Abstract
Introduction
Resistance to azole drugs has been observed in candidiasis due to their long‐term use and poor response to treatment. Resistance to azole drugs in Candida albicans isolates is controlled by several genes including ERG11, CDR1, CDR2, and MDR1. In this study, the expression of the mentioned genes was evaluated in C. albicans isolates susceptible and resistant to fluconazole.
Methods
After identifying the Candida isolates using morphological and molecular methods, the minimum inhibitory concentration (MIC) and drug susceptibility were determined using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) method. RNA was then extracted and cDNA was synthesized from 24 C. albicans isolates from patients with cancer. Then, the mean expressions of these genes were compared in two groups using real‐time polymerase chain reaction (RT‐PCR).
Results
A total of 74 Candida isolates were obtained from the oral cavity of 61 cancer patients with oral candidiasis. After 24 h, 21.6% of the isolates were fluconazole‐resistant, 10.8% were identified as dose‐dependent, and the rest of the isolates (67.6%) were fluconazole‐sensitive. The mean expressions of the CDR1 and MDR1 genes were significantly higher in the resistant isolates than in the sensitive ones. However, the ERG11 and CDR2 genes were not significantly increased in the resistant isolates.
Conclusion
The increased mean expressions of the CDR1 and MDR1 genes had a greater effect on fluconazole resistance among the drug‐resistant strains of C. albicans in chemotherapy patients. It seemed that the accumulation of chemotherapeutic drugs in this organism stimulated some regulatory factors and increased the expression of these two genes and ultimately helped to further increase their expression and resistance to fluconazole.
Keywords: chemotherapy, oral candidiasis, qPCR, resistance genes
Resistance to azole drugs because of long‐term use and poor response to treatment has been observed in candidiasis. Resistance to azole drugs in Candida albicans isolates is controlled by several genes, including ERG11, CDR1, CDR2 and MDR1. In this study, Candida albicans isolates, susceptible and resistant to fluconazole, were evaluated for the expression of the mentioned genes. After identifying Candida isolates using morphological and molecular methods, drug susceptibility was evaluated and MIC was determined. RNA was then extracted and cDNA synthetized from 24 Candida albicans isolated of patients with cancer. Then, the mean expressions of these genes were compared in 2 groups by RT‐PCR. A total of 74 Candida isolates were taken from the oral cavity of 61 cancer patients with oral candidiasis. After 24 h, 21.6% of isolates were fluconazole‐resistant, 10.8% were identified as dose‐dependent and the rest of isolates (67.6%) were fluconazole‐sensitive. The mean expressions of CDR1 and MDR1 genes in resistant isolates were significantly higher than sensitive isolates. However, ERG 11 and CDR2 genes were not significantly increased in resistant isolates. Increased mean expression of CDR1 and MDR1 genes has a greater effect on fluconazole resistance among Candida albicans drug‐resistant strains in chemotherapy patients. It seems that the accumulation of chemotherapeutic drugs in this organism stimulates some regulatory factors and increases the expression of these two genes, and ultimately helps to further increase their expression and resistance to fluconazole. Figure (1): Comparison of mean MDR1.CDRI.CDR2, ERG11 genes expression between fluconazole resistance and sensitive groups. Keywords: oral candidiasis, chemotherapy, qPCR, resistance genes.

1. INTRODUCTION
Oral candidiasis is a common fungal infection in cancer patients and is currently recognized as the most frequent fungal disease in humans. Oral hygiene and early diagnosis are essential in the prevention and treatment of this disease. 1 Nowadays, candidiasis in any clinical form has become a new therapeutic problem especially in HIV patients and those undergoing chemotherapy and radiation therapy. Resistance to azole drugs in oral candidiasis is increasing due to the long‐term use of these drugs and poor response to treatment. 2 , 3 The changes in the target enzymes of azole drugs attributed to the overexpression and mutations of the ERG11 gene have been noted in the study of resistance mechanisms to azole drugs. 4 , 5 However, in a number of studies, the researchers have stated that the overexpression of genes encoding ABC membrane transport proteins (CDR1 and CDR2) is the most frequent mechanism of azole resistance. 6 , 7 , 8 Moreover, the main function of the increase in the expression of the MDR1 gene in fluconazole‐resistant isolates has been determined in several studies. 9 , 10 , 11 , 12 A study (2018) on the combined effects of some (Food and Drug Administration) FDA‐approved oncology drugs and fluconazole on Candida albicans found that some oncology drugs had a negative effect on the antifungal activity of fluconazole, itraconazole, and voriconazole. It seems that these drugs can induce their effects using the mechanism of azole resistance by affecting the genes encoding the peripheral membrane proteins of drug efflux pumps such as MDR1 or ABC (CDR1 and CDR2) and the Erg11 protein. 13 The potential effect of azole drugs on fungal species and the effect of oncology drugs on mammalian host cells depend on the evolutionary similarity and eukaryotic nature of the affected cells. 14
This study aimed to investigate the effects of chemical drugs used in various cancers on the overexpression of the genes (such as ERG11, CDR1, CDR2, and MDR1) involved in fluconazole resistance. To determine whether these genes have an overexpression in C. albicans isolates before exposure to fluconazole as the first line of treatment in Iran for patients with oral candidiasis, the increase in the expressions of these genes in various types of cancer was analyzed.
