Abstract
Despite advances in imaging, image-based vascular systems biology has remained challenging because blood vessel data is often available only from a single modality or at a given spatial scale, and cross-modality data are difficult to integrate. Therefore, there is an exigent need for a multimodality pipeline that enables ex vivo vascular imaging with MRI, CT and optical microscopy of the same sample, while permitting imaging with complementary contrast mechanisms from the whole-organ to endothelial cell spatial scales. To achieve this, we developed ‘VascuViz’ – an easy-to-use method for simultaneous 3D imaging and visualization of the vascular microenvironment using MRI, CT and optical microscopy in the same intact, unsectioned tissue. The VascuViz workflow permits multimodal imaging with a single labeling step using commercial reagents and is compatible with diverse tissue types and protocols. VascuViz’s interdisciplinary utility in conjunction with novel data visualization approaches opens up new vistas in image-based vascular systems biology.
Editor’s summary
VascuViz represents a versatile workflow for multimodal imaging of the vasculature in ex vivo tissue samples across length and resolution scales, paving the way for improved and novel image-based vascular systems biology applications.
Recent advances in imaging methods have made image-based systems biology from the cellular to whole-organ spatial scale a reality1–3. Image-based vascular systems biology involves the acquisition and integration of high-fidelity vasculature related data across spatial scales, and its incorporation in computational models for a holistic understanding of the role of the vascular microenvironment in health and disease. However, this has remained challenging due to the lack of a versatile, multimodality, vascular imaging workflow. This is because labeling blood vessels in preclinical model systems to make them visible in one imaging modality, often precludes the use of other imaging modalities and tissue processing workflows. For example, traditional X-ray or computed tomography (CT)-visible vascular contrast agents4–6 tend to be hydrophobic and polarized, making them invisible on magnetic resonance imaging (MRI), incompatible with immunohistopathology or tissue clearing methods, and therefore difficult to image with complementary imaging methods such as light sheet (LSM) or multiphoton microscopy (MPM). In contrast, the water soluble or fluorescent intravascular labels frequently used for optical imaging7–10 do not contain the contrast moieties necessary to make them visible in MRI or CT images. This limited blood vessel contrast also prevents integration of 3D vascular imaging data acquired using one imaging modality with that acquired using complementary (endogenous or exogenous) contrast mechanisms from the same sample. This includes T1-weighted (T1W) and diffusion-weighted (DW) contrast from MRI, bone contrast from CT, immunohistochemistry or cellular fluorescent protein expression from optical imaging, all of which provide invaluable structural and functional data for vascular systems biology applications. Although many laboratories have reported the development of novel contrast agents11 and imaging techniques7,9 for preclinical vascular applications, these are not ideal for multimodality systems biology workflows. Most vascular contrast agents are optimized for a single imaging modality, tend to require laborious sample preparation4,7,9 or specialized synthesis protocols11, thereby limiting their widespread adoption7,9. While whole brain7–10,12 and whole organ10 vascular mapping techniques have been extensively reported, they often include tissue-destructive methods7,13 that require specialized sectioning and imaging hardware in conjunction with sophisticated image reconstruction techniques to generate 3D maps of the vasculature. Finally, integration of vascular contrasts across imaging modalities and spatial scales spanning several orders of magnitude is often challenging due to the need for externally visible fiducials for image co-registration, and complications that arise from the arboreal topology of the vasculature. Collectively, these hurdles prevent a ‘holistic’ mapping of the vascular microenvironment (VME).
Therefore, we developed an easy-to-use method called “VascuViz” that circumvents the above-mentioned technical challenges and enables multimodality and multiscale 3D imaging of the vasculature in intact, unsectioned tissues using standard sample preparation protocols and commercially available reagents (please see Supplementary Table 1 for a summary of the advantages of VascuViz compared to other vascular imaging workflows). In VascuViz, we combined a water-soluble CT contrast agent (BriteVu®, Scarlet Imaging, UT) with a fluorescently labeled MRI contrast agent (Galbumin-RhodamineB®, BioPAL Inc., MA) to obtain a compound that makes the macro- and microvasculature simultaneously visible in high-resolution imaging with MRI, CT, and optical techniques while also being compatible with conventional tissue processing protocols (e.g. immunohistochemistry and tissue clearing). We demonstrate the widespread utility of VascuViz for vascular systems biology applications by: (i) conducting concurrent high-resolution 3D MRI, CT and optical imaging of the murine vasculature and integrating these multiscale data with complementary endogenous and exogenous image contrasts from the same sample; (ii) showing its compatibility with tissue clearing and conventional histopathology workflows using paraffin-embedded or frozen tissue sections; (iii) exploiting multimodality imaging to characterize the vascular microenvironment in a breast cancer model from the whole-tumor to cancer cell scale; (iv) visualizing multimodality 3D maps of the neurovascular system in the murine brain; (v) simulating hemodynamic contrast from high-resolution 3D neurovascular data and integrating it with cellular scale maps of astrocytic coverage; (vi) multimodality 3D mapping of the vasculature in different murine organ systems; and (vii) mapping in vivo blood flow changes to ex vivo 3D vasculature data acquired from a murine brain. Collectively, we believe these innovations and their broad applicability will make VascuViz invaluable to researchers exploring questions about the VME, and myriad other image-based vascular systems biology applications.
RESULTS:
Vascuviz’s multimodality and multiscale vascular imaging pipeline
Fig. 1 provides an overview of our multimodality, multiscale imaging and visualization (VascuViz) pipeline. First, a GalRh-BVu mixture was prepared by combining the water-soluble CT contrast agent, BriteVu® (Scarlet Imaging, UT) with the fluorescently labeled MRI contrast agent, Galbumin™-Rhodamine-B (BioPAL Inc., MA) (Fig. 1a). Next, intravascular labeling was achieved via transcardial fixation followed by perfusion with the GalRh-BVu mixture (Fig. 1b), after which tissues were excised and immersion fixed overnight. After the GalRh-BVu polymerized, tissues were imaged using endogenous (i.e. due to intrinsic tissue properties) and exogenous (i.e. due to an externally administered label) contrast MRI, CT and optical imaging. MRI images were acquired at 40–100 μm (i.e. macroscopic) spatial resolution (Fig. 1c). Then CT imaging of the same sample was performed at 7.5–9 μm isotropic (i.e. mesoscopic) spatial resolution (Fig. 1d). Next, 10–25 μm tissue sections were prepared and labeled using standard histopathology workflows that included immunohistochemistry (IHC) as well as H&E staining (Fig. 1e). Additionally, thick tissue sections (> 1 mm) were optically cleared (Fig. 1f) and 3D images acquired using MPM or LSM at < 1 μm (i.e. microscopic) spatial resolution (Fig. 1g). Image-based hemodynamic contrasts (e.g. blood flow) were computationally generated (Fig. 1h) using the high-resolution 3D microvascular morphology data. Finally, these multicontrast images at multiple spatial scales were processed and integrated into multilayered 3D data volumes for vascular systems biology applications (Fig. 1i) in the murine brain, kidney, hind limb, an orthotopic breast cancer model, and in bespoke data visualizations.
Fig. 1: Overview of the VascuViz pipeline for multimodality 3D vascular imaging and multiscale data integration:
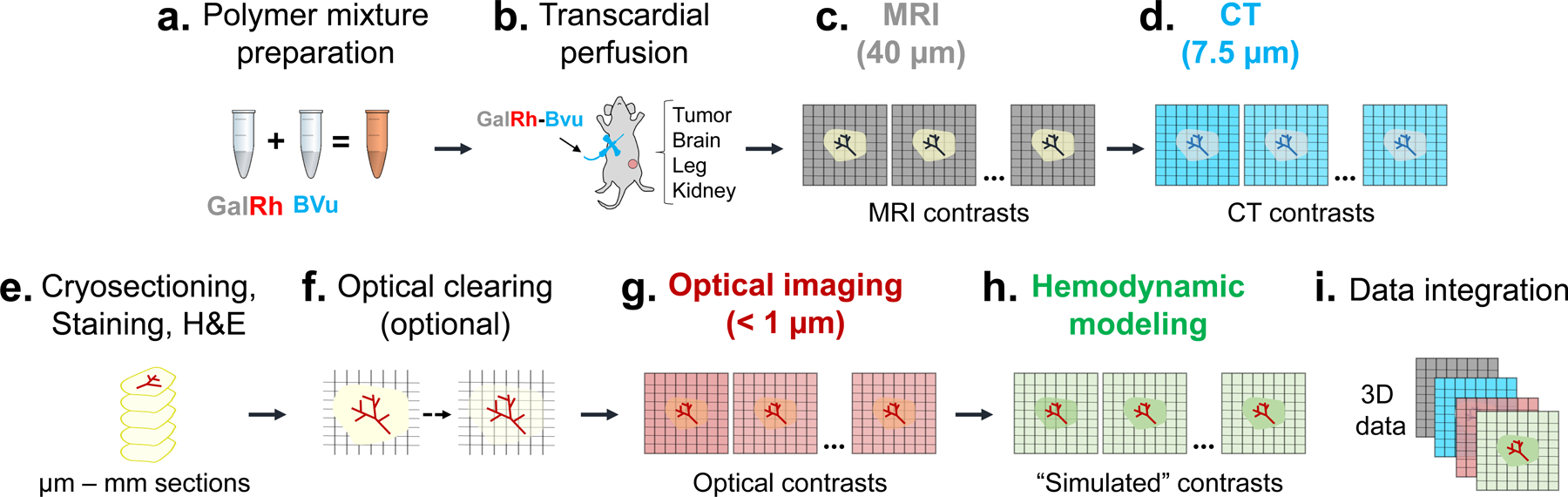
(a) First, the GalRh-BVu mixture was prepared by combining a fluorescently conjugated MRI contrast agent Galbumin™-Rhodamine (GalRh) with a radio-opqaue BriteVu® (BVu) solution (~30 min). (b) Next, vascular labeling was achieved via transcardial perfusion of the GalRh-BVu mixture (~15 min). After perfusion fixation, tumor and healthy tissues (e.g. brain, kidney and hind limb) were excised and immersion fixed overnight. (c) Then, T1-weighted (T1W) and diffusion-weighted (DW) MRI images were acquired at 40 μm and 100 μm isotropic (i.e. macroscopic) spatial resolution, respectively. (d) CT contrasts were acquired from the same sample at 7.5–9 μm isotropic (i.e. mesoscopic) spatial resolution. (e) Then, the sample was sectioned into 10 μm to 1.5 mm slices and immunohistochemical (IHC) or Hematoxylin and Eosin (H&E) staining performed. (f) If required, thicker tissue sections (i.e. > 1 mm) could be optically cleared at this stage. (g) Optical imaging was performed using either 3D light-sheet microscopy (LSM), multiphoton microscopy (MPM) or second harmonic generation (SHG) imaging at 0.5–5 μm lateral and 1–3 μm axial (i.e. microscopic) spatial resolution. (h) Following this step, hemodyamic modeling was performed using high-resolution 3D microvascular network data and additional “simulated” contrasts (e.g. blood flow) generated. (i) Finally, multicontrast data from (c-g) were processed and integrated into 3D data volumes for different vascular systems biology applications.
Concurrent imaging of vasculature and complementary contrasts
To demonstrate the visibility and compatibility of the polymerized GalRh-BVu (i.e. polymer) with other image contrast mechanisms, we performed multicontrast imaging of different tissues with MRI, CT and optical microscopy. Gadolinium-induced shortening of the tissue T1 caused positive enhancement of blood vessel on T1W-MRI (Fig. 2a). The intravascular polymer did not interfere with either endogenous soft tissue (Fig. 2a) or DW-MRI contrast as is evident from the fractional anisotropy (FA) map (Fig. 2b) in which white matter regions in a murine brain (e.g. Corpus Callosum) exhibited elevated FA values (> 0.6) as expected. Moreover, this approach permitted combining these two complementary MRI contrast mechanisms (Fig. 2c). For CT imaging, positive vascular contrast from radio-opaque BVu (Fig. 2d) did not interfere with the X-ray attenuation of bone (Fig. 2e), permitting these two complementary image contrasts to be acquired simultaneously (Fig. 2f). MPM imaging revealed that the polymer bearing vasculature (Fig. 2g) did not hinder cryosectioning or subsequent immunofluorescent labeling of frozen murine brain sections for glial fibrillary acidic protein (GFAP) expression (Fig. 2h). This enabled the concurrent imaging and visualization of GFAP distribution relative to the neurovasculature as shown in Fig 2i. Analogously, stably transduced green fluorescent protein (GFP) expressing MDA-MB-231 breast cancer cells (Fig. 2k, l) and red fluorescent polymer bearing blood vessels (Fig. 2j, l) could be visualized in tissue sections from an orthotopic breast tumor xenograft. In the same tumor sample, one could also image the endogenous second harmonic generation (SHG) signal of collagen fibers (Fig. 2n, o) and fluorescent tumor blood vessels (Fig. 2m, o) without any additional tissue processing. Epifluorescence microscopy images of murine brain sections labeled with antibodies for smooth muscle actin (Fig. 2q, r) and laminin (Fig. 2t, u) in the green channel along with the polymer bearing vasculature in the red channel (Fig. 2p, r, s, u) illustrate the compatibility of VascuViz with conventional immunofluorescence workflows. Finally, the polymer bearing murine tissues could also be optically cleared using the PEGASOS14 method and high-resolution 3D LSM images of the brain vasculature (Fig. 2v), kidney glomeruli (Fig. 2w) and hind limb vasculature (Fig. 2x) acquired. Collectively, these results demonstrate the feasibility of combining VascuViz-induced blood vessel contrast with exogenous and endogenous MRI, CT and optical contrast mechanisms for multimodality and multiscale assessments of the VME in an array of preclinical model systems.
