Abstract
The Coronavirus disease 2019 (COVID19) pandemic has led to a dramatic loss of human life worldwide and caused a tremendous challenge to public health. Immediate detection and diagnosis of COVID19 have lifesaving importance for both patients and doctors. The availability of COVID19 tests increased significantly in many countries, thereby provisioning a limited availability of laboratory test kits Additionally, the Reverse Transcription-Polymerase Chain Reaction (RT-PCR) test for the diagnosis of COVID 19 is costly and time-consuming. X-ray imaging is widely used for the diagnosis of COVID19. The detection of COVID19 based on the manual investigation of X-ray images is a tedious process. Therefore, computer-aided diagnosis (CAD) systems are needed for the automated detection of COVID19 disease. This paper proposes a novel approach for the automated detection of COVID19 using chest X-ray images. The Fixed Boundary-based Two-Dimensional Empirical Wavelet Transform (FB2DEWT) is used to extract modes from the X-ray images. In our study, a single X-ray image is decomposed into seven modes. The evaluated modes are used as input to the multiscale deep Convolutional Neural Network (CNN) to classify X-ray images into no-finding, pneumonia, and COVID19 classes. The proposed deep learning model is evaluated using the X-ray images from two different publicly available databases, where database A consists of 1225 images and database B consists of 9000 images. The results show that the proposed approach has obtained a maximum accuracy of 96% and 100% for the multiclass and binary classification schemes using X-ray images from dataset A with 5-fold cross-validation (CV) strategy. For dataset B, the accuracy values of 97.17% and 96.06% are achieved using multiscale deep CNN for multiclass and binary classification schemes with 5-fold CV. The proposed multiscale deep learning model has demonstrated a higher classification performance than the existing approaches for detecting COVID19 using X-ray images.
Keywords: COVID19, X-ray, Pneumonia, No-findings, FB2DEWT, Multiscale deep CNN
1. Introduction
COVID19, now called severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), has proliferated worldwide. It was first spotted in Wuhan City, Hubei Province, China [1]. After causing nearly 17,205 cases in China [2], [3], it began to proliferate to the Philippines, India, US, UK, Russia, and other countries, eventually causing more than 4, 999, 530 deaths worldwide [2], [4]. Coronavirus is a pathogenic disease that starts by invading the respiratory tract and then guides itself into the cells using an enzyme called furin and ultimately impairs the immune system [5]. Some common symptoms of COVID19 are shortness of breath, chest pain or tightness, a deeper cough, and other breathing difficulties [5], [6]. A molecular test, namely RT-PCR, is used to detect the presence of this pathogen. Even though the RT-PCR test became the standard confirmatory clinical test for detecting COVID19 infection, the limited availability of its test kits and hospital experts created restrictions in diagnosing the infected, who required immediate isolation. The costs of the test kits, especially in developing and underdeveloped countries, are a considerable obstacle for testing in this pandemic. Due to this, chest radiologic examinations using computed tomography (CT) and X-ray as alternatives have become necessary for the early detection of COVID19 [7], [8]. A study on Chest X-ray (CXR) of 88 patients confirmed with COVID19 described the temporal changes of the chest radiological findings throughout the disease course [9]. It concluded that almost half of the patients with COVID19 showed abnormal CXR, and the abnormalities correlated significantly with symptoms. Further studies revealed distinct visual characteristics such as multi-focal, bilateral ground-glass opacities (GGO), and patchy reticular opacities as the most common findings in the CXR of the infected patients [9], [10]. Therefore, these features deem CXRs viable for the analysis and diagnosis of COVID19. CXR is also readily available at most medical centers and has a faster turnaround time during laboratory examination, which adds to its advantages for use.
Artificial Intelligence (AI) is an ever-expanding field and has been proven to show stable and accurate results when analyzing and diagnosing various diseases. With the combination of AI with image analysis, the diagnostic power has been remarkable [11]. Deep learning is one of the fields in AI which has gained importance in the past few years. The image-based classification has advanced to new peaks using hidden layers and deep convolutional networks (CNNs or ConvNets). When we consider the case of COVID19, the use of feature extraction and image classification can be used in the analysis of CXRs [12]. Radiologists, nurses, and hospital staff play a critical role in the diagnosis of COVID19. However, a radiologist can miss these parameters due to the excessive load of patients at the hospitals, which creates fatigue and a need to complete the diagnosis faster and efficiently. Hence, there is a need for the deep learning model for better and faster analysis of X-ray images to detect COVID19.
The residual sections of this paper are organized as follows. The existing methods for the detection of COVID19 using X-ray images are discussed in Section 2. In Section 3, the motivations and the contributions of this paper are written. Section 4 elaborates on the different X-ray datasets used in this study. In Section 5, the proposed approach is described. Section 6 explains the results obtained and the discussions of the results. Finally, Section 7 concludes the paper.
2. Related works
Since the last few decades, various computer-aided detection systems have taken a noticeably significant step, especially in the medical domain. Many applications, particularly detection and diagnosis, pertaining to the medical field have been performed using several AI-based deep learning algorithms. AI has been successful in recent times for the diagnosis of poultry disease [13], osteoporosis [14], breast cancer [15], cardiovascular disease [16], and plant disease [17], respectively. The pandemic has necessitated the need for computer-aided deep learning-based COVID19 detection systems. In this regard, many researchers have developed various machine learning and deep learning models using both X-ray and CT scan images for timely detection. As X-ray tests are cheaper than CT scan tests, it is economical to detect COVID19 using CXR images feasible with higher performance. Afshar et al. have developed a capsule network-based framework (COVIDCAPS) on an X-ray dataset and have arrived at accuracy and specificity of 95.7% and 95.8%, respectively [18]. Such models can efficiently handle the availability of even limited datasets. Similarly, other models have been built using ResNet50, ResNet101, ResNet152, InceptionV3, Inception-ResNetV2 achieving a maximum accuracy of 99.7% produced by ResNet50 for binary classification [19]. Sethy et al., [20] have separated the COVID19 positive cases from the others using the support vector machine (SVM) with learnable features from X-ray images and have successfully achieved an accuracy of 95.38%. Their research has stated that SVM combined with ResNet50 produces fairly superior results.
Furthermore, various researchers have introduced a deep convolutional neural network design on CXR images providing accurate yet practical results [21]. A deep learning model, namely COVIDX-Net, was done on 50 CXR images with 25 COVID19 cases by Hemdan et al. [22]. The model consisted of the architecture of seven deep convolutional neural network (CNN) models. Apostolopoulos et al. have achieved an accuracy of 98.75% and 93.48% for binary (COVID19 and no-finding or normal classes) and multi-class (common pneumonia, COVID19, and normal classes), respectively [23]. Their deep learning model used transfer learning for the classification of 1427 X-ray images. Horry et al. have performed detection through transfer learning using multimodal imaging data [24]. The chosen VGG19 based transfer learning model was fine-tuned with appropriate parameters and reached a precision of 86% for X-ray (three classes), 100% for Ultrasound (three classes), and 84% for CT scans (two classes). The model developed by Ozturk et al. has obtained an accuracy of 98.08% and 87.02% for binary and multi-classes using the DarkCovidNet model [25]. The model has a total of 17 CNN layers with separate filters on each layer. Another hybrid model by Altan and Karasu has been developed by applying 2D curvelet transform on the CXR images, the output coefficients were optimized using CSSA, and the final detection was performed using the EfficientNet-B0 model [26]. Tsiknakis et al. have used transfer learning for the classification of COVID19 and normal X-ray images [27]. They have obtained an overall area under the Receiver Operating Characteristics (ROC) curve as 1. Several deep learning models are used to detect COVID19 patients using CT scans and ultrasound images [27], [28], [29], [30], [31], [32].