2. MATERIALS AND METHODS
2.1. Strains
The samples were obtained from the patients’ oral cavity and tongue using sterile swabs. The mean age of these patients was 59 years. Among the 11 types of cancer, 69% of the isolates were related to leukemia, lymphoma, and liver cancers. Patients with underlying immunodeficiency and diabetics, mentally retarded individuals, and those who had used antifungal drugs in the past 4 weeks were not included in the study. Moreover, patients whose mouth was colonized by Candida (according to the number of colonies on the culture medium and by examining the smear stained with Giemsa) were not included in the study.
This study was conducted on 24 C. albicans isolates (12 sensitive and 12 resistant to fluconazole). These isolates were selected from 74 samples of cancer patients (with 11 types of cancer) undergoing chemotherapy and hospitalized in the Baghaei, Shafa, and Golestan hospitals of Ahvaz, Iran. The isolates were identified using conventional phenotyping methods and polymerase chain reaction‐restriction fragment length polymorphism (PCR‐RFLP). 15 Subsequently, to separate the C. albicans isolates from the C. dubliniensis isolates, a duplex PCR method with CAL and CDU primers was employed on the samples identified as C. albicans during PCR‐RFLP. 16 No case of C. dubliniensis was identified among the C. albicans isolates. The isolates were kept in sterile distilled water until the experiment.
2.2. The evaluation of fluconazole susceptibility using the EUCAST method
To determine the MIC of C. albicans isolates using the EUCAST method, fluconazole was diluted in the range of 0.125–64 mg/ml. In this method, the isolates with the MICs of ≤2 and >4 are considered as sensitive and resistant, respectively. 17
A suspension equivalent to 0.5 McFarland (1–5 × 106 cells per ml) was obtained using a spectrophotometer at a wavelength of 530 nm. Then, 100 µl of the cell suspension was added to each well. 18 Afterward, 100 µl of the serial concentration of drugs prepared in batches of 10 (using RPMI 1640 containing 2% glucose) was added to the wells. The last two wells were used as positive and negative controls. According to the EUCAST protocol, the microplates were incubated at 35°C for 18–24 h. 19 In this approach, the resazurin reagent (Sigma, Germany) was used for the final evaluation and determination of the MIC well. Resazurin was prepared using sterile distilled water in a 0.01% ratio. It was subsequently sterilized using a 0.45‐μm syringe filter. To determine the MIC of fluconazole, the resazurin reagent was diluted with RPMI 1640 medium (1:10 ratio).
To specify the MIC well with this method, the first well in which the blue color of resazurin was observed (without changing to the pink color of resorufin) was considered as the MIC of the drug. 19
2.3. RT‐PCR
2.3.1. RNA isolation
To extract RNA, the isolates were first cultured on Sabouraud dextrose agar (SDA) medium (Merck, Germany) and then incubated for 24 h at 37°C. The RNA extraction steps were performed using the RNeasy Plus Mini Kit (QIAGEN, Germany) according to the kit instructions using completely RNase‐free equipment (4 h in an oven at 200°C). The qualitative assessment of RNA was carried out for all samples using a Nanodrop device (Thermo). The OD A260/A280 nm ratio of 1.9–2 was obtained for all samples, indicating the 90%–100% purity of the nucleic acid.
2.3.2. cDNA synthesis and RT‐PCR
Due to the instability of the extracted RNA, the cDNA synthesis was performed immediately for each sample in two steps using a cDNA synthesis kit (Fermentas) based on the manufacturer's instructions. After standardizing the concentrations of the samples according to the protocol of the cDNA synthesis kit, the volume of RNA used in the cDNA synthesis was determined which ranged from 0.1 ng to 5 µg. The samples were then stored at −20°C.
The RT‐PCR was performed three times for each sample using the primers designed for the ERG11, CDR1, CDR2, and MDR1 genes as well as the ACT1 reference (internal control) gene in Gene Runner software. Notably, one negative control was considered for each gene (without a cDNA pattern) in each run. As a housekeeping gene, ACT1 was used to confirm the PCR reaction in all molecular experiments. 20 The nucleotide sequence of the designed primers is shown in Table 1.
TABLE 1.
The designed primers
| Genes | Primer sequence (5′→3′) | Size (bp) | Accession number |
|---|---|---|---|
| ERG11 | Forward (F) TTGGTGGTGGTAGACATAGATG | 132 | XM_711668.2 |
| Reverse (R) AACTATAATCAGGGTCAGGCAC | |||
| MDR1 | Forward (F) AGTTGCTTGGGGTAGTTCCG | 96 | XM_714072.2 |
| Reverse (R) CTTGCTCTCAACTTTGGTCCG | |||
| CDR1 | Forward (F) TGTTGGGTTGGTCTCGATG | 130 | XM_718116.2 |
| Reverse (R) TCATAACCTGGACCACTTGG | |||
| CDR2 | Forward (F) ATGCCAATGCTGAACCGAC | 154 | XM_718076.2 |
| Reverse (R) AAAGTTGTAGCCAAATTAGCAGC | |||
| ACT1 | Forward (F) ACTGCTTTGGCTCCATCTTCT | 166 | XM_019475182.1 |
| Reverse (R) TGTGGTGAACAATGGATGGAC |
The reaction solutions included 1 μl of cDNA, 0.4 μl of each forward and reverse primer for each gene, and 5 μl of Master Mix Green High Rox. The final reaction volume reached 10 µl by adding 3.2 µl of sterile distilled water.