Fig. 2. Concurrent imaging of GalRh-BVu bearing murine tissues with MRI, CT and optical contrast mechanisms:
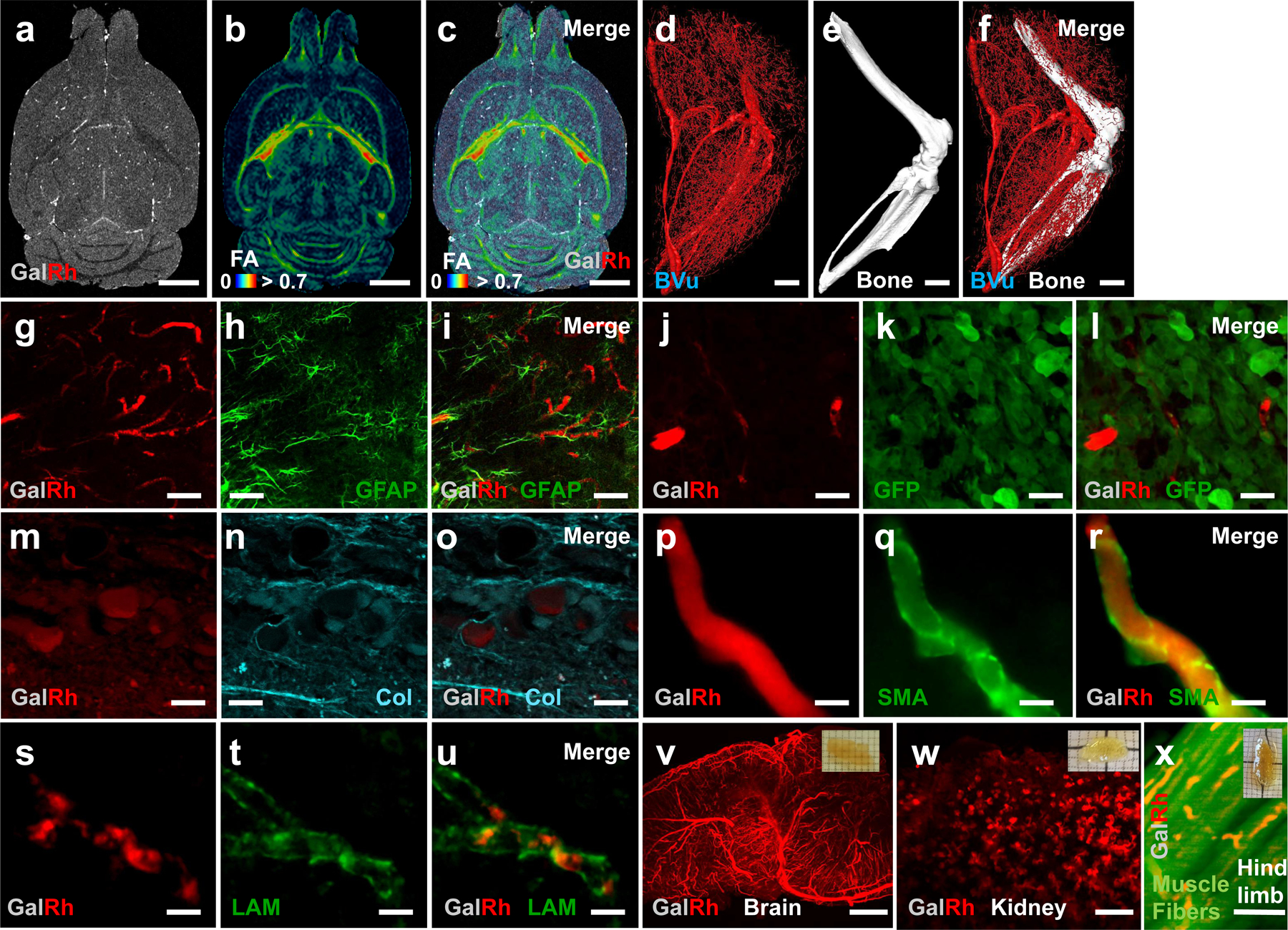
(a) T1W-MRI of a GalRh-BVu bearing murine brain. (b) DW-MRI derived fractional anisotropy (FA) map of the same brain showing elevated (i.e. > 0.6) values in white matter regions. (c) Composite of a-b, illustrating that complementary MRI contrast mechanisms can be simultaneously acquired in the presence of the GalRh-BVu polymer. Simultaneously acquired (d) blood vessel and (e) bone contrast from a GalRh-BVu bearing hind limb using CT. (f) Composite of d-e, showing these contrasts did not interfere with each other. Here, blood vessels are rendered in red and the bone in white. The presence of rhodamine in the polymer made blood vessels fluoresce (red channel) in MPM (g, h, I, j, k, l), epifluroscent microscopy (p-u) and SHG images (m-o). In murine brain tissue sections, the polymer (red channel) did not interfere with fluorescence from other components such as glial fibrillary acidic protein (GFAP) labeled astrocytes (green channel) (h, i), smooth muscle actin (SMA) (q, r) or laminin (LAM) (t, u). In MDA-MB-231 breast tumor sections, the polymer (j, l, m, o) did not interfere with fluorescence from stably transduced green fluorescent protein (GFP) expression of MDA-MB-231 cancer cells (k, l) or the endogenous contrast (cyan channel) from fibrillar collagen (Col) (n, o). The GalRh-BVu bearing tissues could be optically cleared using the PEGASOS protocol as illustrated in the insets of (v-x) for the brain, kidney and hind limb tissues, respectively. High-resolution 3D vasculature data could be acquired using LSM from optically cleared GalRh-BVu bearing (v) brain, (w) kidney and (x) hind limb tissues, and combined with complementary endogenous contrasts such as that from the muscle fibers (x). Scale bars: (a-f, v-x) 1 mm; (g-u) 50 μm. The transparecy of the FA map in (c) was adjusted to enhance the visibility of the soft tissue contrast in the T1W image. (d, f) Blood vessel diameters were scaled by ×1.5 for data visualization. The CNR of the images in (g-i, p-r, v-x) was enhanced by normalizing image intensities to 0.1% of their dynamic range followed by 3D median filtering. Images in (p-x) were enhanced with background subtraction. The upper and lower bounds of the intensity range in merged images (i, l, o, r, u) were adjusted for visualization purposes without any changes to the original data.
Vascuviz is compatible with conventional histopathology workflows
To evaluate the compatibility of the GalRh-BVu polymer with standard histopathology workflows, we performed Hematoxylin and Eosin (H&E) staining on both, paraffin embedded (PE) and frozen tissue sections excised from mice perfused with it. Although paraffin embedding may quench the polymer’s fluorescence, bright-field microscopy of the same sample could be used to detect polymer-bearing blood vessels as shown in Extended Data Fig. 1a–d. The GalRh-BVu polymer appeared dark brown on H&E images as shown for the vascularized tumor rim (Extended Data Fig. 1a, b), and glomeruli (black arrows) and blood vessels of the kidney (Extended Data Fig. 1c, d). In contrast, the polymer’s fluorescence was preserved when H&E staining was performed on frozen tissue sections (Extended Data Fig. 1e–l). The cytoarchitecture of white matter fiber tracts and surrounding tissue were evident in H&E stained regions of the hippocampus (Extended Data Fig. 1e) and the cortex (Extended Data Fig. 1f, g), respectively. H&E staining of tissue sections from a 4T1 breast tumor xenograft (Extended Data Fig. 1i, k) reveal the tumor cytoarchitecture. Extended Data Fig. 1h, j, l show that the red fluorescence from the same polymer-bearing vessels shown in the bright-field images in Extended Data Fig. 1g, i, k was detectable with fluorescence microscopy. These results clearly demonstrated that the vascular contrast agent combination did not interfere with standard histopathology assessments of tissue cytoarchitecture and provided a complementary vascular “tag” for conventional H&E-based histopathological workflows.
Multiscale characterization of the VME in a breast cancer model
We applied our VascuViz pipeline to an orthotopic MDA-MB-231 breast cancer model to enable multicontrast characterization of the vascular microenvironment (VME) from the whole-tumor to cancer cell spatial scale. Multicontrast imaging included the use of macroscopic MRI for blood vessel, soft tissue and cellularity (i.e. necrosis vs. healthy) contrast, mesoscopic CT imaging for vascular contrast, microscopic SHG imaging for contrast from collagen fibers, and MPM for contrast from fluorescent breast cancer cells. The macroscopic MRI data showed the whole-tumor (translucent blue boundary in Fig. 3a) surrounded by large (i.e. 30–114 μm diameter) supporting blood vessels. Co-registration between contrast-enhanced T1W- and DW-MRI enabled simultaneous visualization of the tumor vasculature with fractional anisotropy (FA) (Fig. 3b) and apparent diffusion coefficient (ADC) (Fig. 3c) maps that reflected the cellularity within the vascular microenvironment (VME). The average tumor FA (0.14 ± 0.07) and ADC (0.57 ± 0.18 × 10−3 mm2/s) were lower than that for the surrounding healthy tissue (0.2 ± 0.07 and 1.3 ± 0.34 × 10−3 mm2/s), respectively. Concurrently acquired 3D CT (Fig. 3d) data enabled computation of the inter-vessel distances within the VME as shown by the Euclidian distance map (EDM) in Fig. 3e. The mean and maximum inter-vessel distances within the tumor (80 ± 56.5 μm and 320 μm) were greater than those for surrounding healthy tissue (53 ± 34.8 μm and 150 μm), and complemented the cellularity data (i.e. ADC maps) from DW-MRI (Fig. 3b–c). To directly correlate inter-vessel distance to cellularity, we divided the 3D EDM map into sub-regions corresponding to intervessel distances of 0–50 μm, 51–150 μm and 151–350 μm, respectively (Extended Data Fig. 2a–c). Next, these sub-region boundaries were mapped on to co-registered ADC maps (Extended Data Fig. 2d–f) to facilitate their segmentation into rim and central tumor sub-regions (Extended Data Fig. 2g–h). This approach revealed that ADC for the 151–350 μm EDM regions (5.4 ± 2.2 × 10−4 mm2/s) was significantly elevated (p << 0.001) relative to that for 51–150 μm (3.8 ± 2.4 × 10−4 mm2/s) and 0–50 μm EDM regions (4.3 ± 2.8 × 10−4 mm2/s), respectively (Extended Data Fig. 2i). Collectively, these integrated macroscopic and mesoscopic VME data demonstrated that avascular sub-regions (i.e. regions with elevated intervessel distances) were more necrotic (i.e. exhibited elevated ADC) relative to sub-regions that were well-vascularized (i.e. regions with low intervessel distances) and non-necrotic (i.e. exhibited lower ADC).
Fig. 3: Multicontrast characterization of the vascular microenvironment (VME) in a human breast cancer model.
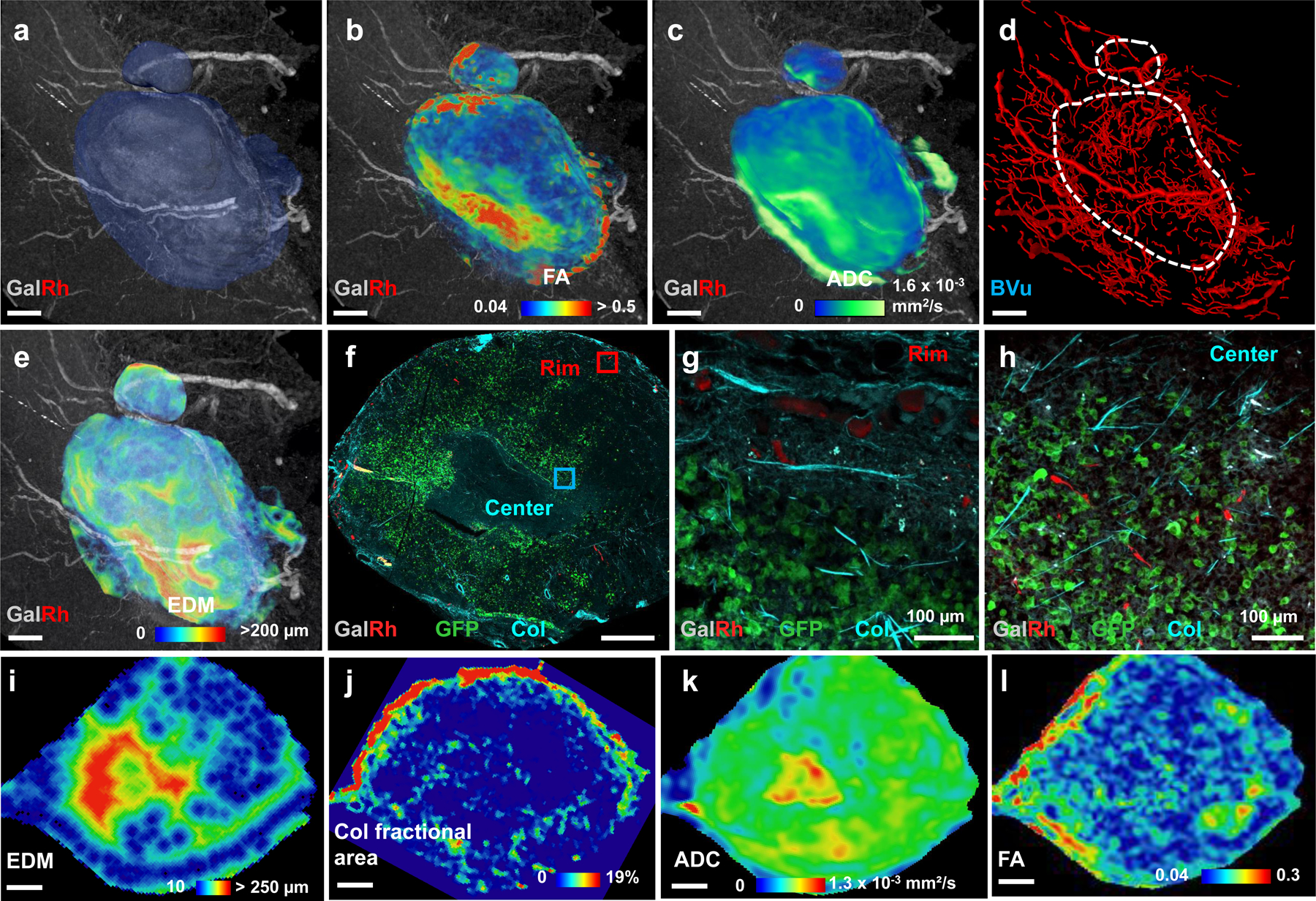
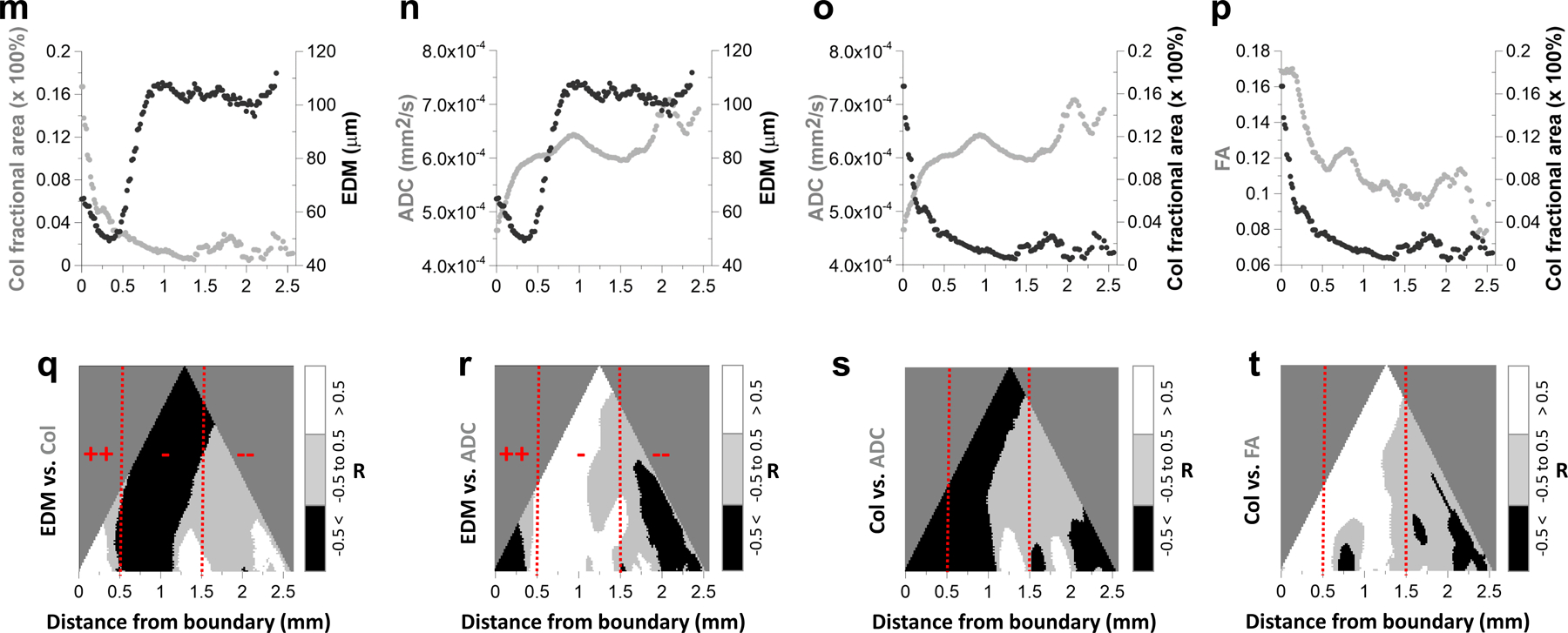
(a) T1W-MRI (40 μm) of the polymer bearing MDA-MB-231 breast tumor xenograft. Here, large (i.e.70–160 μm diameter) feeding blood vessels could be visualized due to the GalRh component of the polymer. Blue region represents the tumor extent. Simultaneous visualization of the vasculature with DW-MRI (100 μm) derived (b) fractional anisotropy (FA) and (c) apparent diffusion coefficient (ADC) maps. (d) Microvascular network (red) of the tumor and surrounding tissue derived from CT (9 μm). Blood vessels are scaled by their mean diameters (9–160 μm). (e) Euclidian distance map (EDM) computed from the CT-derived vascular network in (d) showing the presence of large inter-vessel distances (250–300 μm) within the tumor. (f) MPM-derived MIP image of a 25 μm thick section from the tumor center showed capillaries (red), green fluorescent protein (GFP) expression from MDA-MB-231 cancer cells (green) and fibrillar collagen (Col) from SHG imaging (blue). Here, vascular contrast was due to the presence of rhodamine in the polymer. Representative regions from the tumor rim (red box) and center (blue box) in (f) were magnified (10×) to illustrate the presence of a well-perfused rim (g) in contrast to a relatively avascular tumor center (h). For data integration, each parameter map i.e. EDM (i), Col fractional area (Col Fr Area) (j), ADC (k) and FA (l) was resampled to match the spatial resolution (40 μm) of T1W-MRI and its orientation matched to that of the SHG imaging data. Scatter plots illustrating the mean tumor boundary-to-center profiles of (m) Col Fr Area w.r.t EDM; (n) ADC w.r.t EDM; (o) ADC w.r.t Col Fr Area; and (p) FA w.r.t. Col Fr Area. Hierarchical correlation plots illustrating the spatial correlations of: (q) EDM vs. Col Fr Area; (r) EDM vs. ADC; (s) Col Fr Area vs. ADC; and (t) Col Fr Area vs. FA as a function of distance from the tumor boundary. Correlation coefficients > 0.5 are displayed in white; those from −0.5 to 0.5 in gray; and < −0.5 in black. Potential regions of efficacious drug delivery (++) were within 0–0.5 mm; those with intermediate efficacy of delivery (−) were within 0.5 – 1.5 mm; and regions with potentially poor drug delivery (--) were at distances > 0.5 mm. Red hashed lines indicate 0.5 mm and 1.5 mm from the tumor boundary. Scale bars: 1 mm unless indicated otherwise.