In the past few years, the learnable feature extraction and classification of images have gained much importance in deep learning. Empirical Wavelet Transform (EWT), developed by Gillies [33], is a technique that uses an adaptive wavelet subdivision scheme to create a multiresolution analysis (MRA) of a signal. It is used to design a filter-bank, which is formed based on the segregation of the Fourier spectrum of the non-stationary signals by detecting the boundary points [34]. The evaluated filter-bank is used to compute the modes of a signal or image. The fixed boundary-based EWT has already been used to extract modes from ECG signals [35], [36], [37]. The EWT with fixed boundary points reduces the computational complexity as the detection of the peaks in the Fourier spectrum of the signal is not required [34]. The multiscale convolutional neural network (CNN) developed by considering these modes of ECG signals has shown better performance for the detection of cardiac arrhythmia [34]. The fixed boundary-based EWT has not been explored for image analysis. Therefore, the deep CNN model can be applied in the EWT domain of X-ray images for the automated detection of COVID19.
3. Motivation and contributions
Multiscale deep CNN architecture is adopted in this study for the diagnosis of COVID19. This model essentially takes in information from different modes and then combines it to produce promising results. The modes, or channels, extracted from a particular image are semantically correlated and provide complementary information with respect to each other. Due to the above characteristics, they tend to reflect information that is unique to each other. The proposed model consolidates these heterogeneous modes, producing a more robust diagnosis for the detection of COVID19. The proposed approach uses X-ray images for the classification due to its cost-effectiveness and easy availability. Moreover, CT scans are more expensive and are not readily available due to their availability in only large healthcare centers [27]. The multiscale gated multi-head attention-based CNN model has been used to detect COVID 19 using X-ray and CT scan images [38]. However, the deep CNN model in the empirical wavelet-based multiresolution domain of X-ray images has not been explored to detect COVID19. The novelty of this work is to develop a new multiscale two-dimensional (2D) deep CNN model to detect COVID19 using X-ray images. The significant contributions of this paper are as follows.
-
1.
Fixed boundary-based two-dimensional Empirical Wavelet Transform (FB2DEWT) filter bank is used to extract modes or sub-band images from the CXR images.
-
2.
A novel multiscale deep CNN architecture is proposed to detect COVID19 patients using various modes of the CXR images.
-
3.
The performance of the proposed architecture is compared for both binary and three-class classification of X-ray images for different combinations of the modes.
-
4.
The model is evaluated using two different datasets, consisting of 1125 and 9000 images each, to ensure the robustness of the model.
4. X-ray image datasets
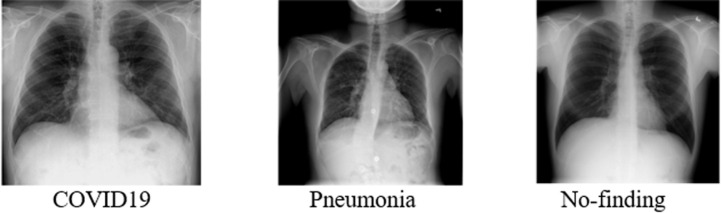
Two publicly available CXR image databases are used in the present work. The first database (dataset A) contains 1125 images, of which 125 were COVID19 images, 500 were pneumonia images, and the remaining 500 were No-Findings images. The 125 COVID19 images are obtained from Cohen’s Covid-CXR dataset, which has been developed by collecting the images from various publications and researchers to maintain the quality of the images [39]. For Pneumonia and No-Findings, 500 images for each class are obtained from Wang et al.’s CXR image dataset [40]. The dataset B contains 9000 images, with the issue of class imbalance in mind; 3000 CXR images are taken for each class of COVID19, No-Findings, and Pneumonia. The COVID19 and no-findings CXR images are obtained from the dataset created by Rahman, T. [41], [42]. He had developed the database of COVID19 CXR images from the Italian Society of Medical and Interventional Radiology (SIRM) COVID-19 database, Novel Coronavirus 2019 Dataset developed by Joseph Paul Cohen and Paul Morrison, and Lan Dao [39]. For Pneumonia, the CXR images are obtained from the large dataset of labeled Optical Coherence Tomography (OCT) and chest X-ray Images developed by Kermany, D. [43]. The X-ray image of each class in dataset A is shown in Fig. 1. The CXR images from the different dataset sources had different resolutions and file formats. For the sake of standardization, all the images in both datasets are converted into Joint Photographic Experts Group (JPEG) format for further processing.
Fig. 1.
CXR images from the dataset A for COVID19, Pneumonia, and No-finding classes.
5. Proposed approach
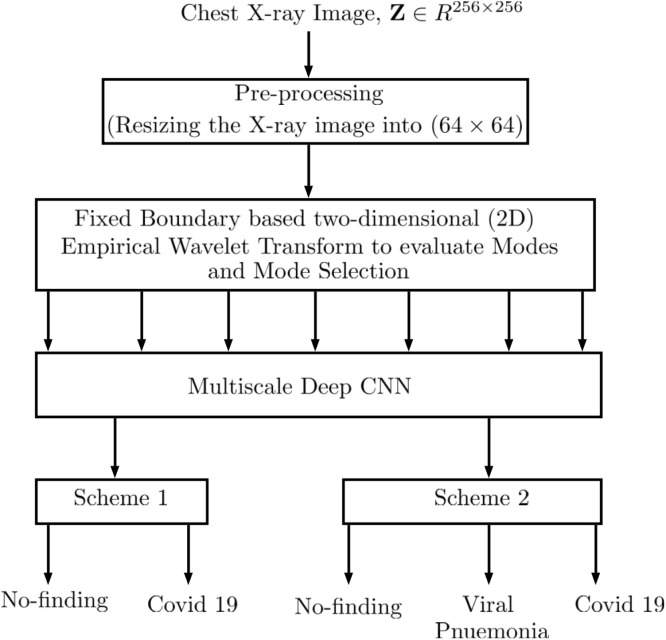
A block diagram of the proposed COVID19 detection model is illustrated in Fig. 2. The approach comprises the pre-processing of the X-ray images. Then, the images are decomposed into seven modes, each containing information unique to each other, using EWT. Further, various combinations of modes are selected randomly as the input to a multiscale deep CNN. These modes are then used as the input to the proposed model for the detection of COVID19. The detection part is split into two schemes. Scheme 1 includes the binary classification where the model considers only two classes, namely COVID19 and No-findings. Scheme 2 is the classification of the image dataset into three classes, namely COVID19, Pneumonia, and No-findings. The above approach was adopted by two different datasets: dataset A (consisting of 1125 images) and dataset B (9000 images).
Fig. 2.
Block-diagram of the proposed approach for the detection of COVID19.
5.1. Pre-processing
The original X-ray images in both databases have the size of 256 × 256. In this work, we have resized the X-ray images of each class to [25]. The image resizing helps to reduce the computational time of the proposed multiscale deep CNN model during training.
