Abstract
Study Objectives
Sleep is an important biological process that is perturbed in numerous diseases, and assessment of its substages currently requires implantation of electrodes to carry out electroencephalogram/electromyogram (EEG/EMG) analysis. Although accurate, this method comes at a high cost of invasive surgery and experts trained to score EEG/EMG data. Here, we leverage modern computer vision methods to directly classify sleep substages from video data. This bypasses the need for surgery and expert scoring, provides a path to high-throughput studies of sleep in mice.
Methods
We collected synchronized high-resolution video and EEG/EMG data in 16 male C57BL/6J mice. We extracted features from the video that are time and frequency-based and used the human expert-scored EEG/EMG data to train a visual classifier. We investigated several classifiers and data augmentation methods.
Results
Our visual sleep classifier proved to be highly accurate in classifying wake, non-rapid eye movement sleep (NREM), and rapid eye movement sleep (REM) states, and achieves an overall accuracy of 0.92 ± 0.05 (mean ± SD). We discover and genetically validate video features that correlate with breathing rates, and show low and high variability in NREM and REM sleep, respectively. Finally, we apply our methods to noninvasively detect that sleep stage disturbances induced by amphetamine administration.
Conclusions
We conclude that machine learning-based visual classification of sleep is a viable alternative to EEG/EMG based scoring. Our results will enable noninvasive high-throughput sleep studies and will greatly reduce the barrier to screening mutant mice for abnormalities in sleep.
Keywords: mouse sleep, inbred mouse strains, sleep states, high-throughput sleep phenotyping, machine learning
Statement of Significance.
We develop a noninvasive sleep state classification system for mice using computer vision. The trained classifier can accurately score wake, non-rapid eye movement sleep, and rapid eye movement sleep states using only the video data. The approach will enable high-throughput automated analysis of sleep states in mice. Validation of key findings will require electroencephalogram/electromyogram recording of sleep.
Introduction
Sleep is a complex behavior that is regulated by a homeostatic process and whose function is critical for survival [1]. Sleep and circadian disturbances are seen in many diseases including neuropsychiatric, neurodevelopmental, neurodegenerative, physiologic, and metabolic disorders [2, 3]. Sleep and circadian functions have a bidirectional relationship with these diseases, in which changes in sleep and circadian patterns can contribute to or be the result of the disease state [4–11]. Even though the bidirectional relationships between sleep and many diseases have been well described, their genetic etiologies have not been fully elucidated. In fact, treatments for sleep disorders are limited because of a lack of knowledge about sleep mechanisms [1]. Rodents serve as a readily available model of human sleep due to similarities in sleep biology and mice, in particular, are a genetically tractable model for mechanistic studies of sleep and potential therapeutics [12–15]. One of the reasons for this critical gap is due to technological barriers that prevent reliable phenotyping of large numbers of mice for assessment of sleep states. The gold standard of sleep analysis in rodents utilizes electroencephalogram/electromyogram (EEG/EMG) recordings. This method is low throughput as it requires surgery for electrode implantation and often requires hand scoring of the recordings. Although new methods utilizing neural networks have started to automate EEG/EMG scoring [16–18], the data generation is still low-throughput. In addition, the use of tethered electrodes limits animal movement potentially altering animal behavior [19].
To overcome this limitation, several noninvasive approaches to sleep analysis have been explored. These include activity assessment through beam break systems, or videography in which certain amount of inactivity is interpreted as sleep [20–24]. Piezo pressure sensors have also been used as a simpler and more sensitive method of accessing activity [25–28]. The latter has been applied toward high throughput studies in the knockout mouse project [29]. These methods only access sleep versus wake and are not able to differentiate wake, rapid eye movement sleep (REM), and non-rapid eye movement sleep (NREM) states. This is critical because activity determination of sleep states can be inaccurate in humans as well as rodents that have low general activity [30]. Other methods to assess sleep states include pulse doppler-based method to assess movement and respiration [31] and whole body plethysmography to directly measure breathing patterns [32]. Both these approaches require specialized equipment. Recently, electric field sensors that detect respiration and other movements have also been used to access sleep states [33]. Both doppler and electric field sensors achieve higher accuracy for 3-state sleep classification. In general, respiration, movement, or posture by themselves are valuable for distinguishing between the three states and we predict that accuracy increases with combined features of movement and respiration.
In our work, to assess wake, NREM, and REM states, we used a video-based method with high resolution and found that information about sleep states is encoded in video data [34]. There are subtle changes in the area and shape of the mouse as it transitions from NREM to REM, likely due to the atonia of the REM stage. This allowed us to discriminate these two states [34]. However, using the methods available at that time, we found low discriminability between the three states. This is because the images we employed were low resolution and we tracked the mouse using an ellipse fit which inherently loses important postural information. Over the past few years, large improvements have been made in the field of computer vision, largely due to advancement in machine learning, particularly in the field of deep learning [35]. We have used these methods to track and determine action of mice [36–38]. Here we use these advanced machine vision methods to greatly improve upon visual sleep state classification. We extract rich features from the video that describe respiration, movement, and posture. These features combine to accurately determine sleep states in mice. This noninvasive video-based method is simple to implement with low hardware investment and yields high-quality sleep state data. The ability to access sleep states reliably, noninvasively, and in a high throughput manner will enable large scale mechanistic studies necessary for therapeutic discoveries. Key findings from this discovery strategy can then be validated in a subset of mice with EEG/EMG recording of sleep states.
Methods
Animal housing, surgery, and experimental setup
Sleep studies were conducted in 16 C57BL/6J (000664) male mice. We also image C3H/HeJ (000659) without surgery for feature inspection. These mice were obtained from Jackson Laboratory at 10–12 weeks of age. All animal studies were performed in accordance with the guidelines published by the National Institutes of Health Guide for the Care and Use of Laboratory Animals and was approved by the University of Pennsylvania Animal Care and Use Committee. The methods employed have been described previously by us [22, 34], and are described briefly here.
Mice were individually housed in an open-top standard mouse cage (6 by 6 inches). The height of each cage was extended to 12 inches prevent mice from jumping out of the cage. This design allowed us to simultaneously assess mouse behavior by video and sleep/wake stages by EEG/EMG recording. Animals were fed water and food ad libitum and were in a 12-hour light/dark cycle. During the light phase, the lux level at the bottom of the cage was 80 lux.
For EEG recording, four silver ball electrodes were placed in the skull; two frontal and two parietotemporal. For EMG recordings, two silver wires were sutured to the dorsal nuchal muscles. All leads were brought subcutaneously to the center of the skull and connected to a plastic socket pedestal (Plastics One) which was fixed to the skull with dental cement. Electrodes were implanted under general anesthesia. Following surgery, animals were given a 10-day recovery period before recording.
To assess how well our video-based algorithm worked when there were dynamic changes in state, we performed studies with intra-peritoneal injections of methamphetamine HCl (Sigma Aldrich). An additional 8 C57BL/6J male singly housed mice were studied with methamphetamine. They had surgery performed as described for insertion of electrodes for recording of EEG and EMG. They were allowed 10 days to recover from surgery before assessment of sleep. Animals were fed water and food ad libitum and were in a 12-hour light/dark cycle. EEG and EMG were recorded simultaneously with video. We recorded an initial 24-hour period of baseline wake and sleep behavior the day prior to the methamphetamine injection. Following the baseline day, animals received two separate intra-peritoneal injections of 1 mg/kg of methamphetamine at the Zeitgeber time (ZT) hours 2 and 6 following lights on. Recording was continued throughout the remainder of the 24-hour period.
EEG/EMG acquisition
For recording of EEG/EMG, raw signals were read using Grass Gamma Software (AstraMed) and amplified (20,000×). The signals were filtered with settings for EEG being low cutoff frequency of 0.1 Hz and high cutoff frequency of 100 Hz. The settings for EMG were a low cutoff frequency of 10 Hz and high cutoff of 100 Hz. Recordings were digitized at 256 Hz samples/second/channel.
Video acquisition
We used a Raspberry Pi 3 model B night vision setup to record high quality video data in both day and night conditions. We utilized the Sainsmart infrared night vision surveillance camera, which is accompanied with infrared light-emitting diodes to illuminate the scene when visible light is absent (SKU:101-40-112). The camera was mounted 18 inches above the floor of the home cage looking down providing a top-down view of the mouse for observation. During the day, video data are color. During the night, video data are monochromatic. We recorded video at 1920 × 1080 pixel resolution and 30 frames per second using the v4l2-ctl capture software. The cost of our video acquisition was approximately $155 ($35 Raspberry Pi3, $ 30 camera, $80 3TB hard drive, $10 power supply).
ideo and EEG/EMG data synchronization
We used computer clock time to synchronize video and EEG/EMG data. The EEG/EMG data collection computer was used as the source clock. At a known time on the EEG/EMG computer, a visual cue was added to the video. The visual cue typically lasted 2–3 frames in the video, suggesting that possible error in synchronization could be at most 100 ms. Since EEG/EMG data are analyzed in 10 s intervals, any possible error in temporal alignment would be negligible.
