Abstract
Background
Coffea arabica L. is an economically important agricultural crop and the most popular beverage worldwide. As a perennial crop with recalcitrant seed, conservation of the genetic resources of coffee can be achieved through the complementary approach of in-situ and ex-situ field genebank. In Ethiopia, a large collection of C. arabica L. germplasm is preserved in field gene banks. Here, we report the whole-genome resequencing of 90 accessions from Choche germplasm bank representing garden and forest-based coffee production systems using Illumina sequencing technology.
Results
The genome sequencing generated 6.41 billion paired-end reads, with a mean of 71.19 million reads per sample. More than 93% of the clean reads were mapped onto the C. arabica L. reference genome. A total of 11.08 million variants were identified, among which 9.74 million (87.9%) were SNPs (Single nucleotide polymorphisms) and 1.34 million (12.1%) were InDels. In all accessions, genomic variants were unevenly distributed across the coffee genome. The phylogenetic analysis using the SNP markers displayed distinct groups.
Conclusions
Resequencing of the coffee accessions has allowed identification of genetic markers, such as SNPs and InDels. The SNPs discovered in this study might contribute to the variation in important pathways of genes for important agronomic traits such as caffeine content, yield, disease, and pest in coffee. Moreover, the genome resequencing data and the genetic markers identified from 90 accessions provide insight into the genetic variation of the coffee germplasm and facilitate a broad range of genetic studies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-022-03449-4.
Keywords: Coffee, Genetic markers, Phylogenetic analysis, Resequencing, Single nucleotide polymorphism
Introduction
Coffee (Rubiaceae) is an important agricultural crop and is mainly grown as a cash crop in several tropical countries [1, 2]. Presently, it is cultivated in more than 80 countries around the globe [3] and serves as a major source of livelihood for smallholder farmers. With only 30% of production consumed domestically, coffee remains an important export commodity [4]. Despite the huge number of species reported in the genus Coffea [5], the primary species utilized for coffee production are Coffea arabica L. and C. canephora Pierre [2, 6]. Arabica coffee is a tetraploid species (2n = 4x = 44) derived from interspecific crosses between C. canephora and C. eugenioides [7]. The cultivation of this crop serves as an important source of income and employment in developing countries of Latin America, Africa, and Asia [8]. Besides, C. arabica L. is valued for its superior beverage quality [9–11] and accounts for about 63% of the global coffee production [11].
Coffea arabica L. is the only coffee species cultivated and exported from Ethiopia [12]. It is the major foreign exchange earner contributing to a quarter of the country’s export earnings [13] and serves as a means of livelihood and employment for an estimated 15 million people [12–14]. The agro-ecology under which coffee grows varies significantly, and the crop is mainly produced in four distinct production systems in Ethiopia; i.e., garden, semi-forest, forest and plantation. Garden coffee is widespread across the country and forest-based (semi-forest and forest) coffee production systems are found in the southeast and southwest parts of the country [14]. Different scholars have reported a high genetic diversity for coffee in Ethiopia, which is of great potential to improve the crop [15–18]. Presently, the global demand for specialty coffee has increased significantly [19]. Hence, developing coffee varieties with high market demand, such as low bean caffeine content, is essential. Ethiopia being the main source of C. arabica L. gene pool, the germplasm found in the country is valuable [16], and could be used for developing varieties with desirable traits [20–22]. Notably, the wild coffee genetic resources are genetically diverse and are believed to possess traits that can be used to improve the cultivated varieties [17, 23]. Specifically, these resources are valuable in light of the projected climate change due to their ability to adapt to environmental change [24]. Despite this fact, the forest coffee gene pool of C. arabica L. is threatened by various factors that could affect the genetic base for the future breeding program [25]. The loss is mainly attributed to deforestation and land-use change [15], climate change [13, 26], and the introduction of new varieties in the coffee forests [27].
Presently, global ex-situ coffee germplasm conservation programs have been implemented in various countries including Ethiopia [28]. Coffee germplasm is conserved as a living tree in the field gene bank due to the recalcitrant nature of the seeds [29]. Likewise, in Ethiopia, the long-term preservation of this important cash crop is achieved by establishing field gene banks. Currently, more than 11,000 coffee accessions collected by random and non-random sampling techniques are maintained in field gene banks in Ethiopia [30]. Evaluating the genetic diversity of the coffee germplasm maintained in field genebank is essential [31], and assists in enhancing the management of the conserved materials. However, the perennial nature of coffee makes the evaluation and breeding work very costly [32]. Hence, developing and using molecular markers in coffee could enhance the development of varieties with desirable traits [33].
Currently, NGS technology has enabled the identification of genetic variation in germplasm collections [34]. Particularly, the availability of reference genome and improvement in the genetic data analysis methods have contributed to advance resequencing studies [35, 36]. Genome resequencing could also help in bridging the knowledge gap between genotype and phenotype and facilitate molecular breeding [37]. Notably, the markers generated from resequencing analysis help to advance the conventional crop breeding approaches [38], and in turn, contribute to shortening the time required to develop new varieties [39].
SNP represent single nucleotide change in DNA sequence and are considered the most abundant form of genetic variation [37, 40]. Presently, SNPs have become the genetic markers of choice in various genetic, ecological and evolutionary studies [41]. Despite the great economic and social importance of C. arabica L., studies with SNP markers are scarce and a small number of SNP markers are available for this species [33]. This study, therefore, aimed to discover the genomic variations in 90 accessions of C. arabica L. by whole-genome resequencing. The genomic data and SNP generated in this study could be of great relevance for undertaking various genetic studies.
