Abstract
Human epidermal growth factor receptor 2 (HER2)‐positive is a particularly aggressive type of the breast cancer. Trastuzumab‐based therapy is a standard treatment for HER2‐positive breast cancer, but some patients are resistance to the therapy. There are no known diagnostic biomarkers to improve the early diagnosis of HER2‐positive breast cancer and the clinical utility of trastuzumab therapy. Using ultrahigh‐performance liquid time of flight mass spectrometry (UPLC‐TOF‐MS)‐based serum metabolomics and multivariate statistical analysis, we investigated and identified the circulating metabolites L‐arginine and arachidonic acid were elevated in trastuzumab‐responsive and trastuzumab‐resistant HER2‐positive breast cancer patients, and increased until reaching their peaks in trastuzumab‐resistant HER2‐positive breast cancer patients. Moreover, an equation for assessing the risk scores based on linear logistic regression models involving L‐arginine and arachidonic acid was created, which was beneficial for revealing metabolic changes in HER2‐positive breast cancer and enhancing current trastuzumab‐based therapy. In summary, we develop serum‐based metabolic biomarkers for diagnosis of HER2‐positive breast cancers and predicts the therapeutic effects of trastuzumab therapy.
Keywords: HER2‐positive breast cancer, metabolic biomarkers, metabolomics, trastuzumab therapy outcomes
Workflow of metabolomics for metabolomic profiling and data interpretation of serum samples from HER2‐positive breast cancer and normal control

1. INTRODUCTION
Currently, breast cancer ranks first in the incidence of female cancer, accounting for 30% of the new female cancer population around the world, and its incidence is increasing year by year. 1 Human epidermal growth factor receptor 2 (HER2)‐positive breast cancer, one type of breast cancer that overexpresses HER2, accounts for approximately 20% of breast cancers and tends to be more aggressive. 2 Previous report have shown that breast cancer with overexpression of the HER2 gene has higher malignancy, earlier recurrence, metastasis and a poor prognosis, which significantly affects the disease‐free survival rate. 2 Although trastuzumab plus adjuvant chemotherapy treatment has markedly enhanced the efficacy of therapy for this type of breast cancer, the high rate of trastuzumab resistance limits its application. 3 , 4 However, there are no known diagnostic biomarkers to improve the early diagnosis of HER2‐positive breast cancer and the clinical utility of trastuzumab therapy.
Cancer‐related liquid biopsy biomarkers can demonstrate the occurrence, progression and prognosis of cancers and are of great value for the early diagnosis of cancers, prediction of treatment response, and prognostic monitoring. 5 Metabolomics has emerged as a powerful analytical tool to provide new discoveries, and modern analysis methods are being used to study metabolic biomarkers related to diseases for clinical applications and to detect their abnormal changes in the living body. 6 As active modulators of gene and protein activity, metabolites have been widely adopted to investigate metabolic mechanisms underlying cancer occurrence, to evaluate treatment efficacy and monitor the prognosis to provide new diagnostic ideas and guide the development of better therapeutic strategies. 7 However, to the authors’ knowledge, untargeted metabolomic investigation of serum metabolites has not been thoroughly conducted.
In our work, we used ultrahigh‐performance liquid time of flight mass spectrometry (UPLC‐TOF‐MS)‐based serum metabolomics and multivariate statistical analysis to investigate the circulating metabolite profiling of HER2‐positive breast cancer. L‐arginine and arachidonic acid were elevated in trastuzumab‐responsive and trastuzumab‐resistant HER2‐positive breast cancer patients, and increased until reaching their peaks in trastuzumab‐resistant HER2‐positive breast cancer patients. Moreover, an equation for assessing the risk scores based on linear logistic regression models involving L‐arginine and arachidonic acid was created, which was beneficial for revealing metabolic changes in HER2‐positive breast cancer and enhancing current trastuzumab‐based therapy. These unique circulating metabolites in serum not only uncover the molecular characteristics of HER2‐positive breast cancer patients but also enable personalized therapy.
2. MATERIALS AND METHODS
2.1. Serum samples
Before surgery or drug therapy, blood specimens were collected at Jiangsu Cancer Hospital (Nanjing, China) between June 2019 and February 2021 after written consent was obtained from all HER2‐positive breast patients and healthy donors who were regarded as normal control subjects. The status of HER2 in all patients was assessed according to standard HER2 testing. All procedures were reviewed and approved by the ethics committee of the hospital and were conducted in accordance with the Declaration of Helsinki. Trastuzumab primary resistance and sensitivity subgroups were defined according to our previous work. 8 Blood samples were drawn from the elbow vein in the fasting state in the morning and stored in ethylenediaminetetraacetic acid vacuum tubes (BD Vacutainer, Franklin Lakes, NJ, USA) and then centrifuged at 1300 g for 10 min at 4°C. The serum was immediately separated and stored at −80°C until analysis.
