Abstract
In order to evaluate the diagnostic yield of a PCR assay for patients with focal complications of brucellosis, we studied by PCR and by conventional microbiological techniques 34 nonblood samples from 32 patients with different focal forms of brucellosis. The samples from patients with brucellosis were paired to an equal number of control samples from the same locations of patients whose illnesses had different etiologies. Thirty-three of the 34 nonblood samples (97%) from the brucellosis patients were positive by PCR, whereas Brucella spp. were isolated from only 29.4% of the conventional cultures. For 11.4% of the patients, the confirmatory serological tests were either negative or showed titers below the diagnostic range. Two patients (6.2%) from the control group, both with tuberculous vertebral osteomyelitis, had a positive PCR result. The brucella PCR of blood from these two patients was also positive, and the two strains of Mycobacterium tuberculosis isolated were analyzed by the brucella PCR, with no evidence of amplification. These results show that the PCR assay is far more sensitive than conventional cultures, and this, coupled with its speed and reduction in risk to laboratory workers, makes this technique a very useful tool for the diagnosis of focal complications of brucellosis.
Brucellosis is a zoonosis widely distributed around the world (8). In humans, brucellosis behaves as a systemic infection with a very heterogeneous clinical spectrum. The disease usually presents as fever with no apparent focus, although in 20 to 40% of cases there are focal forms (7). These focal forms of brucellosis have been described in almost all organs and systems, with the osteoarticular and genitourinary forms being more common and those affecting the heart and the central nervous system being more severe (21).
The diagnosis of focal forms of brucellosis is sometimes difficult, as the yield of conventional cultures of nonblood samples is as low as 10 to 40% among all cases (11, 14). Moreover, as Brucella spp. are slowly growing pathogens, the cultures require prolonged incubation, which can at times lead to excessive delays in diagnosis. Furthermore, serological diagnosis lacks adequate specificity in areas where the disease is endemic, and the results of serology for some slowly evolving focal forms are difficult to interpret (2).
Our group has recently reported that the use of a PCR technique with blood samples provides better results than conventional techniques in the diagnosis of both primary infections and relapses of the disease (15, 16). The aim of the study described here was to evaluate the yield of this PCR technique with nonblood samples from patients with different focal forms of brucellosis compared with that of conventional microbiological techniques.
Patient population.
From January 1997 to December 1999, 34 nonblood samples from 32 patients with different focal complications of brucellosis were studied by PCR assay. Samples came from synovial fluid from eight patients; pus from abscesses from five patients; urine from five patients; cerebrospinal fluid (CSF) from five patients; vertebral or other bone tissue from four patients; sputum from two patients; renal cyst fluid from two patients; and pleural fluid, renal tissue, and thyroid tissue from one patient each. The two patients who had more than one focal form each provided two different samples. All nonblood samples, excluding urine and sputum samples, were obtained by conventional puncture, ultrasound, or computed tomography-guided puncture, depending on the site, or during open surgery.
The diagnosis of brucellosis was established by one of the following criteria: first, isolation of Brucella spp. from blood or any other body fluid or tissue sample or, second, the presence of a compatible clinical picture together with the demonstration of specific antibodies at significant titers or seroconversion. Significant titers were considered to be a Wright's seroagglutination test titer of ≥1/160 or a Coombs antibrucella test titer of ≥1/320.
In all cases, cultures and PCR assays with the sample obtained from the focus of infection, as well as a serological battery including the rose bengal plate agglutination test, Wright's seroagglutination test, and the Coombs antibrucella test, were carried out. For 28 patients (87.5%) two or more sets of blood cultures were done, and for 26 patients (81.25%) PCR was also done with a peripheral blood sample.
Blood cultures were processed in a BACTEC 9240 instrument (Becton Dickinson Diagnostic Instrument Systems, Sparks, Md.) by usual techniques (10), with incubation being maintained for 30 days and with blind subcultures performed on chocolate agar and brucella agar (Biomedics, San Sebastian de los Reyes, Madrid, Spain) after 10, 20, and 30 days. These subcultures were incubated at 37°C in a 5 to 10% CO2 atmosphere for 7 days.