Additionally, the macroscopic MRI and mesoscopic CT data could be integrated with microscopic SHG and MPM measurements for a 25 μm thick tumor section (Fig. 3f) from the same sample. Fig. 3g shows an example of a well-vascularized region of interest (red square in Fig. 3f) from the tumor rim, while Fig. 3h shows a relatively avascular region from the tumor center (cyan square in Fig. 3f). To directly relate macro- and mesoscopic VME parameters to those assessed with microscopic SHG and MPM, we co-registered EDM data and the collagen (Col) fractional area from SHG to the ADC (Fig. 3k) and FA maps (Fig. 3l) as shown in Figs. 3i and 3j, respectively. Next, we computed parameter distributions as a function of their distance from the tumor boundary (Extended Data Fig. 3a–b). These analyses revealed that the mean ADC and EDM were highest near the tumor center (x = 2.5 mm) and lowest at the boundary (x = 0.25mm) (Fig. 3n). FA and Col fractional area (Fig. 3p) profiles showed the opposite trend with the highest values at the boundary and the lowest values near the tumor center. In contrast, both Col fractional area and EDM (Fig. 3m) and ADC and Col fractional area profiles (Fig. 3o) changed in the opposite directions from the tumor boundary to the center. Overall, EDM showed a positive correlation with ADC (R2 = 0.43, p = 0.0084) (Extended Data Fig. 3c) and was inversely correlated with Col fractional area (R2 = 0.35, p = 0.0183) (Extended Data Fig. 3d). A significant correlation was also observed between FA and ADC (R2 = 0.68, p = 0.0002) (Extended Data Fig. 3e) and between FA and Col fractional area (R2 = 0.7, p = 0.0001) (Extended Data Fig. 3f).
To visualize the potential impact of tumor microenvironmental factors on drug transport, we computed 2D “hierarchical correlation” plots using tumor boundary-to-center profiles of intervessel distance (i.e. EDM) vs. Col fractional area (Fig. 3q), EDM vs. ADC (Fig. 3r), Col fractional area vs. ADC (Fig. 3s) and Col fractional area vs. FA (Fig. 3t). These plots helped identify regions of limited drug and nutrient delivery as defined in terms of elevated EDM (i.e. intervessel distance), elevated ADC (i.e. necrosis) and elevated Col fractional area (i.e. extracellular matrix density) – all of which pose barriers to convective and diffusive transport within the tumor microenvironment. For example, near the tumor boundary (i.e. 0–0.5 mm), intervessel distance and Col fractional area were positively correlated as a result of simultaneous decrease in both parameters (Fig. 3m, q), while ADC and intervessel distance (Fig. 3n, r) were negatively correlated. These factors are suggestive of efficacious (denoted by ‘++’) drug delivery in this region due to increased vascular density (i.e. reduced EDM) and a concurrent reduction in collagen fiber density. In contrast, between 0.5–1.5 mm from tumor boundary, one observes that elevated intervessel distance was negatively correlated with Col fractional area (Fig. 3m, q), and positively correlated with ADC (Fig. 3n, r). Simultaneously, a gradual decrease in Col fractional area was negatively correlated with ADC (Fig. 3o, s) and positively correlated with FA (Fig. 3p, t). These factors suggest intermediate (denoted by ‘-’) efficacy of drug transport in this region due to contrasting decreases in vascular density (i.e. elevated EDM) and collagen fiber density. Finally, for regions at distances >1.5 mm from the tumor boundary, the hierarchical correlation plots exhibited further increases in intervessel distance (Fig. 3m) accompanied by a sharp rise in ADC (Fig. 3n), and small increases in Col fractional area (Fig. 3o) and FA (Fig. 3p). Collectively, these observations suggest that this region will be characterized by poor (denoted by ‘--’) drug delivery attributable to necrosis (i.e. elevated ADC) and enhanced barriers to convective (i.e. stagnant Col fractional area) and diffusive transport (i.e. elevated EDM).
Visualizing multimodality data for neurovascular systems biology
We demonstrated the feasibility of using the VascuViz pipeline to generate complementary multiresolution, multimodality 3D data from the murine brain for neurovascular systems biology investigations (Fig. 4). This included anatomical and diffusion-weighted contrast from macroscopic MRI, vascular contrast from mesoscopic CT, and cellular contrast from LSM. Ten representative brain regions were segmented (Fig. 4a) and visualized in 3D: Olfactory Bulb (OB), Cortex (CTX), Caudate Putamen (CP), Thalamus (TH), Amygdala (AG), Hypothalamus (HY), Hippocampus (HP), Striatum (ST), Cerebellum (CB) and Brain Stem (SM) (Fig. 4a). GalRh-BVu contrast-enhanced T1W-MRI data enabled visualization of large blood vessels (i.e. diameters > 40 μm) in different brain regions (Fig. 4a). Fig. 4b shows the entire 3D microvascular network of the same brain derived from CT data acquired at 7.5 μm spatial resolution wherein blood vessels were scaled according to their mean diameters (7.5–180 μm). The overall fractional blood volume (FV) derived from the CT data was 3 ± 1%. Using the CTX, HP, CB and TH as representative examples, we illustrated how VascuViz was used to combine CT contrast at 7.5 μm with complementary DW-MRI contrast at 50 μm (Fig. 4c–t) and LSM contrast at 1.3–5 μm spatial resolution (Fig. 4w–z), in these regions. VascuViz enabled co-registration between T1W-MRI and CT, and facilitated mapping of the microvascular network (at 7.5 μm) in these brain regions as illustrated for the CTX (Fig. 4c), HP (Fig. 4d), TH (Fig. 4e) and CB (Fig. 4f). The gray isosurfaces delineate the regional 3D neuroanatomy from T1W-MRI while the neurovasculature is rendered in red and scaled by its mean diameters. Next, these regional maps were complemented with 3D FA and principal diffusion direction information computed from DW-MRI. We illustrate the integration of these data for the CTX (Fig. 4g–i), CB (Fig. 4j–l), HP (Fig. 4o–q) and TH (Fig. 4r–t). The soft tissue contrast from T1W-MRI (40 μm) was used as the anatomical reference (Fig. 4g–i, j–l, o–q, r–t) with red, blue, green colors representing diffusion along the medial-lateral, rostral-caudal, and dorsal-ventral axes, respectively (Fig. 4i, l, q, t). The creation of these co-registered multiresolution, multicontrast volumes permitted the simultaneous assessment of FA and local FV distributions within the Cornu-Ammonis (CA) and the Dentate Gyrus (DG) fields of the hippocampus (Fig. 4n, u, v). Comparisons of the coefficient of variation (COV) between the CA and DG revealed larger FV heterogeneity in CA (COV=0.43) compared to DG (COV=0.39). This correlated with the larger FA heterogeneity in CA (COV=0.53) compared to DG (COV=0.49). Finally, to facilitate the integration of macroscopic vascular contrasts with complementary LSM contrasts at the cellular scale, a 1.5 mm tissue section from the same brain was optically cleared and LSM data acquired from the highlighted region of interest (white square) from the CTX (Fig. 4g), CB (Fig. 4j) and TH (Fig. 4r) and visualized as maximum intensity projection (MIP) images as shown in Figs. 4m, w, x–z, respectively. Fig. 4x, y, z illustrates how microscopic vascular contrast acquired with LSM can be complemented with GFAP coverage in the thalamus. Collectively, these results demonstrate the utility of VascuViz for generating multicontrast 3D maps of different murine brain regions, and enabling the visualization of complementary information ranging from the spatial scale of the whole brain down to that of individual capillaries, without necessitating additional sectioning or processing of brain tissue.
Fig. 4: Visualization of multimodality 3D data from the murine brain for multiscale neurovascular systems biology.
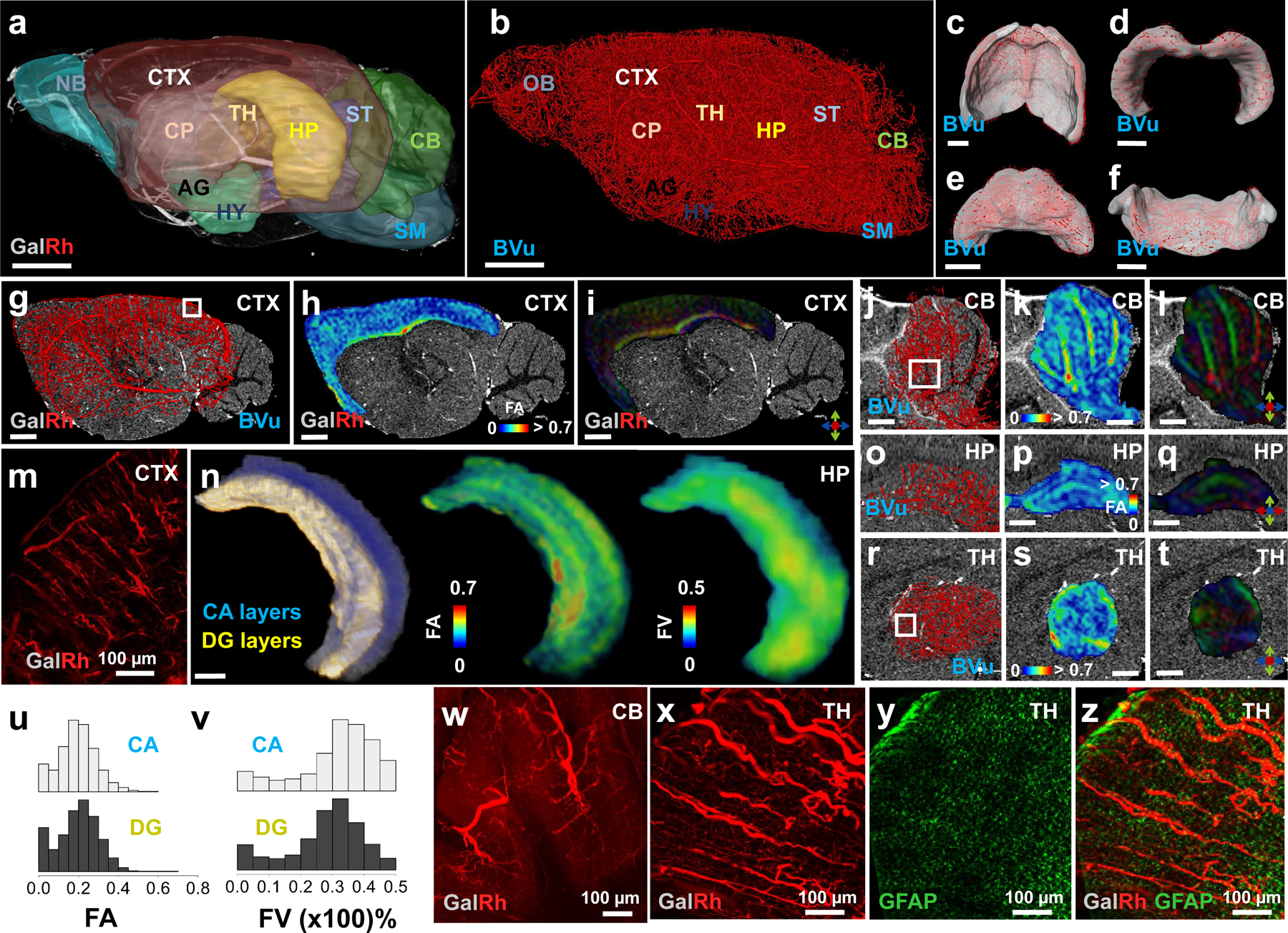
(a) Sagittal 3D view of polymer contrast enhanced T1W-MRI (40 μm) of a murine brain wherein ten representative regions of interest (ROI) are visualized: Olfactory bulb (OB), Cortex (CTX), Caudate Putamen (CP), Thalamus (TH), Amygdala (AG), Hypothalamus (HY), Hippocampus (HP), Striatum (ST), Cerebellum (CB) and Brain stem (SM). GalRh enabled T1-enhancement and visualization of large blood vessels (i.e. diameter > 40 μm). (b) 3D microvascular network of the same brain derived from CT (7.5 μm) wherein blood vessels are scaled by their mean diameter and brain regions identified from (a). CT-derived “regional” microvascular networks are shown for the CTX (c), HP (d), TH (e) and CB (f). Red: vasculature; gray: volumetric ROI of interest obtained from (a). Regional microvascular network data (g, j, o, r) derived from b was combined with DW-MRI-derived FA (h, k, p, s) and principal diffusion directional data (i, q, t) for the CTX in g-i; CB in j-l; HP in o-p and TH in r-t. Diffusion axes in i,l,q,t indicate medial-lateral (red), rostral-caudal (blue) and dorsal-ventral (green) directions. The soft tissue contrast from (a) was used as the anatomical underlay in (g-i, j-l, o-t). (n, u, v) CT-derived local fractional blood volume (FV) data complements MRI data for the right HP: (n) (left) Composite of T1W-MRI and 3D masks for the Cornu-Ammonis (CA) (blue) and Dentate Gyrus (DG) layers (yellow); corresponding maps of the FA (center) and FV (right). Stacked distributions of (u) FA and (v) FV (x 100%) for the CA and DG layers in (n). (m, w, x and y) LSM contrasts for CTX, CB and TH, highlighting complementary optical data visualized for the same tissue as in (a-b). MIP images of the vasculature for: CTX (m) from the ROI in (g), 5 μm; CB (w) from the ROI in (j), 1.3 μm and TH (x) from the ROI in (r), 1.3 μm. (y) MIP image of GFAP expression in TH from the ROI in (r), 1.3 μm. (z) Composite of (x) and (y). Scale bars: 1 mm unless indicated otherwise. The intensity range in (a, c-f, g-t, z) was adjusted for visualization purposes without altering the original data.