RT‐PCR was conducted according to the following temperature program: 95°C for 15 min (one cycle) and 95°C for 15 s and 60°C for 1 min (40 cycles). This was followed by the melt curve program using the ABI Step One device. Finally, the RT‐PCR products were visualized by 2% agarose gel electrophoresis. The measured expression levels of the genes by RT‐PCR were shown in CT units.
The RT‐PCR results were analyzed to investigate the differences in the expressions of the ERG11, CDR1, CDR2, and MDR1 genes in the fluconazole sensitive and resistant groups using the 2−∆∆ CT formula and GraphPad Prism software (version 8). In this relation, the mean cycle threshold (CT) performed in triplicate for each gene was employed. To obtain the ∆CT value, the mean CT of the ACT1 reference gene was subtracted from the mean CTs of the tested CDR1, CDR2, ERG11, and MDR1 genes. Using the 2−∆∆ CT formula, the fold changes in the expressions of the mentioned genes were determined by comparing the observed differences in the ∆CT values.
3. RESULTS
3.1. Testing the antifungal susceptibility of the clinical isolates
To detect the drug resistance genes in the oral candidiasis isolates obtained from individuals undergoing chemotherapy in Ahvaz, 24 C. albicans species including 12 susceptible and 12 resistant isolates were compared. The MIC value was determined using the EUCAST method. The MIC of each sample is shown in Table 2 according to the type of cancer.
TABLE 2.
The MIC of fluconazole against 24 resistant and sensitive Candida albicans isolates as determined by broth microdilution testing (mg/ml)
| Candida albicans isolates | FLC MICs (mg/ml) | Type of cancer | |
|---|---|---|---|
| 1 | 21 | >64 | Liver |
| 2 | 36 | >64 | Gastric |
| 3 | 37 | >64 | Liver |
| 4 | 6 | 64 | Uterine |
| 5 | 49 | 64 | Colon |
| 6 | 23 | 64 | Breast |
| 7 | 16 | >64 | Leukemia |
| 8 | 42 | >64 | Lymphoma |
| 9 | 14 | 64 | Leukemia |
| 10 | 5 | >64 | Lymphoma |
| 11 | 70 | >64 | Uterine |
| 12 | 22 | >64 | Liver |
| 13 | 9 | 0.12 | Lymphoma |
| 14 | 3 | 0.12 | Breast |
| 15 | 11 | 0.25 | Gastric |
| 16 | 13 | 0.25 | Leukemia |
| 17 | 12 | 0.25 | Lymphoma |
| 18 | 33 | 0.25 | Bladder |
| 19 | 34 | 0.06 | Colon |
| 20 | 44 | 0.12 | Liver |
| 21 | 2 | 0.12 | Liver |
| 22 | 18 | 0.25 | Uterine |
| 23 | 4 | 0.5 | Leukemia |
| 24 | 19 | 0.25 | Gastric |
3.2. Gene expression
Using the GraphPad Prism (USA) software, the expression levels of the ERG11, CDR1, CDR2, and MDR1 genes were compared with that of the ACT1 gene. The overexpression of the ERG11, CDR1, CDR2, and MDR1 genes was detected in the fluconazole‐resistant isolates. The most common increase in the expression of genes in the 12 resistant isolates compared to the 12 sensitive ones was related to the MDR1 gene, followed by the CDR1 gene. The mean increase in the expression of these two genes was significant in the drug‐resistant group compared with the sensitive group. According to our results, the highest expression of the MDR1 gene (9/24) among the 12 isolates was observed in isolate No. 5. In addition, an increase in the expression level of this gene was seen in isolates No. 14, 42, 23, and 16. The lowest expression level (0.2) of the MDR1 gene was observed in resistant isolates No. 6 and 49. This gene did not have a significant expression in the sensitive isolates. Finally, the mean expression of the MDR1 gene was significantly higher in the resistant isolates than in the sensitive isolates and the p‐value was < 0.02 (Figure 1).
FIGURE 1.