Materials and methods
Sample preparation and DNA extraction
The C. arabica L. accessions that have been maintained at the Choche germplasm bank of the Ethiopian Biodiversity Institute (Jimma zone, Goma district, southwest Ethiopia) were used in this study. Genomic DNA was isolated from Silica gel dried leaf material of the 90 accessions originally collected from the garden and forest-based coffee production systems using MagicMag Genomic DNA Micro Kit (Sangon Biotech Co. Shanghai, China). The quantity and quality of isolated DNA were checked and analyzed with the NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and 1.0% agarose gel electrophoresis, respectively. Detailed information about each accession included in this study is shown in Additional file 1: Table S1.
Library preparation and sequencing
Paired-end libraries with approximately 350 bp insert sizes were constructed from 1 μg of genomic DNA from each accession using Illumina TruSeq or Nextera (San Diego, CA) kits according to the Illumina manufacturer’s specifications. Whole-genome resequencing was performed for 90 coffee accessions at the Beijing Genomics Institute (Shenzhen, China) using the Illumina Hiseq 2500 Platform (Illumina, San Diego, CA).
Sequence processing and mapping of reads to the reference genome
The filtering of raw reads was accomplished using the FASTQC (version 0.11.3) program. The clean reads were aligned onto the C. arabica L. reference genome using a burrows wheeler aligner (BWA) with default parameters [42]. SAMtools (version 1.3.1) software was used to convert mapping results into the BAM format and filter the unmapped reads [43]. Then, the aligned reads were processed using Piccard tools (http://broadinstitute.github.io/picard/) to remove duplicate reads. The Illumina sequencing reads of each accession is deposited under accession number from SRR17316330 to SRR17316419.
Variant detection, annotation and relationship analysis
The mapped reads were used to detect variants (SNP and InDels) using the Genome Analysis Toolkit (version 3.6) software [44]. The annotation and classification of the genomic variants were performed by SnpEff software [45]. The variants were annotated based on their impact (high, moderate, modifier and low), functional class (synonymous and non-synonymous substitutions) and their genomic regions such as downstream, upstream, exon, intron, intragenic and intergenic region, transcript, 3′ and 5′ untranslated regions (UTRs). DNA substitution mutations (transitions and transversion) and amino acids changes were identified. The whole-genome SNP markers were used to infer the phylogenetic relationship of 90 accessions. A Maximum likelihood (ML) analysis was conducted using the RAxML (version 8.1.2) program [46].
Results
Resequencing 90 accessions of C. arabica L.
Whole-genome resequencing of 90 accessions of C. arabica L. was performed with the Illumina sequencing platform. The genome sequencing generated 6.41billion paired-end reads, with a mean of 71.19 million reads per sample. After removing low-quality reads, high-quality reads of each accession were mapped onto the C. arabica L. reference genome using a BWA aligner. The percentage of reads mapped onto the reference genome varied from 88.93 to 98.11%. The detailed resequencing information was provided in Additional file 2: Table S2. This result indicates that there is a difference in the whole-genome sequences among the studied accessions. A total of 11.08 million variants were identified by mapping the clean reads onto the reference genome. Among these, 9.74 million (87.9%) were SNPs and 1.34 million (12.1%) were InDels (insertions and deletions) (Table 1). The deletions and insertions length observed in the coffee genome ranged from 1 to 46 base pairs.
Table 1.
Type and the number of variants detected in the C. arabica L. genome
| Variant | Total number | % |
|---|---|---|
| SNP | 9,743,804 | 87.9 |
| InDels | ||
| - insertions | 794,963 | 7.2 |
| - deletions | 545,345 | 4.9 |
| Total | 11,084,112 | 100 |
Identification, characterization and annotation of SNPs
The high-quality sequences were used for the identification of SNP, and a total of 9.74 million SNPs were identified in all the 90 coffee accessions. We used the SnpEff [45] program to evaluate the impact and possible effects of the identified SNP could have on the gene. Based on their impact on the coding sequence the SNP were classified into four classes i.e., high, moderate, modifier and low. The analysis revealed a major proportion of SNPs were modifier (97.911% of the SNP with impact on non-coding regions), followed by moderate (1.151% of the SNP could have a non-synonymous substitution), low impact (0.834% of the SNP with synonymous substitution) and the smallest value was recorded for high impact SNP (0.104% of the SNP with disruptive impact on the protein) (Table 2). The distribution of SNPs across genomic regions was compared (Fig. 1). SNPs were most abundant in the intergenic, upstream, downstream, intron and exon of genes, and their proportions are about 29.115, 26.63, 25.721, 5.996 and 2.47%, respectively. A limited number of SNPs were observed in 3’UTR (0.574%) and 5’UTR (0.484). More SNPs were detected in the introns than exons (Fig. 1). Further, the result showed that the majority of genomic variations were located in non-coding regions. The SNP variants were also separated into heterozygous and homozygous, and in all coffee accessions, the number of heterozygous variants was higher than the homogenous ones. The SNP mutations also resulted in a codon modification in genomic regions leading to variations in amino acid sequences. The details of amino acid changes observed in this study are indicated in Additional file 3: Table S3.
Table 2.
The effects of identified SNP on genes as classified by SnpEff program [45]
| Impact class | Count | Percent |
|---|---|---|
| High | 33,443 | 0.104% |
| Moderate | 370,392 | 1.151% |
| Low | 268,283 | 0.834% |
| Modifier | 31,501,326 | 97.911% |
Fig. 1.