Integrating in vivo CBF dynamics with 3D neurovascular data
To further demonstrate the utility of the VascuViz pipeline for neurovascular systems biology applications, we mapped in vivo cerebral blood flow (CBF) changes in response to carbogen gas inhalation assessed with dynamic laser speckle contrast (LSC) imaging (7.7 μm), to ex vivo microvasculature and anatomical data acquired using 3D CT imaging (7.7 μm) (Extended Data Fig. 5 and Supplementary Movie 1). An anesthetized, head-fixed mouse (Extended Data Fig. 5a) was made to breathe room air for 3 min, followed by carbogen (i.e. 95% O2 and 5% CO2) gas for 2 min and room air again for 5 min (Extended Data Fig. 5b) and the functional hyperemic response (i.e. CBF increase due to the carbogen gas perturbation) imaged continuously via a thinned-skull (Extended Data Fig. 5a) using in vivo LSC imaging (Extended Data Fig. 5f–k). Initial 2D intrinsic optical signal (IOS) imaging facilitated the in vivo visualization of small (i.e. diameter < 40 μm), medium (i.e. 40 < dimeter < 60 μm) and large (i.e. diameter > 60 μm) cortical blood vessels (Extended Data Fig. 5c, d and Extended Data Movie 1). Extended Data Fig. 5f–k depicts the in vivo functional hyperemic response in the same blood vessels at representative time points during baseline, carbogen inhalation and recovery phases of the perturbation paradigm. These functional in vivo data revealed an increase in CBF between 3 to 5 minutes in response to carbogen inhalation followed by a gradual return to baseline during the next 5 minutes. A maximum CBF increase of 85% was observed for medium and large blood vessels in contrast to a 45% increase for small blood vessels relative to global mean baseline CBF (Extended Data Fig. 5e). We then euthanized the animal, processed it with the VascuViz workflow (Extended Data Fig. 5l) and imaged the skull and cerebrovasculature with micro-CT and confocal microscopy (< 1 μm). Ex vivo CT imaging facilitated 3D visualization of the cranial anatomy and underlying vasculature (Extended Data Fig. 5m–o), as well as co-registration with in vivo CBF maps via cranial and vascular landmarks. As before, blood vessels were clearly visible at multiple spatial scales ranging from the cortical vascular network (Extended Data Fig. 5m–o) down to individual microvessels (Extended Data Fig. 5p–q) in the same sample. We also created a compelling 4D visualization in which the in vivo functional hyperemic response was mapped on to the 3D anatomical and microvasculature data as shown in Extended Data Fig. 5r–w. In this visualization, one can clearly appreciate the time evolution of the functional hyperemic response to carbogen inhalation, which is even more apparent in the accompanying movie (Supplementary Movie 1). Collectively, these results demonstrate the utility of VascuViz for conducting structural and functional assessments of the neurovasculature by exploiting complementary image contrast derived from the surrounding VME.
Hemodynamic modeling using VascuViz-derived 3D vascular data
We demonstrate the feasibility of generating simulated hemodynamic contrasts (e.g. blood flow) in sub-cortical regions (e.g. thalamus) of the murine brain (shown in Fig. 4) using an “image-based” computational modeling approach. The microvascular network employed for the blood flow simulations is shown in Fig. 5a. It consisted of 12,857 segments and corresponded to a FOV of 300 μm × 300 μm × 100 μm. Microscopic resolution (0.33 μm) LSM data revealed that blood vessel diameters ranged from 1.4–42 μm with a mean value of 3.2 ± 3.1 μm (Fig. 5b). Prior to applying our blood flow model to this data, we validated our approach using a widely used and publicly available dataset derived from a 546 segment rat mesentery microvascular network15 (Extended Data Fig. 6a). We observed excellent agreement between our simulated blood flow rates and discharge hematocrit and those reported in Pries et al15 as indicated by R2 = 0.99 and 0.93, respectively (Extended Data Fig. 6b–c). Our results were also found to satisfy the Fahraeus and Phase Separation effects that are known to be significant in the microcirculation15 (Extended Data Fig. 6e–f). Following validation, we applied our blood flow model to the thalamic microvasculature by imposing a pressure differential ranging from 75 mmHg in inlet vessels to 15 mmHg in outlet vessels and obtained the intravascular pressure distribution shown in Fig. 5c. The resulting blood flow was then visualized on a log scale as shown in Fig. 5d. This image-based simulation resulted in a mean blood flow velocity of 0.7 ± 4.1 mm/s which was within the experimentally observed and simulated ranges reported by others for the murine brain (see Supplementary Table 2 for a summary). Our simulated blood flow rates (nl/min), intravascular pressures (mmHg) and shear stresses (dyne/cm2) were also in excellent agreement with previously reported simulations for the murine brain (Supplementary Table 2).
Fig. 5: Computationally generated hemodynamic contrast from VascuViz-derived 3D neurovascular data.
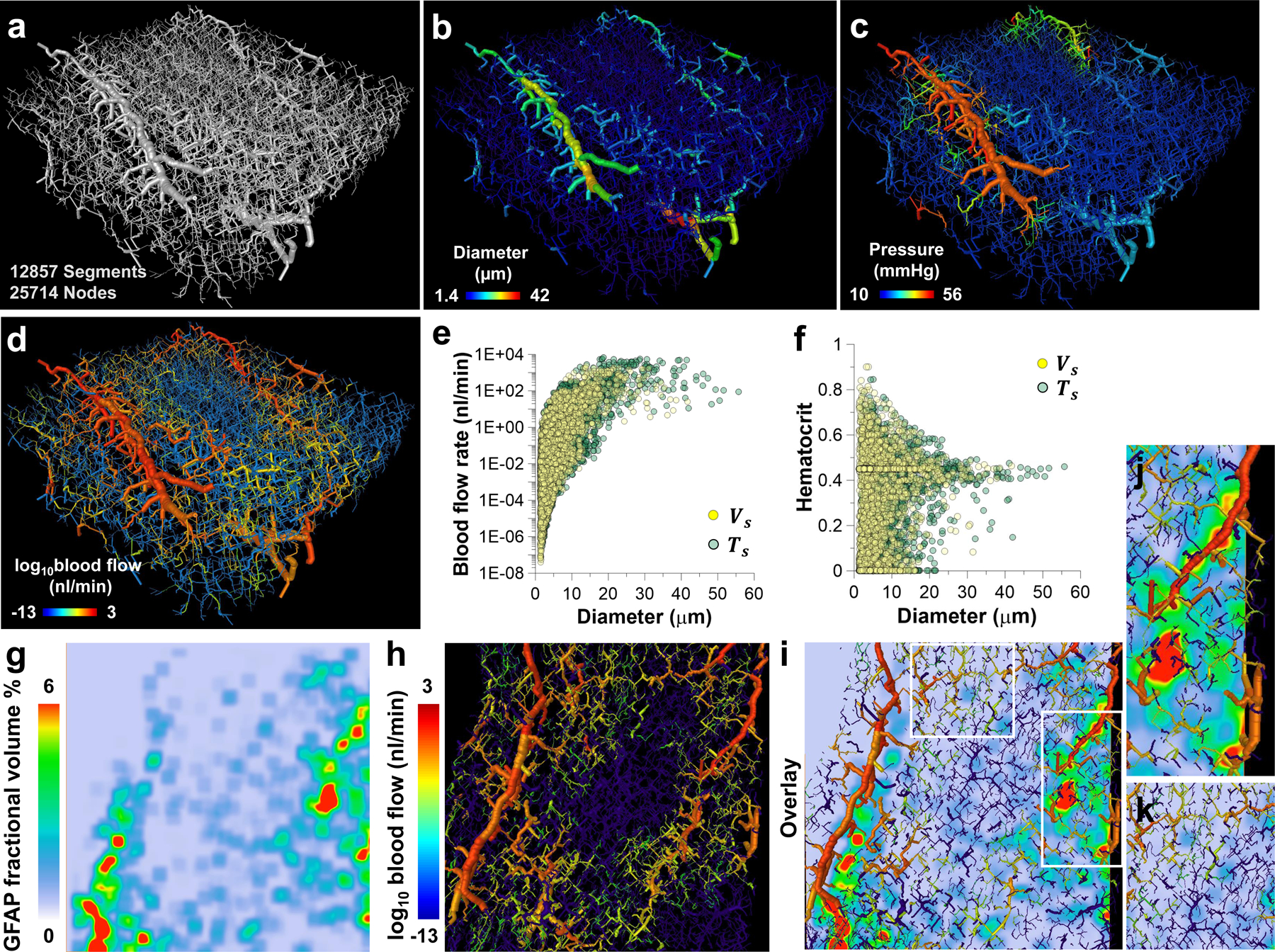
The polymer bearing murine brain tissue section was optically cleared and imaged at a high spatial resolution (0.33 μm) using LSM. (a) A 300 μm × 300μm × 100 μm FOV of the thalamic microvasculature was extracted from the 3D LSM data. (b) Diameter map of the microvascular network in (a), wherein each blood vessel was scaled and color-coded by its mean diameter (1.4–42 μm). (c) Map of the simulated intravascular pressure (mmHg) wherein each blood vessel was scaled by the mean diameter and color-coded by the average pressure across its end points. (d) Map of the simulated blood flow (nl/min) wherein each blood vessel was scaled by the mean diameter and color-coded by the log of the blood flow. Scatter plots of: (e) blood flow (nl/min) vs. blood vessel diameter (μm), and (f) hematocrit vs. blood vessel diameter (μm). Data are shown for two cases: Case I (yellow circles) employed a vessel-based scaling factor ( = 1.25); and Case II (green circles) employed a tissue-based scaling factor (= 1.67) to account for vascular shrinkage due to tissue clearing. These simulated data were complemented with the fractional volume of the GFAP label computed from LSM data (0.33 μm) as shown in (g-k). (g) Map of the LSM-derived fractional GFAP volume (%) corresponding to a z-location 30 μm below the top plane in volume (a); (h) blood flow (nl/min) map corresponding to the same z-location; (i) overlay of the fractional GFAP volume (%) (g) and blood flow (nl/min) (h). White boxes in (i) highlight ROI in which elevated blood flow (nl/min) co-localized with high fraction of GFAP labeling (> 4%), and (j) ROI in which low blood flow (nl/min) co-localized with low fraction of GFAP labeling (< 4%) (k).
In our hemodynamic simulations, we accounted for the vascular shrinkage that results from the use of dehydrating reagents during tissue clearing (Fig. 5a). A vessel shrinkage-based scaling factor () was defined as the ratio of maximum blood vessel diameter measured from the CT image to that measured from LSM data in the same FOV, and was equal to 1.25 (i.e. Case I). We also considered the scenario in which the entire tissue and blood vessels shrank proportionately due to optical clearing (i.e. Case II). To account for this, a second tissue shrinkage-based scaling factor () was defined as the ratio of the brain’s dimensions in the rostral-caudal and dorsal-ventral directions before and after optical clearing. This yielded a mean value of 1.67. Collectively, these two cases represented the lower (1.25) and upper bound (1.67) of blood vessel distortions attributable to tissue handling, processing and dehydration caused by optical clearing. Comparison between the blood flow and hematocrit distributions for Case I () and Case II ( are presented in Fig. 5e–f, respectively. We found that the mean diameter of large vessels (i.e. diameters > 8 μm) in Case II (13 ± 6.3 μm) was slightly greater than those for Case I (12.5 ± 5.1 μm). Consequently, this resulted in an elevated mean blood flow for Case II (7.0 ± 57.4 nl/min) relative to Case I (2.8 ± 18.9 nl/min). Despite these differences, the mean values of all the simulated parameters were within previously reported ranges for the murine brain (see Supplementary Table 3 for a summary). Fig. 5g–j demonstrates the feasibility of combining simulated hemodynamic contrast with complementary contrast (i.e. GFAP expression) acquired with LSM at 0.33 μm. We observed that large blood vessels (i.e. 25–40 μm diameter) within the neurovascular network exhibited elevated blood flow rates (86.7 ± 250.6 nl/min) and co-localized with regions of high GFAP expression (> 4%) (Fig. 5i, j). Analogously, small blood vessels (i.e. 1.4–8 μm diameters) exhibited lower blood flow rates (0.2 ± 0.9 nl/min) and co-localized with regions of low GFAP coverage (< 4%) (Fig. 5i, k). Collectively, these results demonstrate how the VascuViz pipeline enables image-based simulations of multiple hemodynamic contrasts as well as their integration with complementary LSM contrasts at the microscopic scale.
Multimodality 3D vascular mapping in other murine organ systems
We showcase how the VascuViz pipeline can also be used to conduct 3D mapping of the vasculature in other organs besides the murine brain, such as the hind limb (Fig. 6a–g) and kidney (Fig. 6h–m) for which public 3D anatomical atlases are not currently available. The soft tissue contrast from T1W-MRI (40 μm) facilitated slice-by-slice anatomical segmentation and manual annotation of these organs (Fig. 6a, h). The hind limb tissue was segmented into ten major muscle groups: Tibialis Anterior (TA), Extensor Digitorum Longus (EDL), Soleus (SOL), Gluteus Maximus (GA), Biceps Femoris Anterior (BFA), Biceps Femoris Posterior (BFP), Semitendinosus (ST), Rectus Femoris (RF), Vastus Medialis (VM) and Vastus Lateralis (VL) as illustrated in the MRI-derived volume rendering shown in Fig. 6b. High-resolution CT images (9 μm) revealed the 3D architecture of the whole hind limb vasculature (Fig. 6c–e) and bone tissue (Fig. 6d, e). A vascular landmark-based co-registration of the macroscopic MRI and mesoscopic CT data facilitated mapping of the murine hind limb vasculature in different muscle groups (Fig. 6c). Figs. 6f, g demonstrate the feasibility of using 3D LSM (2.6 μm) to map the muscle fiber morphology using endogenous fluorescence contrast from the same sample. Similarly, kidney tissue was segmented into the Cortex (CTX), Medulla (MED) and Renal Pelvis (RP) using the soft tissue contrast from T1W-MRI (60 μm). Next, CT data (Fig. 6i) was co-registered with the T1W-MRI data using the same vascular landmark-based approach to co-register the 3D kidney vasculature (Fig. 6j) from CT with that acquired using T1W-MRI. This approach also enabled the use of tissue masks from T1W-MRI (e.g. renal CTX) to visualize vasculature in a given region of interest, as shown for the renal CTX (Fig. 6k). Finally, we demonstrated the feasibility of using ultra high-resolution nano-CT (2 μm) and LSM to map the kidney glomerular morphology as illustrated in Fig. 6l and Fig. 6m, respectively.