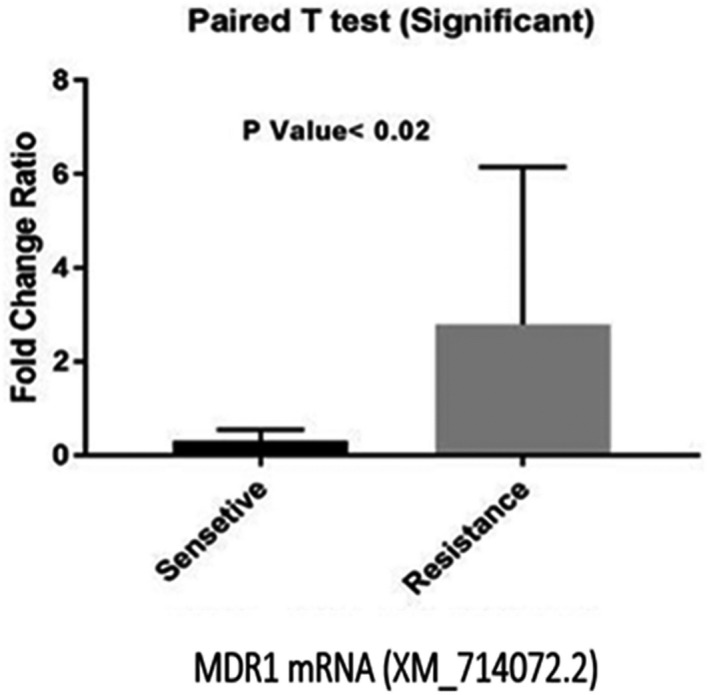
Comparison of mean MDR1 gene expression between fluconazole resistant and sensitive groups
Alternatively, isolate No. 14 showed the highest expression of the CDR1 gene (3.24) among the twelve isolates in the resistant group. Moreover, it was observed that the expression level of this gene had an increase in isolates No. 16 and 42. Among the sensitive isolates, CDR1 had the highest and lowest expression in isolates No. 19 (1.128) and 4 (1.038), respectively. The mean increase in the expression of CDR1 in the resistant group was significantly higher than that of the sensitive group with the p‐value of < 0.03 (Figure 2). The fold changes for the CDR1 and MDR1 genes were calculated to be 1.79 and 9.64, respectively.
FIGURE 2.
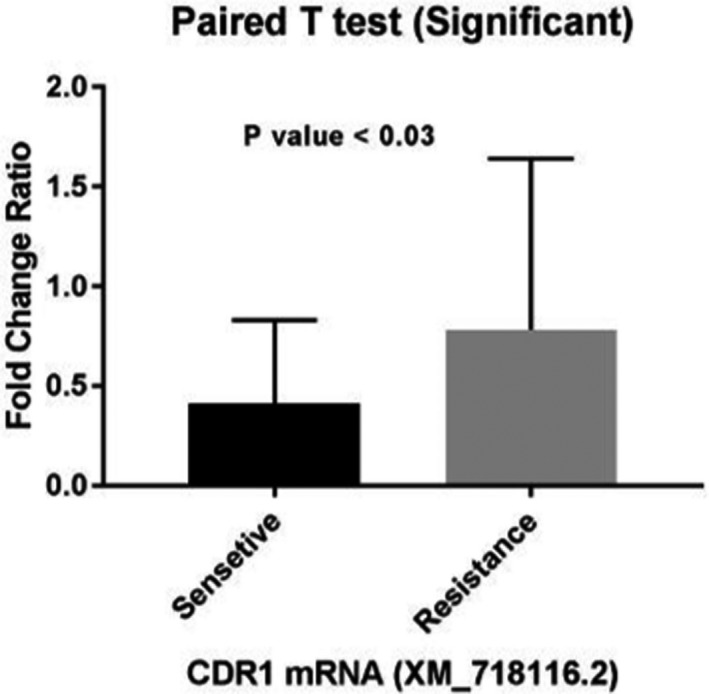
Comparison of mean CDR1 gene expression between fluconazole resistant and sensitive groups
The mean increase in the expression of the ERG11 gene was also observed in the resistant isolates compared with the susceptible ones (Figure 3) so that its highest expression was observed in resistant isolate No. 49 (6.58), followed by isolates No. 6, 70, 16, 23, 5, 42, and 14. Isolate No. 21 showed the lowest expression of the ERG11 gene among the drug‐resistant isolates (0.31). Among the susceptible isolates, the highest expression of ERG11 was found in isolate No. 3 (4.57) and the lowest expression was observed in isolate No. 18 (0.053). Furthermore, no increase in expression was observed in resistant isolates No. 36 and 37 and in sensitive isolates No. 2 and 34. Despite the increase in the mean expression level of ERG11 in the resistant isolates compared with the susceptible ones, there was no significant difference between them given the p‐value of < 0.05. The fold change of this gene was 1.35.
FIGURE 3.