EEG/EMG annotation for training data
We collected 24 h synchronized video and EEG/EMG data for 16 C57BL/6J male mice from the Jackson Laboratory that were 10–12 weeks old. Both the EEG/EMG data and videos were divided into 10 s epoch, each epoch was scored and labeled as REM, NREM, or wake stage based on EEG and EMG signals by trained scorers. A total of 17,700 EEG/EMG epochs were scored by expert humans. Among them, 48.3% ± 6.9% epochs were annotated as wake, 47.6% ± 6.7% as NREM and 4.1% ±1.2% as REM stage. Additionally, we applied SPINDLE’s methods for a second annotation [16]. Similar to human experts, 52% were annotated as wake, 44% as NREM, and 4% as REM. Since SPINDLE annotates 4 s epochs, we joined three sequential epochs to compared to the 10 s epochs and only compare epochs when the three 4 s epochs do not change. When we correlated specific epochs, the agreement between human annotations and SPINDLE was 92% (89% Wake, 95% NREM, and 80% REM).
Data preprocessing
Starting with the video data, we applied a previously described segmentation neural network architecture to produce a mask of the mouse [1]. We annotated 313 frames to train the segmentation network. We applied a 4 × 4 diamond dilation followed by a 5 × 5 diamond erosion filter to the raw predicted segmentation. These routine operations were used to improve segmentation quality. With the predicted segmentation and resulting ellipse fit, we extracted a variety of per-frame image measurement signals in each frame described in Table 1. All these measurements (Table 1) were calculated by applying OpenCV contour functions on the neural network predicted segmentation mask. The OpenCV function we used included fitEllipse, contourArea, arcLength, moments, and getHuMoments. Using all the measurement signal values within an epoch, we derived a set of 20 frequency and time domain features (Table 2). These were calculated using standard signal processing approaches and can be found in our example code (github.com/KumarLabJax/MouseSleep).
Table 1.
Description of per-frame measurements derived from the segmentation and resulting ellipse fit of the segmentation mask of the mouse
| Measurement | Measurement description |
|---|---|
| m00 | Area |
| Perimeter | Perimeter of the mouse silhouette |
| x | Center x-position of ellipse-fit |
| y | Center y-position of ellipse-fit |
| w | Minor axis length for an ellipse-fit |
| l | Major axis length for an ellipse-fit |
| wl_ratio | Width divided by length of minor and major axis of ellipse fit |
| dx | Change in ellipse center x-position |
| dy | Change in ellipse center y-position |
| hu0 | Hu moment 0 |
| hu1 | Hu moment 1 |
| hu2 | Hu moment 2 |
| hu3 | Hu moment 3 |
| hu4 | Hu moment 4 |
| hu5 | Hu moment 5 |
| hu6 | Hu moment 6 |
Table 2.
The transition probability matrix of the sleep stages
| From\to | Wake | NREM | REM |
|---|---|---|---|
| Wake | 97.0% | 3.0% | 0% |
| NREM | 2.4% | 96.5% | 1.1% |
| REM | 10.1% | 4.4% | 85.6% |
Training the classifier
Due to the inherent dataset imbalance, that is, many more epochs of NREM compared to REM sleep, we randomly selected an equal number of REM, NREM, and wake epochs to generate a balanced dataset. We utilized a cross-validation approach to evaluate the performance of our classifier. We randomly selected all epochs from 13 animals from the balanced dataset for training and used imbalanced data from the remaining four animals for testing. The process was repeated 10 times to generate a range of accuracy measurements. This approach allowed us to observe performance on real imbalanced data while taking advantage of training a classifier on balanced data.
Prediction post-processing
We applied a Hidden Markov Model (HMM) approach to integrate larger scale temporal information to enhance prediction quality. The HMM model can correct erroneous predictions made by the classifier by integrating the probability of sleep state transitions and thus obtain more accurate predicted results. The hidden states of the HMM model are the sleep stages, whereas observables come from the probability vector results from the XgBoost algorithm. We computed the transition matrix empirically from the training set sequence of sleep states, then applied the Viterbi algorithm (Viterbi 1967) to infer the most probable sequence of the states given a sequence of the out of bag class votes of the XgBoost. In our cases, the transition matrix is a 3 by 3 matrix T = {S_ij}, here S_ij represents the transition probability from state S_i to state S_j (Table 2).
Classifier performance analysis
Performance was evaluated using metrics of accuracy as well as several metrics of classification performance: precision, recall, and F1 score. Precision is defined as the ratio of epochs classified by both the classifier and the human scorer for a given sleep stage to all of the epochs that the classifier assigned as that sleep stage. Recall is defined as the ratio of epochs classified by both the classifier and the human scorer for a given sleep stage to all of the epochs that the human scorer classified as the given sleep stage. F1 combines precision and recall and measures the harmonic mean of recall and precision. The mean and standard deviation of the accuracy and the performance matrix were calculated from 10-fold cross-validation.
Methamphetamine analysis
Performance of the methamphetamine experiment was conducted by training a classifier using the original 16 animals and predicting on the methamphetamine data. This means that no animals from the methamphetamine experiment were included in the visual classification model we used. We calculated the same metrics as we used in the classifier performance analysis for this dataset as well as summarized hourly time spent in each sleep state. For detecting the effect of methamphetamine, we analyzed after each injection (ZT2 and 6) and compared them to baseline in the same animal.
Results
Experimental design
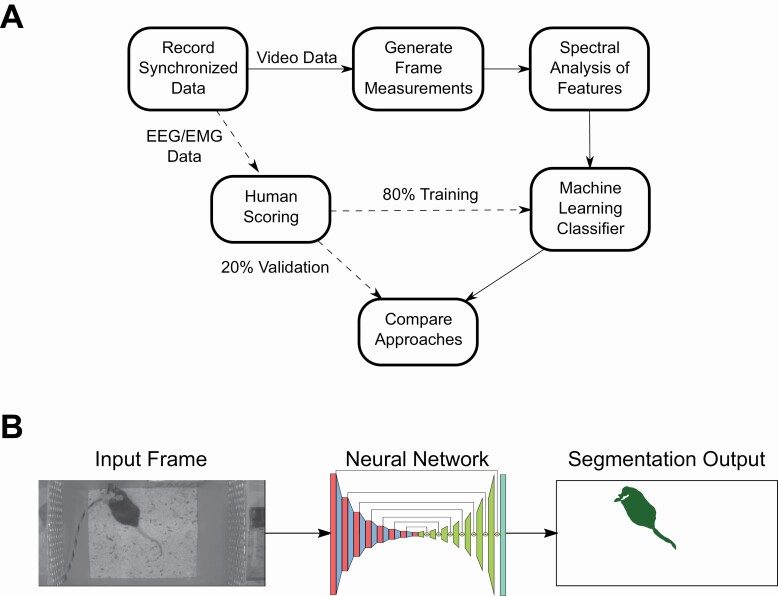
We sought to quantify the feasibility of using exclusively video data to classify mouse sleep states. This entire process is described visually in Figure 1A. We designed an experimental paradigm where we could leverage the current gold standard of sleep state classification, EEG/EMG recordings, as labels for training and evaluating our visual classifier. Overall, we recorded synchronized EEG/EMG and video data in 16 animals (24 h per animal). The data were split into 10 s epochs. Each epoch was hand scored by human experts. Concurrently, we designed features from video data which could be used in a machine learning classifier. These features were built on per frame measurements that describe the animal’s visual appearance in individual video frames (Table 1). We then applied signal processing techniques to the per frame measurements to integrate temporal information to generate a set of features for use in a machine learning classifier (Table 2). Finally, we split the human-labeled dataset by holding out individual animals into training and validation datasets (80:20, respectively). Using the training dataset, we trained a machine learning classifier to classify 10 s epochs of video into three states: wake, sleep NREM, and sleep REM. We used the set of held out animals in our validation dataset to quantify our classifiers performance. When we separate the validation set from the training set, we held out whole animal data which ensured that our classifier generalized well across animals instead of learning to predict well only on the animals it was shown.
Figure 1.
Visual data flow of our experimental pipeline.
(A) A visual description of how we organized data collection, annotation, feature generation, and classifier training. (B) A visual description of frame-level information we use for visual features. We used a trained neural network to produce a segmentation mask of pixels pertaining to the mouse for use in downstream classification.