2.2. Sample extraction and handling
One hundred microliters of serum samples stored at −80°C were taken after slow dissolution at 4°C. Then, 400 μL precooled methanol was added and vortexed for 60 s. After precipitating the protein for 1 h at −20°C, the samples were subjected to centrifugation for 20 min at 16,000 rpm at 4°C. Supernatants were taken and freeze‐dried followed by storage at −80°C until metabolomics analysis.
2.3. Metabolite profile analysis and metabolite identification
The supernatants were separated by a 1290 Infinity LC System with a C18 chromatographic column and then analyzed by an AB SCIEX TripleTOF 5600 mass spectrometer by Clinical Mass Company (Nanjing, China). General conditions were set as follows: column temperature, 40°C; flow rate, 0.4 ml/min; mobile phase A, 0.1% formic acid in water, B: 0.1% formic acid in acetonitrile. The gradient elution procedure was as follows: 0–1 min, B linearly changed from 90% to 70%; 1–19 min, B linearly changed from 70% to 5%; 19–20 min, B was maintained at 5%; 20–25 min, B was maintained at 95%; and the sample was placed in a 4°C autosampler during the entire analysis. Quality control (QC) samples, which were prepared by mixtures of aliquots of samples, were inserted into the sample queue to monitor and evaluate the stability of the system and the reliability of the experimental data. Information‐dependent acquisition (IDA) for both positive and negative ion modes was applied. The collision energy, declustering potential, and ion spray voltage were set to 30, 80, +5200 V for positive mode, while −30, −50 and −5200 V were set for negative mode with the same mass range from 50 to 1700 m/z. The other source parameters, including ion source gas 1, ion source gas 2, curtain gas and drying temperature, were set at 40, 45, 30 psi, and 550°C, respectively.
2.4. Data analysis
The original data were converted into mzML format, and then the MSdial program was applied for peak alignment, retention time correction and peak area extraction. Accurate mass matching and secondary spectrum matching methods to search public databases were performed for metabolite structure identification. After data preprocessing, unsupervised principal component analysis (PCA) analysis and orthogonal partial least squares discriminant analysis (OPLS‐DA) were applied with the MSdial program for multidimensional statistical analysis.
2.5. Metabolic network analysis and metabolic pathway analysis
Through logarithmic conversion and automatic scaling, metabolites with significantly altered MS signal intensities were introduced to the calculation of the Pearson correlation coefficient. Metabolic network analysis based on the correlation was performed using Cytoscape 3.7. In addition, metabolic pathways influenced by metabolites were analyzed using MetaboAnalyst software (http://www.metaboanalyst.ca).
2.6. Statistical processing
Data are expressed as the mean ±standard deviation and were analyzed using SPSS 22.0 (SPSS, Chicago, IL, USA). Continuous data were compared by Student's t test or the Mann–Whitney U test. An equation for assessing the risk scores based on linear logistic regression models was created, and its sensitivity and specificity were assessed by constructing receiver operating characteristic (ROC) curves. p < 0.05 was considered statistically significant.
3. RESULTS
3.1. Demographic and clinical characteristics of enrolled samples
A total of 20 HER2‐positive breast cancer patients and 30 normal controls were recruited. The 30 people in the normal control group were all in the normal range under the available detection methods. Age and sex were not comparable between the HER2‐positive breast cancer patients and normal controls. According to the National Comprehensive Cancer Network (NCCN) guidelines, the pathological grading status of HER2‐positive breast cancer ranges from I to III. In the HER2‐positive breast cancer group, eight patients with disease progression were regarded as trastuzumab resistant, while 12 patients with a pathologically complete response were defined as trastuzumab responsive. Detailed demographic and clinical characteristics of the participants are shown in Table 1.
TABLE 1.