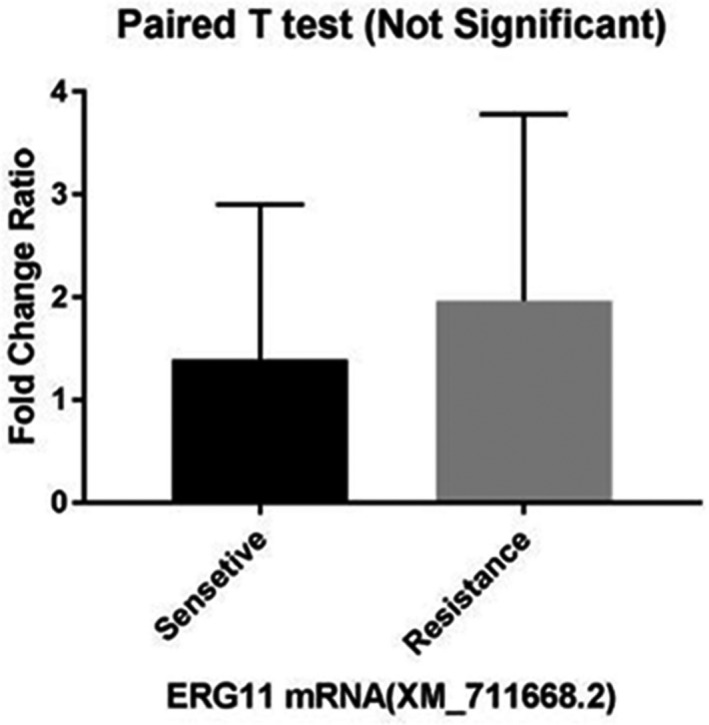
Comparison of mean ERG11 gene expression between fluconazole resistant and sensitive groups
Finally, a review of the expression of the CDR2 gene in the two study groups indicated no significant increase in any group. The highest increase in expression was found in resistant isolate No. 70 (2.96), followed by resistant isolates No. 16 and 14 and sensitive isolate No. 4. There was no increase in the expression of the CDR2 gene in resistant isolates No. 36 and 37. This gene was not expressed in susceptible isolate No. 2. The mean increase in the expression levels of the CDR2 gene in the resistant isolates compared with the sensitive ones was not statistically significant (Figure 4) and the fold change for the CDR2 gene was calculated to be 1.29.
FIGURE 4.
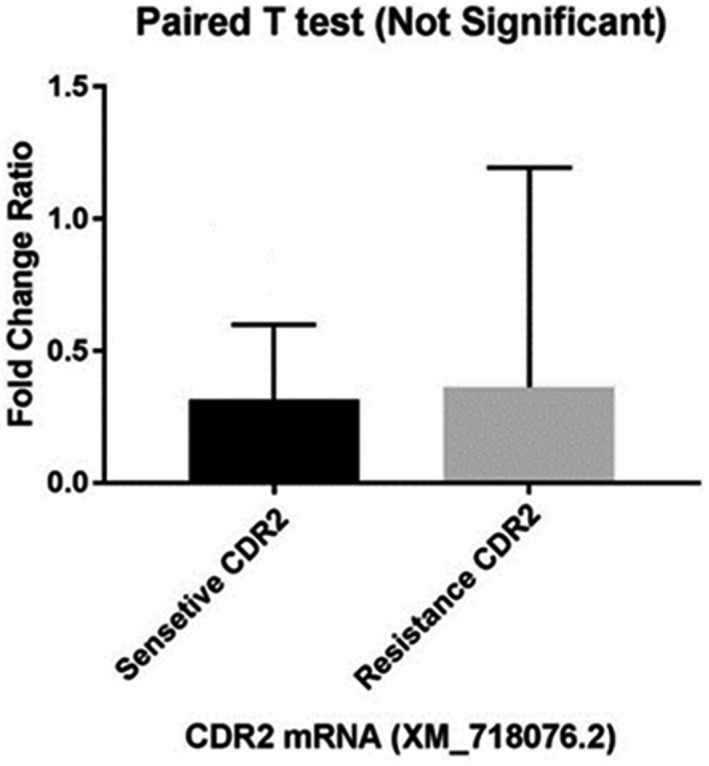
Comparison of mean CDR2 gene expression between fluconazole resistant and sensitive groups
4. DISCUSSION
Oral candidiasis is a common fungal infection in patients with cancer and is currently recognized as the most prevalent fungal disease in humans. 1 In cancer patients, chemotherapy and radiation therapy disrupt the immune system cells and cause neutropenia. As a result, Candida species colonize the mucosal tissues (including the oral cavity) and can eventually enter the bloodstream through the mucosa and lead to candidiasis. Therefore, such patients are at high risk of invasive candidiasis. Today, candidiasis in any of its clinical forms has become a new therapeutic challenge. In addition, resistance to azole drugs with their long‐term use and poor response to the treatment of oral candidiasis are also increasing. 2 , 3
Fluconazole is an antifungal triazole drug which is used as the first line of systemic treatment in patients undergoing chemotherapy. Furthermore, fluconazole is widely used to prevent fungal infections in neutropenic patients with malignancy. 21 , 22 Various reports have shown that over the past few decades, the susceptibility of albicans and non‐albicans species to fluconazole has gradually decreased. 23 , 24 In a study conducted on patients undergoing chemotherapy, the resistance of Candida species to fluconazole was reported to be 47.2%, with the highest resistance observed in the C. albicans isolates. The increased resistance of C. albicans species to fluconazole has been reported by several researchers. 25 , 26 Identifying the reason for the resistance of clinical isolates of C. albicans to azole drugs will lead to more appropriate treatment strategies in the future.