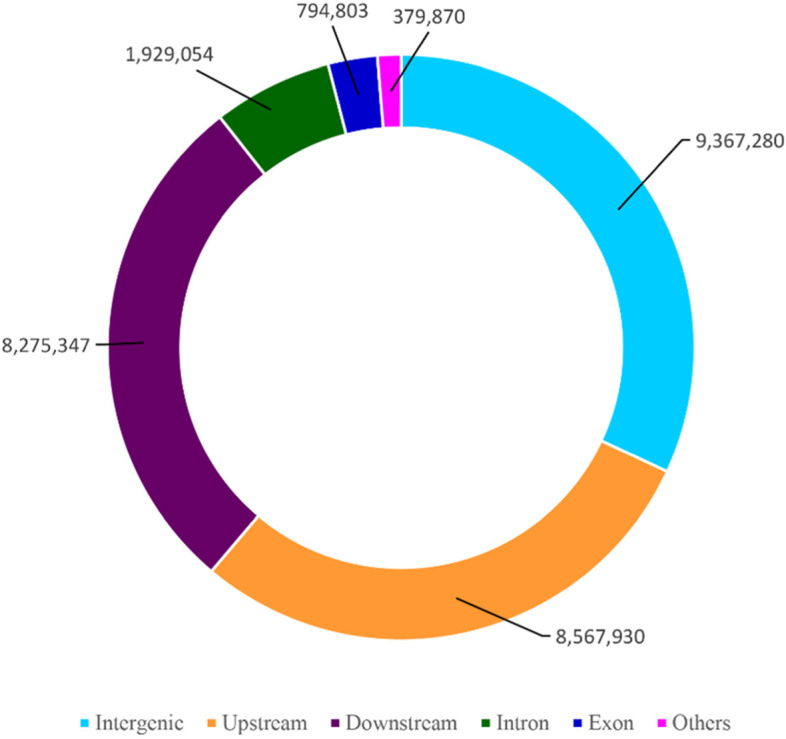
The distribution of the identified SNP in the different genomic regions (Intergenic, Upstream, Downstream, Intron, Exon and other) of the C. arabica L. genome
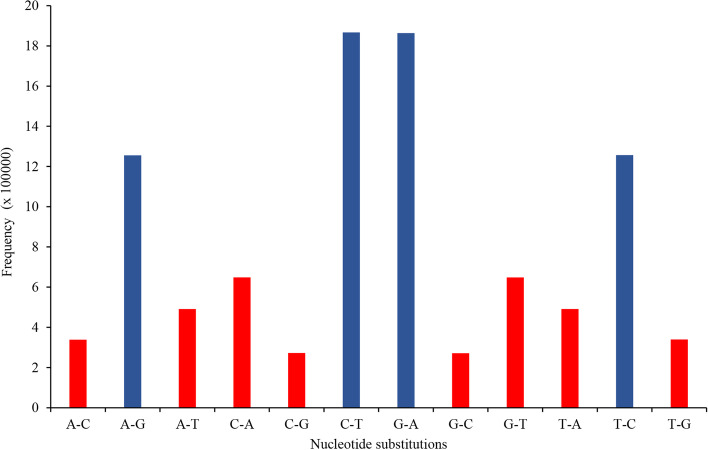
Based on the nucleotide substitutions, the SNPs identified in the coffee genome were classified into two classes, namely, transitions (A/G and C/T) and transversions (A/C, C/G, A/T, G/T). The total transitions and transversions detected were 204,665,968 and 103,693,961, with a transitions/transversions (Ts/Tv) ratio of 1.97. The transition frequency of C/T was more than G/A. The transversion frequency of C/A was higher, similarly within transitions; the C/T transition was higher in number (Additional file 4: Table S4 and Fig. 2). The SNP found in the coding region are of two types i.e., synonymous and non-synonymous. A total of 215,712 synonymous SNPs were detected in the coding sequence of the coffee genome (Additional file 4: Table S4). Often, synonymous SNPs do not affect the normal function of genes.
Fig. 2.
Transition and transversion SNPs detected in C. arabica L. genome (Transitions (A-G, C-T) indicated with red color; Transversions (C-G, A-C, G-T, A-T) indicated with blue color
Relationship analysis
We inferred the phylogenetic relationships of the 90 accessions of C. arabica L. by constructing a Maximum-likelihood phylogenetic tree using the whole-genome SNP markers. The analysis revealed four major clusters, and each cluster also sub-divided into sub-clades. Forest-based accessions sampled from southwest Ethiopia were distributed in different clusters, while those sampled from the southeast part clustered together except MHSF4. Cluster I (Black), contained six accessions, of which two were collected from the northwest, while four were collected from the southwest. Cluster II (Purple) consisted of accessions collected from the southwest and the north part of the country. Cluster III (Blue) mainly consisted of accessions sampled from the southwest parts of the country i.e., GUG5, YCG2 and GNG4 sampled from the country’s north, south and southeastern parts, clustered together. Almost all of the south and southeast accession were clustered in group IV (Green). The garden coffee accessions collected from similar geographical areas consistently clustered together in the same group (e.g., TGG1, TGG3 and TGG5; ZPG2, ZPG4 and ZPG5; LAG3, LAG4 and LAG5; KOG1, KOG2 and KOG3; BEG1, BEG4 and BEG5; WLG1, WLG2 and WLG3; JIG2, JIG3 and JIG5; MKG1, MKG2 and MKG3, and ISG1, ISG2 and ISG3). Only, GNG, GUG, YCG, SAG, GMG and WEG garden coffee samples showed exceptional grouping patterns (Fig. 3). This suggests that the SNP markers identified in this study have the potential to give insights into the evolutionary relationship of the coffee accessions.
Fig. 3.