All nonblood samples were cultured onto blood and chocolate agar media, MacConkey agar, and brucella agar. The samples of urine, sputum, and abscess pus were also inoculated onto a modified Thayer-Martin medium (Biomedics). The plates were incubated in a 10% CO2 atmosphere at 37°C for at least 7 days. If growth appeared, the suspected colonies were identified by colonial morphology; Gram staining; oxidase, catalase, and urease tests; and positive agglutination with specific antiserum. All isolated strains were sent to the National Brucellosis Reference Laboratory in Valladolid, Spain, for definitive identification and biotyping. The serological tests were all performed by previously described techniques (1).
DNA extraction.
All samples destined for PCR study were maintained at −20°C until processing. The volume was varied depending on the type of sample. Samples from the different tissues, synovial fluid, and purulent sample collections were resuspended in 1 ml of erythrocyte lysis solution (320 mM saccharose, 5 mM MgCl2, 1% Triton X-100, 10 mM Tris HCl [pH 7.5]), mixed, and centrifuged at 15,000 × g for 2 min. The supernatant was discarded, and the pellet was washed with Milli-Q water and centrifuged as described above. This washing with water was repeated until the pellet lost all reddish coloring. Purification and precipitation of DNA were performed as reported previously (16). DNA was extracted from urine and CSF samples by boiling.
PCR assay.
Once the DNA was extracted, the amplification process was performed by a previously described technique (16). Briefly, this consisted of amplification of a 223-bp fragment from the gene coding for the synthesis of an immunogenic membrane protein of Brucella abortus BCSP31, described by Mayfield et al. (13). This protein, with a molecular mass of 31 kDa, is specific to the Brucella genus and is present in all its biovars. The B4 and B5 primers described by Baily et al. (3) were used in the amplification process. Positive controls based on DNA from B. abortus B-19 were included in all the tests, as were negative controls, which contained all the elements of the reaction except DNA. A sample was considered positive when DNA with a molecular mass expected for the amplified product was fluorescent in the presence of ethidium bromide after 2% agarose gel electrophoresis.
To guarantee the reliability of the results and detect any possible contamination, all the samples were processed in duplicate. The test was considered positive if the signal from the amplified fragment was clearly visible in both samples. In order to study the specificity of the technique, all the samples from patients with brucellosis were paired to an equal number of samples from controls with lesions at the same site as the patients but with a different etiology. The organisms from the control group were isolated from synovial fluid (Staphylococcus aureus from five patients and Neisseria meningitidis, Neisseria gonorrhoeae, and Staphylococcus epidermidis from one patient each), liver or splenic abscesses (Escherichia coli from three patients and Streptococcus intermedius and Pseudomonas aeruginosa from one patient each), urine (E. coli from three patients and Klebsiella pneumoniae and Proteus mirabilis from one patient each), CSF (Streptococcus pneumoniae from two patients, and N. meningitidis, Mycobacterium tuberculosis, and S. epidermidis from one patient each), vertebral osteomyelitis tissue (M. tuberculosis from three patients and S. aureus from one patient), sputum (S. pneumoniae and Branhamella catarrhalis), renal cyst fluid (P. mirabilis and E. coli), pleural fluid (K. pneumoniae), renal tissue (P. mirabilis), and thyroid tissue (Streptococcus agalactiae).
Statistical analysis.
Data were analyzed with the help of SPSS 8.0 for Windows. Sensitivity, specificity, and positive and negative predictive values were calculated. Likelihood ratios and their 95% confidence intervals were calculated with the Roc Curve Analyzer program, version 6, as described by Centor (6).
Of the 32 patients included in the study, 26 (81.2%) were male and 6 (18.7%) were female. The mean age of the group was 48.9 ± 17.3 years (range, 19 to 81 years). For 17 patients (53.1%) the diagnosis of brucellosis was established by isolating the pathogen in blood cultures or in cultures of other samples. For the remaining 15 patients (46.8%), the diagnosis was made by clinical-serological means. All the strains isolated were identified as Brucella melitensis. Relevant clinical data and the sites of the different focal forms are presented in Table 1.