5.2. FB2DEWT for X-ray image decomposition and mode selection
In this work, we have considered the FB2DEWT to decompose each X-ray image X(, ) into modes as shown in Fig. 3. In the first step of two-dimensional EWT (2DEWT), the pseudo-polar Fast Fourier Transform (FFT) of the input image is evaluated and it is denoted as [44]. After evaluating the pseudo-polar FFT, the average spectrum is computed as follows [44]:
| (1) |
where ‘m’ is denoted as the number of phase angles. The boundary points can be evaluated using any of the boundary detection techniques such as local maxima based, local minima based, scale-space, etc. [33] using the average spectrum, . In this study, we have considered the fixed boundary points such as [, , , , , ] [0.5, 1, 1.5, 2, 2.5, 3] in the average spectrum to decompose each X-ray image into modes [45]. Once the boundary points are assigned, the 2D empirical Littlewood–Paley and Mayer wavelets are used to evaluate both 2D scaling and 2D wavelet functions [44]. The empirical 2D scaling function is written as Eq. (2) in Box I [44].
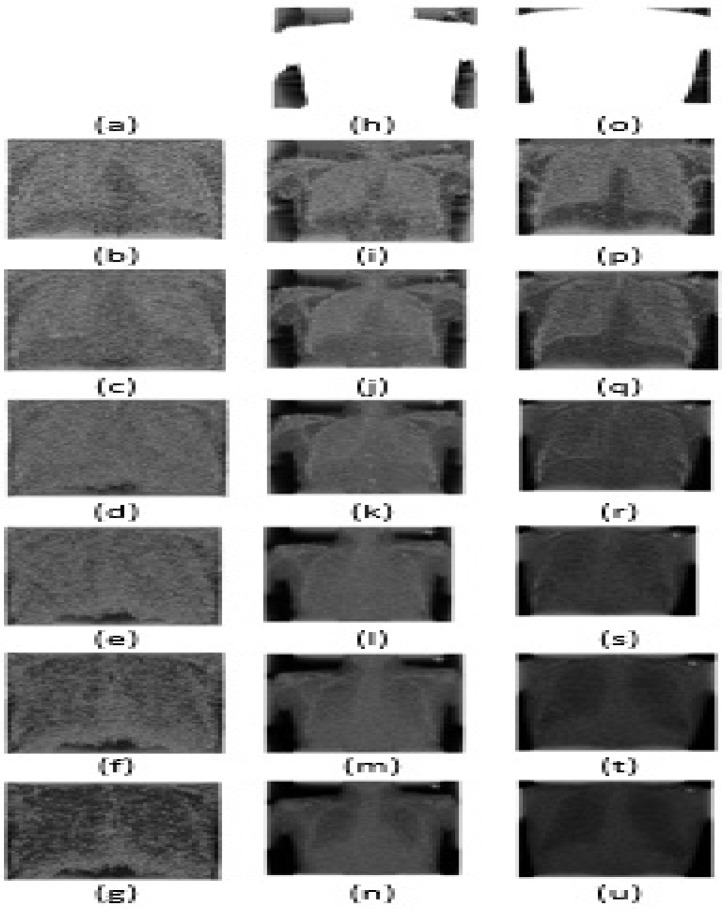
Fig. 3.
The output of seven modes of a CXR image from dataset A obtained using fixed boundary based EWT for all the classes (COVID19, Pneumonia, No findings): (a) COVID19 mode-1. (b) COVID19 mode-2. (c) COVID19 mode-3. (d) COVID19 mode-4. (e) COVID19 mode-5. (f) COVID19 mode-6. (g) COVID19 mode-7. (h) Pneumonia mode-1. (i) Pneumonia mode-2. (j) Pneumonia mode-3. (k) Pneumonia mode-4. (l) Pneumonia mode-5. (m) Pneumonia mode-6. (n) Pneumonia mode-7. (o) No-findings mode-1. (p) No-findings mode-2. (q) No-findings mode-3. (r) No-findings mode-4. (s) No-findings mode-5. (t) No-findings mode-6. (u) No-findings mode-7.
Box I.
| (2) |
The empirical 2D scaling function is used to evaluate the first mode of the X-ray image with the foundry range as [0, K1]. Similarly, the remaining other modes of X-ray images are computed using empirical 2D wavelet function. This empirical 2D wavelet function is given as Eq. (3) in Box II [44].
Box II.
| (3) |
Where are the index for the boundary points. Similarly, the parameter ‘’ is interpreted as mode number and it is given as . The boundary ranges evaluated from the boundary points for the design of 2D wavelet functions are given as [, ], [, ], [, ], [, ], and [, ], respectively. As six boundary points are considered as prior, hence we can evaluate 7 modes from the X-ray image. For obtaining the last modes, the wavelet function is written as Eq. (4) in Box III [44].
Box III.
| (4) |
The boundary range for the seventh mode is given as [, ]. The first mode is evaluated by considering the 2D scaling function is given as follows [44]:
| (5) |
Similarly, the second to seventh modes are computed as follows:
| (6) |
Where is denoted as the mode. The ( (, )) (, ) is termed as the pseudo-polar FFT of the X-ray image [44]. Similarly, the F−1 is called the inverse pseudo-polar FFT. The modes evaluated using the FB2DEWT of X-ray images for COVID19, pneumonia, and no-finding classes are shown in Figs. 3(a), (d), (g), (j), (m), (p), (s), 3(b), (e), (h), (k), (n), (q), (t), and 3(c), (f), (i), (l), (o), (r), (u), respectively. The X-ray image information for each class is divided into different modes based on the frequency range. These modes of X-ray images can be used as input to the deep CNN model for the detection of COVID19.
5.3. Multi-scale deep CNN
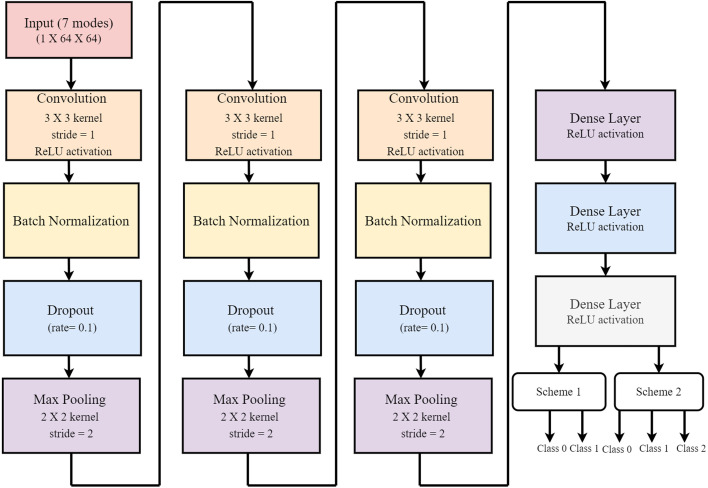
Although deep neural networks eliminate the need for dedicated feature extraction, information aggregation from different modalities can improve the network’s predictive ability. Therefore, while designing the deep model used in this study, multiscale deep CNN is adopted [34]. A unique classification model is developed in this work to diagnose the COVID19 using two different datasets, containing 1125 images and 9000 images. All the learnable features, from a combination of different modes, are combined and merged at a fully connected layer to predict the output. The robustness of the proposed multiscale deep CNN model is evaluated using two datasets. In this deep CNN model, the modes are fed to the different convolution layers as channels individually. Each mode is passed through 14 layers (block) individually and then merged with additional 4 layers. Each block has one input layer followed by a convolution layer, batch normalization, dropout, and max-pooling layer, and the same setup is repeated three times in a successive form. These blocks are then merged using a concatenate layer, subsequently following two dense layers and one softmax layer for the correct diagnosis. Each layer parameter and the feature map size are shown in the deep CNN architecture block diagram in Fig. 4. The feature map of the th convolution layer is computed as [46],
| (7) |
where is the output feature map from the convolution layer, represents the 2-dimensional kernel that evaluates the feature map, corresponds to the feature map output of the previous layer. p and r denote the size of the 2D kernel (K), is the bias, and corresponds to the ReLU activation. Followed by the 2D convolution layer is the batch normalization (BN) layer. This layer helps standardize the inputs as well as accelerates training [47]. This is essentially done to put our output data into a known scale, mainly between 0 and 1. In many cases, this can also provide some regularization to reduce generalization error. BN is computed by normalizing the output obtained from Eq. (7) [46],
| (8) |
where is the new value of a single component, is the mean within the respective batch and is the standard deviation within a batch. After the Batch normalization layer, the dropout layer is used for the proposed multiscale deep CNN model [47]. The function of the dropout operation is to decorrelate the weights in order to prevent the neurons from converging to the same goal (overfitting) [47]. After the dropout layer, the pooling layer feature map is evaluated as [46],
| (9) |
where . Max[•] is the maximum value of the elements in the feature map for the given range. One block consists of the above computation successively done three times. Later the blocks of all the modes are combined, and the output of the merged layers is flattened to formulate a feature vector (V). In our model, the output layer consists of three neurons, each representing one class, and the output is evaluated, using a softmax function, as [46],
| (10) |
where is the weight vector of the neuron of the output layer, and v is the feature vector for the previous fully connected layer. The proposed model is trained using 70 epochs for dataset A and 50 epochs for dataset B for both the schemes (binary and multiclass). Both the datasets use the 5-fold cross-validation (CV) approach to infer the robustness of the developed model, while the addition of holdout validation is adopted for dataset B [47]. The Hyperparameters of the proposed deep CNN model for dataset A and dataset B are given in Table 1. To compare the performances of the multiscale deep CNN models with different combinations of modes, we have used accuracy, precision, recall, and F1-score measures, which are calculated from the confusion matrix [48]. These metrics are calculated over 70 epochs with 5-folds cross-validation for dataset A and 50 epochs with both holdout validation and 5-folds CV for dataset B [48]. The validation and training accuracies are maximum, with epochs being 50 and 70 for dataset A and dataset B, respectively.