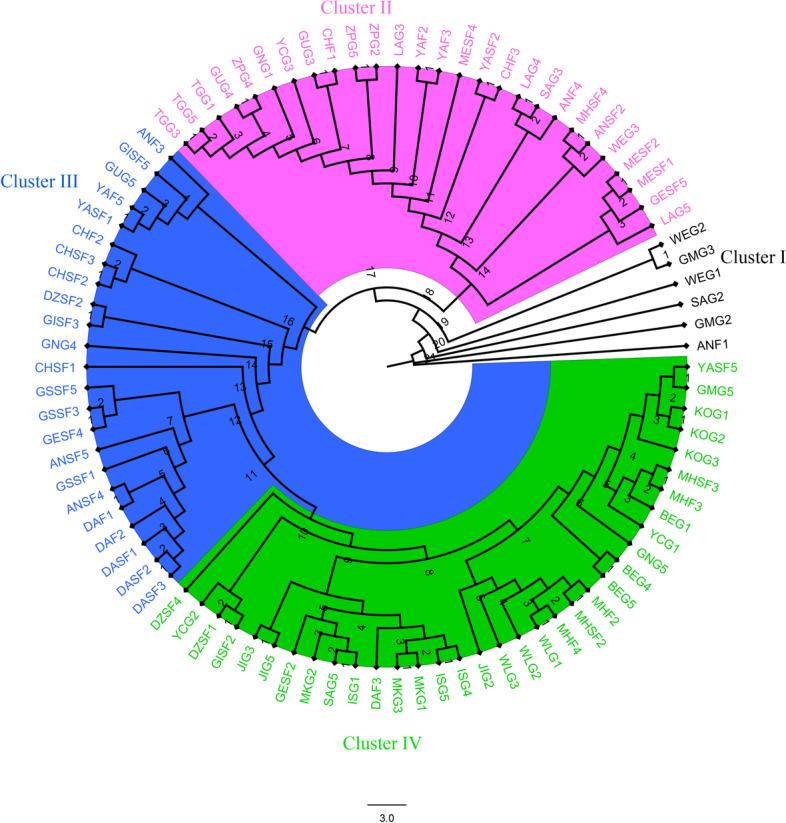
Maximum-likelihood (ML) phylogeny of C. arabica L. accessions inferred from RAxML using SNPs detected in whole-genome resequencing data. The accessions within different clades are highlighted with different colors
Discussion
Whole-genome sequence diversity
Presently, whole-genome resequencing is the most convenient approach for genome-wide SNP identification. It provides information of great relevance for crop genetics and breeding [35]. Besides, characterization of genome-wide DNA variation can help to understand the trait-genotype associations [37, 47]. In this study, we performed the whole-genome resequencing analysis of coffee, in which diverse accessions sampled from different geographic regions and production systems were included. The percentage of reads mapped onto the reference genome varied from 88.93 to 98.11% indicating the good quality of the data generated. The observed variation in the mapping rate could be attributed to the difference between the sequenced accessions and the reference genome. SNP and InDels identification using the C. arabica L. nuclear, mitochondrial and chloroplast genomes yielded a total of 9.74 million SNPs and 1.34 million InDels across 90 accessions. These variants were heterogeneously distributed across the eleven chromosomes of C. arabica L. Such uneven dispersal of variants has also been reported in other plant species such as in Solanum melongena and C. canephora [35, 48].
The variant identified by the resequencing study could be used for various studies including marker-assisted selection, genome-wide association studies, phylogenetic and diversity analyses [49]. SNPs also considered a valuable genetic marker, and often associated with the gene or trait of interest. Because of this, they are widely used genetic markers to identify genes responsible for traits of agricultural importance [33, 36]. A genome-wide association study was performed in coffee to identify genomic regions associated with lipid, cafestol and kahweol contents in green coffee beans [50]. The study discovered SNP located inside or near candidate genes related to metabolic pathways of these chemical compounds. SNP markers have also been utilized in coffee genetic diversity and population structure analysis [49]. In other studies, SNP markers associated with pathways of caffeine and trigonelline biosynthesis were reported [50]. Further, Tran et al. [51] identified 1444 non-synonymous SNPs associated with caffeine content using the draft genome of C. arabica L. Among these, the Kyoto Encyclopedia of Genes and Genome pathways analysis discovered 11 SNPs that have direct associations with genes encoding enzymes involved in caffeine biosynthesis pathways. These studies demonstrated the importance of SNP in identifying the genetic basis of traits of interest. The resequencing efforts of [52], identified SNPs that are potentially responsible for bacterial wilt disease in the Capsicum annuum. Furthermore, [53] highlighted that whole-genome resequencing is critical for the development of molecular markers. In this regard, marker development from variants identified from sequencing data has been done successfully for various agronomical important traits [54]. Thus, the SNP identified in the present study could be a valuable source of new allelic variations to advance coffee genomic research and germplasm improvement programs. Further, analyzing the diversity in the coffee genome could help uncover variants that could be used to better understand the genetic basis of agriculturally important traits.
SNP analysis
Phenotypic variations in crop plants are the result of variation at the DNA level [55]. SNP is an abundant form of genetic variation and may cause phenotypic diversity among individuals [36]. Based on the nucleotide substitution SNP is generally classified as transitions and transversions [56]. In this study, the number of SNP with transitions was two-fold higher than SNP with transversions. G/A and C/T transitions were observed in equal numbers. The transition/transversion ratio was 1.97, which indicates transitions were the most frequent mutations similar to the findings from previous reports. For instance, [57, 58] reported a Ts/Tv ratio of 2.0 and 2.4 in rice, respectively. Generally, transversion SNP mutations have a high potential to alter the amino acid sequence of proteins than transitions [59]. Our study indicated that most of the detected SNP were located in intergenic and non-coding regions of the coffee genome. This suggests that these SNPs do not affect the gene functions. Similar results have also been reported in the previous studies on coffee, tea, potato and rice [31, 38, 59, 60].
The variant effects that might affect the protein-coding sequences include: synonymous/non-synonymous amino acid replacement, start codon gains or losses, stop codon gains or losses [45]. In this study, only 0.11% of SNP with high impact effects on genes were detected in the coffee genome. These SNPs might disturb the proper functioning of genes ultimately affecting the enzyme activity. Generally, the non-synonymous SNP in the coding sequence are disruptive and result in gene product change [36, 59, 60]. Hence, the identification of SNP found in genes, and analysis of their effects on phenotype could help to understand their impact on gene function and contribute to crop improvement programs [36]. In a previous study, stop gain, splice donor variant, intron variant, and splice acceptor variant were reported as disruptive variant effects in coffee affecting the proper functioning of genes [31]. Hence, the determination of the genomic location of variants contributes to identification of the genetic region responsible for trait variations.