TABLE 1.
Clinical features of patients with brucellosis
| Characteristic | Value |
|---|---|
| Mean age (yra) | 48.9 (19–81) |
| Mean duration of symptoms (wka) | 18.3 (1–102) |
| Fever (no.b of patients) | 30 (93.7) |
| Location of the focal forms (no.b of patients) | |
| Peripheral arthritisc | 8 (23.5) |
| Hepatic or splenic abscess | 5 (14.7) |
| Orchiepididymitis | 5 (14.7) |
| Neuromeningesd | 5 (14.7) |
| Osteomyelitise | 4 (11.8) |
| Pneumonia | 2 (5.8) |
| Other focal complicationsf | 5 (14.7) |
Range is given in parentheses.
Percentage(s) is given in parentheses.
Knee arthritis, six patients; coxitis, two patients.
Meningitis, three patients; meningoencephalitis, two patients.
Vertebral osteomyelitis, three patients; femur osteomyelitis, one patient.
Infected renal cysts two patients; empyema, pyelonephritis, and suppurative thyroiditis, one patient each.
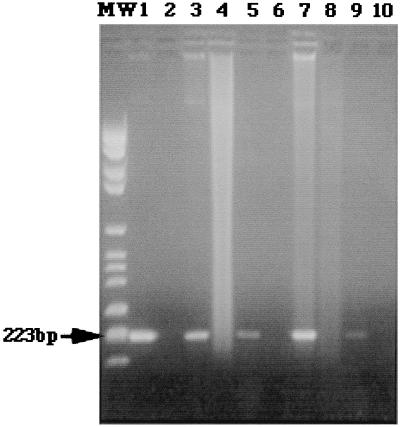
Thirty-three of the 34 (97%) nonblood samples from the patients with brucellosis had a positive PCR result, whereas Brucella spp. were isolated from cultures of the same samples from only 29.4% of the patients. Clear visualization of the PCR-amplified fragments was possible for all positive samples (Fig. 1). The only false-negative PCR result corresponded to a sample of synovial fluid for which culture was positive. This sample was mistakenly preserved with heparin. Seventy-five percent of the patients whose samples were culture negative had previously received antibiotic treatment for other suspected diagnoses not related to brucellosis. There was no difference in the sensitivity of the PCR between individuals in the treated group and those who had not received antimicrobial therapy prior to sample extraction.
FIG. 1.
Agarose gel electrophoresis and ethidium bromide staining. Lane MW, DNA ladder (223 bp); lane 1, positive control (B. abortus B-19 DNA); lane 2, a sample to which no DNA was added; lane 3, synovial fluid from a brucellosis patient with knee arthritis; lane 4, synovial fluid from a patient with knee arthritis due to S. aureus; lane 5, urine sample from a patient with orchiepididymitis due to B. melitensis; lane 6, urine sample from a patient with E. coli pyelonephritis; lane 7, sample of pus from a liver abscess due to B. melitensis; lane 8, sample of pus from a liver abscess due to E. coli; lane 9, CSF from a brucellosis patient with meningitis; lane 10, CSF from a patient with meningitis due to M. tuberculosis. The photocomposition of the figure was obtained from the original Polaroid films with a ScanJet IIcx scanner (Hewlett-Packard, Corvallis, Oreg.) After the initial image was scanned and saved as a tagged image file format file, the file was opened in Adobe Photoshop (Adobe System, Inc., Seattle, Wash.).
The rose bengal test was positive for 91.2% of the patients. Wright's seroagglutination test and the Coombs antibrucella test had titers within the diagnostic range for 73.5 and 88.5% of the patients, respectively. Both confirmatory tests were negative or showed titers below the diagnostic range for 11.4% of patients.
