Abstract
CTX-M-14 β-lactamase was identified in a stool isolate of Shigella sonnei and in blood isolates of Escherichia coli (one isolate) and Klebsiella pneumoniae (two isolates) from different parts of Korea. The amino acid sequence differed by one amino acid from CTX-M-9 (Ala-231→ Val) and was identical to that of β-lactamases recently found in China and Japan.
Because resistance is more than locally relevant with increasingly mobile populations and since unique resistance mechanisms may evolve anywhere antibiotics are used, we have investigated several unusually resistant clinical isolates from Korea.
One strain of Escherichia coli and two strains of Klebsiella pneumoniae that had high levels of resistance to cefotaxime were isolated from the blood of patients in Seoul National University Children's Hospital in 1995 and 1996. One strain of Shigella sonnei isolated from a pediatric patient on Cheju Island in 2000 also had a high level of cefotaxime resistance. By disk susceptibility testing, the strains were resistant to amoxicillin, cephalothin, and cefotaxime but were susceptible to ceftazidime, cefoxitin, and amoxicillin-clavulanic acid. Isoelectric focusing showed that the four strains produced a β-lactamase with an isoelectric point (pI) of 8.0. PCR with SHV-specific primers (3) was negative for all strains. Cefotaxime resistance was transferred by conjugation (9) from K. pneumoniae strain 95151 along with a plasmid of 160 kb to E. coli J53 Azir (met pro azide resistant) to produce E. coli J53 Azir/pMG267. The β-lactamase gene was cloned from plasmid pMG267 with EcoRI as an 8-kb insert into vector plasmid pBC SK (Stratagene, La Jolla, Calif.) carrying chloramphenicol resistance to produce plasmid pMG268. For sequencing, a Tn7-based transposon carrying a kanamycin resistance gene was inserted into purified pMG268 using the GPS-1 Genome Priming System-1 kit (New England BioLabs, Beverly, Mass.), and the resulting derivative was introduced into E. coli DH10B (Gibco BRL, Rockville, Md.) by electroporation. After selection with 50 μg of kanamycin per ml and 30 μg of chloramphenicol per ml, colonies were screened for loss of resistance to 100 μg of ampicillin per ml. In ampicillin-susceptible colonies, the transposon was assumed to have been inserted into the β-lactamase gene. With primers (primerN and primerS) that matched nucleotides at the extremities of the inserted transposon, cycle sequencing (Perkin-Elmer Cetus, Norwalk, Conn.) was initiated and continued by primer walking until both DNA strands were analyzed.
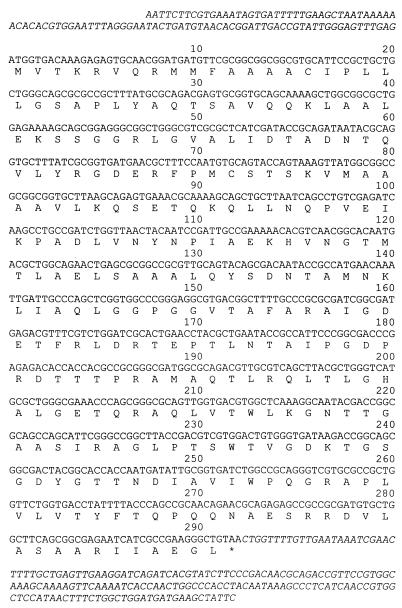
The open reading frame of the bla gene was 876 bp, and it encoded a protein with 291 amino acid residues (Fig. 1). The deduced amino acid sequence had one amino acid difference from that of the CTX-M-9 β-lactamase discovered in Spain (10), a change from Ala to Val at position 231. A BLAST search (2) indicated that the 99 nucleotides upstream from blaCTX-M-14 differed by only one base from a sequence upstream from the gene for Toho-2 β-lactamase, another member of the CTX-M family (4). The DNA from nucleotides 882 to 1055 downstream from the bla gene was 99% identical to the sequence of transposable element IS903 in kanamycin-resistant transposon Tn2680 (7).
FIG. 1.
The nucleotide and deduced amino acid sequences of the CTX-M-14 β-lactamase identified in four strains found in Korea. The upstream and downstream sequences are shown in italics. ∗, termination codon.