Phylogenetic analysis
SNP markers have been employed for analyzing the phylogenetic relationships and differences between genotypes [61]. In this study, the phylogenetic relationship of the 90 coffee accessions was analyzed using whole-genome SNP markers. The analysis revealed four major groups, comprising several sub-clades. Clusters I, II and III mainly contained accessions from the southwest. It is also noted that coffee accessions collected from southwest Ethiopia were found in different clusters. Specifically, the grouping of accessions collected from the southwest regions in different groups could explain coffee accessions that originated in that region had a broad genetic base. In another study, Spinoso-Castilillo et al. [11] reported that the SNP markers generated by DArTseq technology separated the 87 accessions of Coffea spp. into five distinct groups. Further, Silvestrini et al. [62] reported that even if coffee accessions had originated in the same localities, there was a possibility of separating genetically by the domestication process due to human selection activity. The finding also supports earlier reports that suggested southwest Ethiopia as the center of origin and diversity of C. arabica L. [8, 12, 17, 63].
Most of the garden coffee accessions collected from the north parts of the country are grouped with southwest forest-based accessions. Whereas, the majority of garden coffee accessions sampled from southern parts of the country are grouped with southeast forest-based accession. Delsuc et al. [64] reported that phylogenetic analysis is one of the tools that could help understand the evolutionary relationships of crop plants. Hence, the grouping of garden coffee accessions sampled from different areas with southwest and southeast forest-based accessions shows the ancestor of these accessions probably originated in the southwest and southeast. The garden coffee accessions collected from similar geographical areas consistently clustered together in the same group. These accessions also formed a sub-clade within a forest-based accession clade.
A recent study by Benti et al. [12] on the Ethiopian commercial C. arabica L. varieties also found the grouping of varieties into different clusters regardless of their geographic origin. The clustering pattern found in this study could indicate the presence of a high level of genetic diversity within coffee accessions sampled from the same geographic origin. Furthermore, the garden coffee accessions sampled from the neighboring regions were consistently grouped with few exceptions (Fig. 3). This might have been attributed to gene flow between adjacent populations or the garden coffee farms probably established from seeds obtained from the same source. The clustering of southwest coffee accessions with the south and southeast population was previously reported [8, 65, 66]. Moreover, Mishra et al. [67] noted that coffee accessions collected from the same geographical origin in Ethiopia did not cluster together. All these studies indicate that the C. arabica L. germplasm found in Ethiopia has a broad genetic base, and is valuable in developing varieties that could sustain global coffee production.
Conclusion
The availability of reference genomes and the continuous improvements of genetic data analysis methods are fostering resequencing studies. In this study, we performed the resequencing of coffee accessions, which has allowed the identification of genetic markers, such as SNPs and InDels. The SNPs discovered in this study might contribute to the variation in important pathways of genes for important agronomic traits such as caffeine content, yield, disease, and pest in coffee. Moreover, the genome resequencing data and the genetic markers identified from 90 accessions provide insight into the genetic variation of the coffee germplasm and facilitate a broad range of genetic studies.
Supplementary Information
Additional file 1: Table S1. The details of the analyzed 90 accession of C. arabica L.
Additional file 2: Table S2. Summary of the C. arabica L. whole-genome sequencing data.
Additional file 3: Table S3. Amino acid changes identified by SnpEff software. Rows are reference amino acids and columns are changed amino acids. E.g., Row 'A' column 'E' indicates how many 'A' amino acids have been replaced by 'E' amino acids.
Additional file 4: Table S4. Summary count of SNPs with effects on the genome.
Acknowledgments
The authors would like to thank the Ethiopian Biodiversity Institute for providing the leaf material used in this research. Jara Negash and his colleagues highly acknowledged for their assistance during sample collection. Boniface N. Karega also deserves special thanks for reading and providing valuable comments on the draft version of the manuscript.
Field study permissions
Ethiopian Biodiversity Institute (EBI) approved this study (EBI 71/150042/2018).
Authors’ contributions
QF.W., GW. H and Y.M designed the experiment. Y.M performed the experiment. Y.M., K.T. and JK. S. analyzed the data. Y.M. wrote the manuscript. K.T., JK. S. and X.D revised the manuscript. QF. W and GW.H. secured the research fund and provided useful advice. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the Hubei Provincial Key Research and Development Program (2020BHB022), the National Natural Science Foundation of China (31961143026, 31970211), International Partnership Program of Chinese Academy of Sciences (151853KYSB20190027), and the Sino-Africa Joint Research Center, CAS (SAJC202101).
Availability of data and materials
The data generated and analyzed during this study are included in this article, its supplemental information files and the sequence data have been deposited into NCBI sequence read archive under submission identifier SUB10821951 with the Bio project identifier PRJNA790687.
Declarations
Study protocol must comply with relevant institutional, national, and international guidelines and legislation.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhang D, Vega FE, Solano W, Su F, Infante F, Meinhardt LW. Selecting a core set of nuclear SNP markers for molecular characterization of Arabica coffee (Coffea arabica L.) genetic resources. Conserv Genet Resour. 2021;13:329–335. doi: 10.1007/s12686-021-01201-y. [DOI] [Google Scholar]
- 2.Vidal RO, Mondego JMC, Pot D, Ambrósio AB, Andrade AC, Pereira LFP, et al. A high-throughput data mining of single nucleotide polymorphisms in Coffea species expressed sequence tags suggests differential homeologous gene expression in the allotetraploid Coffea arabica. Plant Physiol. 2010;154(3):1053–1066. doi: 10.1104/pp.110.162438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musoli P, Cubry P, Aluka P, Billot C, Dufour M, De BF, et al. Genetic differentiation of wild and cultivated populations : diversity of Coffea canephora Pierre in Uganda. Genome. 2009;52:634–646. doi: 10.1139/G09-037. [DOI] [PubMed] [Google Scholar]
- 4.ICO. The value of coffee. Sustainability, inclusiveness, and resilience of the coffee global value chain. Coffee development report. 2020. International Coffee Organization.