Per frame features
We applied computer vision techniques to extract detailed visual measurements of the mouse in each frame. The first computer vision technique that we used is segmentation of the pixels pertaining to the mouse versus background pixels (Figure 1B). We trained a segmentation neural network as an approach that operates well in dynamic and challenging environments such as light and dark conditions as well as the moving bedding seen in our arenas [1]. Segmentation also allowed us to remove the EEG/EMG cable emanating from the instrumentation on head of each mouse so that it does not affect the visual measurements with information about the motion of the head. Our segmentation network predicts pixels that are only the mouse and as such the measurements are only based on mouse motion and not the motion of the wire connected to the mouse’s skull. We annotated frames randomly sampled from all videos to achieve this high-quality segmentation and ellipse fit using a previously described network [36] (Figure 1B). The neural network required only 313 annotated frames to achieve good performance segmenting the mouse. We show example performance of our segmentation network by coloring pixels predicted as not mouse with red and pixels predicted as mouse as blue on top of the original video (Video 1). Following the segmentation, we calculated 16 measurements from the neural network predicted segmentation that describes the shape and location of the mouse (Table 1). These include, major, minor length, and ratio of the mouse from an ellipse fit that describes the mouse shape. We extracted the location of the mouse (x, y) and change in x, y (dx, dy) for the center of the ellipse fit. We also calculated the area of the segmented mouse (m00), perimeter, and 7 Hu image moments that are rotationally invariant (HU0-6) [2]. Hu image moments are numerical descriptions of the segmentation of the mouse through integration and linear combinations of central image moments [3].
Time-frequency features
Next, we used these per frame features to carry out time and frequency-based analysis in each 10 s epoch. This allowed us to integrate time information by applying signal processing techniques. For each per frame feature in an epoch, we extracted six time-domain features: kurtosis, mean, median, std, max, min of each signal and 14 frequency domain features: kurtosis of power spectral density, skewness of power spectral density, mean power spectral density for 0.1–1 Hz, 1–3 Hz, 3–5 Hz, 5–8 Hz, 8–15 Hz, total power spectral density, max, min, average, and standard deviation of power spectral density (Table 3). These resulted in 320 total features (16 measurements × 20 time-frequency features).
Table 3.
Time and frequency features extracted from the per frame measurements in Table 1
| Label | Description | Domain | |
|---|---|---|---|
| k | Kurtosis of raw signal | Time domain | 1 |
| k_psd | Kurtosis of power spectral density | Frequency domain | 2 |
| s_psd | Skewness of power spectral density | Frequency domain | 3 |
| MPL_1 | Mean power spectral density (0.1–1 HZ) | Frequency domain | 4 |
| MPL_3 | Mean power spectral density (1–3 HZ) | Frequency domain | 5 |
| MPL_5 | Mean power spectral density (3–5 HZ) | Frequency domain | 6 |
| MPL_8 | Mean power spectral density (5–8 HZ) | Frequency domain | 7 |
| MPL_15 | Mean power spectral density (8–15 HZ) | Frequency domain | 8 |
| Tot_PSD | Total power spectral density | Frequency domain | 9 |
| Max_PSD | Max power spectral density | Frequency domain | 10 |
| Min_PSD | Min power spectral density | Frequency domain | 11 |
| Ave_PSD | Average power spectral density | Frequency domain | 12 |
| Std_PSD | Standard deviation of power spectral density | Frequency domain | 13 |
| Ave_Signal | Average raw signal | Time domain | 14 |
| Std_Signal | Standard deviation of raw signal | Time domain | 15 |
| Max_Signal | Max raw signal | Time domain | 16 |
| Min_Signal | Max raw signal | Time domain | 17 |
| TOP_SIGNAL | Frequency that corresponds to MAX_PSD | Frequency domain | 18 |
| MED_SIGNAL | Median raw signal | Time domain | 19 |
| MED_PSD | Median power spectral density | Frequency domain | 20 |
The analysis results in 320 total features for each 10 s epoch.
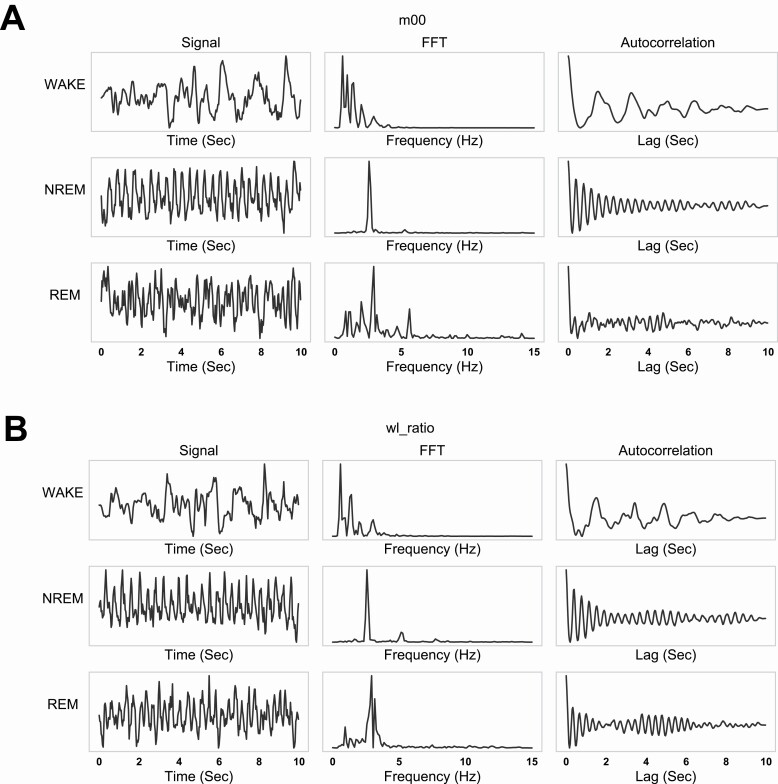
We visually inspected these spectral window features to determine if they vary between wake, REM, and NREM states. Figure 2, A and B show representative epoch examples of m00 (area, Figure 2A) and wl_ratio (width-length ratio of ellipse major and minor axis, Figure 2B) features that vary in time and frequency domain for wake, NREM, and REM state. The raw signals for m00 and wl_ratio show clear oscillation in NREM and REM states (Figure 1, A and B, left) which can be seen in the fast Fourier transform (FFT) (Figure 1, A and B, middle) and autocorrelation (Figure 1, A and B, right). We observed a single dominant frequency present in NREM epochs and a wider peak in REM. Additionally, the FFT peak frequency varied slightly between NREM (2.6 Hz) and REM (2.9 Hz) and in general we observed more regular and consistent oscillation in NREM epochs than REM. Thus, an initial examination of our features revealed differences between the sleep states and provided confidence that useful metrics are encoded in our features for use in a visual sleep classifier.
Figure 2.
Examples of selected signals in time and frequency domain within one epoch.
(A) the leftmost column shows m00 (area of the segmentation mask) for the wake, NREM, REM states; the middle column gives the FFT of the corresonding signals;the rightmost column shows the auto-correlation of the signals. (B) wl_ratio in time and frequency domain, similar to panel (A).
Breathing rate
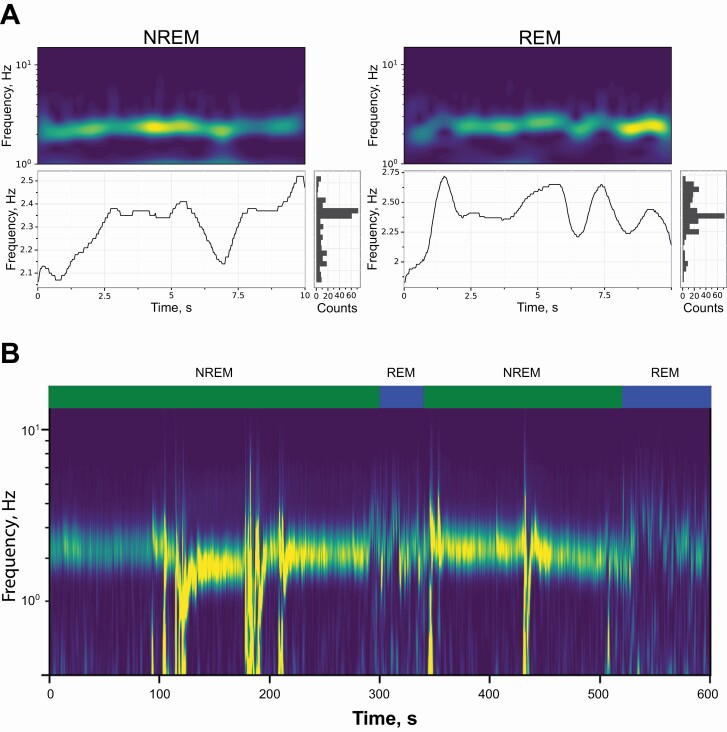
Previous work in both humans and rodents has demonstrated that breathing and movement varies between sleep stages [39–43]. In our examination of m00 and wl_ratio features, we discovered a consistent signal between 2.5 and 3 Hz that appears as a ventilatory waveform (Figure 2). An examination of the video revealed that changes in body shape and changes in chest size due to breathing were visible and may be captured by our time-frequency features. To visualize this signal, we carried out continuous wavelet transform (CWT) spectrogram for the wl_ratio feature (Figure 3A, top). To summarize data from these CWT spectrograms, we identified the dominant signal in the CWT (Figure 3A, bottom), and a histogram of dominant frequencies in the signal (Figure 3A, bottom right). From this histogram, we calculated the mean and variance of the frequencies contained in the dominant signal.