Detailed demographics of enrolled participants
| Characteristics | Normal control (n = 30) | Breast cancer (n = 20) |
|---|---|---|
| Age‐years | ||
| Median (range) | 51 (42–67) | 52 (47–65) |
| HER2 status‐no. (%) | ||
| IHC(3+) | 14 | |
| IHC(2+) and FISH(+) | 6 | |
| Estrogen receptor status (%) | ||
| Positive | 8 | |
| Negative | 12 | |
| No. of metastatic sites | ||
| 1 | 12 | |
| 2 | 4 | |
| 3 | 1 | |
| ≥4 | 0 | |
| Clinical stage | ||
| I | 1 | |
| II | 9 | |
| III | 10 | |
| Site of metastasis (%) | ||
| Lymph node | 20 | |
| Lung | 3 | |
| Pleura | 1 | |
| Bone | 2 | |
| Liver | 0 | |
| Brain | 0 | |
| Others | 0 | |
| Treatment regimens | ||
| Paclitaxel + trastuzumab | 9 | |
| Docetaxel + trastuzumab | 11 | |
| Trastuzumab resistant | 8 | |
| Trastuzumab responsive | 12 | |
3.2. Metabolomics workflow
Figure 1 briefly represents the entire workflow of the metabolomics research. We collected serum from HER2‐positive breast cancer patients and normal controls. The metabolomics products were extracted from the serum and analyzed by UPLC‐TOF‐MS. In general, characteristic features were consistently measured in all serum samples, including ESI+ and ESI− modes. The interpretation of these data was carried out through a set of bioinformatics tools. First, PCA and OPLS‐DA were applied to analyze the metabolomics and abundance of each sample of MS‐related results. The difference in metabolic features between HER2‐positive breast cancer and normal controls was carried out to provide a total view. Volcano maps and heat maps were generated from statistically significant extracted metabolic features and dysregulated metabolites, which met the criteria of downregulation to fold change <0.67 or fold change >1.5, VIP > 1.0 and p value < 0.05. Then, we conducted metabolic network analysis through correlation and pathway enrichment to investigate the biological significance. L‐Arginine and arachidonic acid were upregulated in trastuzumab‐responsive and trastuzumab‐resistant HER2‐positive breast cancer patients, and increased until reaching their peaks in trastuzumab‐resistant HER2‐positive breast cancer patients. A diagnostic risk equation was then created based on their linear regression models, and their correlation with the occurrence of HER2‐positive breast cancer and clinical trastuzumab therapy outcome data were assessed.
FIGURE 1.
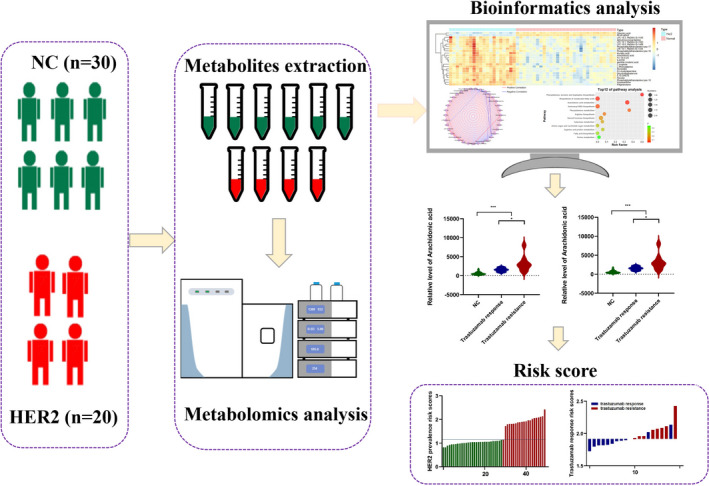
Workflow of metabolomics for metabolomic profiling and data interpretation of serum samples from HER2‐positive breast cancer and normal control
3.3. Global overview of serum metabolites in HER2‐positive breast cancer
First, we performed PCA on all 50 serum samples and found a significant difference between HER2‐positive breast cancer and the control cases. QC analysis was performed during the PCA process to ensure stability and reliability. As shown in Figure 2A,B, the PCA score chart clearly proves the reliability of the metabolomics platform used. Subsequently, through OPLS‐DA (a high‐performance tool for multivariate statistical analysis that can be monitored in real‐time), we found differences in the metabolic features between HER2‐positive and normal samples. As shown in Figure 2C,D, there was a clear separation between HER2‐positive and normal tissues through the ESI+ and ESI− modes, respectively. The quality of these OPLS‐DA models was counted through internal cross‐validation, and the applicability of the parameters (R2Y) and the predictive ability of the model (Q2) were calculated. R2Y with 0.988 and Q2 with 0.957 in ESI+ mode and R2Y with 0.987 and Q2 with 0.959 in ESI+ mode were determined, and the difference between the two was <0.2, suggesting that the calculation model was not overfit. All of these results showed that there was indeed a significant difference in metabolomics between HER2‐positive breast cancer and normal controls.
FIGURE 2.