Fig. 4.
Proposed multiscale deep CNN architecture developed for the detection of COVID19 (scheme 1: class 0: No-finding, class 1: Covid19, scheme 2: class 0: No-finding, class 1: Viral pneumonia, class 2: COVID19).
Table 1.
Hyperparameters table for the proposed multiscale deep CNN for the classification of COVID19.
| Hyperparameters | Values for dataset A | Values for dataset B |
|---|---|---|
| COVID19 instances | 125 | 3000 |
| Pneumonia instances | 500 | 3000 |
| No-Findings instances | 500 | 3000 |
| Learning rate | 0.001 | 0.001 |
| Batch size | 64 | 64 |
| Epochs | 70 | 50 |
| Optimizer | Adam | Adam |
| Loss function | Sparse categorical cross-entropy | Sparse categorical cross-entropy |
In our study, both multi-class and binary classification schemes are performed using the CXR dataset. For multi-class classification, three classes such as COVID19 (cases having COVID19), pneumonia (cases having pneumonia but not COVID19), and no-Findings (cases that do not have COVID19 or Pneumonia) are formulated [26], [31]. The positives and negatives of the classification change concerning the class that we are referencing. For example, if the precision of pneumonia is to be determined, the positives will be the pneumonia cases, and the negatives will be the Non-Pneumonia (which includes the COVID19 and the no-Finding cases). Here, true positive (TP) and true negative (TN) will be the number of correctly diagnosed Pneumonia and Non-Pneumonia cases, respectively. In contrast, false-positive (FP) and false-negative (FN) will represent the number of incorrectly diagnosed Pneumonia and Non-Pneumonia cases, respectively [26], [31]. Similarly, the performance indices of all three classes are calculated. For binary classification, two classes of COVID19 and no-Findings were formed. In this, the positives and negatives were assigned to COVID19 and no-Finding (non-COVID19) cases, respectively. Hence, TP and TN represent the number of correctly diagnosed COVID19 and non-COVID19 infections, respectively. At the same time, FP and FN represent the number of incorrectly diagnosed COVID and non-COVID19 infections, respectively. An ANOVA test is performed to determine the statistical significance of the obtained results using the proposed model [47].
6. Results and discussions
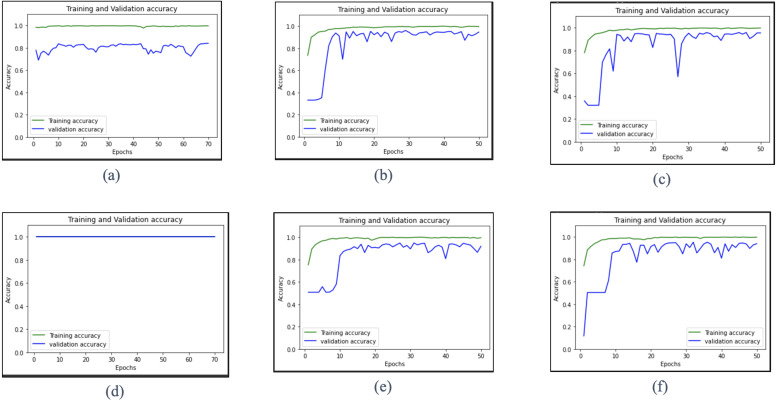
This section presents the results obtained using the proposed multi-scale deep CNN model with the modes extracted from the FB2DEWT based decomposition of CXR images for both datasets. The proposed model based on two datasets, two classifying instances, and two CV parameters gave us six different sets of results. These results are given as multi-class and binary classification of dataset A with 5-fold CV, multi-class and binary classification of dataset B with 5-fold CV, and multi-class and binary classification of dataset B with hold-out validation, respectively. The hold-out cross-validation for dataset A is not considered because the size was considerably small, and the presence of a class imbalance would have fetched over-fitted results. The accuracy versus epoch plots obtained based on the training and validation of the multi-scale deep CNN models for all six different sets of results are shown in Fig. 5. Here, for the 5-fold CV, we have shown the accuracy vs. epoch plot for one fold. It has been observed that the training accuracy values are obtained as 100% for both binary and multi-class classification tasks using the proposed multi-scale deep CNN models with different combinations of modes. From Fig. 5 (d), it is seen that both training and validation accuracy values are obtained as 100% at each epoch.
Fig. 5.
Accuracy versus epoch graphs evaluated using multi-scale deep CNN for training and validation data of the best results obtained from the 6 models: (a) multi-class classification of dataset A with 5-fold CV using mode 3 and mode 4. (b) multi-class classification of dataset B with 5-folds using mode 4, mode 5 and mode 6. (c) multi-class classification of dataset B with hold-out validation using mode 4, mode 5 and mode 6. (d) binary classification of dataset A with 5-fold CV using mode 1 and mode 2. (e) binary classification of dataset B with 5-fold CV using all seven modes. (f) binary classification of dataset B with hold-out validation using the first six modes.