Figure 3.
Breathing singal extraction from video.
(A) Exemplar spectral analysis plots for REM and NREM epochs. The continuous wavelet transform spectral response (top) and associated dominant signal (bottomleft), and a histogram of the dominant signal (bottom right). NREM epochs typically show a lower mean and standard deviation than REM epochs. (B) Inspecting a larger time scale of epochs shows that the NREM signal is stable until a bout of REM. Dominant frequencies are typical mouse breathing rate frequencies.
Previous work has demonstrated that C57BL/6J mice have a breathing rate of 2.5–3 Hz during NREM state [43, 44]. Examination of a long bout of sleeping (10 min), which include both REM and NREM, showed that the wl_ratio signal is more prominent in NREM than REM, although it was clearly present in both (Figure 3B). Additionally, the signal varies more within the 2.5–3.0 Hz range while in the REM state. This is because the REM state causes higher and more variable breathing rate than the NREM state. We also observed low-frequency noise in this signal in the NREM state due to larger motion of the mouse such as adjusting their sleeping posture. This suggests that the wl_ratio signal is capturing the visual motion of the mouse abdomen.
Breathing rate validation
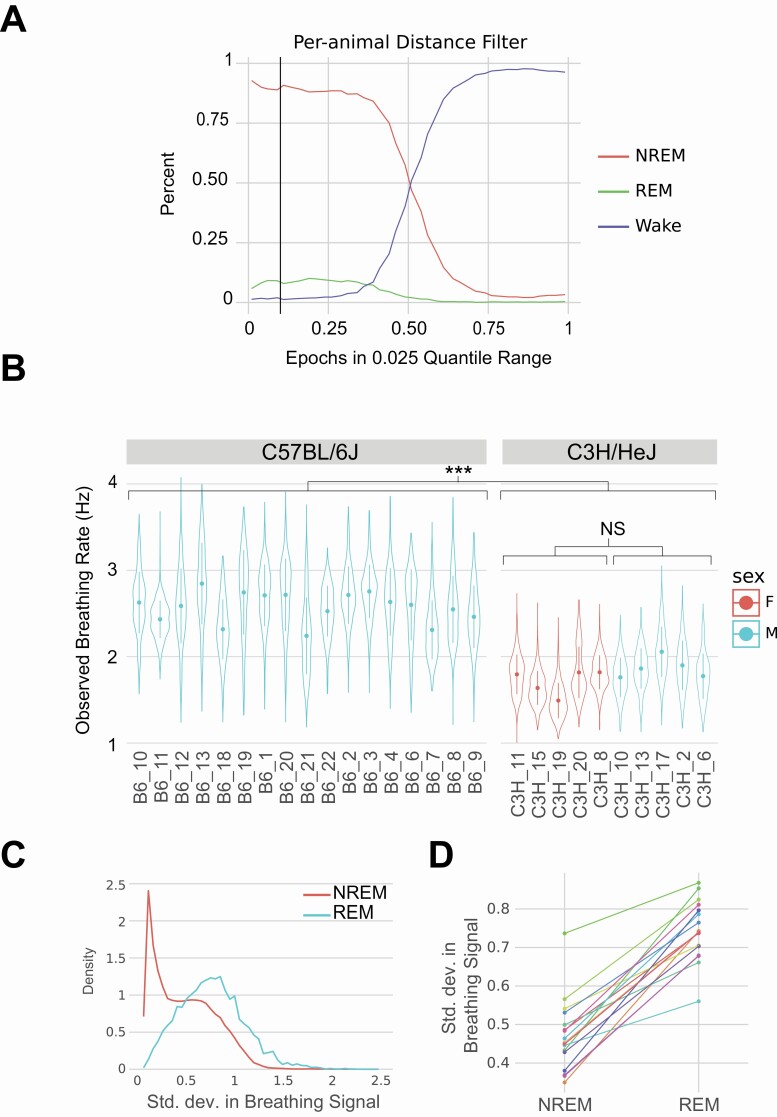
To confirm that the signal we observe in REM and NREM epochs for m00 and wl_ratio features is abdominal motion and correlates with breathing rate, we carried out a genetic validation test. C3H/HeJ mice have been previously demonstrated to have a wake breathing frequency that is approximately 30% less than that of C57BL/6J mice, ranging from 4.5 versus 3.18 Hz [45], 3.01 versus 2.27 Hz [46], and 2.68 versus 1.88 Hz [47] for C57BL/6J and C3H/HeJ, respectively. We video-recorded un-instrumented C3H/HeJ (5M/5F) and applied classical sleep/wake heuristic of movement (distance traveled) [22] to identify sleep epochs. We conservatively selected epochs with in lowest 10% quantile for motion. We used annotated C57BL/6J EEG/EMG data to confirm that our movement-based cutoff was able to accurately identify sleep bouts. Using the EEG/EMG annotated data for the C57BL/6J mice, we found that this cutoff primarily identifies NREM and REM epochs (Figure 4A). Epochs selected in our annotated data consists of 90.2% NREM, 8.1% REM, and 1.7% wake epochs. Thus, as expected, this mobility-based cutoff method correctly distinguished between sleep/wake and not REM/NREM. From these low motion sleep epochs, we calculated the mean value of the dominant frequency in the wl_ratio signal. We selected this measurement due to its sensitivity to chest area motion. We plotted the distribution of this measurement for each animal and observed similar distributions between animals within the same strain. For instance, C57BL/6J animals have an oscillation range from mean frequency of 2.2 to 2.8 Hz, while C3H/HeJ range from 1.5 to 2.0 Hz, in which C3H/HeJ breathing rates are approximately 30% less than of C57BL/6J. This is a statistically significant difference between the two strains, that is, C57BL/6J and C3H/HeJ (p < .001) (Figure 4B) and similar in range to previously reported data for breathing rate [45–47]. Thus, using this genetic validation method, we conclude that the signal we observe correlates strongly with breathing rate.
Figure 4.
Validation of breathing signal in video data for wl_ratio measurement.
(A) The mobility cutoff we use to select for sleeping epochs in C57BL/6J versus C3H/HeJ breathing rate analysis. At the 10% quantile cutoff (black line), epochs below this threshold consist of 90.2% NREM, 8.1% REM, and 1.7% wake. (B) Strain comparison of dominant frequency observed in sleeping epochs. (C) Using the C57BL/6J annotated epochs, we observe a higher standard deviation in dominant frequency in REM state than NREM state. (D) This increase in standard deviation is consistent across all animals.
In addition to overall changes in breathing frequency due to genetics, breathing during sleep has been shown to be more organized and with less variance during NREM than REM in both humans and rodents [26, 48]. We hypothesized that the breathing signal we detected would show greater variation in REM than NREM epochs. We determined whether there are changes in variation of CWT peak signal in epoch across REM and NREM states in the EEG/EEG annotated C57BL/6J data. Using only the C57BL/6J data, we partition the epochs by NREM and REM states and observed the variation in the CWT peak signal (Figure 4C). NREM states showed a smaller standard deviation of this signal while the REM state has a wider and higher peak. The NREM state appeared to comprise of multiple distributions, possibly indicating sub-divisions of the NREM sleep state [49]. To confirm that this odd shape of the NREM distribution is not an artifact of combining data from multiple animals, we also plotted data for each animal and show that each animal increased its standard deviation from NREM to REM state (Figure 4D). We also observed that individual animals show this long-tailed NREM distribution. Both these experiments indicate that the signals we observe are in fact a breathing rate signal. Knowing this, we expected good classifier performance.
Classification
Finally, we trained a machine learning classifier to predict sleep state using the 320 visual features. For validation, we held out entire animal’s data to avoid any bias that may be introduced by correlated data within a video. For calculation of training and test accuracy, we carried out 10-fold cross-validation by shuffling which animals were held out. We created a balanced dataset (see Methods section) and compared multiple classification algorithms, including XgBoost, Random Forest, multilayer perceptron (MLP), logistic regression, singular value decomposition (SVD), and observed a wide variety of performances between classifiers (Table 4). XgBoost and Random Forest both achieved good accuracies in the held-out test data. However, the Random Forest algorithm achieves 100% training accuracy, indicating that it overfits the training data. Overall, the best performing algorithm is the XgBoost classifier.