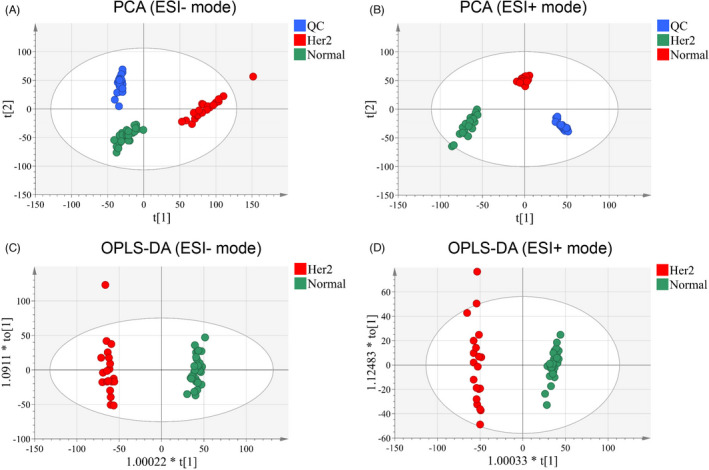
Multivariate statistical analysis results. PCA score plot of the analysis in ESI (−) mode (A) and ESI (+) mode (B). OPLS‐DA score plot of the analysis in ESI (−) mode (C) and ESI (+) mode (D)
3.4. Discovery of metabolites in the serum of HER2‐positive breast cancer
To reveal the serum metabolites in HER2‐positive patients, high‐confidence metabolites that contribute to HER2‐positive breast cancer were identified and confirmed. Among these metabolic characteristics, we distinguished the differences in metabolic features under the criteria of fold change >1.5 and p < 0.05. In addition, a VIP value >1.0, which was calculated by OPLS‐DA scoring, was selected as a significantly different metabolic feature for analysis. Subsequently, metabolic features with significant differences were identified (Figure 3A,B). Using a public metabolite library, the refined significant metabolic features were then searched to confirm the identity of 24 metabolites (Table 2), including 2 upregulated and 22 downregulated metabolites in the HER2‐positive breast cancer group. Hierarchical clustering analysis also revealed a different metabolic pattern between HER2‐positive breast cancer patients and normal controls (Figure 3C).
FIGURE 3.
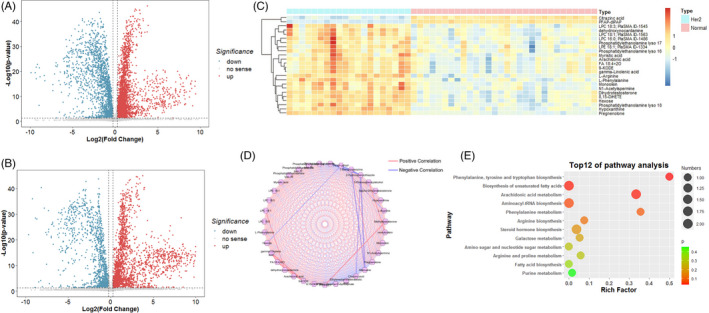
Representative Volcano plot (fold change >1.5 and p value < 0.05) in ESI (+) mode (A) and ESI (+) mode (B) metabolomics data. (C) Representative heat map of significant different metabolites (fold change >1.5, VIP > 1 and p value < 0.05). (D) Metabolic network analysis. (E) Metabolic pathway analysis
TABLE 2.
Differential metabolites identified from metabolomics profiling
| Metabolite name | HER2 | NC | FC | VIP | Mode |
|---|---|---|---|---|---|
| Dihydrotestosterone | 1601 | 680 | 2.35 | 1.12 | POS |
| Hypoxanthine | 2008 | 532 | 3.78 | 1.40 | POS |
| Pregnenolone | 1849 | 697 | 2.65 | 1.24 | POS |
| Monoolein | 1775 | 268 | 6.61 | 1.19 | POS |
| N1‐Acetylspermine | 1621 | 337 | 4.81 | 1.09 | POS |
| L‐Arginine | 1516 | 29 | 51.58 | 1.44 | POS |
| Citrazinc acid | 648 | 1850 | 0.35 | 1.34 | POS |
| 8,15‐DiHETE | 1919 | 631 | 3.04 | 1.69 | NEG |
| 9‐KODE | 1770 | 669 | 2.65 | 1.34 | NEG |
| Arachidonic acid | 2232 | 573 | 3.89 | 1.69 | NEG |
| dehydroxynocardamine | 1430 | 849 | 1.68 | 1.03 | NEG |
| FA 18:4+2O | 1738 | 670 | 2.59 | 1.30 | NEG |
| gamma‐Linolenic acid | 1822 | 635 | 2.87 | 1.39 | NEG |
| Hexose | 1501 | 698 | 2.15 | 1.33 | NEG |
| L‐Phenylalanine | 1285 | 782 | 1.64 | 1.03 | NEG |
| LPC 16:0; PlaSMA ID‐1486 | 1369 | 827 | 1.66 | 1.01 | NEG |
| LPC 18:1; PlaSMA ID‐1563 | 1458 | 838 | 1.74 | 1.07 | NEG |
| LPC 18:3; PlaSMA ID‐1545 | 1853 | 689 | 2.69 | 1.28 | NEG |
| LPE 18:1; PlaSMA ID‐1334 | 1645 | 870 | 1.89 | 1.03 | NEG |
| Myristic acid | 1681 | 748 | 2.25 | 1.08 | NEG |
| Phosphatidylethanolamine lyso 16 | 1802 | 810 | 2.22 | 1.22 | NEG |
| Phosphatidylethanolamine lyso 17 | 1553 | 867 | 1.79 | 1.01 | NEG |
| Phosphatidylethanolamine lyso 18 | 1486 | 674 | 2.20 | 1.30 | NEG |
| PFAP‐diPAP | 276 | 1583 | 0.17 | 1.83 | NEG |
Abbreviations: FC, Fold change; NC, Normal control; NEG, Negative; POS, Positive; VIP, Variable important in projection.