Resistance to fluconazole can be due to several factors including the increased expressions of the ERG11, CDR1, CDR2, and MDR1 genes. In the study of the mechanisms of resistance to azoles, the changes in the target enzyme of azole drugs have been discussed and attributed to the overexpression of and mutations in the ERG11 gene. 4 , 5 The expression level of ERG11 in C. albicans varies significantly in the presence and absence of fluconazole. The overexpression of ERG11 in azole‐resistant isolates has been observed in several investigations. 27 , 28
However, some researchers have concluded that there is no clear relationship between the expression levels of ERG11 and increased fluconazole resistance. 29 In this study, the expression level of ERG11 was 50% and 67% in 24 clinical isolates of C. albicans which were sensitive and resistant to fluconazole, respectively. The increased expression of this gene in half of the sensitive isolates showed a poor correlation with fluconazole resistance. Salari et al. 30 (2015) also compared two sensitive and resistant groups of C. albicans isolated from HIV patients. They concluded that the increased expression of the ERG11 gene was not associated with resistance to azoles.
In previous studies, researchers have stated that the overexpression of genes encoding ABC membrane‐transport proteins (CDR1 and CDR2) is the most frequent mechanism of azole resistance. 6 , 7 , 8 , 27 Indeed, the overexpression of ABC transporter genes (CDR1 and CDR2), which encode Cdr1p and Cdr2p plasma membrane proteins, has been accepted as an important factor in the resistance of Candida isolates to fluconazole. In the current study, the mean increase in the expression of CDR1 and CDR2 in the resistant isolates has been compared with that of the sensitive isolates. The mean expression of the CDR1 gene in the resistant isolates was higher than that of the sensitive isolates. This difference was statistically significant. Although the mean expression of the CDR2 gene in the resistant isolates was shown to be higher than that of the sensitive isolates, no statistically significant difference was observed in this regard.
Our results were similar to those of Holmes et al. 10 who evaluated the increased expression of genes encoding Cdr1p and Cdr2p in 18 fluconazole‐resistant clinical isolates of C. albicans. They showed that Cdr1p had a more effective role than Cdr2p in increasing resistance to fluconazole in these isolates. Similarly, the higher importance of Cdr1p in the resistance of C. albicans isolates to fluconazole has been demonstrated by Nakamura et al. and Sanglard et al. 31 , 32 The studies of Holmes, Hiller, Morschhäuser, Park, and Perea et al. 9 , 10 , 11 , 12 have revealed the essential role of the increase in the expression of MDR1 in fluconazole‐resistant isolates. In the current study, the expression of MDR1 depending on the MFS superfamily encoding membrane transport proteins was typically increased only in the resistant isolates and the difference in the expression between the two groups was statistically significant. Moreover, among the four genes under study, MDR1 showed the highest increase in expression among the fluconazole‐resistant isolates. In a study by Hiller et al. 9 on the increase in the expression of the MDR1 gene in the clinical isolates of C. albicans, the relationship between the expression level of this gene and the excretion of toxins in the cell was examined. They found that the expression of MDR1 was directly associated with the level of resistance to toxins.
In a 2018 study on the combined effect of some FDA‐approved oncology drugs and fluconazole on C. albicans, it was found that several oncology drugs had a negative impact on the antifungal activity of fluconazole, itraconazole, and voriconazole. The combination of the oncology drugs with the mentioned azoles increased the antifungal resistance of C. albicans in vitro. These drugs are prescribed for the treatment of breast cancer, myeloid leukemia, lymphoma, prostate cancer, bladder cancer, and sarcoma. It appears that oncology drugs can induce their effect using the azole resistance mechanism, affecting the genes encoding the drug efflux proteins such as MDR1 or ABC (CDR1 and CDR2), and influencing the Erg11 protein. 13 Given that the C. albicans isolates of the present study were obtained from patients with various cancers, a closer examination of the resistant isolates and the type of cancer revealed that isolates No. 5, 14, and 42, obtained from patients with lymphoma and leukemia, had the highest expression of the MDR1 gene. On the other hand, the increased expression of the CDR1 gene was observed only in isolates No. 14, 16, and 42 from patients with lymphoma and leukemia. Therefore, in this study, it is likely that the oncology drugs prescribed to treat patients with leukemia and lymphoma were a factor in increasing the mean expression of the MDR1 and CDR1 genes and in decreasing the absorption of the antifungal drug fluconazole in patients undergoing chemotherapy. One of the mechanisms for increasing the expression of the MDR1 and CDR1 genes is the regulatory factor MRR1 which increases the expressions of the MDR1 and CDR1 genes in conditions caused by the accumulation of chemical drugs, toxins, benomyl, etc. MRR1 is not involved in the regulation of the CDR2 and ERG11 genes. 33 , 34 It seems that the use of chemotherapy drugs in the patients in this study stimulated the MRR1 regulatory factor of the MDR1 and CDR1 genes, leading to a greater effect on increasing the expressions of these two genes.
Recent studies have shown that cell excretion associated with ABC and MDR1 superfamilies is an important mechanism of resistance in C. albicans, C. glabrata, and C. auris. Alternatively, it has been shown that the deletion of the CDR1 gene reduces fluconazole resistance up to sixfold, whereas the deletion of the CDR2 and MDR1 genes reduces resistance to fluconazole by 1.5‐ and 2‐fold, respectively. 35 Therefore, in the present study, it appears that the overall increase in the expression of the CDR1 gene had a greater effect on the fluconazole resistance of the drug‐resistant C. albicans species in patients undergoing chemotherapy.