To sequence the β-lactamase genes of the other strains, they were amplified using primers designed for a CTX-M-type gene. The primers used for amplification were CTX-1 (5′-CGCTTTATGCGCAGACGA) and CTX-2 (5′-GATTCTCGCCGCTGAAGC). The PCR amplification mixture was denatured at 95°C for 30 s and annealed at 58°C for 1 min, and the chain was extended at 72°C for 1 min in a thermal cycler (Perkin-Elmer Cetus). This cycle was repeated 35 times. The nucleotide sequencing analyses were performed with the BigDye Terminator Cycle Sequencing Ready Reaction kit and ABI 377 automated sequencer (Applied Biosystems, Inc., Foster City, Calif.). Primers CTX-1, CTX-2, and CTX-3 (5′-TCAAAGGCAATACGACCG) were used for sequencing. The nucleotide sequences of the genes from the other Korean strains (K. pneumoniae 96062, E. coli 960402, and S. sonnei 153) were identical to those of K. pneumoniae 95151.
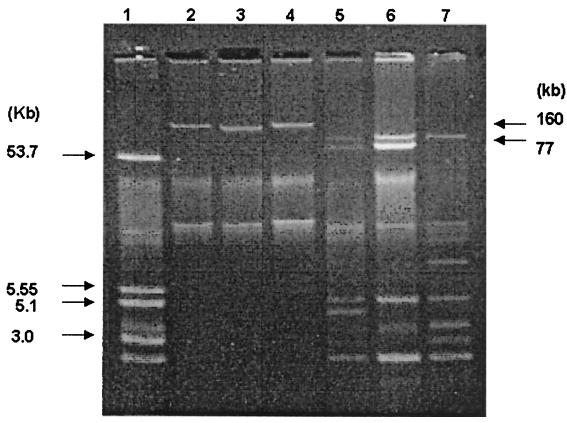
To compare the resistance plasmids of the four strains, plasmid analyses were performed. Cefotaxime resistance was transferred by conjugation at frequencies of 10−3 to 10−4 per donor from each strain. Plasmids from strains 96062 (pMG269) and 960402 (pMG270) mediated resistance to β-lactams, streptomycin, and sulfonamide, whereas plasmid pMG267 carried resistance to amikacin, chloramphenicol, gentamicin, streptomycin, sulfonamide, tetracycline, and trimethoprim as well as to β-lactams. S. sonnei 153, however, transferred cefotaxime resistance alone. Plasmids from the transconjugants were extracted and separated by electrophoresis (6). Transconjugants from strains 960402, 96062, and 95151 each had one plasmid with a size of about 160 kb. S. sonnei 153 contained nine plasmids of different sizes, one of which was a plasmid of 77 kb (pMG271) that was most likely responsible for cefotaxime resistance (Fig. 2).
FIG. 2.
Plasmid agarose gel electrophoresis. Lanes: 1, plasmid size standards from E. coli strain V517 (5); 2, E. coli J53 Azir/pMG270; 3, E. coli J53 Azir/pMG269; 4, E. coli J53 Azir/pMG267; 5, S. sonnei 153; 6, S. sonnei 153 × E. coli J53 Azir transconjugant selected with cefotaxime; 7, S. sonnei 153 × E. coli J53 Azir transconjugant selected with ampicillin. The lack of cefotaxime resistance suggests that the cefotaxime resistance gene was carried on the missing 77-kb plasmid.
The MICs of the β-lactams were determined using Etest strips (AB Biodisk, Dalvägen, Sweden). Similar values were found for all strains (Table 1). The strains all had high levels of resistance to cefotaxime but much less resistance to ceftazidime or aztreonam. The MICs of amoxicillin decreased more than eightfold for each strain when the drug was combined with clavulanic acid.
TABLE 1.
Antimicrobial susceptibility of clinical isolates, transconjugants, and strains with cloned genesa
| Strain | MIC (μg/ml) of:
|
||||||
|---|---|---|---|---|---|---|---|
| AMX | AMC | ATM | CAZ | CEF | CTX | FOX | |
| K. pneumoniae 95151 | >256 | 12 | 12 | 4 | >256 | >256 | 4 |
| E. coli J53/pMG267 | >256 | 8 | 8 | 3 | >256 | >256 | 6 |
| E. coli DH10B/pMG268 | >256 | 4 | 8 | 3 | >256 | >256 | 8 |
| K. pneumoniae 96062 | >256 | 8 | 12 | 2 | >256 | >256 | 4 |
| E. coli J53/pMG269 | >256 | 8 | 12 | 3 | >256 | >256 | 4 |
| E. coli 960402 | >256 | 12 | 6 | 2 | >256 | >256 | 6 |
| E. coli J53/pMG270 | >256 | 12 | 2 | 1.5 | >256 | >256 | 4 |
| S. sonnei 153 | >256 | 24 | 6 | 1 | >256 | >256 | 2 |
| E. coli J53/pMG271 | >256 | 24 | 6 | 1 | >256 | >256 | 2 |
Abbreviations: AMX, amoxicillin; AMC, amoxicillin-clavulanic acid; ATM, aztreonam; CAZ, ceftazidime; CEF, cephalothin; CTX, cefotaxime; FOX, cefoxitin.