3.5. Metabolic network mapping and pathway enrichment analysis
To further identify and discover the biological significance behind these aberrant metabolites, we conducted a correlation‐based metabolic network analysis, which means that metabolites are analyzed in pairs to discover the relationship between their abundances, thereby inferring potential biological explanations. We used Cytoscape and MetScape to visualize the potential biological relationships between potentially active metabolites. In this ring network (Figure 3D), each node represents the significant changes in metabolites; the connecting line between every two nodes represents the correlation index, where a red line indicates a positive correlation and a blue line indicates a negative correlation. The wider the line, the higher the correlation index. From the results of the molecular network, we concluded that there is a strong internal correlation between the metabolites of metabolic disorders, which reveals that these metabolites have special potential biological significance in HER2‐positive breast cancer. In addition, we used the KEGG library to reveal special potential biological significance. Assuming there were 11 metabolic pathways, their significant P values were all less than 0.05. The most important metabolic pathways are phenylalanine, tyrosine, tryptophan biosynthesis, biosynthesis of unsaturated fatty acids, arachidonic acid metabolism, and others (Figure 3E).
3.6. Risk scores for HER2‐positive breast cancers based on linear logistic regression models
We further analyzed the differential expression of metabolites in trastuzumab‐responsive and trastuzumab‐resistant groups. L‐arginine and arachidonic acid were significantly different between the trastuzumab‐responsive and trastuzumab‐resistant groups, while no significant differences in other metabolites were found (Figure 4A). To reveal the diagnostic performance of these two circulating metabolites, ROC analysis was carried out, and the results (Figure 4B) showed that the ROC curve for L‐arginine had an AUC of 1, and the ROC curve for arachidonic acid had an AUC of 0.95. According to the intensity of these two metabolites, the following formula was calculated for each patient: Ln (risk score) = 0.2234 × Ln (L‐arginine) + 0.0475 × Ln (arachidonic acid) − 1.0025, obtained using logistic regression models. The risk score of the HER2‐positive group was significantly higher than that of the normal control group (Figure 4C). The AUC of the risk score for HER2‐positive patient occurrence was determined to be 1 (Figure 4D). Then, all participants were discriminated into low‐ and high‐risk score groups according to the threshold point cutoff value (1.1492). Based on the exploration of the association between risk score distribution and patient prevalence, the rate of patients in the high‐risk subtype was obviously higher than that in the low‐risk group (Figure 4E,F).
FIGURE 4.
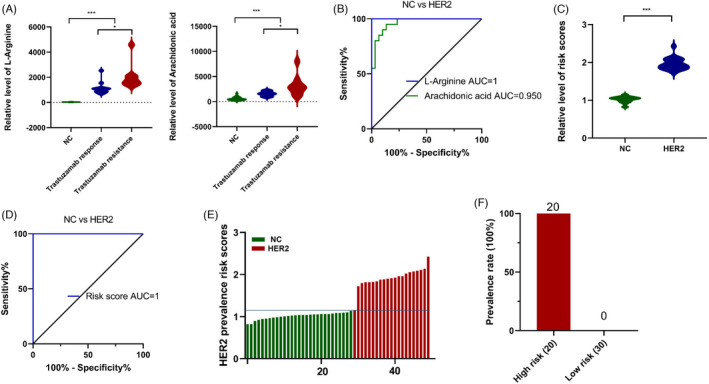
Development of risk score for HER2‐positive breast cancer using linear logistic regression models. (A) Statistical analysis of L‐arginine and arachidonic acid between normal control, trastuzumab responsive and resistant group. (B) ROC curves were created to evaluate the diagnostic power of L‐arginine and arachidonic acid. (C) Statistical analysis of risk score between HER2‐positive breast cancer and normal control. (D) ROC analysis of risk score for HER2‐positive breast cancer. (E) The distribution of risk score. (F) The prevalence of HER‐positive breast cancer in high‐risk group and low‐risk group. (*, p < 0.05; **, p < 0.01; ***, p < 0.001)
3.7. An advanced diagnosis panel for trastuzumab resistance
Given that L‐arginine and arachidonic acid were upregulated in the trastuzumab resistance group, ROC analysis was performed, and the results (Figure 5A) showed that the AUC for L‐arginine was 0.896 and that for arachidonic acid was 0.823 for trastuzumab resistance. According to the logistic regression formula, the risk score of trastuzumab resistance was significantly higher than that of the normal control group (Figure 5B). The sensitivity and specificity of the risk score for trastuzumab resistance were 84.9% and 89.7%, respectively, with an AUC of 0.896 (Figure 5C). Then, the HER2‐positive breast cancer patients were divided into low‐ and high‐risk score groups according to the threshold point cutoff value (1.9159). Based on the investigation of the relationship between the risk score distribution and trastuzumab response status, the rate of patients in the high‐risk subtype was obviously higher than that in the low‐risk group (Figure 5D). Notably, HER2‐positive patients with higher risk scores were all in the trastuzumab‐resistant group (Figure 5E). Further investigation revealed that the low‐risk group had a better objective response rate (ORR) (66.67%, 52/78) than the high‐risk group (27.78%, 10/36) (Figure 5F).