Two patients from the control group (6.2%), both with tuberculous vertebral osteomyelitis, had positive PCR assay results. The blood of two patients, a 25-year-old shepherd with a 14-year history of type I diabetes mellitus and poor metabolic control and a 38-year-old farmer, also had positive PCR assay results. The patients were treated surgically and had favorable resolutions of their infections with conventional tuberculostatic therapy plus doxycycline at 100 mg/12 h per os for 3 months. However, neither of them demonstrated a serological response by the rose bengal test, the seroagglutination test, or the Coombs antibrucella test during the following 12 months. In order to rule out a cross-reaction with Brucella spp., the two strains of M. tuberculosis isolated from these patients were analyzed by the brucella PCR assay, and both were negative by the assay. Table 2 shows the diagnostic yields of the PCR versus those of the conventional cultures.
TABLE 2.
Diagnostic yield of PCR assay with nonblood samplesa
| Parameter | Percent (95% CI)
|
|
|---|---|---|
| PCR assay | Culture | |
| Sensitivity | 97.1 (93.1–99.8) | 29.4 (18.5–40.2) |
| Specificity | 94.1 (88.5–99.7) | 100 |
| PPV | 94.3 (88.8–99.8) | 100 |
| NPV | 97.0 (92.9–99.8) | 58.6 (46.9–70.3) |
| Positive LR | 16.5 (5.1–59.7) | 20.1 (2.2–196.7) |
| Negative LR | 0.03 (0.01–0.16) | 0.71 (0.54–0.83) |
Abbreviations: CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; positive LR, positive likelihood ratio; negative LR, negative likelihood ratio.
Almost one-third of all patients with brucellosis present with focal complications, either initially or during the course of the disease (7). The focal complications can affect any organ or system, which explains why these patients are not always seen by infectious disease specialists, so that many other medical and surgical specialists are involved in the care of these patients (17, 19).
Even when there is a high degree of suspicion, the diagnosis of brucellosis is not always easy. Blood cultures lack sufficient sensitivity; and serological tests, although more sensitive, are not wholly specific, may be difficult to interpret in areas where the disease is endemic, and may be negative during the early stages of the disease (2, 20). Diagnosis of focal forms of brucellosis can occasionally be difficult, not only because the physician may not consider it as the cause of the clinical picture but also because even when it is considered the yields of blood and nonblood sample cultures for patients with these forms are low (7, 12, 14).
This is the first report to date that has analyzed the yield of a PCR assay for the diagnosis of focal complications of human brucellosis. Our results show that the sensitivity of the PCR assay was far superior to that of conventional culture, which was positive for only 29.4% of cases. This low yield of culture is similar to the 28.5% yield reported by others for a series of brucella meningitis or the 23.8% reported for brucella spondylitis (12, 14). The explanation for this low yield of conventional culture appears to us to be related more to the lack of suspicion of brucellosis and the widespread use of antimicrobial therapy in patients with no etiological diagnosis than to the technical difficulty of isolating Brucella spp. from clinical samples. In the present study, 75% of patients whose cultures were negative had received different antibiotic treatments for various diagnostic suspicions.
The high degree of sensitivity of the PCR assay, even for patients who had previously received antibiotic therapy, seems to be related to the high degree of detection capacity of the technique. We have estimated in previous studies that the detection capacity is about 10 to 100 fg of DNA (16). Such a small amount of DNA, which is the equivalent of that from just 2 to 20 cells, can be expected to be present in any clinical sample from a patient with an active infection. Moreover, PCR is able to amplify intramacrophage pathogens or pathogens which are damaged or nonviable as a result of previous treatment and which would be impossible to isolate in conventional cultures.
Despite the use of a wide battery of serological tests, 11.4% of the patients with brucellosis in the present study failed to show titers within the diagnostic range. In fact, this figure would have risen to 26.5% if we had used just the seroagglutination test, which is the serological test most commonly used by any physician faced with a possible diagnosis of brucellosis. The low degree of sensitivity of the seroagglutination test for the focal forms of the disease is well known and is due to the longer duration of the clinical picture in these patients. The immune response to brucella infection is characterized by an initial rise in immunoglobulin M (IgM) agglutinating antibody titers that is followed within a few weeks by a switch to IgG antibodies, with the latter being detected better by other serological tests, such as the 2-mercaptoethanol agglutination test, the Coombs antibrucella test, an IgG enzyme-linked immunosorbent assay, or complement fixation (2, 20).