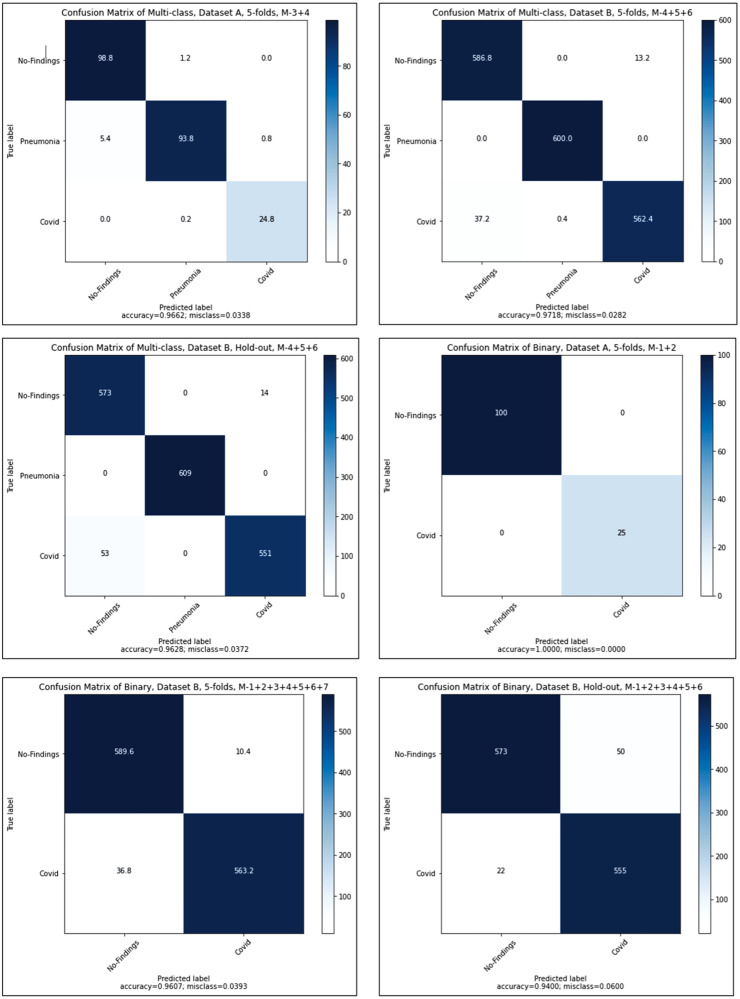
Fig. 6 shows the confusion matrix plots for multi-scale deep CNN model with hold-out validation and 5-fold CV for different combinations of the modes of X-ray images. It is observed from these results that for all cases, the accuracy values are obtained as more than 0.94. For the binary-classification scheme (No-finding versus COVID19), the multi-scale deep CNN model has achieved 100% accuracy using the combination of mode 1 and mode 2 of X-ray images. In the below two tables, the notations such as ACC denotes the testing accuracy, PRE represents the precision, REC shows the recall or sensitivity of the model, SPE denotes the specificity, and F1 represents the obtained f1-score from the proposed model. Table 2 shows the multi-class classification performances of our model for different combinations of modes extracted from the obtained CXR datasets. It is observed that the combination of mode 3 and mode 4, has achieved the best performance for COVID19 and Pneumonia detection (testing accuracy of 0.96) in dataset A using 5-fold CV. It is also noticed that the combination of mode 4, mode 5, and mode 6 of X-ray images has produced the maximum accuracy values of 0.97 and 0.96 using 5-fold CV and hold-out validation, respectively, when the model is evaluated using dataset B. It can also be seen from the table that as the number of modes considered for the multi-scale CNN model decreases, the testing accuracy vaguely increases. The mode selection criteria helped select the discriminative local information of X-ray images at different frequency ranges, thereby increasing the classification performance of multi-scale deep CNN for the multi-class classification task. The maximum time for execution for the multi-scale deep CNN model is 4205.29 s for the multi-class classification task.
Fig. 6.
Confusion matrices of the best results obtained from the 6 models.
Table 2.
Classification performance of multi-scale deep CNN for the multi-class classification task.
| Multi-class classification of dataset A (5-fold cross-validation) |
Multi-class classification of dataset B (5-fold cross-validation) |
Multi-class classification of dataset B (Hold-out cross-validation) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Modes | ACC | PRE | REC | F1 | ACC | PRE | REC | F1 | ACC | PRE | REC | F1 |
| M-1+2 | 0.93 | 0.94 | 0.94 | 0.93 | 0.94 | 0.94 | 0.94 | 0.94 | 0.93 | 0.93 | 0.93 | 0.93 |
| M-3+4 | 0.96 | 0.96 | 0.96 | 0.96 | 0.95 | 0.95 | 0.96 | 0.96 | 0.93 | 0.93 | 0.93 | 0.93 |
| M-5+6 | 0.90 | 0.87 | 0.95 | 0.90 | 0.96 | 0.96 | 0.96 | 0.96 | 0.89 | 0.89 | 0.89 | 0.89 |
| M-6+7 | 0.86 | 0.88 | 0.89 | 0.86 | 0.95 | 0.95 | 0.95 | 0.95 | 0.92 | 0.92 | 0.92 | 0.92 |
| M-1+2+3 | 0.83 | 0.89 | 0.86 | 0.86 | 0.95 | 0.96 | 0.95 | 0.95 | 0.93 | 0.93 | 0.93 | 0.93 |
| M-4+5+6 | 0.87 | 0.91 | 0.90 | 0.89 | 0.97 | 0.97 | 0.97 | 0.97 | 0.96 | 0.96 | 0.96 | 0.96 |
| M-5+6+7 | 0.95 | 0.95 | 0.96 | 0.95 | 0.94 | 0.94 | 0.94 | 0.94 | 0.92 | 0.93 | 0.92 | 0.92 |
| M-1+2+3+4 | 0.78 | 0.87 | 0.82 | 0.82 | 0.93 | 0.94 | 0.93 | 0.93 | 0.95 | 0.95 | 0.95 | 0.95 |
| M-4+5+6+7 | 0.84 | 0.90 | 0.87 | 0.87 | 0.92 | 0.94 | 0.92 | 0.93 | 0.93 | 0.94 | 0.93 | 0.93 |
| M-1+2+3+4+5 | 0.88 | 0.91 | 0.89 | 0.89 | 0.95 | 0.96 | 0.95 | 0.95 | 0.75 | 0.85 | 0.74 | 0.71 |
| M-3+4+5+6+7 | 0.85 | 0.90 | 0.87 | 0.87 | 0.90 | 0.93 | 0.90 | 0.91 | 0.93 | 0.94 | 0.94 | 0.93 |
| M-2+3+4+5+6+7 | 0.86 | 0.90 | 0.88 | 0.88 | 0.93 | 0.91 | 0.93 | 0.94 | 0.87 | 0.88 | 0.87 | 0.87 |
| M-1+2+3+4+5+6+7 | 0.74 | 0.79 | 0.80 | 0.79 | 0.86 | 0.93 | 0.86 | 0.89 | 0.96 | 0.96 | 0.96 | 0.96 |
On the other hand, Table 3 shows the performance of the multi-scale deep CNN model for binary classification tasks using different combinations of modes. As can be seen, the model’s overall performance for binary classification exceeds that of the multi-class classification task. As perceived in the multi-class classification task, there is no specific trend for the binary classifications, and the accuracies remain consistent with the change in the number of modes considered. Combining all seven modes with multi-scale deep CNN has achieved a remarkable performance of training and testing accuracy of 1. The multi-scale deep CNN has shown a precision of 1, the sensitivity of 1, and the F1-score of 1 in detecting COVID19 using modes of the X-ray images from dataset A with a 5-fold CV. While testing with dataset B, an accuracy of 0.96 is obtained using multi-scale deep CNN with all seven modes for 5-fold CV. For holdout validation, the accuracy value of the proposed model is obtained as 0.93 using the combination of the first six modes of X-ray images. The maximum time taken for a model to execute is 1969.33 s for binary classification.
Table 3.