Fig. 6: Multimodality 3D mapping of the vasculature in the murine hind limb and kidney.
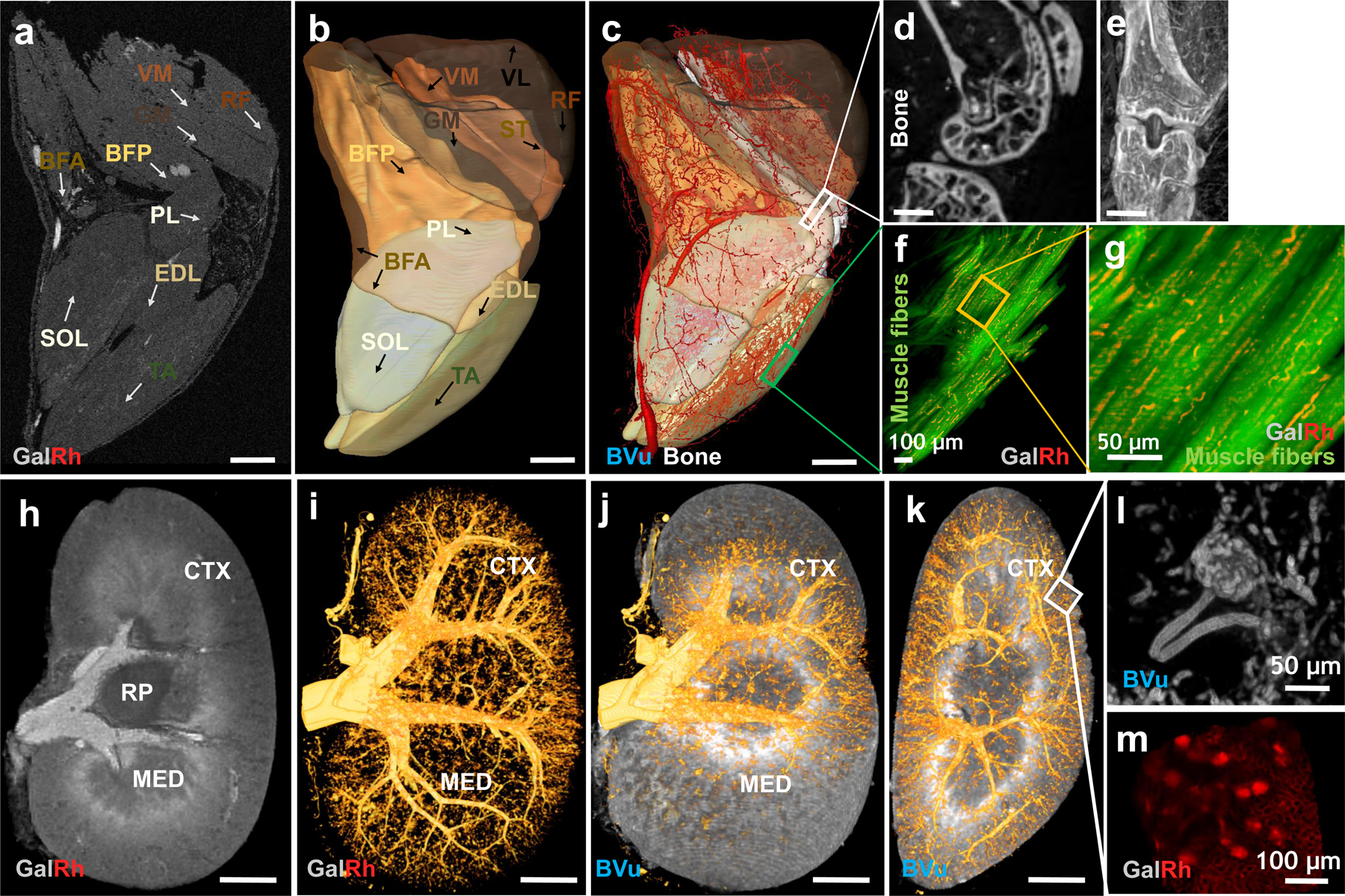
Ten major muscle groups in the hind limb were manually segmented using T1W-MRI data: Tibialis Anterior (TA), Extensor Digitorum Longus (EDL), Soleus (SOL), Gluteus Maximus (GA), Biceps Femoris Anterior (BFA), Biceps Femoris Posterior (BFP), Semitendinosus (ST), Rectus Femoris (RF), Vastus Medialis (VM) and Vastus Lateralis (VL). Three major regions in the kidney were also manually segmented using T1W-MRI data: Cortex (CTX), Medulla (MED) and Renal Pelvis (RP). (a) Axial T1W-MRI (40 μm) of the hind limb showing cross sections of eight muscle groups within the FOV (except the VL and ST). Blood vessels appeared bright due to GalRh while the bone appeared dark in T1W-MRI. (b) All ten muscle groups visualized in 3D using transparent isosurface rendering. (c) Co-registered T1W-MRI (40 μm) and CT (9 μm) data enabled simultaneous visualization of the vasculature in all the muscle groups along with contrast from the bone. Blood vessel diameters were scaled by ×1.5 for data visualization. These data could be complemented with high-resolution bone morphology data from CT (18 μm) (d-e) as well as (f-g) LSM derived optical contrast (0.6 μm) in which muscle fibers (green channel) and capillaries (red channel) were visible. (h-m). Macro- to microscopic scale 3D mapping of the vasculature in a polymer bearing murine kidney. (h) Sagittal T1W-MRI (40 μm) shows the Renal Cortex (CTX), Medulla (MED), Renal Pelvis (RP) and large renal blood vessels (i.e. diameter > 40 μm) within the FOV. (i) High-resolution (7.5 μm) CT-derived microvascular network in the same kidney. (j) Co-registered data from (h) and (i) enabled visualization of the vasculature in the renal CTX as shown in (k). These data could be complemented with glomeruli data acquired using ultrahigh-resolution (2 μm) nano-CT (i) or (1.2 μm) LSM of an optically cleared sagittal kidney tissue section (1 mm thickness) (m). The positive glomerular contrast was due to BVu in nano-CT and GalRh in LSM acquired images, respectively. Scale bars: 1 mm unless indicated otherwise. The intensity range was adjusted in merged images (b, c, j, k) for visualization purposes without any changes to the original data.
METHODS:
Preparation of the GalRh-BVu mixture
To prepare the GalRh-BVu mixture, one part of the BriteVu® CT contrast agent (Scarlet Imaging, UT) was mixed with 4.5 parts of distilled water and 2% w/v BriteVu Enhancer® (Scarlet Imaging, UT). The solution was warmed to 60–70°C before removing it from the heating plate and adding the Galbumin™-Rhodamine B, a fluorescently conjugated MRI contrast agent (BioPAL Inc., MA) at a concentration of 0.96 mg/ml. The final GalRh-BVu mixture was maintained at ~40–50°C until it was ready to be perfused into the animal.
Vascular labeling with GalRh-BVu
All animal experiments were conducted in accordance with an approved Johns Hopkins University Animal Care and Use Committee (JHU ACUC) protocol. Mice were housed in barrier caging, under standard dark/light cycle, ambient temperature and humidity conditions in the Johns Hopkins University animal facility, which is accredited by the American Association for the Accreditation of Laboratory Animal Care, and meets the National Institutes of Health standards as set forth in the “Guide for the Care and Use of Laboratory Animals”. The GalRh-BVu mixture was injected transcardially into the vasculature of three anesthetized orthotopic MDA-MB-231 tumor bearing NCr nu/nu mice, one healthy NCr nu/nu mouse and one orthotopic 4T1-BALB/C tumor bearing mouse. Please see the accompanying protocols document for a detailed perfusion procedure.
MRI
Please see the accompanying protocols document for pre-imaging sample preparation steps. Samples were imaged on a 9.4T vertical bore MRI scanner (Bruker BioSpin Corp, Billerica, MA) using either a 10- or 20-mm RF volume coil and ParaVision (v5.1) MRI software. T1-weighted (T1W) images were acquired using a 3D-FLASH sequence with a flip angle = 30°, TE/TR = 4.2/40 ms, 4 averages, and 40 μm isotropic spatial resolution. Diffusion tensor imaging (DTI) data were acquired using a 3D diffusion-weighted (DW) GRASE sequence16 with TE/TR = 32/800 ms, 12 echoes per excitation, 2 averages, diffusion gradient duration/separation = 2.8/10 ms, and 16 directions with a b-value = 1700 s/mm2 and 100 μm isotropic spatial resolution, zero-padded to 50 μm during reconstruction.
CT Imaging
Please see the accompanying protocols document for pre-imaging sample preparation steps. CT (CT) imaging was performed on a Skyscan 1275 scanner (Bruker, USA) using the following acquisition parameters: 0.5 mm aluminum filter, 55 kVp, 145 μA, 335 ms exposure time, 0.2 rotation step, 3 averages. The brain and kidney samples were imaged at 7.5 μm, and the hind limb and tumor samples at 9 μm isotropic spatial resolution, respectively. 3D CT image reconstruction was performed using the NRecon software (v1.7.0.4). Ultra-high spatial resolution (2 μm) data of the kidney glomeruli were acquired and reconstructed using the nano-CT Xradia Versa 520 scanner at the Biotechnology Resource Center Imaging Facility at Cornell University using the following parameters: 120 kVp, 145 μA, 0.2 rotation step, 3 averages.
Immunofluorescent labeling and histology
Please see the accompanying protocols document for pre-labeling tissue preparation steps. The polymer bearing brain tissue sections were cut onto silanized slides for immunofluorescent labeling. Astrocytes were labeled with Alexa Flour 488 conjugated monoclonal GFAP (GA5) mouse antibody (dilution 1:50, Cell Signaling, Danvers, MA), vasculature associated smooth muscle with FITC-conjugated α-smooth muscle actin antibody (dilution 1:50, Sigma-Aldrich, St. Louis, MO), blood vessel endothelium with anti-rabbit laminin antibody (dilution 1:100, Sigma-Aldrich, St. Louis, MO). The secondary antibody used was goat anti-rabbit IgG Alexa Fluor 488 (10 μg/ml, ThermoFisher, MA) to complement rhodamine emission from the GalRh-BVu polymer. H&E staining on paraffin-embedded tissues was conducted at the Johns Hopkins Pathology Core facility using their standard pathology workflow.
Tissue clearing
Please see the accompanying protocols document for details of the tissue clearing. The polymer bearing murine brain, kidney and hind limb samples (1–2 mm) were optically cleared using the PEGASOS method14.
Ex vivo optical imaging
Tissue sections (< 20 μm) were imaged on a Nikon ECLIPSE-TS100 microscope (Nikon Instruments Inc., NY) at 2×, 10× and 40× magnification with the appropriate filters for detecting immunofluorescence.
Multiphoton Microscopy (MPM)
Tumor and brain tissue sections (25–50 μm) were imaged on an Olympus laser scanning FV1000 MPE multiphoton microscope. Incident laser light at 860 nm was used for acquiring the second harmonic generation (SHG) images from tumor collagen (Col) fibers as well as for the two-photon excitation of GFP from cancer cells, GFAP from brain astrocytes, and rhodamine fluorescence in blood vessels. The SHG signal from collagen fibers was detected at 420–460 nm, GFP/GFAP fluorescence at 495–540 nm, and rhodamine fluorescence at 575–630 nm, respectively. The 5 mm × 5 mm tumor section was imaged at 1 μm spatial resolution with a 3 μm z-interval using a 10× objective while a 25× lens was used to acquire multiple 500 μm × 500 μm fields-of-view from the tumor rim and core at 0.1 μm spatial resolution with a z-interval of 2 μm. Similarly, a 200 μm × 200 μm fields-of-view were acquired for the brain sample at 0.1 μm spatial resolution with a z-interval of 3 μm.
Light Sheet Microscopy (LSM)
Optically cleared brain, hind limb and kidney tissue sections (> 1 mm) were imaged on a light sheet microscope (Ultramicroscope II, LaVision BioTec, Germany) using benzyl benzoate-PEG14 as the mounting medium. Excitation wavelengths were 488 nm for Alexa 488 fluorophore and 561 nm for the GalRh-BVu polymer. The corresponding emission wavelengths detected were between 525–550 nm and 620–660 nm, respectively. 3D images of the brain vasculature were acquired at the following spatial resolutions using 1 μm z-step size: cortex, 5 μm; cerebellum, 0.65 μm; and thalamus vessels and their astrocytic coverage, 0.33–0.56 μm. Similarly, 3D images of the glomeruli in the kidney cortex and hind limb vasculature were acquired at 0.6 μm lateral resolution.
In vivo optical functional imaging
To demonstrate the compatibility of the VascuViz technique with in vivo functional imaging workflows, a 6-week-old, homozygous Nu/J male mouse (Jackson Laboratories, Bar Harbor, ME) was maintained under 1% isoflurane (Iso Flo, Cat. No. 06–8550-2/R1) anesthesia mixed with air using a Vapomatic Model 2 vaporizer (AM Bickford, Inc., NY) and administered at 1.5 L/min via a nose cone. Next, a thinned-skull cranial window was surgically prepared according to17 after which the animal’s head was secured in a custom-designed stereotaxic frame. Following this, in vivo optical imaging was conducted using a benchtop imaging system18 at high spatial (7.7 μm) and temporal (5 s) resolution. This included imaging of total hemoglobin (HbT) absorption or cerebral blood volume (CBV) via intrinsic optical signal (IOS) contrast imaging under 570 ± 5 nm illumination and cerebral blood flow (CBF) via laser speckle contrast (LSC) imaging under 632.8 nm illumination, respectively. First, baseline IOS and LSC data were acquired for 3 min under room air inhalation. Then, the animal was subjected to carbogen (95% oxygen/5% carbon dioxide) gas inhalation for 2 min followed by room air inhalation for another 5 minutes (i.e. recovery phase) with dynamic LSC data being continuously acquired. Following in vivo imaging, the animal was perfused with the vascular contrast agent combination of GalFITC-BVu (BioPAL Inc., MA) at a concentration of 0.45 mg/ml and imaged ex vivo using 3D CT and confocal microscopy as described above.
Segmentation of vasculature from 3D CT and LSM/MPM data
Blood vessels were segmented from 3D images using the Interactive Learning and Segmentation toolkit Ilastik (v 1.3)19 (Heidelberg, Germany) as described in the accompanying protocols document.
CT-derived EDM map
The Euclidian distance map (EDM) was calculated from CT imaging data using the 26-voxel neighborhood chamfer metric in Amira™ (v5.4).
Computation of fractional blood volume and fractional area maps
To calculate the fractional blood volume for each brain region, a 3D binary vessel mask was generated by implementing a logical AND operation between the binary tissue mask derived from T1W-MRI and the co-registered CT data. Next, using the “material statistics” feature in Amira™ (v5.4), we calculated the fractional blood volume for each brain region as the ratio of the total number of blood vessel voxels from CT to the total number of tissue voxels from T1W-MRI. The mean fractional blood volume of the whole brain was the average fractional blood volume of the ten brain regions.