The specificity of the PCR can be considered good, although the existence of two patients with false-positive results requires a search for an explanation. Previous studies, as well as our own results obtained by PCR with the B4 and B5 primers, have demonstrated the high degree of specificity of the technique with a large panel of microorganisms. Only the DNA from Ochrobactrum spp., a pathogen very closely related phylogenetically to Brucella spp., has been amplified with these primers (5). Cross-reactivity with M. tuberculosis has not been reported before. The two patients with false-positive results had habitual contact with cattle, and the suspensions of the pure cultures of these strains of M. tuberculosis failed to amplify. This led us to think more of a coinfection than of a false-positive result. In fact, coinfection with Mycobacterium and Brucella has been reported in our area, where the prevalence of infections with both of these organisms is high (9). In addition, in situ immunohistochemical studies have demonstrated that active effector cells in granulomas from patients with tuberculosis expressed high levels of cytokines such as tumor necrosis factor alpha, interleukin-6, and gamma interferon, which might result in activation of other latent infections (4). It is possible, then, that severe tuberculosis in these two patients led to the subclinical reactivation of a previous brucella infection.
Finally, in addition to the high yield of the PCR assay for the diagnosis of focal complications in patients with brucellosis, other important aspects of the technique make it especially attractive for use with patients with these types of complications. First, PCR is fast, providing results in 24 h, which is much less than the time required for conventional methods to rescue a fastidious microorganism such as a Brucella sp. Second, the technique almost completely obviates the necessity for direct handling of the pathogen, thus drastically reducing the risk of infection of laboratory personnel (18). Third, the sample can be stored at −20°C until processing, thus enabling it to be collected by any physician and processed immediately or else stored and safely sent to another laboratory if necessary.
In conclusion, the high degrees of sensitivity and specificity of the PCR assay, together with its speed, versatility in sample handling, and risk reduction for laboratory personnel, make this technique a very useful tool for the diagnosis of focal complications of brucellosis.
Acknowledgments
This work received financial support from the Inter-Ministerial Commission for Science and Technology (CICYT) and the European Commission (grant IFD97-0539).
We thank Isolde Gornemann and Ian Johnstone for comments and help with the English language version of the text.
REFERENCES
- 1.Alton G G, Jones L M, Pietz D E. Laboratory techniques in brucellosis. 2nd ed. Geneva, Switzerland: World Health Organization; 1975. [PubMed] [Google Scholar]
- 2.Ariza J, Pellicer T, Pallarés R, Foz A, Gudiol F. Specific antibody profile in human brucellosis. Clin Infect Dis. 1992;14:131–140. doi: 10.1093/clinids/14.1.131. [DOI] [PubMed] [Google Scholar]
- 3.Baily G G, Kranhn J B, Drasar B S, Stoker N G. Detection of Brucella melitensis and Brucella abortus by DNA amplification. J Trop Med Hyg. 1992;95:271–275. [PubMed] [Google Scholar]
- 4.Barnes P F, Lu S, Abrams J S, Wang E, Yamamura M, Modin R L. Cytokine production at the side of disease in human tuberculosis. J Immunol. 1993;61:3482. doi: 10.1128/iai.61.8.3482-3489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casañas M C, Queipo-Ortuño M I, Rodriguez-Torres A, Orduña A, Colmenero J D, Morata P. Specificity of a polymerase chain reaction assay of a target sequence on the 31-kilodalton Brucella antigen DNA used to diagnose human brucellosis. Eur J Clin Microbiol Infect Dis. 2001;20:127–131. doi: 10.1007/pl00011242. [DOI] [PubMed] [Google Scholar]