Classification performance of the multi-scale deep CNN for the binary classification task.
| Binary-class classification for dataset A (5-fold cross-validation) |
Binary-class classification for dataset B (5-fold cross-validation) |
Binary-class classification for dataset B (Hold-out cross-validation) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Modes | ACC | PRE | REC | F1 | SPE | ACC | PRE | REC | F1 | SPE | ACC | PRE | REC | F1 | SPE |
| M-1+2 | 1 | 1 | 1 | 1 | 1 | 0.92 | 0.93 | 0.92 | 0.92 | 0.88 | 0.89 | 0.9 | 0.89 | 0.89 | 0.91 |
| M-1+2+3 | 0.99 | 0.99 | 0.99 | 0.99 | 1 | 0.95 | 0.95 | 0.95 | 0.95 | 0.96 | 0.87 | 0.89 | 0.87 | 0.87 | 0.96 |
| M-1+2+3+4 | 0.99 | 0.99 | 0.98 | 0.98 | 1 | 0.93 | 0.93 | 0.93 | 0.93 | 0.95 | 0.92 | 0.92 | 0.92 | 0.92 | 0.91 |
| M-1+2+3+4+5 | 0.98 | 0.99 | 0.97 | 0.98 | 1 | 0.95 | 0.95 | 0.94 | 0.95 | 0.98 | 0.93 | 0.93 | 0.93 | 0.93 | 0.94 |
| M-1+2+3+4+5+6 | 0.99 | 0.99 | 0.98 | 0.98 | 1 | 0.94 | 0.94 | 0.94 | 0.94 | 0.97 | 0.93 | 0.94 | 0.94 | 0.94 | 0.91 |
| M-1+2+3+4+5+6+7 | 1 | 1 | 1 | 1 | 1 | 0.96 | 0.96 | 0.96 | 0.93 | 0.98 | 0.93 | 0.93 | 0.93 | 0.93 | 0.95 |
The objective of this work is to develop a multiscale deep CNN model for the detection of COVID 19 using X-ray images. Both the datasets used for the COVID19 classification in our study are customized as mentioned in Section 4. Hence, all the datasets considered for the diagnosis of the disease in the comparison study are different from the dataset taken for the proposed model. The summary of the comparison of the proposed work with existing techniques for the detection of COVID19 using X-ray images is shown in Table 4. It can be observed from the table that most of the models have adopted ResNet, Inception, VGG19 based transfer learning algorithms to detect COVID19 using X-ray images. In [18], the suggested model achieved an accuracy of 95.7% for the binary classification scheme as COVID19 vs. No-finding. In [19], authors have considered various standard models for their classification and have concluded that the ResNet50 has produced the highest performance among the other four used models. In [20], authors have successfully detected the presence of COVID19 using the ResNet50 plus SVM model and obtained an accuracy of 95.33% using X-ray images. The paper implies that SVM is more robust than other transfer learning approaches. In [22], COVIDX-Net achieves an accuracy of 90%. This model includes seven different architectures, such as VGG19 and MobileNet, and other models. Using transfer learning with convolutional neural networks [23], authors have extracted significant information from the X-ray images and obtained an accuracy of 96.78% for binary classification tasks. The DarkCovidNet model has obtained an accuracy of 98.08% for binary classification and 87.02% for multiclass classification tasks [25]. A rapid and high accuracy of 99.69% was observed in [26] using a new hybrid model consisting of 2D curvelet transform and chaotic optimization algorithm. The suggested transfer learning methodology (Inception-V3) trained on the ImageNet database was used for the effective diagnosis of COVID19 [27]. The model achieves an overall area under ROC of 1 for the binary classification task. The models mentioned above are compared with the proposed multi-scale deep CNN model, for multiclass and binary classification tasks [47]. Table 5, Table 6 show the ANOVA results of multi-class and binary-class classification tasks. Similarly, in Fig. 7, we have shown the graph of the computed ANOVA results.
Table 4.
Comparison of algorithms of the existing methods for the automated diagnosis of COVID19.
| Ref. no. | Method | Classification scheme | Accuracy | Recall/sensitivity | CV |
|---|---|---|---|---|---|
| Proposed model | COVID-19 vs. Pneumonia vs. No-findings (Dataset A) |
96% | 96.67% | 5-Fold | |
| COVID-19 vs. No-findings (Dataset A) |
100% | 100% | 5-Fold | ||
| COVID-19 vs. Pneumonia vs. No-findings (Dataset B) |
97.17% | 97.17% | 5-Fold | ||
| COVID-19 vs. No-findings (Dataset B) |
96.06% | 96% | 5-Fold | ||
| [18] | COVID-CAPS | COVID vs. non-COVID | 95.7% | 90% | Hold out |
| [19] | InceptionV3 | COVID-19 vs. No-finding | 96.2% | 97.1% | 5-Fold |
| ResNet50 | COVID-19 vs. No-finding | 96.1% | 91.8% | 5-Fold | |
| ResNet101 | COVID-19 vs. No-finding | 96.1% | 78.3% | 5-Fold | |
| ResNet152 | COVID-19 vs. No-finding | 93.9% | 65.4% | 5-Fold | |
| Inception-ResNetV2 | COVID-19 vs. No-finding | 94.2% | 83.5% | 5-Fold | |
| [20] | ResNet50 plus SVM | COVID-19 vs. Pneumonia vs. No-finding | 95.33% | 95.33% | Hold out |
| [21] | VGG-19 | COVID-19 vs. Pneumonia vs. No-finding | 82.24% | 83% | Hold out |
| ResNet-50 | COVID-19 vs. Pneumonia vs. No-finding | 90.67% | 90.6% | Hold out | |
| COVID-Net | COVID-19 vs. Pneumonia vs. No-finding | 93.34% | 93.3% | Hold out | |
| [22] | COVIDX-Net | COVID-19 vs. No-finding | 90% | 90% | Hold out |
| [23] | Transfer learning with convolutional neural networks | COVID-19 vs. Pneumonia vs. No-finding | 94.72% | 98.66% | 10-Fold |
| [25] | DarkCovidNet | COVID-19 vs. Pneumonia vs. No-finding | 87.02% | 85.35% | 5-Fold |
| COVID-19 vs. No-finding | 98.08% | 95.13% | 5-Fold | ||
| [26] | EfficientNet-B0 | COVID-19 vs. Pneumonia vs. No-finding | 95.24% | 93.61% | Hold out |
| 2D curvelet transform-EfficientNet-B0 | COVID-19 vs. Pneumonia vs. No-finding | 96.87% | 95.68% | Hold out | |
| [27] | Inception-V3 | COVID-19 vs. Pneumonia vs. No-finding | 85% | 94% | 5-Fold |
Table 5.
ANOVA test for multiclass classification from the comparison table.
| ANOVA table | |||||
|---|---|---|---|---|---|
| Source | SS | df | MS | F | Prob > F |
| Columns | 641.989 | 6 | 106.998 | 11.21 | 2.276 |
| Error | 267.31 | 28 | 9.547 | ||
| Total | 909.299 | 34 | |||
Table 6.
ANOVA test for binary classification from the comparison table.
| ANOVA table | |||||
|---|---|---|---|---|---|
| Source | SS | df | MS | F | Prob > F |
| Columns | 2107.95 | 3 | 702.652 | 52.46 | 1.682 |
| Error | 214.3 | 16 | 13.394 | ||
| Total | 2322.25 | 19 | |||
Fig. 7.
ANOVA test statistics plot for the multi-class and binary classification tasks. (a) ANOVA test graph for the comparison between the proposed model and the other models for multiclass classification, (b) ANOVA test graph for the comparison between the proposed model and the other models for binary classification.