FIGURE 5.
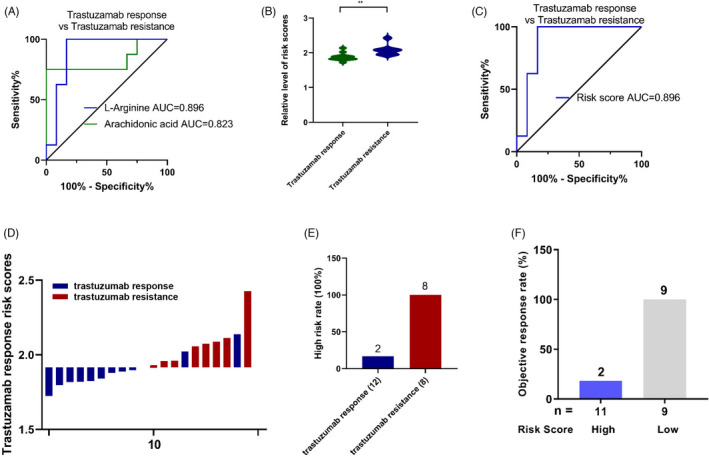
Development of risk score for trastuzumab resistance using linear logistic regression models. (A) ROC curves were created to evaluate the diagnostic power of L‐arginine and arachidonic acid for trastuzumab resistance. (B) Statistical analysis for distribution of risk score between trastuzumab responsive and resistant group (**, p < 0.01). (C) ROC analysis of risk score for trastuzumab resistance. (D) The distribution of risk score between trastuzumab responsive and resistant group. (E) High‐risk rate in trastuzumab responsive and resistant group. (F) Objective response rate in high and low‐risk group. (*, p < 0.05; **, p < 0.01; ***, p < 0.001)
4. DISCUSSION
The overall goal of this study was to delineate the unique serum metabolic biomarkers of HER2‐positive breast cancer patients. By using the UPLC‐TOF‐MS method platform to analyze the serum samples of 20 women with HER2‐positive breast cancer and 30 normal women in the control group, we detected a series of significantly changed metabolites that were associated with HER2‐positive breast cancer, covering a wide range of metabolic classes. Finally, from archived trastuzumab response data from HER2‐positive breast cancer patients, we discovered 2 metabolites and developed a risk score that can predict trastuzumab resistance status. Through comprehensive data interpretation, we acquired a better understanding of the metabolic features in HER2‐positive breast cancer and trastuzumab therapy status.
The emergence of omics methods is effectively accelerating predictive, treating and personalized therapeutics. 9 Metabolites more directly reflect and are linked closer to the phenotype of the pathology than genes and proteins. 10 Focusing on metabolite differences and the discovery of characteristic metabolites can be a shortcut and supplement the gene and protein level omics methods, and could also efficiently explain the mechanisms underlying various phenotypic variations. Thus, metabolomics screening is considered to be an effective, money‐saving, and noninvasive option.
Each breast cancer subtype has inherent molecular features and metastatic potential, and its natural heterogeneity results in a high degree of difference in prognosis and the clinical response to available drugs, even for patients with a similar diagnosis, histology, and disease stages. 11 Therefore, accurate determination of the molecular subtype of breast cancer is very important for personalized treatment. In fact, there is evidence that compared with patients with mismatched therapies, patients who receive the correct molecularly matched therapy have a higher overall response rate, less treatment failure, and higher survival rates. 12 Clinically, liquid biopsy procedures and subsequent histopathological analysis are usually used to study the molecular and genetic information of cancer cells to diagnose and differentiate breast cancer and classify it into subtypes. 13 This analysis technique is invasive and time‐consuming. Therefore, there is an urgent need for a noninvasive, fast, and accurate analysis method for distinguishing different breast cancer subtypes.