Additionally, we used custom MATLAB® (vR2017A) code to generate local fractional blood volume maps (FV) from binary 3D CT vascular data. To do this, we replaced each voxel with the vascular fraction computed within a sliding volume of 20×20×20 voxels. The FV varied from 0–1, where 0 indicated no blood vessel voxels detected within the window, and 1 indicated that the window consisted only of blood vessel voxels. A similar approach was employed to generate the GFAP fractional volume map from the 3D LSM vascular data. This included a preprocessing step to eliminate stripe artifacts in LSM images by applying an fast Fourier transform (FFT) based structural filter in FIJI (v2.1). For the 2D data, MIP images from SHG and MPM data (co-registered with MRI and CT data) were used to generate maps of the Col fractional area and vascular fractional area.
DW-MRI derived ADC, FA and 3D direction-encoded color maps
Fractional anisotropy (FA), apparent diffusion coefficient (ADC) and 3D direction-encoded color (DEC) maps were computed at 50 μm isotropic spatial resolution from DW-MRI data using DTI-Studio software20 (www.mristudio.org) as described previously16.
Boundary-to-center profiles of VME parameters
First, 2D binary mask of the tumor was generated using FIJI (v2.1). Then, a distance map was generated from the binary mask using custom MATLAB® code (vR2017A) having a set of contours that were parallel to the shape of the boundary as shown by white points in Extended Data Fig. 3a, b. Next, a set of radial lines that were oriented normal to each contour were generated as shown by the dashed red lines in Extended Data Fig. 3a, b. Using a grid of 150 (radial)×210 (azimuthal) points mapped on to each parameter map (Extended Data Fig. 3a, b), a mean boundary-to-center profile was calculated and plotted using the Grapher™ 12 software.
Hierarchical correlation plots for identifying VME niches
To visualize the relationship between boundary-to-center profiles of EDM, Col fractional area, ADC and FA, we generated hierarchical correlation plots using custom MATLAB® code (vR2017A). First, for each parameter’s profile, combinations of all possible sub-profiles were computed by varying the number of data points in each. For example, the Pearson correlation coefficient computed and displayed at level 1 (i.e. at the apex of the hierarchical correlation plot), corresponded to that for the entire profile (e.g. N data points) for each variable, while correlation coefficients displayed at level 2 (i.e. just below the apex) corresponded to that between two sub-profiles comprised of (N-1) data points for each variable, and so on. In this plot, Pearson correlation coefficients < −0.5 were color coded black, those between −0.5 to 0.5 color coded grey, and those > 0.5 color coded white. Thus, the hierarchical correlation plot provided a visualization of the spatial correlation (i.e. at discrete distances from the tumor boundary) between variable pairs that characterized the VME. Moreover, in each plot we identified zones of efficacious (++), intermediate (−) or poor (−) drug transport within the tumor based on the spatial correlations between these image-derived parameters.
Image-based hemodynamic modeling
Reconstruction of the microvascular network:
A GalRh-BVu polymer bearing murine brain section (1.5 mm) was optically cleared and imaged at microscopic spatial resolution (0.33 μm) over a FOV of 300 μm × 300 μm × 100 μm using LSM. First, vascular segmentation and skeletonization were performed as described above. Then, the “filament editor” tool in Amira™ (v5.4) was employed to visualize and remove isolated vessel segments at the domain boundary to yield a fully connected microvascular network that comprised of 12,900 nodes and 12,899 vessel segments. We observed that a total of 64 nodes (i.e. 0.4%) were of the 4th degree. As there was insufficient information available on the location of vascular trifurcations in the murine brain, we converted these 4th degree nodes into bifurcating nodes according to the following geometric rules: 4th degree nodes that were connected to at least one first degree node were converted to 3rd degree nodes by removing the boundary segment from the network. This step was equivalent to removing isolated segments at the boundary as described earlier. Fourth degree nodes that only had 3rd degree connections were converted into bifurcations by inserting a new node between the 4th degree node and one of its neighboring nodes, and then connecting this new node with its two neighbors. These steps enabled us to generate a fully connected microvascular graph suitable for computational hemodynamic simulations.
Scaling of blood vessel diameters and lengths:
To account for tissue and vascular distortions due to optical clearing, we computed two geometric scaling factors for each blood vessel segment. First, we estimated the vessel-based scaling factor (Vs) using the CT measurements (7.5 μm) of the murine thalamus from which a small FOV was also acquired using LSM (0.33 μm). The ratio between the maximum diameters observed in CT data to that observed in LSM data was measured to be 1.25. Next, a tissue-based scaling factor (Ts) was estimated as the ratio between the dimensions of the tissue sample before and after optical clearing in the rostral-caudal (1.67±0.06) and dorsal-ventral (1.65±0.17) directions. These two measurements helped define the lower (1.25) and upper (1.67) bounds for the geometric scaling factor for the microvascular network.
Boundary conditions:
To assign boundary conditions for blood flow simulations, we first needed to define the inlets and outlets of the microvascular network. All nodes that were < 8 μm and only had one connection (i.e. boundary nodes) were assigned zero flow according to21. Next, from the remaining vessel segments (i.e. 8–50 μm in diameter), inlets or outlets were identified as those segments with diameters larger than the mean diameter of 12 μm. Then, to distinguish inlets from outlets, additional criteria were imposed based on the known microvascular topology of the murine brain and other rodent models22,23. For example, inlets (i.e. arterioles) were defined as having more branches and smaller mean diameter (15.1 μm) than outlets (i.e. venules) (15.7 μm)22. These criteria resulted in the identification of 14 inlet and 10 outlet nodes. The ratio of the number of inlets/outlets (1.4:1) in the microvascular network was in agreement with observations from other rodent models23. Following this classification, constant pressure boundary conditions were prescribed at all inlets (75 mmHg) and outlets (15 mmHg)21.
Blood flow model:
We adapted our recently reported blood flow model24 for this data as follows: our model was based on the 1D formulation of Poiseuille’s law and accounted for nonlinear rheological effects of blood flow25. To solve for pressure at all the blood vessel junctions and subsequently blood flow rate in each vessel segment under the above-mentioned boundary conditions, a system of linear equations was derived to satisfy mass conservation at all the interior vessel nodes. To solve for hematocrit in each vessel segment, we employed the in vivo viscosity law25. The pressure and hematocrit values were iteratively computed for each vessel segment until convergence was reached24. To validate our hemodynamic model, we applied our blood transport model to a publicly available 546 segment rat mesentery microvascular network15 according to24. We then computed the correlation between our simulated blood flow rates, discharge hematocrit and discharge to tube hematocrit ratio and those reported in Pries et al15. Additionally, we determined if the simulated fractional erythrocyte flow vs. fractional blood flow distributions obtained using our approach exhibited the phase separation effect reported by Pries et al15.
Integrating ex vivo 3D data from MRI, CT and optical imaging
Tumor data:
We first segmented the tumor based on its soft tissue contrast on T1W-MRI images. ADC and FA maps were co-registered to the T1W-MRI data using a rigid body transformation in DiffeoMap (www.mristudio.org)20. Vascular CT data and CT-derived EDM maps were co-registered to the T1W-MRI data via a vascular landmark-based co-registration approach26 using Amira™ (v5.4). To co-register MPM data with MRI data, the tumor sample was embedded in an agarose block (Extended Data Fig. 4a) prior to CT imaging. This enabled us to match the sample’s orientation before CT imaging to that after MPM. Before sectioning of the tumor sample, the location of the cutting plane relative to the tumor center was visually aligned with the T1W-MRI data as illustrated in Extended Data Fig. 4a. This orientation was preserved throughout the entire cutting (Extended Data Fig. 4b) and cryosectioning (Extended Data Fig. 4c) process with the help of directional annotations on the sample.
MPM images were pre-processed using FIJI (v2.1)27.Maximum intensity projection (MIP) images were calculated from vascular, Col and GFP image z-stacks (~50 μm in thickness), respectively. Each intensity histogram was normalized to 0.1% of its dynamic range followed by smoothing with a 2D median filter (radius = 2 pixels), and visual alignment of the MPM data to the MRI data. This alignment was possible because VascuViz made the vasculature concurrently visible in the MRI, CT and MPM images. Finally, the MPM image was co-registered to the target MRI slice using an affine transformation based on a normalized mutual information approach in Amira™ (v5.4). To correlate MPM contrasts with those from MRI and CT, we used the average boundary-to-center profiles for each parameter as described previously.
Brain data:
We first computed a nonlinear registration based on diffeomorphic metric mapping29 between the DW-MRI data and a reference anatomical atlas29 in MRI-Studio20 to generate 3D labels representing ten distinct brain regions of interest. The label assignments were validated by visual inspection and regional boundaries edited for accuracy using the segmentation toolbox in Amira™ (v5.4). To map the Cornu Ammonis (CA) and the Dentate Gyrus (DG) within the hippocampus, hippocampal data from the Australian Mouse Brain Mapping Consortium (AMBMC, www.imaging.org.au/AMBMC) were co-registered to the T1W-MRI data in Amira™ (v5.4) using a normalized mutual information based affine transformation. Next, DW-MRI derived FA, 3D direction- encoded color (DEC) map data and vascular CT data were co-registered to T1W-MRI data as described above. The 3D spatial graph representing the neurovasculature was manually aligned with 3D T1W-MRI data by using the large blood vessels as internal landmarks.
Hind limb and kidney data:
We first manually segmented ten muscle groups in the hind limb based on soft tissue contrast from T1W-MRI using Amira™ (v5.4), and annotated each muscle by matching our data with previously reported data on muscle anatomy30. Next, vascular CT data were co-registered to T1W-MRI data via a landmark based registration approach that employed fiducials placed on large blood vessels (> 40 μm)26 and bone. This enabled the 3D mapping and visualization of the hind limb vasculature in ten different muscle groups as well as around the bone. To combine LSM contrasts with the 3D vasculature, the Tibialis Anterior (TA) muscle was isolated, optically cleared and imaged to visualize muscle fibers and capillaries at 2.6 μm. Similarly, to perform 3D mapping of the vasculature in kidney, ROI such as the cortex, medulla and renal pelvis were segmented based on soft tissue contrast from T1W-MRI data using Amira™ (v5.4) that was followed by co-registration between vascular CT data and T1W-MRI data as described above. Then, blood vessels were segmented from the vascular CT images and combined with the cortical ROI using a logical AND operation to generate a cortex specific 3D vascular tree.
Integration of dynamic in vivo CBF data with ex vivo vascular CT data
Data pre-processing:
First, 2D cerebral blood flow (CBF) maps were computed from laser speckle contrast (LSC) data as described previously32. Next, a background mask was generated using ilastik (v 1.3)19 (Heidelberg, Germany) and subtracted from the CBF maps to enhance the vascular CNR. Next, IOS data was used to quantify in vivo vascular morphology as described in the previous section. Ex vivo vascular and bone masks were also derived from 3D CT images using ilastik (v 1.3). Finally, %ΔCBF was computed for each vessel w.r.t a global mean baseline CBF (i.e. % ΔCBF = (CBF − CBFbaseline)/CBFbaseline×100) to enable visualization of the functional hyperemic response.
Co-registration of in vivo to ex vivo data:
2D maps of dynamic CBF data were co-registered to CT-derived 3D bone and neurovascular data via a landmark-based registration approach that employed fiducials placed on blood vessels 26 and cranial landmarks (e.g. the bregma). These combined data resulted in a 4D image volume that was then converted to a movie of the dynamic functional hyperemic response to carbogen gas inhalation by using the “5D Explorer” plugin in Amira (v 2020, Visage Imaging, San Diego, CA, USA).
Statistical analysis
Pearson correlations between boundary-to-center EDM profiles, ADC, FA and Col fractional area were calculated using NCSS (v11) statistical software (NCSS, Kaysville UT). A two-tailed, nonparametric Mann–Whitney U-test was used (α = 0.05) to determine if there were any significant differences between the ADC distributions corresponding to the different EDM ranges. To demonstrate that the polymer did not interfere with other fluorescent moieties, optical microscopy was used to acquire multiple fields of view (FOV) from GalRh-BVu perfused tissue samples. The MDA-MB-231 breast tumor sample (FOV = 2) brain tissue (FOV = 1) were imaged with MPM. The brain sample was also imaged with epifluorescence microscopy (FOV = 2). Finally, to demonstrate the compatibility of the VascuViz workflow with tissue clearing, a GalRh-Bvu perfused and PEGASOS cleared brain was imaged using LSM (FOV = 3).
Data Availability
The authors declare that all the data supporting the findings of this study are included within the paper and its Extended Data and supplementary information files. This includes the availability of imaging data for: (i) the murine brain in Figs. 4 and 5, Supplementary Fig. 5–7 and Supplementary Tables 2 and 3; and Supplementary Movie 1, (ii) the breast tumor xenograft in Fig. 3 and Extended Data Figs. 2 and 3; (iii) the kidney and hind limb in Fig. 6. Finally, IHC and H&E data are available in Fig. 2 and Extended Data Fig. 1. The mouse hippocampal data that was used for the labeling of the CA and DG layers can be accessed freely from the Australian Mouse Brain Mapping Consortium (AMBMC) weblink: www.imaging.org.au/AMBMC.
Code Availability
MATLAB® code used in the manuscript will be made available upon reasonable request from the corresponding author.
DISCUSSION:
Image-based vascular systems biology necessitates a versatile multimodality imaging approach that enables acquisition of vascular data in a single tissue from the macroscopic whole organ scale to the mesoscopic vascular network scale, down to the microscopic scale of individual cells without requiring additional sample processing for each complementary imaging method employed. To achieve this, we developed a method called VascuViz that enables concurrent multimodality, multiscale imaging and 3D visualization of the vascular microenvironment (VME) using MRI, CT and optical microscopy in intact, unsectioned tissues.
There have been recent reports of elegant tissue preparation and imaging approaches for microvascular applications in a range of preclinical models5,7,9,10,33–35. However, many of these techniques are not suitable for multimodality imaging workflows (as summarized in Supplementary Table 1) as they require specialized sample preparation protocols5,7,9,10,33 (e.g. resin embedding, cell labeling, optical clearing, decalcification) or involve tissue sectioning prior to imaging, or both7,13. The use of unique or bespoke vascular tags and labels (discussed in detail below) often precludes the use of complementary imaging methods due to their deleterious effects on endogenous and exogenous tissue contrast, and tissue sectioning hampers subsequent 3D imaging and histopathological analyses. Collectively, this limits the utility of such approaches in multimodality imaging workflows. In contrast, some non-destructive techniques34 do enable high-resolution 3D vascular imaging in tissue samples, but are often limited by their need for specialized hardware. For example, high resolution X-ray synchrotron imaging33,34 requires a particle accelerator to generate high energy X-rays, which limits its accessibility and widespread use in multimodality imaging workflows.
As shown, VascuViz overcomes these limitations and enables multimodality, multiscale, 3D imaging and visualization of the vasculature with MRI, CT and LSM/MPM in intact, unsectioned tissues. The systemic perfusion of the entire animal during the VascuViz protocol ensured that the vasculature within any organ system could be imaged, and integrated with images of the same sample acquired with complementary (endogenous or exogenous) image contrast mechanisms without the need for additional tissue preparation or imaging hardware. VascuViz expedited multiscale imaging workflows because it did not involve relabeling of the blood vessels prior to multimodality imaging, immunohistochemistry or optical clearing. It also enabled the incorporation of high-fidelity 3D microvascular data in computational biology applications, such as hemodynamic simulations of blood flow in tissues. Finally, VascuViz is applicable for imaging healthy or pathologic vasculature in any preclinical disease model.