Table 4.
The accuracy of the model on dataset used for constructing the model (training accuracy) and the accuracy of the samples on the examples the model has not seen (test accuracy)
| Classifer | Training accuracy | Test accuracy |
|---|---|---|
| XgBoost | 0.875 | 0.852 |
| Random Forest | 1.000 | 0.857 |
| Neural network | 0.635 | 0.696 |
| SVM | 0.597 | 0.564 |
Significantly lower accuracy in the training set implies overfitting.
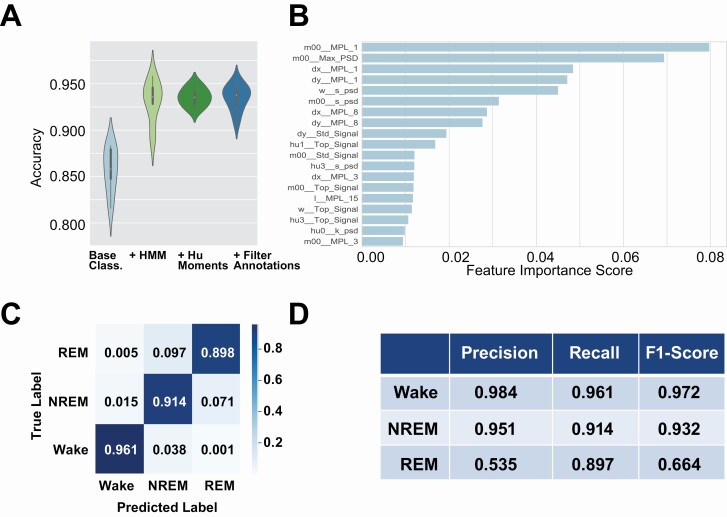
Transitions between wake, NREM, and REM states are not random and generally follow expected patterns. For instance, generally wake transitions to NREM which then transitions to REM sleep. The HMM is an ideal candidate to model the dependencies between the sleep states. The transition probability matrix and the emission probabilities in a given state are learned using the training data. We observed that by adding HMM model, the overall accuracy improved by 7% (Figure 5A, HMM) from 0.839 ± 0.022 to 0.906 ± 0.021.
Figure 5.
Classifier performance metrics.
(A) shows the performance of our classifier. We compare performance at different stages of our classifier, starting with the XgBoost classifier, adding an HMM model, increasing features to include 7 Hu moments, and integrating SPINDLE annotations to improve epoch quality. We can see the overall accuracy improves by adding each of these steps. (B) The top 20 most important features for the classifier. (C) Confusion matrix obtained from 10-fold cross validation. (D) Precision-recall table.
To enhance classifier performance, we adopted Hu moment measurements from segmentation for inclusion in input features for classification [50]. These image moments are numerical descriptions of the segmentation of the mouse through integration and linear combinations of central image moments. The addition of Hu moment features achieved a slight increase in overall accuracy and increased robustness of classifier through decreased variation in cross-validation performance (Figure 5A, Hu moments) from 0.90 6 ± 0.021 to 0.913 ± 0.019.
Even though the EEG/EMG scoring was performed by human trained experts, there is often disagreement between trained annotators [22]. Indeed, two experts only generally agree between 88% and 94% of the time for REM and NREM [22]. We used a recently published machine learning method to score our EEG/EMG data to complement data from human scorers [16]. We compared annotations between SPINDLE and human annotation and found that these two annotations agree in 92% of all epochs. We then used only epochs with both the human and machine-based methods agreed as labels to train our visual classifier. Training a classifier using only epochs where SPINDLE and humans agree added an additional 1% increase in accuracy (Figure 5A, filter annotations). Thus, our final classifier is able to achieve a three-state classification accuracy of 0.92 ± 0.05.
We investigated the most important features used in classification and discover that area of the mouse and motion measurements are most important (Figure 5B). This makes sense because motion is the only feature used in binary sleep-wake classification algorithms. Additionally, three of the top five features are low frequency (0.1–1.0 Hz) power spectral densities (Figure 2, A and B, FFT column). We note that wake epochs have the most power in low frequencies, REM has low power in low frequencies, and NREM has the least power in low-frequency signals.
Using our highest performing classifier, we observed good performance (Figure 5C). Rows in the matrix represent sleep stated assigned by the human scorer, while columns represent stages assigned by the classifiers. Wake has the highest accuracies of the classes at 96.1% accuracy. By observing the off-diagonals of the matrix, our classifier performed better at distinguishing wake from either sleep state than between the sleep states. This shows that distinguishing REM from NREM is a difficult task.
An average of 0.92 ± 0.05 overall accuracy was achieved in our final classifier. The prediction accuracy for wake stage is 0.97 ± 0.01, with average precision recall rate of 0.98. The prediction accuracy for NREM stage is 0.92 ± 0.04, with average precision recall rate of 0.93. The prediction accuracy for REM stage is around 0.88 ± 0.05, with average precision-recall rate of 0.535. The lower precision-recall rate for REM is largely due to a very small percentage of epochs that are labeled as REM stage (4%).
In addition to the prediction accuracy, we showed performance metrics including precision, recall, F1 score to evaluate the model (Figure 5D) from the 10-fold cross-validation. Given the imbalanced data, precision-recall were better metrics for classifier performance [51, 52]. We also used precision measures, the proportion of positive items that was correctly predicted, while recall measures the proportion of actual positives that was identified correctly. F1 score is the weighted average of precision and recall.
Here TP, TN, FP, and FN are true positives, true negatives, false positives, and false negatives, respectively.
We also attempted a variety of data augmentation approaches to improve classifier performance. The proportion of the different sleep states in 24 h is severely imbalanced (WAKE 48%, NREM 48%, and REM 4%). The typical augmentation techniques used for time series data include jittering, scaling, rotation, permutation, and cropping. These methods can be applied in combination with each other. In a recent study, it was shown that the classification accuracy could be increased by augmenting the training set by combining four data augmentation techniques [53]. However, the features we extract from the time series depend on the spectral composition and therefore we decided to use a dynamic time warping based approach to augment the dataset for improving the classifier [54]. The results of applying this data augmentation are shown in Supplementary Figure S1 and Supplementary Table S1. This data augmentation approach did not improve classifier performance and was not pursued further. In addition to data augmentation, we also considered using smaller epoch sizes, since two epoch sizes (4 and 10 s) are commonly used in sleep research. However, we observed decreased performance when training our classifier with 4 s epoch data (Supplementary Figure S2).
Our classifier is exceptional for both the wake and NREM states. However, the poorest performance was noted for REM stage, which has a precision of 0.535 and the F1 of 0.664. Most of the misclassified stages were between NREM and REM. As REM state is the minority class (only 4% of the dataset), even a relatively small false positive rate will cause a high number of false positives which will overwhelm the rare true positives. For instance, 9.7% of REM bouts were incorrectly identified as NREM by the visual classifier, and 7.1 of the predicted REM bouts are actually NREM (Figure 5C). These misclassification errors seem small, but can disproportionately affect the precision of the classifier due to the imbalance between REM and NREM. Despite this, our classifier is also able to correctly identify 89.7% of REM epochs present in the validation dataset.
Within the context of other existing alternatives to EEG/EMG recordings, our model performs exceptionally. We report performances elsewhere in literature to provide context of our model performance (Table 5). We note that each of these performances uses different datasets with different characteristics. Notably, the piezo system is evaluated on a balanced dataset which may present higher precision due to reduced possible false positives. Our approach outperforms all approaches for Wake and NREM state prediction. REM prediction is a more difficult task for all approaches. Of the machine learning approaches, our model achieves the best accuracy.
Table 5.
Performance comparison across published approaches
| Approach | Wake | NREM | REM | Overall | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | Accuracy cc. | Precision | Recall | Accuracy | Precision | Recall | Accuracy | |
| Video (mice) [34] | 0.767 | |||||||||
| Doppler (rats) [31] | 0.916 | 0.898 | 0.834 | 0.851 | 0.852 | 0.917 | 0.697 | 0.718 | 0.615 | 0.844 |
| Piezo (mice) [26] | 0.91 | 0.841 | 0.9 | 0.831 | 0.717 | 0.81 | 0.834 | 0.815 | 0.66 | 0.787 |
| Electric field* (mice) [33] | 0.938 | 0.943 | 0.943 | 0.834 | 0.94 | |||||
| Ours (mice) | 0.961 | 0.984 | 0.961 | 0.914 | 0.951 | 0.914 | 0.898 | 0.535 | 0.897 | 0.92 |
Bold indicates best performing approach for each metric.