A number of studies have explored the possibility of using metabolite profiling as a biomarker for early diagnosis, cancer characterization and clinical outcome prediction. Body fluids such as human saliva, urine, serum and plasma have been re‐emphasized. These are important sources for the discovery of potential biomarkers. Therefore, we analyzed and summarized the metabolic profile that may represent systemic metabolic abnormalities in HER2‐positive breast cancer patients. 14 However, so far, due to the high degree of heterogeneity shown by breast cancer, from histology to clinical prognosis, early recurrence, high risk of metastasis progression or low response rate to treatment and relatively low survival rates, personalized treatment methods based on highly accurate markers or proven targets have been unable to achieve the desired results.
After collecting, processing, and analyzing their samples, we found that the metabolites in the serum of HER2‐positive breast cancer patients were significantly different from those in normal volunteers. Subsequently, from the perspective of confirmation, we found that there were 24 significantly different metabolites to further understand the characteristics and treatment of HER2‐positive breast cancer patients. It is worth noting that our research samples were collected from patients who had just been diagnosed, and they had no interference from any drugs or surgery. Our data were in accordance with previous studies showing that a series of altered metabolites play a role in the development of HER2‐positive breast cancer. 15 We discovered the following new metabolites for the first time: lipids and amino acids.
Our results also showed that the levels of L‐arginine were increased in the HER2‐positive breast cancer and trastuzumab‐resistant groups. L‐arginine, an amino acid naturally found in red meat, poultry, and others, is necessary for making proteins and is commonly found in serum. It has generally been acknowledged that L‐arginine, a fundamental metabolite, plays a role in nutrition and the urea cycle, followed by an explosion of research after the discovery of the biological function of nitric oxide synthesis. 16 L‐arginine is also associated with the occurrence and development of hypertension and atherothrombosis because L‐arginine improves nitric oxide bioactivity. 17 , 18 L‐arginine and its metabolites have also been shown to be novel diagnostic metabolic markers of the pathological progression of kidney disease. 19 Moreover, L‐arginine synthesis is not sufficient for the high nutritional needs of cancer cells, forcing them to rely on an extracellular supply of arginine. 20 L‐arginine has been shown to be correlated with the regulation of immune responses, both innate and adaptive immunity, which have been found to be associated with trastuzumab resistance in our previous work, suggesting a close relationship between the level of L‐arginine and trastuzumab resistance. 21 Arachidonic acid, a polyunsaturated fatty acid present in the phospholipids of membranes of the body's cell, is necessary for the function of the immune system. 22 Other research revealed the arachidonic acid metabolic pathway in breast cancer metastasis with an emphasis on arachidonic acid as a novel therapeutic target. 23 The effect of arachidonic acid signaling pathways on the promotion of drug‐resistant breast cancer has been disclosed, demonstrating the role of arachidonic acid signaling pathways in the development of drug resistance. 24
Notably, the small sample size in the present study limited the robustness of the equation we established for assessing the risk scores, and thus, further validation in a larger sample size is required. In brief, we successfully identified the nontargeted metabolite profile of HER2‐positive breast cancers and found that L‐arginine and arachidonic acid were enhanced in the trastuzumab‐resistant group. Using these biomarkers, we created an equation for assessing the risk scores, which effectively distinguished the normal control, trastuzumab‐responsive and trastuzumab‐resistant groups. In conclusion, the serum metabolites we identified were beneficial to the diagnosis of HER2‐positive breast cancers and trastuzumab therapy outcomes.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
Jin‐Hai Tang and Li Li conceived of and designed the experiments; Changfei Mao and Min Wang. performed the experiments; Changfei Mao analyzed the data and Li Li wrote the manuscript.
ACKNOWLEDGMENTS
The National Key Research and Development Program of China (2016YFC0905900), National Natural Science Foundation of China (81872365) and Jiangsu Provincial Key Research Development Program (BE2019731) to Dr. Tang are gratefully acknowledged. Grant from Jiangsu Research Hospital Association for Precision Medication (JY202023) are gratefully acknowledged. The authors would also like to thank American Journal Experts for proofreading the article.
Mao C, Wang M, Li L, Tang J‐H. Circulating metabolites serve as diagnostic biomarkers for HER2‐positive breast cancer and have predictive value for trastuzumab therapy outcomes. J Clin Lab Anal. 2022;36:e24212. doi: 10.1002/jcla.24212
Changfei Mao and Min Wang contributed equally to this work.