- 5.Davis AP, Tosh J, Ruch N, Fay MF, Museum NH, Road C, et al. Growing coffee : Psilanthus (Rubiaceae) subsumed on the basis of molecular and morphological data ; implications for the size, morphology, distribution and evolutionary history of Coffea. Bot J Linn Soc. 2011;167:357–377. doi: 10.1111/j.1095-8339.2011.01177.x. [DOI] [Google Scholar]
- 6.Cui L, Hanika K, Visser RGF. Improving pathogen resistance by exploiting plant susceptibility genes in coffee (Coffea spp.). Agronomy. 2020;10(12):1928. 10.3390/agronomy10121928.
- 7.Lashermes P, Combes MC, Robert J, APT, D’Hont A, Charrie A. Molecular characterization and origin of the Coffea arabica L. genome. Mol Gen Genet. 1999;261:259–266. doi: 10.1007/s004380050965. [DOI] [PubMed] [Google Scholar]
- 8.Anthony F, Bertrand B, Quiros O, Wilches A, Lashermes P, Berthaud J, et al. Genetic diversity of wild coffee (Coffea arabica L.) using molecular markers. Euphytica. 2001;118(1):53–65. doi: 10.1023/A:1004013815166. [DOI] [Google Scholar]
- 9.Jingade P, Huded AK, Kosaraju B, Kumar M. Diversity genotyping of Indian coffee (Coffea arabica L.) germplasm accessions by using SRAP markers. J Crop Improv. 2019;33(3):1–19. doi: 10.1080/15427528.2019.1592050. [DOI] [Google Scholar]
- 10.Tran HTM, Slade Lee L, Furtado A, Smyth H, Henry RJ. Advances in genomics for the improvement of quality in coffee. J Sci Food Agri. 2016;96:3300–3312. doi: 10.1002/jsfa.7692. [DOI] [PubMed] [Google Scholar]
- 11.Spinoso-Castillo JL, Escamilla-Prado E, Aguilar-Rincón VH, et al. Genetic diversity of coffee (Coffea spp.) in Mexico evaluated by using DArTseq and SNP markers. Genet Resour Crop Evol. 2020;67:1795–1806. 10.1007/s10722-020-00940-5.
- 12.Benti T, Gebre E, Tesfaye K, Berecha G, Lashermes P, Kyallo M, et al. Genetic diversity among commercial arabica coffee (Coffea arabica L.) varieties in Ethiopia using simple sequence repeat markers. J Crop Improv. 2021;35(2):147–168. doi: 10.1080/15427528.2020.1803169. [DOI] [Google Scholar]
- 13.Moat J, Williams J, Baena S, Wilkinson T, Gole TW, Challa ZK, et al. Resilience potential of the Ethiopian coffee sector under climate change. Nat Plants. 2017;3. 10.1038/nplants.2017.81. [DOI] [PubMed]
- 14.ECFF. Coffee: Ethiopia’s gift to the world. Environment and Coffee Forest Forum. Addis Ababa; 2015.
- 15.Legesse A. Assessment of coffee (Coffea arabica L.) genetic erosion and genetic resources management in Ethiopia. Int J Agric Ext. 2019;07(03):223–229. doi: 10.33687/ijae.007.03.2972. [DOI] [Google Scholar]
- 16.Benti T. Progress in Arabica coffee breeding in Ethiopia : achievements, challenges and prospects. Int J Sci Basic Appl Res. 2017;33(2):15–25. [Google Scholar]
- 17.Tesfaye K, Govers K, Bekele E, Borsch T. ISSR fingerprinting of Coffea arabica throughout Ethiopia reveals high variability in wild populations and distinguishes them from landraces. Plant Syst Evol. 2014;300(5):881–897. doi: 10.1007/s00606-013-0927-2. [DOI] [Google Scholar]
- 18.Labouisse J, Kotecha S. Preserving diversity for specialty coffees. A focus on production systems and genetic resources of Arabica coffee in Ethiopia. 2008. [Google Scholar]
- 19.Schuit P, Moat J, Gole TW, Challa ZK, Torz J, MacAtonia S, et al. The potential for income improvement and biodiversity conservation via specialty coffee in Ethiopia. PeerJ. 2021;9:e10621. https://doi.org/10.7717/peerj.10621. [DOI] [PMC free article] [PubMed]
- 20.Mehrabi Z, Lashermes P. Protecting the origins of coffee to safeguard its future. Nat Plants. 2017;3:16209. doi: 10.1038/nplants.2016.209. [DOI] [PubMed] [Google Scholar]
- 21.van der Vossen H, Bertrand B, Charrier A. Next generation variety development for sustainable production of arabica coffee (Coffea arabica L.): a review. Euphytica. 2015;204(2):243–256. doi: 10.1007/s10681-015-1398-z. [DOI] [Google Scholar]
- 22.Hein L, Gatzweiler F. The economic value of coffee (Coffea arabica) genetic resources. Ecol Econ. 2006;60(1):176–185. doi: 10.1016/j.ecolecon.2005.11.022. [DOI] [Google Scholar]
- 23.Moat J, Gardens RB, Gole TW, Ababa A, Davis AP, Gardens RB. Least concern to endangered : applying climate change projections profoundly influences the extinction risk assessment for wild Arabica coffee. Glob Change Biol. 2019;25:390–403. doi: 10.1111/gcb.14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiwuka C, Goudsmit E, Douma JC, Bellanger L, Crouzillat D, Stoffelen P, et al. Genetic diversity of native and cultivated Ugandan Robusta coffee (Coffea canephora Pierre ex A. Froehner ): climate influences, breeding potential and diversity conservation. PLoS One 2021;16(2):e0245965. 10.1371/journal.pone.0245965. [DOI] [PMC free article] [PubMed]
- 25.Krishnan S. Current status of coffee genetic resources and implications for conservation. CAB Rev. 2013;8(16). 10.1079/PAVSNNR20128016_2013. [DOI]
- 26.Davis AP, Gole TW, Baena S, Moat J. The impact of climate change on indigenous Arabica coffee (Coffea arabica): predicting future trends and identifying priorities. PLoS One. 2012;7(11):e47981. doi: 10.1371/journal.pone.004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aerts R, Geeraert L, Berecha G, Hundera K, Muys B, De KH, et al. Conserving wild Arabica coffee : emerging threats and opportunities. Agriculture, Ecosyst Environ. 2017;237:75–79. doi: 10.1016/j.agee.2016.12.023. [DOI] [Google Scholar]
- 28.Bramel P, Krishnan S, Horna D, Lainoff B, Montagnon C. Global Conservation Strategy for Coffee Genetic Resources. 2017. [Google Scholar]
- 29.Zhou L, Vega FE, Tan H, Lluch AER, Meinhardt LW, Fang W, et al. Developing single nucleotide polymorphism (SNP) markers for the identification of coffee germplasm. Trop Plant Biol. 2016. 10.1007/s12042-016-9167-2.