*Electric field approach uses human annotation, not a machine learning algorithm.
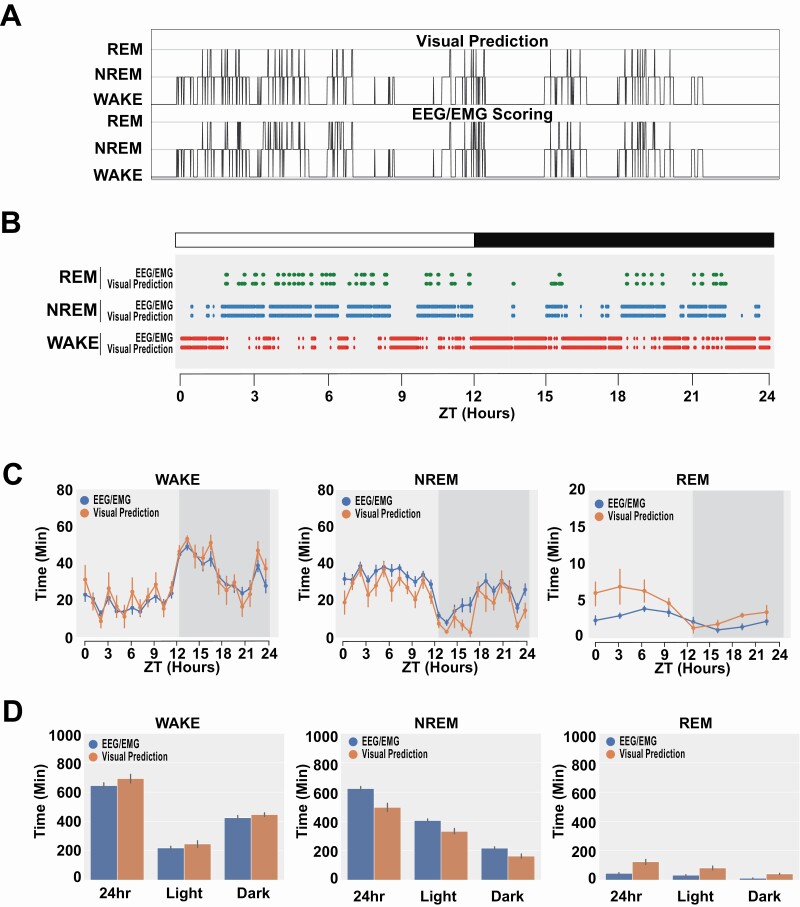
Figure 6, A and B display visual performance comparisons of our classifier to manually scored by a human expert (hypnogram). The x axis is time, consisting of sequential epochs, and the y axis corresponds to the three stages. For each subfigure, the top panel represent the human scoring results and the bottom panel represent the scoring results of the classifier. The hypnogram shows accurate transitions between stages along with frequency of isolated false positives (Figure 6A). We also plot data obtained by human scorers from EEG/EMG data and our visual classifier scoring for a single animal over 24 h (Figure 6B) as well as for the remaining animals in the validation dataset (Supplementary Figure S3). The raster plot shows exceptional global correlation between classification of state (Figure 6B). We then compare all C57BL/6J animals between human EEG/EMG scoring and our visual scoring (Figure 6, C and D). We observe high correlation across all states and conclude that our visual classifier scoring results are consistent with human scorers. Finally, we conduct full bout analysis to compare EEG/EMG scores and our visual prediction classifier (Supplementary Figure S4). These results in tandem with the 24-hour plots (Supplementary Figure S3) suggest that errors in classification are uniform over all data and do not skew sleep-related behavioral measurements. There are notably only two exceptions to this, that is, animal ID 21_3’s longest NREM bout and animal ID 18_4’s longest REM bout (Supplementary Figure S4). These errors represent an overall minority of the validation data. We inspected the segments when these longer bouts are predicted by our visual system and discovered that the wire used for EEG/EMG recording became twisted and is obscuring the mouse more than most other video segments, which would cause a drastic shift in features.
Figure 6.
Validation of visual scoring.
(A) Hypnogram of visual scoring and EEG/EMG scoring. (B) 24-Hour EEG/EMG scored sleep stage (top) and visual classifier predicted stage (bottom) for a mouse (B6J_7). (C, D) Comparison of human and visual scoring across all C57BL/6J mice show high concordance between the two methods. Data plotted in 1 h bin across 24 h (C) and in 24 or 12 h periods (D).
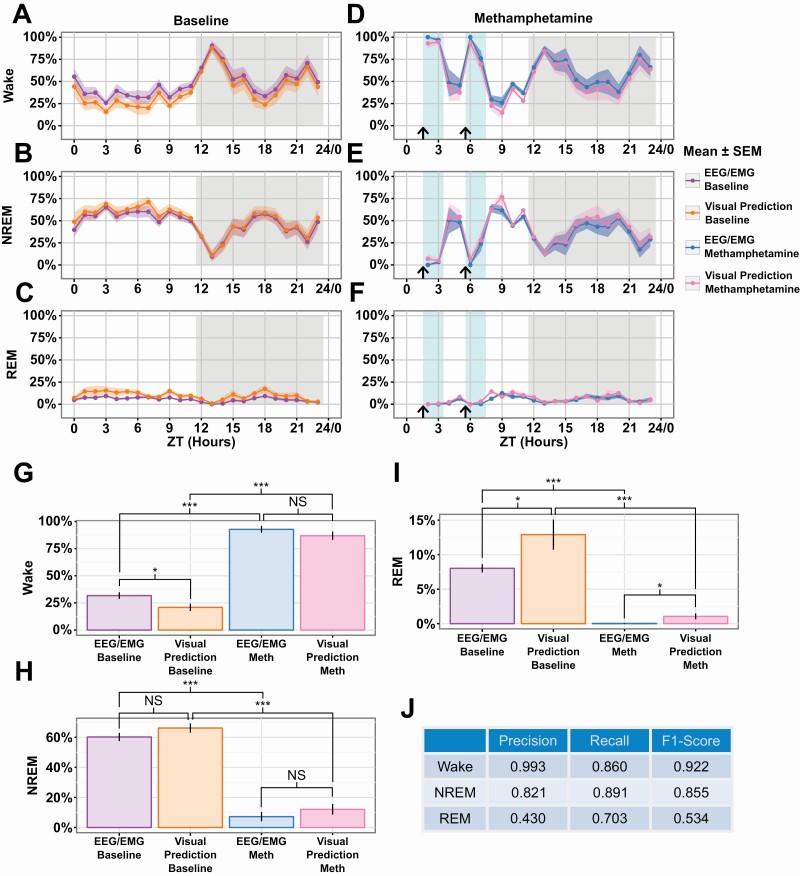
We conducted an experiment with a drug perturbation to validate our system further, that is, did we capture dynamic changes in sleep state. We observed mice both preinjection and postinjection of methamphetamine because it has previously been shown to affect sleep/wake behavior [55]. We observe that our classifier is robust to the drug perturbation and can accurately predict both the baseline and post-injection data (Figure 7A–F). Using EEG/EMG annotations, we detect sleep state differences for the first 2 h after each injection (Supplementary Figure S5). Additionally, we compared the 2 h after each injection and detected differences between baseline and injection for both EEG/EMG annotations as well as our visual prediction (Figure 7G–I). Finally, we present the precision-recall and F1 scores for this entire experiment and find that they are within expected variation based on our held-out validation set (Figures 5D and 7J). These results show that our classifier can accurately be extended to detect drug-induced sleep perturbation experiments without modification.
Figure 7.
Scoring comparison of methamphetamine treatment.
Effect of methamphetamine treatment observed with EEG/EMG is preserved using visual classification (A–C). Animals were treated twice (arrow). Comparison of time spent per hour in the Wake (A), NREM (B), and REM (C) states for the baseline of the methamphetamine experiment (n = 8 animals). (D–F) Comparison of the response to methamphetamine in the Wake (D), NREM (E), and REM (F) states (n = 6 animals, blue box). (G–I) Comparison of ZT hours 2, 3, 6, and 7 between baseline and post methamphetamine injection for Wake (G), NREM (H), and REM (I). Hours compared are marked with a light blue background in panels (A–F). (J) Precision-recall table. Data is shown as Mean ± SEM in (A–I). p value (* ≤ .05, *** ≤ .001).
Overall, the visual sleep state classifier is able to accurately identify sleep states using only visual data. Inclusion of HMM, Hu moments, and highly accurate labels, improve performance, whereas data augmentation using dynamic time warping and motion amplification did not improve performance.