Contributor Information
Li Li, Email: muzichen_1118@126.com.
Jin‐Hai Tang, Email: jhtang@njmu.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. [DOI] [PubMed] [Google Scholar]
- 2. Loibl S, Gianni L. HER2‐positive breast cancer. Lancet. 2017;389(10087):2415‐2429. [DOI] [PubMed] [Google Scholar]
- 3. Pinto AC, Ades F, de Azambuja E, Piccart‐Gebhart M. Trastuzumab for patients with HER2 positive breast cancer: delivery, duration and combination therapies. Breast. 2013;22(Suppl 2):S152‐155. [DOI] [PubMed] [Google Scholar]
- 4. Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2‐positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2011;9(1):16‐32. [DOI] [PubMed] [Google Scholar]
- 5. Marrugo‐Ramirez J, Mir M, Samitier J. Blood‐based cancer biomarkers in liquid biopsy: a promising non‐invasive alternative to tissue biopsy. Int J Mol Sci. 2018;19(10):2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gowda GA, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Metabolomics‐based methods for early disease diagnostics. Expert Rev Mol Diagn. 2008;8(5):617‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frezza C. Metabolism and cancer: the future is now. Br J Cancer. 2020;122(2):133‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang T, Fu Z, Zhang Y, Wang M, Mao C, Ge W. Serum proteomics analysis of candidate predictive biomarker panel for the diagnosis of trastuzumab‐based therapy resistant breast cancer. Biomed Pharmacother. 2020;129:110465. [DOI] [PubMed] [Google Scholar]
- 9. Chen R, Snyder M. Promise of personalized omics to precision medicine. Wiley Interdiscip Rev Syst Biol Med. 2013;5(1):73‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17(7):451‐459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prat A, Pineda E, Adamo B, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24(Suppl 2):S26‐35. [DOI] [PubMed] [Google Scholar]
- 12. Zhang C, Zhang C, Wang Q, Li Z, Lin J, Wang H. Differences in stage of cancer at diagnosis, treatment, and survival by race and ethnicity among leading cancer types. JAMA Netw Open. 2020;3(4):e202950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alimirzaie S, Bagherzadeh M, Akbari MR. Liquid biopsy in breast cancer: a comprehensive review. Clin Genet. 2019;95(6):643‐660. [DOI] [PubMed] [Google Scholar]
- 14. Armitage EG, Ciborowski M. Applications of metabolomics in cancer studies. Adv Exp Med Biol. 2017;965:209‐234. [DOI] [PubMed] [Google Scholar]
- 15. Fan Y, Zhou X, Xia TS, et al. Human plasma metabolomics for identifying differential metabolites and predicting molecular subtypes of breast cancer. Oncotarget. 2016;7(9):9925‐9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morris SM Jr. Arginine metabolism revisited. J Nutr. 2016;146(12):2579S‐2586S. [DOI] [PubMed] [Google Scholar]
- 17. Gokce N. L‐arginine and hypertension. J Nutr. 2004;134(10 Suppl):2807S‐2811S;discussion 2818S–2819S. [DOI] [PubMed] [Google Scholar]
- 18. Loscalzo J. L‐arginine and atherothrombosis. J Nutr. 2004;134(10 Suppl):2798S‐2800S; discussion 2818S–2819S. [DOI] [PubMed] [Google Scholar]
- 19. Popolo A, Adesso S, Pinto A, Autore G, Marzocco S. L‐Arginine and its metabolites in kidney and cardiovascular disease. Amino Acids. 2014;46(10):2271‐2286. [DOI] [PubMed] [Google Scholar]
- 20. Al‐Koussa H, El Mais N, Maalouf H, Abi‐Habib R, El‐Sibai M. Arginine deprivation: a potential therapeutic for cancer cell metastasis? A Review. Cancer Cell Int. 2020;20:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bronte V, Zanovello P. Regulation of immune responses by L‐arginine metabolism. Nat Rev Immunol. 2005;5(8):641‐654. [DOI] [PubMed] [Google Scholar]
- 22. Tallima H, El Ridi R. Arachidonic acid: physiological roles and potential health benefits ‐ a review. J Adv Res. 2018;11:33‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Borin TF, Angara K, Rashid MH, Achyut BR, Arbab AS. Arachidonic acid metabolite as a novel therapeutic target in breast cancer metastasis. Int J Mol Sci. 2017;18(12):2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hammamieh R, Sumaida D, Zhang X, Das R, Jett M. Control of the growth of human breast cancer cells in culture by manipulation of arachidonate metabolism. BMC Cancer. 2007;7:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