Conventional vascular contrast agents and intravital labeling methods have not been amenable to multimodality imaging workflows due to the following limitations. The synthesis of many vascular contrast agents is optimized for maximal contrast-to-noise ratio (CNR) with a single imaging modality or a given contrast mechanism, such as radiopaque compounds for CT, Gadolinium chelates for MRI, and fluorescent probes for optical imaging, which limits their use with complementary imaging modalities. Recently, a few dual modality (e.g. MRI/optical or CT/optical) preclinical vascular contrast agents such as Galbumin-Rhodamine (Biopal, Inc., MA) and Pu4ii (vasQtech, Zurich, Switzerland) have become commercially available. However, these too have been optimized for initial imaging with MRI or CT followed by fluorescence microscopy and are invisible to other imaging modalities. Additionally, the physiochemical properties of vascular contrast agents such as their lipophilicity11, water insolubility6, polarization upon polymerization6, mismatched viscosity for vascular filling33, toxicity36, burdensome handling36 have limited their use for multimodality imaging pipelines that culminate in immunohistopathology workflows. Some vascular contrast agents11 have been shown to leak from perfused organs even in combination with other agents, preventing their use in tissue clearing protocols and subsequent imaging with LSM/MPM. Finally, other characteristics such as the low CNR of intravital labeling methods (e.g. intravital lectins, anti-CD31 antibodies) in comparison with lumen-filing gels, and the similarity of X-ray attenuation coefficients of radiopaque compounds (e.g. Microfil®) to bone necessitate additional sample preparation steps37 such as combining vascular labeling with bone decalcification37, combining two or more antibody labels35, and specialized image processing routines35 for vascular data extraction.
Here we demonstrated that our GalRh-BVu combination significantly simplified the multimodality vascular imaging workflow in preclinical models by obviating the need to relabel blood vessels differently for each modality. Also, one did not need to expend resources on quality control during synthesis of the polymer’s components or on reproducibility of the conjugated fluorophores since they are commercially available. In contrast to the lengthy sample preparation times required for intravascular resins (typically 4 days)4 or knife-edge-scanning-microscopy (KESM) protocols (typically 1–3 weeks)13, the tissues bearing polymerized GalRh-BVu could be concurrently imaged with commercially available MRI, CT or 2D/3D optical microscopy systems within 24 hours of fixation, and also be successfully integrated and detected in routine histopathology workflows based on paraffin-embedded or frozen tissue sections.
We also demonstrated the compatibility of GalRh-BVu polymer with tissue clearing workflows that enabled submicron resolution imaging of the vasculature with LSM/MPM. Moreover, Extended Data Fig. 7 shows successful LSM imaging of a GalRh-BVu polymerbearing and optically cleared brain eleven months after sample preparation. This confirms the long-term stability of the polymer and its associated contrast moieties within the vasculature for up to a year. In the future, VascuViz could be adapted to be compatible with tissue clearing protocols geared towards specific organ systems such as the kidney68 or muscle69. The GalRh-BVu polymer enabled comprehensive mapping of the VME by enabling the integration of 3D vasculature with complementary contrast mechanisms acquired from the same tissue at spatial scales ranging from the macroscopic whole-organ level (e.g. DW-MRI) to the microscopic or cellular level (e.g. collagen fibers, cancer cells). To the best of our knowledge, this is the first study to report a multimodality, multiscale pipeline for image-based vascular systems biology that is compatible with such a plethora of ex vivo imaging methods (please see Supplementary Table 1 for a summary of these capabilities and a comparison with other techniques reported in the literature).
We demonstrated the utility of VascuViz for multicontrast characterization of the vascular microenvironment (VME) in an orthotopic MDA-MB-231 breast cancer xenograft. At the whole-tumor (i.e. macroscopic) scale, our observations of intratumoral necrosis were consistent with similar DW-MRI based measurements in breast tumor xenografts38–40 that showed decreased FA and elevated ADC in these regions. Additionally, analysis at the vascular network (i.e. mesoscopic) scale exhibited a strong positive correlation between elevated ADC and EDM, suggestive of necrosis at the tumor center41,42. These observations were consistent with necrosis resulting from diffusion- or perfusion-limited hypoxia resulting from an abnormal, angiogenic and heterogenous tumor vasculature43,44. The lack of GFP-expressing tumor cells within the tumor center assessed at the microscopic scale using MPM further corroborated these observations.
Tumor boundary-to-center profiles facilitated comparisons between different image-derived VME parameters across spatial scales. For example, Col fractional area profiles revealed the greatest fiber density at the tumor boundary and lower fiber density towards the center. While heterogeneous distribution of collagen fiber density between peripheral and central regions has been observed in different preclinical tumor models45, the elevated peripheral collagen fiber density in our preclinical breast cancer model is consistent with the well-known invasiveness of the MDA-MB-231 breast cancer cell line. Our whole-tumor analysis showed an inverse correlation between intervessel distance (i.e. EDM) and Col fractional area. These observations are consistent with elevated measurements of microvascular and collagen fiber density at the tumor boundary in contrast to the tumor center reported in other preclinical studies of breast cancer45. Analogously, at the whole-tumor level we observed a positive correlation between FA and Col fractional area profiles, consistent with previous studies implicating the role of collagen fibers in modulating the directionality of water diffusion in a similar breast cancer model41. Finally, we also observed a negative correlation between ADC and Col fractional area profiles, which might be indicative of the presence of a hypoxic phenotype characterized by large intervessel distances, decreased collagen fiber density and elevated ADC at the tumor center41,46.
Efficacious drug and nutrient delivery within the VME requires both perfusive transport in the blood, and diffusive or convective transport into the surrounding extracellular matrix47. In solid tumors, these mechanisms are often hindered by the abnormal architecture of the vasculature and/or density of extracellular matrix components (e.g. collagen fibers)47,48. VascuViz made it possible to visualize the mechanistic barriers to drug transport in the tumor microenvironment by spatially correlating empirical measurements of intervessel distance from CT, collagen fractional area from multiphoton microscopy and ADC from DW-MRI using hierarchical correlation plots. At a local level (i.e. at discrete distances from tumor boundary), this approach revealed that peripheral tumor regions tended to be zones of efficacious delivery, while regions of poor delivery were located closer to the tumor center. In the future, one could utilize this approach to visualize the response to cancer therapies that target the vasculature and reduce intervessel distance49 (e.g. antiangiogenic therapy) or alleviate collagen-mediated solid stress48,50 (e.g. collagenase therapy) in order to enhance intratumoral drug delivery. DW-MRI derived maps of ADC and FA could act as surrogate markers of Col fractional area to further aid in such characterizations41 and have already been used as therapeutic biomarkers in cancer40. Furthermore, such VME data could be incorporated in image-based computational models24 of drug delivery to elucidate the role of other microenvironmental factors such as interstitial fluid pressure and fluid flow on the efficacy of different therapeutic strategies. Collectively, these data demonstrate how VascuViz can provide mechanistic insights into the barriers to intratumoral drug transport via the integration of multiscale, multimodality imaging data and image-based hemodynamic modeling.
A number of studies using MRI, positron emission tomography (PET) and ultrasound (US) imaging have demonstrated that fractional blood volume (FV) can serve as a metric of healthy brain function6,51,52. Therefore, our ability to directly quantify FV from high-resolution 3D CT data is useful for the structural and functional characterizations of the vasculature in the brain53 as well as other organs52 (e.g. kidney, heart). Our FV estimates for the murine brain obtained from CT were in agreement with other CT-based measurements reported in the literature53. Although one cannot quantify the contribution of capillaries at 7.5 μm isotropic spatial resolution, other high-resolution imaging studies have shown that they contribute to less than 1.5% of the FV in the murine brain54. Our multiscale assessment of the murine hippocampus showed that DW-MRI derived fractional anisotropy (FA), which is sensitive to underlying microstructure, was more heterogeneous than CT-derived FV which represents vascular morphology. This heterogeneity of FA within the hippocampus could be due to the presence of microscopic features such as axons and other cellular components that are known to influence DW-MRI contrast55. Nonetheless, one could use VascuViz to obtain LSM-derived microvasculature distributions for comparison with DW-MRI data to better understand and model the sensitivity of DW-MRI to the underlying microvasculature55. Moreover, high-resolution microvasculature data could also be incorporated in physiologically accurate biophysical models that elucidate the effect of changes in microvascular geometry on MRI contrast mechanisms56, which remains an underexplored area of research. This would be especially useful for characterizing microvascular changes in applications such as brain tumors6, stroke35 and Alzheimer’s disease57 in which the vascular phenotype is altered.
We demonstrated the feasibility of integrating in vivo functional imaging data with VascuViz derived ex vivo imaging contrasts in the murine brain. The ability to co-register dynamic functional imaging data (e.g. changes in CBF in response to a carbogen gas challenge) with the underlying microvascular network and 3D bone contrast illustrates the applicability of VascuViz for functional and physiological assessments of the neurovascular microenvironment (Supplementary Movie 1). One could envision using VascuViz to integrate structural and functional imaging data (e.g. in vivo oxygen saturation58, vascular permeability59) to assess vascular remodeling associated with bone regeneration or wound healing60 in preclinical models. In the future, the generation of such integrated 4D (i.e. 3D + time) data could provide a more complete characterization of the structural and functional changes in the VME due to disease progression or treatment40,45,60.
We showcased the utility of VascuViz in yielding high-fidelity 3D microvascular data from deep (i.e. sub-cortical) regions of the murine brain for “image-based” hemodynamic simulations. Since our image-based hemodynamic modeling approach is modality-agnostic, it would work with high-fidelity 3D vascular network data derived from either lightsheet microscopy of optically cleared samples, or high-resolution CT or multiphoton microscopy data from uncleared tissues. Previous simulations of murine brain blood flow have been performed for the cortex using microvascular network measurements from two-photon microscopy that are limited to a depth of ~600 μm. In this study, we demonstrated the feasibility of acquiring high-resolution (0.33 μm) high-fidelity 3D microvascular network data from deep murine brain regions such as the thalamus, and incorporating it in the generation of additional hemodynamic contrast. We presented two approaches for scaling blood vessel diameters and lengths to account for either vascular or gross tissue shrinkage resulting from the use of dehydrating agents during optical clearing of the samples. We found that in both cases, while the simulated hemodynamic data were within the ranges reported in the literature (Supplementary Table 2), the mean perfusion estimated from the vessel-based scaling approach (13 ml/min/100g) was lower than that for tissue-based scaling approach (40 ml/min/100g) as well as the ranges reported (40–165 ml/min/100g) for the murine cortex61,62. This may be due to our choice of zero flow boundary conditions at the capillaries21. When conducting hemodynamic simulations, obtaining a unique blood flow solution for a microvascular network requires the specification of complete boundary conditions, i.e. experimental perfusion or pressure measurements at all inlets and outlet vessels as well as discharge hematocrit measurements at all inlet vessels. Large microvascular networks can have hundreds to thousands of blood vessels as inlets and outlets. Unfortunately, such measurements are rare for murine tissues because it is challenging to acquire in vivo data over a large 3D field of view (e.g. ~ 500 μm × 500 μm × 500 μm), for hundreds to thousands of vessel segments at capillary spatial resolution. While some measurements have been made in mesentery15,63 and cortical21,64 vascular networks, boundary condition measurements for a sub-cortical network are even more challenging since most in vivo optical imaging techniques are limited to a depth < 1 mm from the surface of the brain65. Therefore, to more accurately model tissue perfusion one could optimize inlet/outlet pressures and shear rates with respect to target values assessed from in vivo blood flow measurements 66. To this end, one could also use VascuViz to integrate in vivo functional imaging data (e.g. blood flow measurements with laser speckle imaging or multiphoton microscopy) with high-resolution ex vivo vasculature data acquired from the same tissue. Since glial cells have been shown to play a major role in regulating cerebral blood flow67, we also demonstrated the feasibility of combining simulated blood flow data with astrocytic coverage at the microscopic scale assessed with LSM (0.33 μm). Consistent with their role in modulating perfusion67, we observed that large blood vessels (i.e. 25–40 μm diameter) exhibiting elevated blood flow rates co-localized with regions of high astrocytic coverage in contrast to smaller blood vessels (i.e. 4–8 μm diameter) exhibiting lower blood flow rates. Collectively, VascuViz enabled the integration of microvascular data into image-based hemodynamic models and facilitated comparisons between mesoscopic and microscopic aspects of the circulation. We expect this image-based hemodynamic modeling approach to be adaptable to investigations of the structural and function of the circulation in in other tissues such as the retina, placenta, heart, liver and lung.
We developed VascuViz to be a multimodality imaging pipeline that exploits the strengths of a wide range of ex vivo imaging approaches and made it applicable to any organ-system and preclinical disease model. Although the use of commercially available imaging reagents lower the barriers to accessibility of the imaging equipment and reproducibility regarding data acquisition and integration, some imaging systems (e.g. light sheet microscopes) might not be as widely available as others (e.g. MRI and CT). However, this would not limit the VascuViz’s utility as it permits adaptation of the imaging workflow. For example, since blood vessels are simultaneously visible in each imaging modality, the absence of a modality would not hinder VascuViz’s ability to integrate vascular contrast with complementary contrasts at other spatial scales. We were able to demonstrate the strengths of CT imaging for mapping the 3D macro-and microvasculature in whole-organs, and its utility in multimodal image co-registration via the use of vascular fiducials instead of surface-based fiducials. Moreover, VascuViz provides a range of options for optical imaging of the GalRh-BVu perfused samples ranging from conventional 2D epifluorescence microscopy to 3D multiphoton microscopy, second harmonic generation imaging and light sheet microscopy (LSM). Finally, the VascuViz protocol does not limit the size or volume of the tissues that can be imaged Sample volumes can range from a tissue sub-region (e.g. a muscle group as shown in Fig. 6f–g) to whole organs (e.g. the hind limb in Fig. 6a–g or the brain in Fig. 4), or an entire organism (e.g. whole-mouse). As long as the sample can be accommodated within the gantry of the imaging system being employed, one can acquire and integrate multimodality and multiscale imaging data using VascuViz. As demonstrated here, one could envision the widespread integration of the VascuViz workflow with in vivo functional imaging data (e.g. fMRI, PET, ultrasound) acquired from the same sample in the near future.
In this paper, we have presented a novel multimodality, multiscale vascular imaging pipeline using MRI, CT and optical imaging with an intravascular contrast agent mixture, GalRh-BVu. This method dubbed VascuViz, has direct applications in image-based vascular systems biology of the brain, kidney and hind limb as well as preclinical disease models such as breast cancer. In addition to enabling high-resolution vascular mapping in intact unsectioned tissues, our method provides for the first time, the ability to integrate vascular data with multiple image contrast mechanisms and spatial scales ranging from individual endothelial cells to the whole-organ. It is our hope that these advances in preclinical vascular imaging in conjunction with the novel visualization approaches presented here will open up new vistas for image-based systems biology of the vasculature, and help answer important questions in the broader field of microcirculation and its role in health and disease.
Extended Data
Extended Data Fig. 1. The GalRh-BVu polymer did not interfere with histopathological workflows.
