqPath polymerase chain reaction. J Infect Dis. 2021;223:1666‐1670. doi: 10.1093/infdis/jiab082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Washington NL, White S, Barrett KMS, Cirulli ET, Bolze A, Lu JT. S Gene Dropout patterns in SARS‐CoV‐2 tests suggest spread of the H69del/V70del mutation in the US. medRxiv. 2020. doi: 10.1101/2020.12.24.20248814:2020.12.24.20248814 [DOI] [Google Scholar]
- 10. Artesi M, Bontems S, Göbbels P, et al. A recurrent mutation at Position 26340 of SARS‐CoV‐2 is associated with failure of the E Gene quantitative reverse transcription‐PCR utilized in a commercial dual‐target diagnostic assay. J Clin Microbiol. 2020;58:e01598‐20. doi: 10.1128/JCM.01598-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ziegler K, Steininger P, Ziegler R, Steinmann J, Korn K, Ensser A. SARS‐CoV‐2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 2020;25:2001650. doi: 10.2807/1560-7917.ES.2020.25.39.2001650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tegally H, Wilkinson E, Giovanetti M, et al. Detection of a SARS‐CoV‐2 variant of concern in South Africa. Nature. 2021;592:438‐443. doi: 10.1038/s41586-021-03402-9 [DOI] [PubMed] [Google Scholar]
- 13. Roquebert B, Trombert‐Paolantoni S, Haim‐Boukobza S, et al. The SARS‐CoV‐2 B.1.351 lineage (VOC β) is outgrowing the B.1.1.7 lineage (VOC α) in some French regions in April 2021. Euro Surveill. 2021;26:2100447. doi: 10.2807/1560-7917.ES.2021.26.23.2100447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mwenda M, Saasa N, Sinyange N, et al. Detection of B.1.351 SARS‐CoV‐2 variant Strain–Zambia, December 2020. MMWR Morb Mortal Wkly Rep. 2021;70(8):280‐282. 10.15585/mmwr.mm7008e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. CDC . Centers for Disease Control and Prevention, novel coronavirus (2019‐nCoV) real‐time RT‐PCR diagnostic panel. CDC; 2020. [Google Scholar]
- 17. Zhen W, Manji R, Smith E, Berry GJ. Comparison of four molecular in vitro diagnostic assays for the detection of SARS‐CoV‐2 in nasopharyngeal specimens. J Clin Microbiol. 2020;58:e00743‐20. doi: 10.1128/JCM.00743-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liotti FM, Menchinelli G, Marchetti S, et al. Evaluation of three commercial assays for SARS‐CoV‐2 molecular detection in upper respiratory tract samples. Eur J Clin Microbiol Infect Dis. 2021;40:269‐277. doi: 10.1007/s10096-020-04025-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Broeders S, Huber I, Grohmann L, et al. Guidelines for validation of qualitative real‐time PCR methods. Trends Food Sci Technol. 2014;37:115‐126. doi: 10.1016/j.tifs.2014.03.008 [DOI] [Google Scholar]
- 20. Park M, Won J, Choi BY, Lee CJ. Optimization of primer sets and detection protocols for SARS‐CoV‐2 of coronavirus disease 2019 (COVID‐19) using PCR and real‐time PCR. Exp Mol Med. 2020;52:963‐977. doi: 10.1038/s12276-020-0452-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fung B, Gopez A, Servellita V, et al. Direct comparison of SARS‐CoV‐2 analytical limits of detection across seven molecular assays. J Clin Microbiol. 2020;58:e01535‐20. doi: 10.1128/JCM.01535-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Braunstein GD, Schwartz L, Hymel P, Fielding J. False positive results with SARS‐CoV‐2 RT‐PCR tests and how to evaluate a RT‐PCR‐positive test for the possibility of a false positive result. J Occup Environ Med. 2021;63(3):e159. doi: 10.1097/JOM.0000000000002138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mayers C, Baker K. Impact of false‐positives and false‐negatives in the UK's COVID‐19 RT‐PCR testing programme. Government Office for Science; 2020. www.gov.uk/government/publications/gos‐impact‐of‐false‐positives‐and‐negatives‐3‐june‐2020 [Google Scholar]
- 24. Bullard J, Dust K, Funk D, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020;71:2663‐2666. doi: 10.1093/cid/ciaa638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Basile K, McPhie K, Carter I, et al. Cell‐based culture of SARS‐CoV‐2 informs infectivity and safe de‐isolation assessments during COVID‐19. Clin Infect Dis. 2020;73:e2952‐e2959. doi: 10.1093/cid/ciaa1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim MC, Cui C, Shin KR, et al. Duration of culturable SARS‐CoV‐2 in hospitalized patients with Covid‐19. N Engl J Med. 2021;384:671‐673. doi: 10.1056/NEJMc2027040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuan J, Kou S, Liang Y, Zeng J, Pan Y, Liu L. Polymerase chain reaction assays reverted to positive in 25 discharged patients with COVID‐19. Clin Infect Dis. 2020;71:2230‐2232. doi: 10.1093/cid/ciaa398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID‐19. Nat Med. 2020;26:672‐675. doi: 10.1038/s41591-020-0869-5 [DOI] [PubMed] [Google Scholar]
- 29. Fang FC, Naccache SN, Greninger AL. The laboratory diagnosis of coronavirus disease 2019‐ frequently asked questions. Clin Infect Dis. 2020;71:2996‐3001. doi: 10.1093/cid/ciaa742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wollschläger P, Todt D, Gerlitz N, et al. SARS‐CoV‐2 N gene dropout and N gene Ct value shift as indicator for the presence of B.1.1.7 lineage in a commercial multiplex PCR assay. Clin Microbiol Infect. 2021;27:1353.e1‐1353.e5. doi: 10.1016/j.cmi.2021.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tab S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