- 30.Labouisse JP, Bellachew B, Kotecha S, Bertrand B. Current status of coffee (Coffea arabica L.) genetic resources in Ethiopia: implications for conservation. Genet Resour Crop Evol. 2008;55:1079–1093. doi: 10.1007/s10722-008-9361-7. [DOI] [Google Scholar]
- 31.Huang L, Xiaoyang W, Dong Y, Long Y, Hao C, Yan L, et al. Resequencing 93 accessions of coffee unveils independent and parallel selection during Coffea species divergence. Plant Mol Biol. 2020. 10.1007/s11103-020-00974-4. [DOI] [PubMed]
- 32.Ferra V, Fanelli H, Giovanni C, Luı G, Fritsche-neto R. The effect of bienniality on genomic prediction of yield in Arabica coffee. Euphytica. 216:101. 10.1007/s10681-020-02641-7.
- 33.Sousa TV, Caixeta ET, Alkimim ER, Fernando M, Resende R De, Zambolim L. Population structure and genetic diversity of coffee progenies derived from Catuaí and Híbrido de Timor revealed by genome-wide SNP marker. Tree Genet Genomes. 2017;13(124). 10.1007/s11295-017-1208-y.
- 34.Van Treuren R, Van Hintum TJL. Next-generation gene banking: plant genetic resources management and utilization in the sequencing era. Plant Genet Resour Characterisation Util. 2014;12(3):298–307. doi: 10.1017/S1479262114000082. [DOI] [Google Scholar]
- 35.Gramazio P, Yan H, Hasing T, Vilanova S, Prohens J, Bombarely A. Whole-genome resequencing of seven eggplant (Solanum melongena) and one wild relative (S. incanum) accessions provides new insights and breeding tools for eggplant enhancement. Front. Plant Sci. 2019;10:1–17. doi: 10.3389/fpls.2019.01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huq A, Akter S, Sup I, Hoy N, Kim T, Jin Y, et al. Identification of functional SNPs in genes and their effects on plant phenotypes. J Plant Biotechnol. 2016;43:1–11. doi: 10.5010/JPB.2016.43.1.1. [DOI] [Google Scholar]
- 37.Guo L, Gao Z, Qian Q. Application of resequencing to rice genomics, functional genomics and evolutionary analysis. Rice. 7(1):4. 10.1186/s12284-014-0004-7. [DOI] [PMC free article] [PubMed]
- 38.Xia E, Tong W, Hou Y, An Y, Chen L, Wu Q, et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol Plant. 2020;13(7):1013–1026. doi: 10.1016/j.molp.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka N, Shenton M, Kawahara Y, Kumagai M, Sakai H, Kanamori H, et al. Whole-genome sequencing of the NARO world rice core collection (WRC) as the basis for diversity and association studies. Plant Cell Physiol. 2020;61(5):922–932. doi: 10.1093/pcp/pcaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mammadov J, Aggarwal R, Buyyarapu R, Kumpatla S. SNP markers and their impact on plant breeding. Int J Plant Gen. 2012;2012. 10.1155/2012/728398. [DOI] [PMC free article] [PubMed]
- 41.Tang W, Wu T, Ye J, Sun J, Jiang Y, Yu J, et al. SNP-based analysis of genetic diversity reveals important alleles associated with seed size in rice. BMC Plant Biol. 2016;16(1):1–11. doi: 10.1186/s12870-016-0779-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aaron MK, Hanna M, Banks E, Sivachenko A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms. SnpEff Fly. 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Han X, Zhang Y, Li D, Wei X, Ding X, et al. Deep resequencing reveals allelic variation in Sesamum indicum. MBC Plant Biol. 2014;14. 10.1186/s12870-014-0225-3. [DOI] [PMC free article] [PubMed]
- 48.Anagbogu CF, Bhattacharjee R, Ilori C, Tongyoo P, Dada KE, Muyiwa AA, et al. Genetic diversity and re-classification of coffee (Coffea canephora Pierre ex A. Froehner) from south western Nigeria through genotyping-by-sequencing-single nucleotide polymorphism analysis. Genet Resour Crop Evol. 2019;66(3):685–696. doi: 10.1007/s10722-019-00744-2. [DOI] [Google Scholar]
- 49.Alkimim ER, Caixeta ET, Sousa TV, da Silva FL, Sakiyama NS, Zambolim L. High-throughput targeted genotyping using next-generation sequencing applied in Coffea canephora breeding. Euphytica. 2018;214:50. doi: 10.1007/s10681-018-2126-2. [DOI] [Google Scholar]
- 50.Tran HTM, Furtado A, Alberto C, Vargas C, Smyth H, Lee LS, et al. SNP in the Coffea arabica genome associated with coffee quality. Tree Genet Genomes. 2018;14(72). 10.1007/s11295-018-1282-9.