From Table 5, Table 6 results, it is observed that the p values for multi-class and binary classification schemes are obtained as less than 0.001, proving that the null hypothesis () is true. The difference in the classification results of existing methods and proposed multi-scale deep CNN model are significant for both binary and multiclass based COVID classification schemes. The computational complexity of the proposed multiscale CNN is evaluated as follows. The model described in the study (as shown in Fig. 4) consists of three convolution layers, three batch normalization layers, three dropout layers, and three pooling layers for a single mode. The respective layers for each mode are combined through three dense layers. The computational complexity of the three convolution layers is calculated as [49], where and are defined as the spatial dimensions of the convolution layer out of the three layers. In this study, we are considering the 1D convolution layer, so =1. Similarly, and are the spatial dimensions of the kernel of the convolution layer. As the 1D kernel is considered in this study, so is equal to 1. The number of inputs and output feature maps of the convolution layer is denoted as and respectively. The computational complexities of a single batch normalization layer, dropout layer, and pooling layer are , , and respectively. where represents the size of the input to batch normalization layer, represents the size of the input to the dropout layer and represents the size of the input to the pooling layer. Similarly, for the dense layer, the computational complexity is calculated as [49]. Here and are the size of the weight matrix in the dense layer and is the number of output neurons in the dense layer. The overall computational complexity of the model depends on the number of modes considered for the evaluation. Thereby, the computational complexity of the proposed deep CNN model for a single-mode is calculated as follows
Compared to VGG16, ResNet-based models, the proposed multi-scale CNN model has lower computational complexity due to less convolution and dense layers. The advantages of the proposed COVID detection work are as follows:
-
1.
Fixed boundary-based EWT is used as a pre-processing step for the mode extraction of the medical images.
-
2.
A new deep CNN architecture is projected in the multiscale domain for the diagnosis of COVID19.
-
3.
The proposed model may assist medical experts in the correct diagnosis of the virus.
The proposed model can also be used for the diagnosis of COVID19 using CT scans. However, CT is a more expensive process and is not readily available as it is used in only larger health care centers [25]. Moreover, there is no point in conducting CT scans for mild cases since one may find patches in CT scans even if the subject is asymptomatic [50]. For such cases, X-rays would be the optimal method for diagnosis. The subjects who are detected positive by this model can be further tested using appropriate medical techniques without delay. In addition, those who are tested negative by the model can be restricted from undergoing RT-PCR thereby eliminating the problem of medical kit shortage. Furthermore, the proposed COVID19 detection system can be implemented on a cloud-based platform for automated patient monitoring and helps in the immediate reinstatement of the affected patients.
7. Conclusions
In this paper, an intelligent healthcare model has been developed to detect COVID19 using EWT and multiscale deep CNN. Our model has been constructed using two different publicly available datasets, where dataset A consists of 1125 images and dataset B consists of 9000 images. The model has been compiled using 5-fold CV for dataset A and both 5-fold CV and holdout validation for dataset B. The X-ray Ray images have been first split into seven modes using the FB2DEWT filter-bank. A deep CNN model coupled with the extracted modes of an image has been used to detect COVID19 disease. The proposed model has been successfully able to differentiate COVID19 from viral pneumonia and no findings yielding the accuracy, precision, recall, and F1 score values of 0.96, 0.97, 0.99, and 0.98 respectively in the multiclass classification model and 1 for all the above four performance indices in the binary classification model for dataset A with 5-fold CV. For dataset B, the model has produced accuracy, precision, recall, and F1 score values of all 0.97 for multiclass classification task and 0.96 all for binary classification task using 5-fold CV. Similarly, for holdout validation with dataset B, the accuracy, precision, recall, and F1 score values achieved are 0.96 for multiclass classification and 0.93, 0.94, 0.94, and 0.94 for binary classification. Furthermore, an ANOVA test has been performed to manifest no significant difference between the proposed model and the other novel architectures, thus demonstrating the robustness of the model. The shortcoming of this model is that the obtained results are a cause of random combinations of the extracted modes. Therefore, the model has to be executed with all the combinations to obtain the desired result. In the future, evolutionary computing algorithms can be used to automatically select modes of X-ray images for the multiscale deep CNN to detect COVID19 disease.
CRediT authorship contribution statement
Neha Muralidharan: Data curation, Implementation, Software, Writing – original draft. Shaurya Gupta: Data curation, Implementation, Software, Writing – original draft. Manas Ranjan Prusty: Visualization, Investigation, Validation, Reviewing and editing. Rajesh Kumar Tripathy: Conceptualization, Methodology, Supervision, Validation, Reviewing and editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
All authors have read and agreed to the published version of the manuscript.
References
- 1.Shaukat N., Ali D.M., Razzak J. Physical and mental health impacts of COVID-19 on healthcare workers: A scoping review. Int. J. Emerg. Med. 2020;13(1):40. doi: 10.1186/s12245-020-00299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.2021. Coronavirus in China may have come from bats: Studies—ETHealthWorld, ethealthworld.com. https://health.economictimes.indiatimes.com/news/diagnostics/coronavirus-in-china-may-have-come-from-bats-studies/73923178. (Accessed 18 February 2021) [Google Scholar]
- 3.2021. COVID-19 coronavirus epidemic has a natural origin—ScienceDaily. https://www.sciencedaily.com/releases/2020/03/200317175442.htm. (Accessed 18 February 2021) [Google Scholar]
- 4.Coronavirus update (live) 2021. 453,562 Cases and 2,441,360 deaths from COVID-19 virus pandemic - Worldometer. https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1?%22%20%5Cl%20%22countries. (Accessed 18 February 2021). 110. [Google Scholar]
- 5.Coleman C.M., Frieman M.B. Coronaviruses: Important emerging human pathogen. J. Virol. 2014;88(10):5209–5212. doi: 10.1128/jvi.03488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., Zhang L.J. Coronavirus disease 2019 (COVID-19): A perspective from China. Radiology. 2020;296(2):E15–E25. doi: 10.1148/radiol.2020200490. (Accessed 18 February 2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandra T.B., Verma K., Singh B.K., Jain D., Netam S.S. Coronavirus disease (COVID-19) detection in chest X-ray images using majority voting based classifier ensemble. Expert Syst. Appl. 2021;165 doi: 10.1016/j.eswa.2020.113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rousan L.A., Elobeid E., Karrar M., Khader Y. Chest X-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulm. Med. 2020;20(1):245. doi: 10.1186/s12890-020-01286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong W., Agarwal P.P. Chest imaging appearance of COVID-19 infection. Radiol. Cardiothorac. Imaging. 2020;2(1) doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J., Xue K., Zhang K. Current status and future trends of clinical diagnoses via image-based deep learning. Theranostics. 2019;9(25):7556–7565. doi: 10.7150/thno.38065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekeroglu B., Ozsahin I. Detection of COVID-19 from chest X-ray images using convolutional neural networks. SLAS Technol. 2020;25(6):553–565. doi: 10.1177/2472630320958376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okinda C., Nyalala I., Korohou T., Okinda C., Wang J., Achieng T., Wamalwa P., Mang T., Shen M. A review on computer vision systems in monitoring of poultry: A welfare perspective. Artif. Intell. Agric. 2020;4:184–208. doi: 10.1016/j.aiia.2020.09.002. [DOI] [Google Scholar]