5. CONCUSIONS
Detecting the mechanism of resistance to azole compounds and identifying the Candida isolates in clinical laboratories are of high importance. In the present study, the increase in the mean expression of the MDR1 gene followed by that of the CDR1 gene among the drug‐resistant C. albicans species were found to be statistically significant in the subjects undergoing chemotherapy compared with those of the susceptible group.
Since the overexpression of these genes occurred exclusively on the isolates from patients with different types of cancer and under treatment with various chemical drugs, it is suggested that more studies be done on chemical drugs and the type of cancer to identify the mechanisms that increase the overexpression of the MDR1 and CDR1 genes. In addition, according to the results of this study, it is suggested that in future studies, the overexpression of the MRR1 gene which has a regulatory role in these genes be investigated in patients undergoing chemotherapy.
CONFLICT OF INTEREST
There are no financial conflicts of interest to disclose.
AUTHOR CONTRIBUTIONS
Mahnaz Fatahinia was involved in the study design, the interpretation of the data of the study, and the final editing of the manuscript. Mehrnoush Maheronnaghsh contributed to all steps of the experimental work, data analysis, and preparation of the manuscript draft. Parvin Dehghan contributed to the collection and preparation of clinical samples. Ali Teimoori was involved in the study design and the interpretation of the data of the study.
ACKNOWLEDGEMENTS
We would like to thank the Infectious and Tropical Diseases Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran, for their cooperation in this study.
Maheronnaghsh M, Teimoori A, Dehghan P, Fatahinia M. The evaluation of the overexpression of the ERG‐11, MDR‐1, CDR‐1, and CDR‐2 genes in fluconazole‐resistant Candida albicans isolated from Ahvazian cancer patients with oral candidiasis. J Clin Lab Anal. 2022;36:e24208. doi: 10.1002/jcla.24208
Funding information
This work was supported by a grant (No. OG‐9733) from Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
DATA AVAILABILITY STATEMENT
Data are available on request from the authors.
REFERENCES
- 1. Al‐Abeid HM, Abu‐Elteen KH, Elkarmi AZ, Hamad MA. Isolation and characterization of Candida spp. in Jordanian cancer patients: prevalence, pathogenic determinants, and antifungal sensitivity. Jpn J Infect Dis. 2004;57(6):279‐284. [PubMed] [Google Scholar]
- 2. Kauffman CA. Candiduria. Clin Infect Dis. 2005;41(Supplement_6):S371‐S376. [DOI] [PubMed] [Google Scholar]
- 3. Schelenz S, Abdallah S, Gray G, et al. Epidemiology of oral yeast colonization and infection in patients with hematological malignancies, head neck and solid tumors. J Oral Pathol Med. 2011;40(1):83‐89. [DOI] [PubMed] [Google Scholar]
- 4. Hargrove TY, Friggeri L, Wawrzak Z, et al. Structural analyses of Candida albicans sterol 14α‐demethylase complexed with azole drugs address the molecular basis of azole‐mediated inhibition of fungal sterol biosynthesis. J Biol Chem. 2017;292(16):6728‐6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marichal P, Koymans L, Willemsens S, et al. Contribution of mutations in the cytochrome P450 14α‐demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology. 1999;145(10):2701‐2713. [DOI] [PubMed] [Google Scholar]
- 6. Chen L, Xu Y, Zhou C, Zhao J, Li C, Wang R. Overexpression of CDR1 and CDR2 genes plays an important role in fluconazole resistance in Candida albicans with G487T and T916C mutations. J Int Med Res. 2010;38(2):536‐545. [DOI] [PubMed] [Google Scholar]
- 7. Jia X‐M, Ma Z‐P, Jia Y, et al. RTA2, a novel gene involved in azole resistance in Candida albicans. Biochem Biophys Res Comm. 2008;373(4):631‐636. [DOI] [PubMed] [Google Scholar]
- 8. Perea S, López‐Ribot JL, Kirkpatrick WR, et al. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high‐level fluconazole resistance isolated from human immunodeficiency virus‐infected patients. Antimicrob Agents Chemother. 2001;45(10):2676‐2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hiller D, Sanglard D, Morschhäuser J. Overexpression of the MDR1 gene is sufficient to confer increased resistance to toxic compounds in Candida albicans. Antimicrob Agents Chemother. 2006;50(4):1365‐1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holmes AR, Lin Y‐H, Niimi K, et al. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole‐resistant Candida albicans clinical isolates. Antimicrob Agents Chemother. 2008;52(11):3851‐3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morschhäuser J, Barker KS, Liu TT, Blaß‐Warmuth J, Homayouni R, Rogers PD. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 2007;3(11):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park S, Perlin DS. Establishing surrogate markers for fluconazole resistance in Candida albicans. Microb Drug Resist. 2005;11(3):232‐238. [DOI] [PubMed] [Google Scholar]