- 51.Tran HTM, Ramaraj T, Furtado A, Lee LS, Henry RJ. Use of a draft genome of coffee (Coffea arabica) to identify SNPs associated with caffeine content. Plant Biotechnol J. 2018;16:1756–1766. doi: 10.1111/pbi.12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang YJ, Ahn YK, Kim KT, Jun TH. Resequencing of Capsicum annuum parental lines (YCM334 and Taean) for the genetic analysis of bacterial wilt resistance. BMC Plant Biol. 2016;16(1):1–9. doi: 10.1186/s12870-016-0931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee J, Izzah NK, Jayakodi M, Perumal S, Joh HJ, Lee HJ, et al. Genome-wide SNP identification and QTL mapping for black rot resistance in cabbage. BMC Plant Biol. 2015;15(1):1–11. doi: 10.1186/s12870-015-0424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramakrishna G, Kaur P, Nigam D, Chaduvula PK, Yadav S, Talukdar A, et al. Genome-wide identification and characterization of InDels and SNPs in Glycine max and Glycine soja for contrasting seed permeability traits. BMC Plant Biol. 2018;18(1):1–15. doi: 10.1186/s12870-018-1341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones N, Ougham H, Thomas H, Pašakinskienë I. Markers and mapping revisited : finding your gene. New Phytol. 2009;183:935–966. doi: 10.1111/j.1469-8137.2009.02933.x. [DOI] [PubMed] [Google Scholar]
- 56.Pavlopoulos GA, Oulas A, Iacucci E, Sifrim A, Moreau Y, Schneider R, et al. Unraveling genomic variation from next generation sequencing data. BioData Min. 2013;6(13). [DOI] [PMC free article] [PubMed]
- 57.Subbaiyan GK, Waters DLE, Katiyar SK, Sadananda AR, Vaddadi S, Henry RJ. Genome-wide DNA polymorphisms in elite indica rice inbreds discovered by whole-genome sequencing. Plant Biotechnol J. 2012;10(6):623–634. doi: 10.1111/j.1467-7652.2011.00676.x. [DOI] [PubMed] [Google Scholar]
- 58.Subudhi PK, Shankar R, Jain M. Whole-genome sequence analysis of rice genotypes with contrasting response to salinity stress. Sci Rep. 2020;10(1):1–13. doi: 10.1038/s41598-020-78256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, Colleoni C, Zhang J, Liang Q, Hu Y, Ruess H, et al. Genomic analyses yield markers for identifying agronomically important genes in potato. Mol Plant. 2018;11(3):473–484. doi: 10.1016/j.molp.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 60.Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X, et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol. 2012;30(1):105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- 61.Shavrukov Y, Suchecki R, Eliby S, Abugalieva A, Kenebayev S, Langridge P. Application of next-generation sequencing technology to study genetic diversity and identify unique SNP markers in bread wheat from Kazakhstan. BMC Plant Biol. 2014;14(1):1–13. doi: 10.1186/s12870-014-0258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silvestrini M, Junqueira MG, Favarin AC, Guerreiro-Filho O, Mirian PM, Silvarolla MB, Colombo CA. Genetic diversity and structure of Ethiopian, Yemen and Brazilian Coffea arabica L. accessions using microsatellites markers. Genet Resour Crop Evol. 2007;54:1367–1379. doi: 10.1007/s10722-006-9122-4. [DOI] [Google Scholar]
- 63.Aerts R, Berecha G, Gijbels P, Hundera K, Van Glabeke S, Muys B, et al. Genetic variation and risks of introgression in the wild Coffea arabica gene pool in south-western Ethiopian. Evol Appl. 2012:243–52. 10.1111/j.1752-4571.2012.00285.x. [DOI] [PMC free article] [PubMed]
- 64.Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6. 10.1038/nrg1603. [DOI] [PubMed]
- 65.Mekbib Y, Saina JK, Tesfaye K, Eshetu G, Hu G. Chloroplast genome sequence variations and development of polymorphic markers in Coffea arabica. Plant Mol Biol Rep. 2020;38:491–502. doi: 10.1007/s11105-020-01212-3. [DOI] [Google Scholar]
- 66.Aga E, Bekele E, Bryngelsson T. Inter-simple sequence repeat (ISSR) variation in forest coffee trees (Coffea arabica L.) populations from Ethiopia. Genetica. 2005;124:213–214. doi: 10.1007/s10709-005-1484-6. [DOI] [PubMed] [Google Scholar]
- 67.Mishra MK, Nishani S, Gowda M, Padmajyothi D, Suresh N, Sreenath H, et al. Genetic diversity among Ethiopian coffee (Coffea arabica L.) collections available in Indian gene bank using sequence-related amplified polymorphism markers. Plant Breed Seed Sci. 2014;70(1):29–40. doi: 10.1515/plass-2015-0011. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The details of the analyzed 90 accession of C. arabica L.
Additional file 2: Table S2. Summary of the C. arabica L. whole-genome sequencing data.
Additional file 3: Table S3. Amino acid changes identified by SnpEff software. Rows are reference amino acids and columns are changed amino acids. E.g., Row 'A' column 'E' indicates how many 'A' amino acids have been replaced by 'E' amino acids.
Additional file 4: Table S4. Summary count of SNPs with effects on the genome.
Data Availability Statement
The data generated and analyzed during this study are included in this article, its supplemental information files and the sequence data have been deposited into NCBI sequence read archive under submission identifier SUB10821951 with the Bio project identifier PRJNA790687.