- 14.Tu K.N., Lie J.D., Wan C.K.V., Cameron M., Austel A.G., Nguyen J.K., Van K., Hyun D. Osteoporosis: A review of treatment options. Pharm. Ther. 2018;43(2):92–104. [PMC free article] [PubMed] [Google Scholar]
- 15.Waks A.G., Winer E.P. Breast cancer treatment: A review. JAMA. 2019;321(3):288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 16.S. Dimmeler. Cardiovascular disease review series. EMBO Mol. Med. 2011;3(12):697. doi: 10.1002/emmm.201100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh V., Sharma N., Singh S. A review of imaging techniques for plant disease detection. Artif. Intell. Agric. 2020;4:229–242. doi: 10.1016/j.aiia.2020.10.002. [DOI] [Google Scholar]
- 18.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recognit. Lett. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021:1–14. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sethy P.K., Behera S.K., Ratha P.K., Biswas P. 2020. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. https://www.preprints.org/manuscript/202003.0300/v2. (Accessed 18 February 2021) [Google Scholar]
- 21.Wang L., Lin Z.Q., Wong A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. COVIDX-net: A framework of deep learning classifiers to diagnose COVID-19. X-ray Images, Cs, Eess. arxiv:2003.11055. (Accessed 18 February 2021) [Google Scholar]
- 23.Apostolopoulos I.D., Mpesiana T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horry M.J., Chakraborty S., Paul M., Ulhaq A., Pradhan B., Saha M., Shukla N. COVID-19 detection through transfer learning using multimodal imaging data. IEEE Access. 2020:8–149824. doi: 10.1109/access.2020.3016780. PubMed:149808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altan A., Karasu S. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsiknakis N., Trivizakis E., Vassalou E.E., Papadakis G.Z., Spandidos D.A., Tsatsakis A., Sánchez-García J., López-González R., Papanikolaou N., Karantanas A.H., Marias K. Interpretable artificial intelligence framework for COVID-19 screening on chest X-rays. Exp. Ther. Med. 2020;20(2):727–735. doi: 10.3892/etm.2020.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajinikanth V., Dey N., Raj A.N.J., Hassanien A.E., Santosh K.C., Raja N.S.M. 2020. Harmony-search and Otsu based system for coronavirus disease (COVID-19) detection using lung CT scan images. Arxiv:2004.03431 [Cs, Eess, q-Bio]. arXiv:2004.03431. (Accessed 18 February 2021) [Google Scholar]
- 29.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., Cao K., Liu D., Wang G., Xu Q., Fang X., Zhang S., Xia J., Xia J. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: Evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva P., Luz E., Silva G., Moreira G., Silva R., Lucio D., Menotti D. COVID-19 detection in CT images with deep learning: A voting-based scheme and cross-datasets analysis. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2020:1–8. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 32.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. 2020. Deep learning-based detection for COVID-19 from chest CT using weak label. MedRxiv. (2020).03.12.20027185. [DOI] [Google Scholar]
- 33.J. Gilles. Empirical wavelet transform. IEEE Trans. Signal Process. 2013;61(16):3999–4010. doi: 10.1109/TSP.2013.2265222. [DOI] [Google Scholar]
- 34.Panda R., Jain S., Tripathy R.K., Acharya U.R. Detection of shockable ventricular cardiac arrhythmias from ECG signals using FFREWT filter-bank and deep convolutional neural network. Comput. Biol. Med. 2020;124 doi: 10.1016/j.compbiomed.2020.103939. [DOI] [PubMed] [Google Scholar]
- 35.Tripathy R.K., Bhattacharyya A., Pachori R.B. Localization of myocardial infarction from multi-lead ECG signals using multiscale analysis and convolutional neural network. IEEE Sens. J. 2019;19(23):11437–11448. doi: 10.1109/JSEN.2019.2935552. [DOI] [Google Scholar]
- 36.Kumar R., Saini I. Empirical wavelet transform based ECG signal compression. IETE J. Res. 2014;60(6):423–431. doi: 10.1080/03772063.2014.963173. [DOI] [Google Scholar]
- 37.Tripathy R.K., Bhattacharyya A., Pachori R.B. A novel approach for detection of myocardial infarction from ECG signals of multiple electrodes. IEEE Sens. J. 2019;19(12):4509–4517. doi: 10.1109/JSEN.2019.2896308. [DOI] [Google Scholar]
- 38.Hong G., Chen X., Chen J., Zhang M., Yumeng Ren, Zhang X. A multi-scale gated multi-head attention depthwise separable CNN model for recognizing COVID-19. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-97428-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen J.P., Morrison P., Dao L. 2020. COVID-19 image data collection. ArXiv Prepr, arXiv:2003.11597. [Google Scholar]
- 40.X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri, R.M. Summers, Chestx-ray8: Hospital-scale chest X-ray database and benchmarks on weakly supervised classification and localization of common thorax diseases, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 2097–2106.
- 41.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Al-Emadi N.A., Reaz M.B.I., Islam M.T. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 42.Rahman T., Khandakar A., Qiblawey Y., Tahir A., Kiranyaz S., Abul Kashem S.B., Islam M.T., Maadeed S.Al, Zughaier S.M., Khan M.S., Chowdhury M.E.H. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kermany D., Zhang K., Goldbaum M. 2018. Large dataset of labeled optical coherence tomography (OCT) and chest X-ray images. Mendeley Data, 3. [DOI] [Google Scholar]
- 44.Gilles J., Tran G., Osher S. 2D Empirical transforms, wavelets, ridgelets, and curvelets revisited. SIAM J. Imaging Sci. 2014;7(1):157–186. doi: 10.1137/130923774. [DOI] [Google Scholar]
- 45.Panda R., Jain S., Tripathy R.K., Acharya U.R. Detection of shockable ventricular cardiac arrhythmias from ECG signals using FFREWT filter-bank and deep convolutional neural network. Comput. Biol. Med. 2020;124 doi: 10.1016/j.compbiomed.2020.103939. [DOI] [PubMed] [Google Scholar]
- 46.Madhavan S., Tripathy R.K., Pachori R.B. Time-frequency domain deep convolutional neural network for the classification of focal and non-focal EEG signals. IEEE Sens. J. 2020;20(6):3078–3086. doi: 10.1109/jsen.2019.2956072. [DOI] [Google Scholar]
- 47.Maheshwari D., Ghosh S.K., Tripathy R.K., Sharma M., Acharya U.R. Automated accurate emotion recognition system using rhythm-specific deep convolutional neural network technique with multi-channel EEG signals. Comput. Biol. Med. 2021;134 doi: 10.1016/j.compbiomed.2021.104428. [DOI] [PubMed] [Google Scholar]
- 48.Prusty M.R., Jayanthi T., Velusamy K. Weighted-SMOTE: A modification to SMOTE for event classification in sodium cooled fast reactors. Prog. Nucl. Energy. 2017;100:355–364. doi: 10.1016/j.pnucene.2017.07.015. [DOI] [Google Scholar]
- 49.Radhakrishnan T., Karhade J., Ghosh S.K., Muduli P.R., Tripathy R.K., Acharya U.R. AFCNNet: Automated detection of AF using chirplet transform and deep convolutional bidirectional long short term memory network with ECG signals. Comput. Biol. Med. 2021;137 doi: 10.1016/j.compbiomed.2021.104783. [DOI] [PubMed] [Google Scholar]
- 50.Dailyhunt . Guleria Warns Against Risk of Cancer. Hindustan Times — DailyHunt; 2021. 1 CT scan is equivalent to 300 chest X-rays. https://m.dailyhunt.in/news/india/english/hindustan+times-epaper-httimes/1+ct+scan+is+equivalent+to+300+chest+x+rays+guleria+warns+against+risk+of+cancer-newsid-n276402280?ss=wsp&s=pa. [Google Scholar]