- 6.Centor R M. Estimating confidence intervals of likelihood ratios. Med Decision Making. 1992;12:229–233. doi: 10.1177/0272989X9201200309. [DOI] [PubMed] [Google Scholar]
- 7.Colmenero J D, Reguera J M, Martos F, Sanchez-de Mora D, Delgado M, Causse M, Martín-Farfán A, Juarez C. Complications associated with Brucella melitensis infection: a study of 530 cases. Medicine (Baltimore) 1996;75:195–211. doi: 10.1097/00005792-199607000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Corbel M J. Brucellosis: an overview. Emerg Infect Dis. 1997;3:313–321. doi: 10.3201/eid0302.970219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giner P, El-Amrani A, Corrales A J, Guijarro R, Sanchez-Palencia J A, Jiménez-Alonso J. Simultaneous isolation of Brucella melitensis and Mycobacterium tuberculosis in pleural empyema. Enferm Infec Microbiol Clin. 1990;8:595. [PubMed] [Google Scholar]
- 10.Hausler W J, Moller N P, Holcomb L A. Brucella. In: Lennette E H, Balows A, Hausler W J Jr, Shadomy H J, editors. Manual of clinical microbiology. 4th ed. Washington, D.C.: American Society for Microbiology; 1984. pp. 382–386. [Google Scholar]
- 11.Khateeb M I, Araj G F, Majeed S A, Lulu A R. Brucella arthritis: a study of 96 cases in Kuwait. Ann Rheum Dis. 1990;49:994–998. doi: 10.1136/ard.49.12.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lifeso R M, Harder E, McCorkell S J. Spinal brucellosis. J Bone Joint Surg Br Vol. 1985;67:345–351. doi: 10.1302/0301-620X.67B3.3997939. [DOI] [PubMed] [Google Scholar]
- 13.Mayfield J E, Bricker B J, Godfrey H, Crosby R M, Knight D J, Halling S M, Balinsky D, Tabatabai L B. The cloning and nucleotide sequence of a gene coding for an immunogenic Brucella abortus protein. Gene. 1988;63:1–9. doi: 10.1016/0378-1119(88)90540-9. [DOI] [PubMed] [Google Scholar]
- 14.McLean D R, Russell N, Khan Y. Neurobrucellosis. Clinical and therapeutic features. Clin Infect Dis. 1992;15:582–590. doi: 10.1093/clind/15.4.582. [DOI] [PubMed] [Google Scholar]
- 15.Morata P, Queipo-Ortuño M I, Reguera J M, García-Ordoñez M A, Pichardo C, Colmenero J D. Posttreatment follow-up of brucellosis by PCR assay. J Clin Microbiol. 1999;37:4163–4166. doi: 10.1128/jcm.37.12.4163-4166.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Queipo-Ortuño M I, Morata P, Ocón P, Manchado P, Colmenero J D. Rapid diagnosis of human brucellosis by peripheral blood PCR assay. J Clin Microbiol. 1997;35:2927–2930. doi: 10.1128/jcm.35.11.2927-2930.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samra Y, Shaked M, Altman G. Brucellosis: difficulties in diagnosis and a report on 38 cases. Infection. 1993;11:310–312. doi: 10.1007/BF01641353. [DOI] [PubMed] [Google Scholar]
- 18.Yagupsky P, Peled N, Riesenberg K, Banai M. Exposure of hospital personnel to Brucella melitensis and occurrence of laboratory-acquired disease in an endemic area. Scand J Infect Dis. 2000;32:31–35. doi: 10.1080/00365540050164182. [DOI] [PubMed] [Google Scholar]
- 19.Young E J. Human brucellosis. Rev Infect Dis. 1983;5:821–842. doi: 10.1093/clinids/5.5.821. [DOI] [PubMed] [Google Scholar]
- 20.Young E J. Serologic diagnosis of human brucellosis: analysis of 214 cases by agglutination test and review of the literature. Rev Infect Dis. 1991;13:359–372. doi: 10.1093/clinids/13.3.359. [DOI] [PubMed] [Google Scholar]
- 21.Young E J. An overview of human brucellosis. Clin Infect Dis. 1995;21:283–290. doi: 10.1093/clinids/21.2.283. [DOI] [PubMed] [Google Scholar]