Discussion
Sleep disturbances are a hallmark of numerous diseases and high-throughput studies in model organisms are critical for discovery of new therapeutics [1–3]. Sleep studies in mice are challenging to conduct at scale due to the time investment for conducting surgery, recovery time, and scoring of recorded EEG/EMG signals. We propose a system which provides a low-cost alternative to EEG/EMG scoring of mouse sleep behavior. This alternative will enable researchers to conduct larger-scale sleep experiments that would have been previously cost prohibitive. Previous systems have been proposed to conduct such experiments but have only been shown to adequately distinguish between wake and sleep states. Our system builds on these approaches and can also distinguish the sleep state into REM and NREM states. We argue that this is particularly useful for high throughput analysis of sleep, for example, in multiple lines of mutant mice. Findings from this approach in discovery would then be validated in only a small subset of these lines by EEG/EMG recording of sleep. Thus, our approach is not proposed as an alternative to EEG/EMG recording of sleep but rather a high throughput approach to discovery.
The system we designed utilized sensitive measurements of mouse movement and posture during sleep. We show that our system detects features that correlate with mouse breathing rates using analysis of video behavior. It provides assessments of wake and sleep and importantly the substages of sleep, that is, NREM and REM sleep. Previously published systems that attempt to use noninvasive sleep scoring include plethysmography [32] or piezo systems [26, 28]. Additionally, we show that based on our features, our system may be capable of identifying subclusters of NREM sleep epochs. This could shed additional light on the structure of mouse sleep.
While our system consists of low-cost, off-the-shelf parts, and is accurate and scalable, it is not without limitation. To achieve the sensitive measurements, our system relies upon high-resolution video relatively close to a mouse. These measurements are, therefore, vulnerable to feature shifts when the mouse is obscured from camera view. Processing and classification of video data require data storage and graphics processing units (GPUs) for data analysis resources. While our system outperforms other similar scalable systems, our REM recall performance is not sufficient to fully replace the gold standard of EEG/EMG. Instead, we consider the advantages of this system are for extending sleep analyses to more broad experimental paradigms while still reserving EEG/EMG for confirming sleep state differences.
Our breathing rate measurements are limited to when the mouse is immobile. But this is not an issue for how we use these measurements since assessments of breathing rate and its variability are done when the mouse is immobile during sleep. We have validated the measurement of breathing rate using comparisons of breathing rate in two mouse strains known to have different breathing frequencies. To fully validate the measurement as directly measuring breathing, a follow-up experiment of video inside a plethysmograph system would be necessary. To the best of our knowledge, this is not feasible to carry out given the size of the current widely used mouse plethysmograph system. We do not believe that this is required since we are not proposing that this is a method to measure breathing. Rather, we have extracted a feature that provides data to determine sleep states.
The current study is limited to isogenic C57BL/6J mice, which is the mouse reference strain. As proof of principle, this is the first step and future work needs to extend these methods to diverse strains as well as genetically heterogeneous populations such as the Diversity Outcross [56, 57] and Collaborative Cross [58]. This is technically feasible since we have applied our segmentation and tracking methods to diverse mouse strains [36]. Application of our approach to multiple mouse strains will introduce a variety of challenges for our machine learning approach such as the impact of variability in body size, coat color, and breathing rate which need to be explored and re-validated. During feature selection, we prioritized nondimensional features to reduce the impact of this variability. Notably, the genetic difference between resting breathing rates may pose a challenge for our visual sleep classifier which could be addressed by selecting different frequency domain features.
While we provide reliable estimates of baseline sleep, we do not assess sleep homeostasis. This is typically done studying increase in delta power during recovery sleep following a period of sleep deprivation [59]. While our technique does not directly address this, there are other approaches to studying the propensity to sleep that provide information on sleep drive. Our analysis provides data not only in amounts of sleep and its stages, but also bout duration. During recovery sleep following a period of sleep deprivation, NREM sleep becomes more consolidated with longer bouts [60]. There are also interventions that can be used that assess difference in sleep propensity. Veasey et al. describe a murine equivalent of the multiple sleep latency test to assess time to fall asleep during multiple nap opportunities during the lights-on period [61]. Latency to sleep can be assessed by patterns of activity and inactivity [56]. Progressive decrease in latency to sleep during these multiple naps is a measure of sleep propensity that is heritable, as revealed by studies in the founder mice for the Collaborative Cross [56] and in Diversity Outbred mice [56, 57]. Thus, assessment of mouse behavior with video analysis can be conducted during this type of intervention.
Extensions of this approach to genetically diverse mouse strains will enable high throughput studies that are needed for mapping genetic architecture of sleep, as well as application to interventional studies for better sleep therapeutics. In addition to extension to genetically diverse populations, video-based sleep assay has the exciting possibility of examining sleep in mice housed in groups thereby elucidating social genetic effects [62]. Tethered EEG/EMG approaches have forced analysis of sleep states in isolated animals but video-based assays have the potential to distinguish multiple animals and track each for sleep state determination [63, 64]. Our noninvasive method also allows mice to be evaluated for sleep over long time periods, potentially through the lifetime of the animal, and in group-housed environments. Combined, these will enable aging studies and studies that require longitudinal monitoring of animals, such as those to model Alzheimer’s disease [4, 65]. This is particularly salient in interventional studies where sleep disruption serves as a biomarker for disease progression [11]. In conclusion, we present a high-throughput, noninvasive, computer vision-based method for sleep state determination in mice that will be of utility to the research community.
Supplementary Material
Acknowledgments
We thank members of the Kumar Lab for helpful advice and Taneli Helenius for editing. We thank JAX Information Technology team members Edwardo Zaborowski, Shane Sanders, Rich Brey, David McKenzie, and Jason Macklin for infrastructure support.
Funding
This work was funded by The Jackson Laboratory Directors Innovation Fund, National Institute of Health DA041668 (NIDA), DA048634 (NIDA) (VK), and HL094307 (NHLBI) (AIP).
Disclosure Statement
None declared.
Data Availability
All code are available at Kumar Lab Github at https://github.com/KumarLabJax/MouseSleep.
Feature data from experiments are available on Zenodo at https://zenodo.org/record/5180680.
References
- 1. Webb JM, et al. Recent advances in sleep genetics. Curr Opin Neurobiol. 2021;69:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scammell TE, et al. Neural circuitry of wakefulness and sleep. Neuron. 2017;93(4):747–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allada R, et al. Unearthing the phylogenetic roots of sleep. Curr Biol. 2008;18(15):R670–R679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Green TRF, et al. The bidirectional relationship between sleep and inflammation links traumatic brain injury and Alzheimer’s disease. Front Neurosci. 2020;14:894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Firth J, et al. A meta-review of “lifestyle psychiatry”: the role of exercise, smoking, diet and sleep in the prevention and treatment of mental disorders. World Psychiatry. 2020;19(3):360–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benjamin SE. Sleep in patients with neurologic disease. Continuum (Minneap Minn). 2020;26(4):1016–1033. [DOI] [PubMed] [Google Scholar]
- 7. Ashton A, et al. Disrupted sleep and circadian rhythms in schizophrenia and their interaction with dopamine signaling. Front Neurosci. 2020;14:636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Freeman D, et al. Sleep disturbance and psychiatric disorders. Lancet Psychiatry. 2020;7(7):628–637. [DOI] [PubMed] [Google Scholar]
- 9. Eacret D, et al. Bidirectional relationship between opioids and disrupted sleep: putative mechanisms. Mol Pharmacol. 2020;98(4):445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krystal AD. Sleep therapeutics and neuropsychiatric illness. Neuropsychopharmacology. 2020;45(1):166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carter P, et al. Sleep and memory: the promise of precision medicine. Sleep Med Clin. 2019;14(3):371–378. [DOI] [PubMed] [Google Scholar]
- 12. Mackiewicz M, et al. Functional genomics of sleep. Respir Physiol Neurobiol. 2003;135(2-3):207–220. [DOI] [PubMed] [Google Scholar]