- 13. Butts A, Reitler P, Ge W, Fortwendel JR, Palmer GE. Commonly used oncology drugs decrease antifungal effectiveness against Candida and Aspergillus species. Antimicrob Agents Chemother. 2018;62(7): e00504‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thakur JK, Arthanari H, Yang F, et al. A nuclear receptor‐like pathway regulating multidrug resistance in fungi. Nature. 2008;452(7187):604‐609. [DOI] [PubMed] [Google Scholar]
- 15. Roudbary M, Roudbarmohammadi S, Bakhshi B, Farhadi Z. Relation of ALS 1 and ALS3 genes and fluconazole resistance in Candida albicans isolated from vaginal candidacies. Int J Mol Clin Microbiol. 2012;2(2):170‐174. [Google Scholar]
- 16. Ahmad S, Khan Z, Asadzadeh M, Theyyathel A, Chandy R. Performance comparison of phenotypic and molecular methods for detection and differentiation of Candida albicans and Candida dubliniensis. BMC Infect Dis. 2012;12(1):1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Testing ECoAS . Clinical Breakpoints and Dosing of Antibiotics. Version; 2020. [Google Scholar]
- 18. Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST) . EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin Microbiol Infect. 2008;14(4):398‐405. [DOI] [PubMed] [Google Scholar]
- 19. Repp KK, Menor SA, Pettit RK. Microplate Alamar blue assay for susceptibility testing of Candida albicans biofilms. Med Mycol. 2007;45(7):603‐607. [DOI] [PubMed] [Google Scholar]
- 20. Jin L, Cao Z, Wang Q, et al. MDR1 overexpression combined with ERG11 mutations induce high‐level fluconazole resistance in Candida tropicalis clinical isolates. BMC Infect Dis. 2018;18(1):1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ghiasian SA, Aghamirian MR, Eshghi GR. Nosocomial candiduria in critically Ill patients admitted to intensive care units in Qazvin, Iran. Avicenna Journal of. Clin Microbiol Infect. 2014;1(2):21622. [Google Scholar]
- 22. Vardakas KZ, Michalopoulos A, Falagas ME. Fluconazole versus itraconazole for antifungal prophylaxis in neutropenic patients with haematological malignancies: a meta‐analysis of randomised‐controlled trials. Br J Haematol. 2005;131(1):22‐28. [DOI] [PubMed] [Google Scholar]
- 23. Rodrigues CF, Silva S, Henriques M. Candida glabrata: a review of its features and resistance. Eur J Clin Microbiol Infect Dis. 2014;33(5):673‐688. [DOI] [PubMed] [Google Scholar]
- 24. Shokouhi T, Bandalizadeh Z, Hedayati MT, Mayahi S. In vitro antifungal susceptibility of Candida species isolated from oropharyngeal lesions of patients with cancer to some antifungal agents. Jundishapur J.Microbiol. 2011;4(12):19‐26. [Google Scholar]
- 25. Kumar CPG, Sundararajan T, Menon T, Venkatadesikalu M. Candidosis in children with onco‐hematological diseases in Chennai, south India. Jpn J Infect Dis. 2005;58(4):218. [PubMed] [Google Scholar]
- 26. Maheronnaghsh M, Tolouei S, Dehghan P, Chadeganipour M, Yazdi M. Identification of Candida species in patients with oral lesion undergoing chemotherapy along with minimum inhibitory concentration to fluconazole. Adv Biomed Res. 2016;5(1):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goldman GH, da Silva Ferreira ME, dos Reis ME, et al. Evaluation of fluconazole resistance mechanisms in Candida albicans clinical isolates from HIV‐infected patients in Brazil. Diagn Microbiol Infect Dis. 2004;50(1):25‐32. [DOI] [PubMed] [Google Scholar]
- 28. Ribeiro MA, Paula CR. Up‐regulation of ERG11 gene among fluconazole‐resistant Candida albicans generated in vitro: is there any clinical implication? Diagn Microbiol Infect Dis. 2007;57(1):71‐75. [DOI] [PubMed] [Google Scholar]
- 29. White TC, Holleman S, Dy F, Mirels LF, Stevens DA. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob Agents Chemother. 2002;46(6):1704‐1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salari S, Khosravi A, Mousavi S, Nikbakht‐Brojeni G. Mechanisms of resistance to fluconazole in Candida albicans clinical isolates from Iranian HIV‐infected patients with oropharyngeal candidiasis. J Mycol Med. 2016;26(1):35‐41. [DOI] [PubMed] [Google Scholar]
- 31. Nakamura K, Niimi M, Niimi K, et al. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrob Agents Chemother. 2001;45(12):3366‐3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40(10):2300‐2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mogavero S, Tavanti A, Senesi S, Rogers PD, Morschhäuser J. Differential requirement of the transcription factor Mcm1 for activation of the Candida albicans multidrug efflux pump MDR1 by its regulators Mrr1 and Cap1. Antimicrob Agents Chemother. 2011;55(5):2061‐2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schillig R, Morschhäuser J. Analysis of a fungus‐specific transcription factor family, the Candida albicans zinc cluster proteins, by artificial activation. Mol Microbiol. 2013;89(5):1003‐1017. [DOI] [PubMed] [Google Scholar]
- 35. Tsao S, Rahkhoodaee F, Raymond M. Relative contributions of the Candida albicans ABC transporters Cdr1p and Cdr2p to clinical azole resistance. Antimicrob Agents Chemother. 2009;53(4):1344‐1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available on request from the authors.