A search of GenBank detected two as yet unpublished β-lactamase gene sequences identical to that found in the Korean isolates: one in an E. coli isolate from the People's Republic of China (accession number AF252622) and the other in an E. coli isolate from Japan (accession number AF311345). The prevalence of this enzyme in these countries is not known. In our study, we identified strains of different genera producing CTX-M-14 β-lactamase in different parts of the country over a 5-year period, suggesting dissemination throughout Korea. This β-lactamase is also the first CTX-M-type extended-spectrum β-lactamase (ESBL) to be identified in Korea.
In 2000, an epidemic of S. sonnei gastroenteritis occurred on Cheju Island. During the outbreak, several strains of S. sonnei resistant to expanded-spectrum cephalosporins were isolated, including strain 153, which was studied here. The widespread use of ceftriaxone during the epidemic may be the reason for the emergence of Shigella species that produced the CTX-M-type ESBL. Until now there have been few reports of ESBL-producing Shigella species. In India, an SHV-11-producing Shigella dysenteriae strain was isolated (1), and in Japan, a Shigella flexneri strain which produced a metallo-β-lactamase has been identified (8). This is the third report of ESBL-producing Shigella species strains and the first report of such a strain producing an enzyme of the CTX-M type.
Acknowledgments
This work was supported in part by a Merit Review award from the VA/DoD Collaborative Research Program on Mechanisms of Emerging Pathogens.
REFERENCES
- 1.Ahamed J, Kundu M. Molecular characterization of the SHV-11 β-lactamase of Shigella dysenteriae. Antimicrob Agents Chemother. 1999;43:2081–2083. doi: 10.1128/aac.43.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim J, Kwon Y, Pai H, Kim J W, Cho D T. Survey of Klebsiella pneumoniae strains producing extended-spectrum β-lactamases: prevalence of SHV-12 and SHV-2a in Korea. J Clin Microbiol. 1998;36:1446–1449. doi: 10.1128/jcm.36.5.1446-1449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma L, Ishii Y, Ishiguro M, Matsuzawa H, Yamaguchi K. Cloning and sequencing of the gene encoding Toho-2, a class A β-lactamase preferentially inhibited by tazobactam. Antimicrob Agents Chemother. 1998;42:1181–1186. doi: 10.1128/aac.42.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macrina F L, Kopecko D J, Jones K R, Ayers D J, McCowen S M. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978;1:417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- 6.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 7.Mollet B, Iida S, Arber W. An active variant of the prokaryotic transposable element IS903 carries an amber stop codon in the middle of an open reading frame. Mol Gen Genet. 1985;199:534–536. doi: 10.1007/BF00330770. [DOI] [PubMed] [Google Scholar]
- 8.O'Hara K, Haruta S, Sawai T, Tsunoda M, Iyobe S. Novel metallo beta-lactamase mediated by a Shigella flexneri plasmid. FEMS Microbiol Lett. 1998;162:201–206. doi: 10.1111/j.1574-6968.1998.tb12999.x. [DOI] [PubMed] [Google Scholar]
- 9.Pai H, Lyu S, Lee J H, Kim J, Kwon Y, Kim J W, Choe K W. Survey of extended-spectrum β-lactamases in clinical isolates of Escherichia coli and Klebsiella pneumoniae: prevalence of TEM-52 in Korea. J Clin Microbiol. 1999;37:1758–1763. doi: 10.1128/jcm.37.6.1758-1763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabaté M, Tarragó R, Navarro F, Miró E, Vergés C, Barbé J, Prats G. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob Agents Chemother. 2000;44:1970–1973. doi: 10.1128/aac.44.7.1970-1973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]