- 13. Mavanji V, et al. Sleep and obesity: a focus on animal models. Neurosci Biobehav Rev. 2012;36(3):1015–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kelly JM, et al. Mammalian sleep genetics. Neurogenetics. 2012;13(4):287–326. [DOI] [PubMed] [Google Scholar]
- 15. Toth LA, et al. Animal models of sleep disorders. Comp Med. 2013;63(2):91–104. [PMC free article] [PubMed] [Google Scholar]
- 16. Miladinović Đ, et al. SPINDLE: end-to-end learning from EEG/EMG to extrapolate animal sleep scoring across experimental settings, labs and species. PLoS Comput Biol. 2019;15(4):e1006968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamabe M, et al. MC-SleepNet: large-scale sleep stage scoring in mice by deep neural networks. Sci Rep. 2019;9(1):15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barger Z, et al. Robust, automated sleep scoring by a compact neural network with distributional shift correction. PLoS One. 2019;14(12):e0224642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang X, et al. Telemetric recording of sleep and home cage activity in mice. Sleep. 2002;25(6):691–699. doi: 10.1093/SLEEP/25.6.677. [DOI] [PubMed] [Google Scholar]
- 20. Brown LA, et al. Simultaneous assessment of circadian rhythms and sleep in mice using passive infrared sensors: a user’s guide. Curr Protoc Mouse Biol. 2020;10(3):e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fisher SP, et al. Rapid assessment of sleep-wake behavior in mice. J Biol Rhythms. 2012;27(1):48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pack AI, et al. Novel method for high-throughput phenotyping of sleep in mice. Physiol Genomics. 2007;28(2):232–238. [DOI] [PubMed] [Google Scholar]
- 23. Brown LA, et al. COMPASS: continuous open mouse phenotyping of activity and sleep status. Wellcome Open Res. 2016;1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singh S, et al. Low-cost solution for rodent home-cage behaviour monitoring. PLoS One. 2019;14(8):e0220751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Flores AE, et al. Pattern recognition of sleep in rodents using piezoelectric signals generated by gross body movements. IEEE Trans Biomed Eng. 2007;54(2):225–233. [DOI] [PubMed] [Google Scholar]
- 26. Mang GM, et al. Evaluation of a piezoelectric system as an alternative to electroencephalogram/ electromyogram recordings in mouse sleep studies. Sleep. 2014;37(8):1383–1392. doi: 10.5665/sleep.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Donohue KD, et al. Assessment of a non-invasive high-throughput classifier for behaviours associated with sleep and wake in mice. Biomed Eng Online. 2008;7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yaghouby F, et al. Noninvasive dissection of mouse sleep using a piezoelectric motion sensor. J Neurosci Methods. 2016;259:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Joshi SS, et al. Noninvasive sleep monitoring in large-scale screening of knock-out mice reveals novel sleep-related genes. Neuroscience 2019. doi: 10.1101/517680 [DOI] [Google Scholar]
- 30. Tang X, et al. Home cage activity and behavioral performance in inbred and hybrid mice. Behav Brain Res. 2002;136(2):555–569. [DOI] [PubMed] [Google Scholar]
- 31. Zeng T, et al. Automated determination of wakefulness and sleep in rats based on non-invasively acquired measures of movement and respiratory activity. J Neurosci Methods. 2012;204(2):276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bastianini S, et al. Accurate discrimination of the wake-sleep states of mice using non-invasive whole-body plethysmography. Sci Rep. 2017;7:41698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kloefkorn H, et al. Noninvasive three-state sleep-wake staging in mice using electric field sensors. J Neurosci Methods. 2020;344:108834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McShane BB, et al. Assessing REM sleep in mice using video data. Sleep. 2012;35(3):433–442. doi: 10.5665/sleep.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Raghu M, et al. A survey of deep learning for scientific discovery. ArXiv:200311755 Cs Stat. 2020. http://arxiv.org/abs/2003.11755. Accessed March 13, 2021
- 36. Geuther BQ, et al. Robust mouse tracking in complex environments using neural networks. Commun Biol. 2019;2:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Geuther BQ, et al. Action detection using a neural network elucidates the genetics of mouse grooming behavior. bioRxiv. 2020. doi: 10.1101/2020.10.08.331017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheppard K, et al. Gait-level analysis of mouse open field behavior using deep learning-based pose estimation. bioRxiv. https://www.biorxiv.org/content/10.1101/2020.12.29.424780v1. Accessed March 13, 2021. [DOI] [PMC free article] [PubMed]
- 39. Stradling JR, et al. Changes in ventilation and its components in normal subjects during sleep. Thorax. 1985;40(5):364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gould GA, et al. Breathing pattern and eye movement density during REM sleep in humans. Am Rev Respir Dis. 1988;138(4):874–877. [DOI] [PubMed] [Google Scholar]
- 41. Douglas NJ, et al. Respiration during sleep in normal man. Thorax. 1982;37(11):840–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kirjavainen T, et al. Respiratory and body movements as indicators of sleep stage and wakefulness in infants and young children. J Sleep Res. 1996;5(3):186–194. [DOI] [PubMed] [Google Scholar]
- 43. Friedman L, et al. Ventilatory behavior during sleep among A/J and C57BL/6J mouse strains. J Appl Physiol (1985). 2004;97(5):1787–1795. [DOI] [PubMed] [Google Scholar]
- 44. Fleury Curado T, et al. Sleep-disordered breathing in C57BL/6J mice with diet-induced obesity. Sleep. 2018;41. doi: 10.1093/sleep/zsy089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Berndt A, et al. Comparison of unrestrained plethysmography and forced oscillation for identifying genetic variability of airway responsiveness in inbred mice. Physiol Genomics. 2011;43(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Groeben H, et al. Heritable differences in respiratory drive and breathing pattern in mice during anaesthesia and emergence. Br J Anaesth. 2003;91(4):541–545. [DOI] [PubMed] [Google Scholar]
- 47. Breathing-Rate-Jan-2019.pdf. https://4e0msbd6u0p3nnihfrzedkd8-wpengine.netdna-ssl.com/wp-content/uploads/2019/01/Breathing-Rate-Jan-2019.pdf. Accessed January 10, 2021.
- 48. Terzano MG, et al. The cyclic alternating pattern as a physiologic component of normal NREM sleep. Sleep. 1985;8(2):137–145. doi: 10.1093/sleep/8.2.137 [DOI] [PubMed] [Google Scholar]
- 49. Katsageorgiou VM, et al. A novel unsupervised analysis of electrophysiological signals reveals new sleep substages in mice. PLoS Biol. 2018;16(5):e2003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hu MK. Visual pattern recognition by moment invariants. IRE Trans Inf Theory. 1962;8(2):179–187. doi: 10.1109/TIT.1962.1057692 [DOI] [Google Scholar]
- 51. Powers DMW. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. ArXiv201016061 Cs Stat. 2020. http://arxiv.org/abs/2010.16061. Accessed January 10, 2021.
- 52. Saito T, et al. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One. 2015;10(3). doi: 10.1371/journal.pone.0118432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rashid KM, et al. Times-series data augmentation and deep learning for construction equipment activity recognition. Adv Eng Inform. 2019;42:100944. doi: 10.1016/j.aei.2019.100944 [DOI] [Google Scholar]
- 54. Fawaz HI, et al. Data augmentation using synthetic data for time series classification with deep residual networks. ArXiv180802455 Cs. 2018. http://arxiv.org/abs/1808.02455. Accessed January 10, 2021.
- 55. Kitahama K, et al. Strain differences in amphetamine sensitivity in mice. Psychopharmacology. 1979;66:189–194. [DOI] [PubMed] [Google Scholar]
- 56. Keenan BT, et al. High-throughput sleep phenotyping produces robust and heritable traits in Diversity Outbred mice and their founder strains. Sleep. 2020;43(5): doi: 10.1093/sleep/zsz278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Churchill GA, et al. The Diversity Outbred mouse population. Mamm Genome. 2012;23(9-10):713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Swanzey E, et al. Mouse genetic reference populations: cellular platforms for integrative systems genetics. Trends Genet. 2021;37(3):251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Franken P, et al. The homeostatic regulation of sleep need is under genetic control. J Neurosci. 2001;21(8):2610–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Franken P, et al. Genetic determinants of sleep regulation in inbred mice. Sleep. 1999;22(2):155–169. doi: 10.1093/SLEEP/22.2.155. [DOI] [PubMed] [Google Scholar]
- 61. Veasey SC, et al. Murine Multiple Sleep Latency Test: phenotyping sleep propensity in mice. Sleep. 2004;27(3):388–393. doi: 10.1093/sleep/27.3.388. [DOI] [PubMed] [Google Scholar]
- 62. Baud A, et al. Genetic variation in the social environment contributes to health and disease. PLoS Genet. 2017;13(1):e1006498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ohayon S, et al. Automated multi-day tracking of marked mice for the analysis of social behaviour. J Neurosci Methods. 2013;219(1):10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pereira TD, et al. SLEAP: multi-animal pose tracking. bioRxiv. 2020. doi: 10.1101/2020.08.31.276246 [DOI] [Google Scholar]
- 65. Roh JH, et al. Disruption of the sleep-wake cycle and diurnal fluctuation of β-amyloid in mice with Alzheimer’s disease pathology. Sci Transl Med. 2012;4(150):150ra122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code are available at Kumar Lab Github at https://github.com/KumarLabJax/MouseSleep.
Feature data from experiments are available on Zenodo at https://zenodo.org/record/5180680.