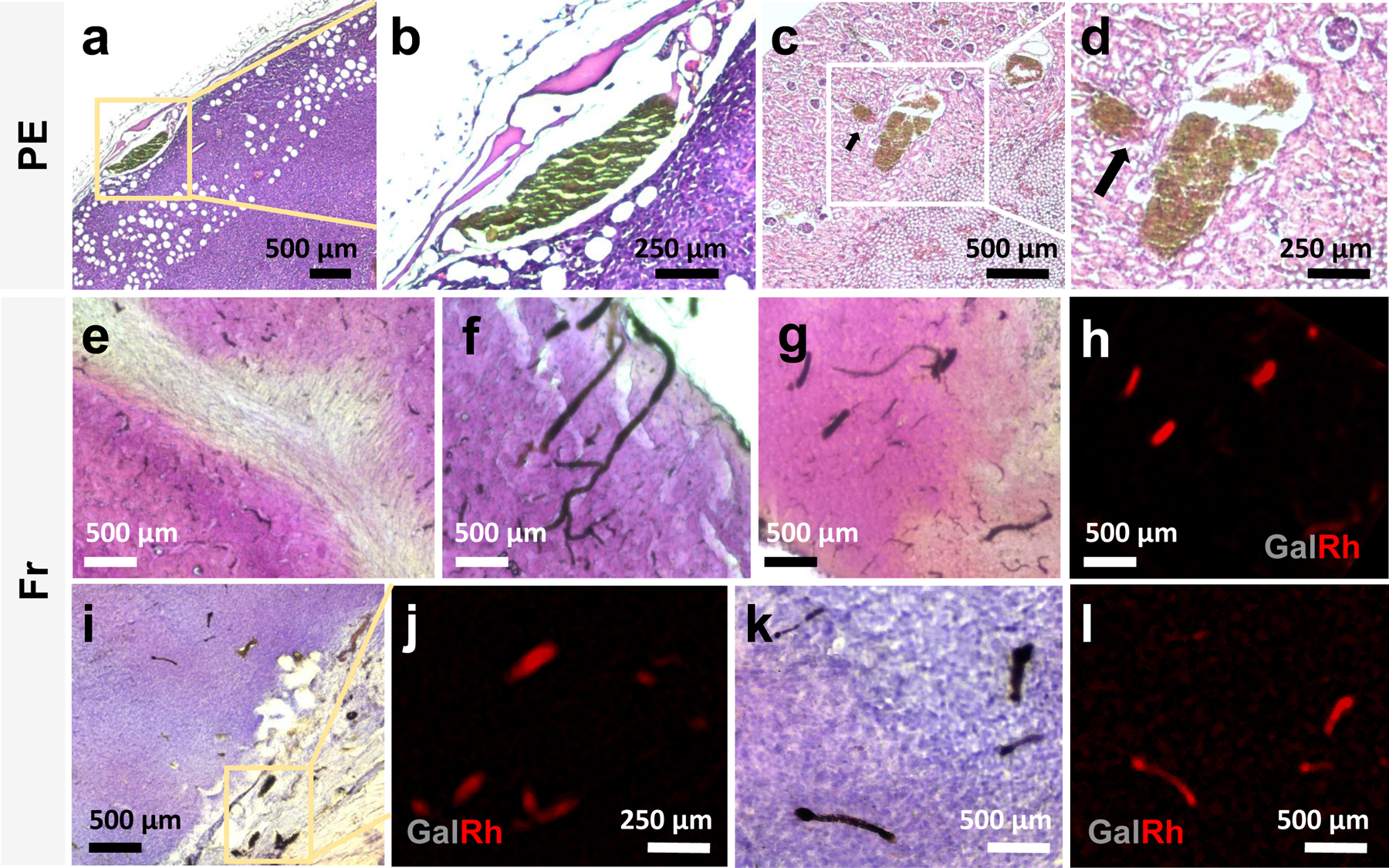
The GalRh-BVu polymer was compatible with H&E staining of paraffin-embedded (PE) tissues as shown for a 4T1 tumor xenograft sample (a-b), and a kidney sample (c-d). The intravascular polymer appeared dark brown on H&E images as seen in (b) the tumor rim vasculature from (a), and (d) renal cortex vasculature from (c). Black arrows point to a perfused glomerulus in (c-d). The polymer also did not interfere with H&E staining of frozen tissues as shown for the murine hippocampus (e) and cortex (f-g). In H&E labeled images, the cytoarchitecture of the hippocampus and cortex could also be complemented with the vascular visibility of the GalRh-BVu polymer (e-g). The GalRh-BVu polymer bearing blood vessels in (g) could also be imaged using fluorescence microscopy as shown in (h). Similarly, tissue cytoarchitectural alterations seen in a H&E stained 4T1 tumor xenograft sample (i, k) could be complemented with the vascular visibility of the GalRh-BVu polymer in bright field (i, k) and fluorescence microscopy images (j, l). N.B. The brightness and contrast of H&E images were adjusted for visualization purposes without any changes to the original data.
Extended Data Fig. 2. Combining Euclidean distance maps derived from CT with ADC maps derived from DW-MRI in a human breast cancer model.
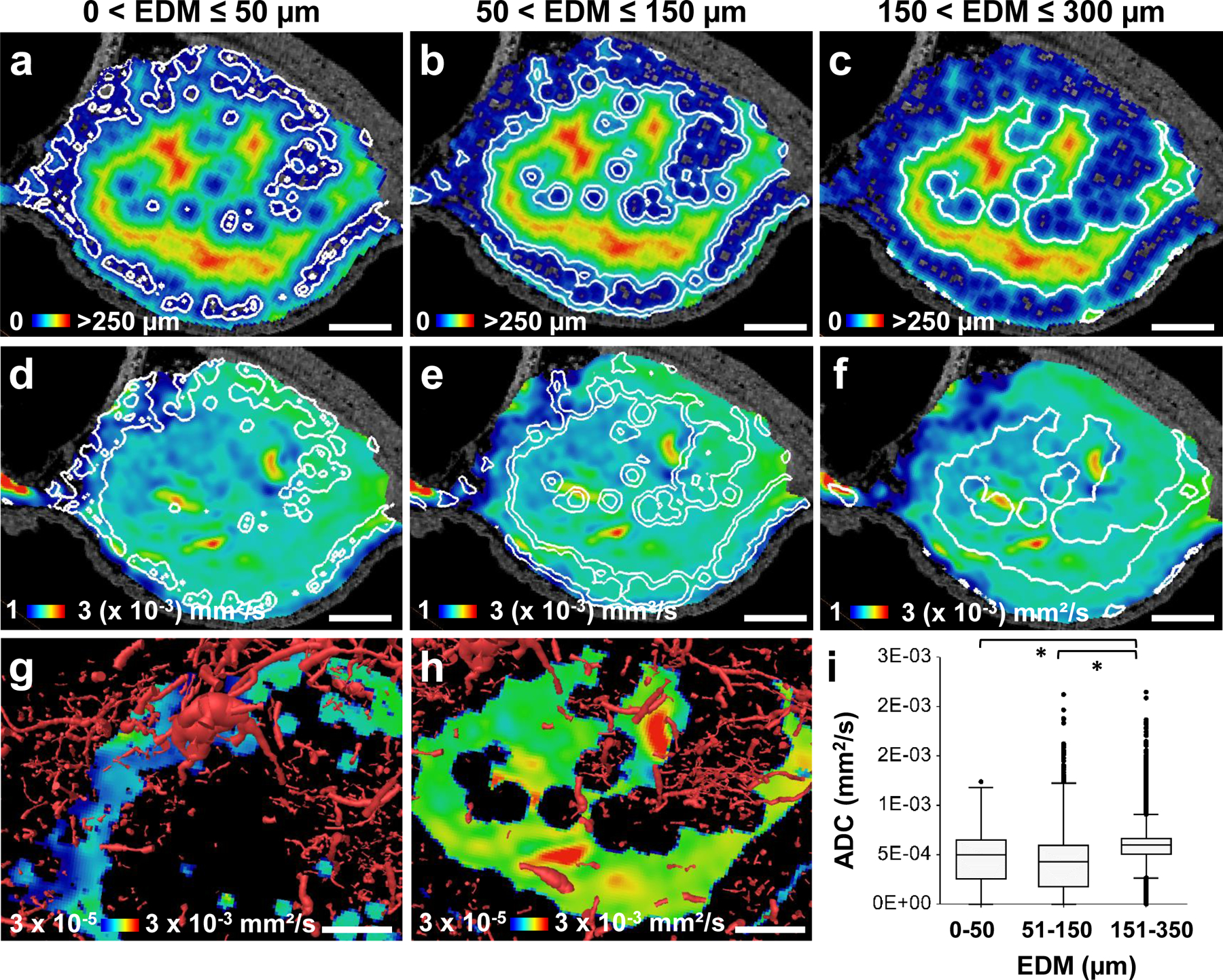
(a-c) A 2D section from the 3D Euclidian distance map (EDM) is shown with an overlay of white contour lines to highlight regions within inter-vessel distance ranges 0–50 μm (a), 51–150 μm (b) and 151–350 μm (c), respectively. Soft tissue contrast from T1W-MRI data of the same region was employed as the underlay image in (a-c). The contour lines corresponding to the inter-vessel distance ranges shown in (a-c) were mapped on to co-registered apparent diffusion coefficient (ADC) maps derived from DW-MRI (d-f), respectively. Rim and central tumor sub-regions from (d) and (f) were selected and visualized with volume rendered tumor blood vessels (red) derived from CT (g-h). Low ADC regions (blue-green) co-localized with regions with a high density of tumor vasculature (g), while high ADC regions (yellow-red) co-localized with regions exhibiting low vessel density (h). (i) Box and whisker plots of whole-tumor ADC distributions showed that 151–350 μm EDM regions were significantly different (p << 0.001) from those for 51–150 μm EDM regions and 0–50 μm EDM regions using a two-tailed Mann-Whitney U-test test at alpha = 0.05. The box and whisker plots corresponding to 0 – 50 μm, 50–150 μm and 150–350 μm EDM regions show the median, interquartile range (IQR) and the data within the Q1–1.5IQR and Q3+1.5IQR range. The upper and lower bounds of the displayed intensity range for the merged images shown in (a-f) were adjusted for visualization purposes without any changes to the original data.
Extended Data Fig. 3. Correlation between tumor boundary-to-center profiles for MRI-, CT- and optical imaging-derived vascular microenvironmental (VME) parameters in a human breast cancer model.
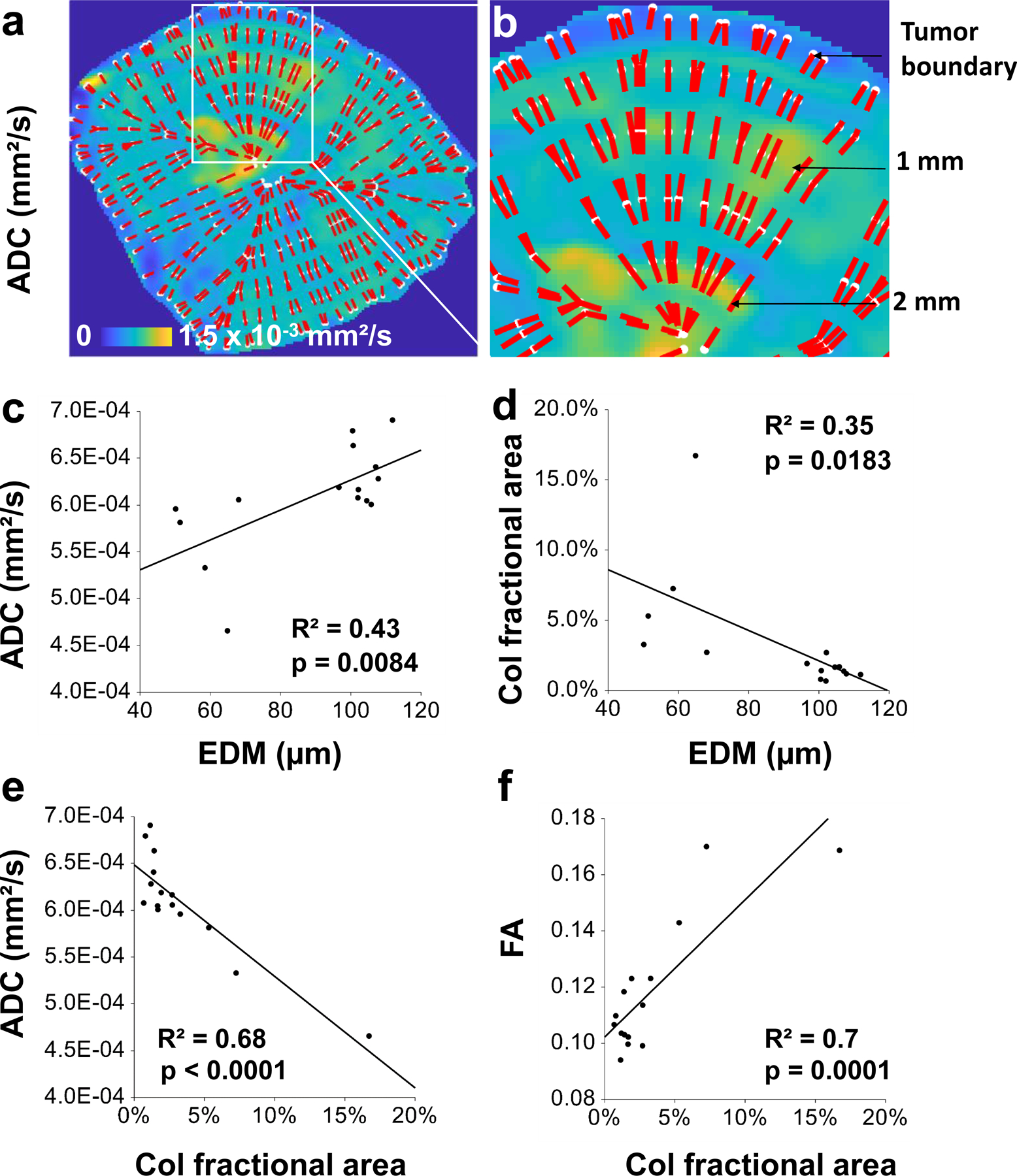
(a-b) For each VME parameter map (e.g. ADC), we overlaid a 2D grid of points along azimuthal (white points) and radial directions (dashed red lines). An enlarged view of the grid is shown in (b) wherein black arrows point to white contours that are located at the tumor boundary and at 1 mm and 2 mm normal to it. Next, boundary-to-center profiles were calculated along each dashed red line and an average radial profile generated for each VME parameter map. (c) ADC correlated with EDM (R2= 0.43, p = 0.0084). (d) Collagen (Col) fractional area correlated inversely with EDM (R2= 0.35, p = 0.0183). (e) FA correlated inversely with ADC (R2= 0.68, p < 0.0001). (f) FA correlated with Col fractional area (R2=0.7, p = 0.0001). The Pearson correlation coefficients between each variable pair shown in (a-d) was significant.
Extended Data Fig. 4. Steps for matching sample orientation between CT and MPM imaging.
(a) The sample was embedded in an agarose block prior to CT imaging and directional annotations made. Before sectioning the sample for MPM, the location of the cutting plane was determined relative to the tumor center by matching the embedded sample with its T1W-MRI image, as shown in (b). Red hatched line indicate where the sample was cut. This orientation was then preserved with the help of the directional annotations shown in (c) during cutting, and (d) during cryosectioning.
Extended Data Fig. 5. Creating a 4D (i.e. 3D + time) visualization by mapping the temporal dynamics of the in vivo functional hyperemic response to 3D ex vivo neurovascular and anatomical data.
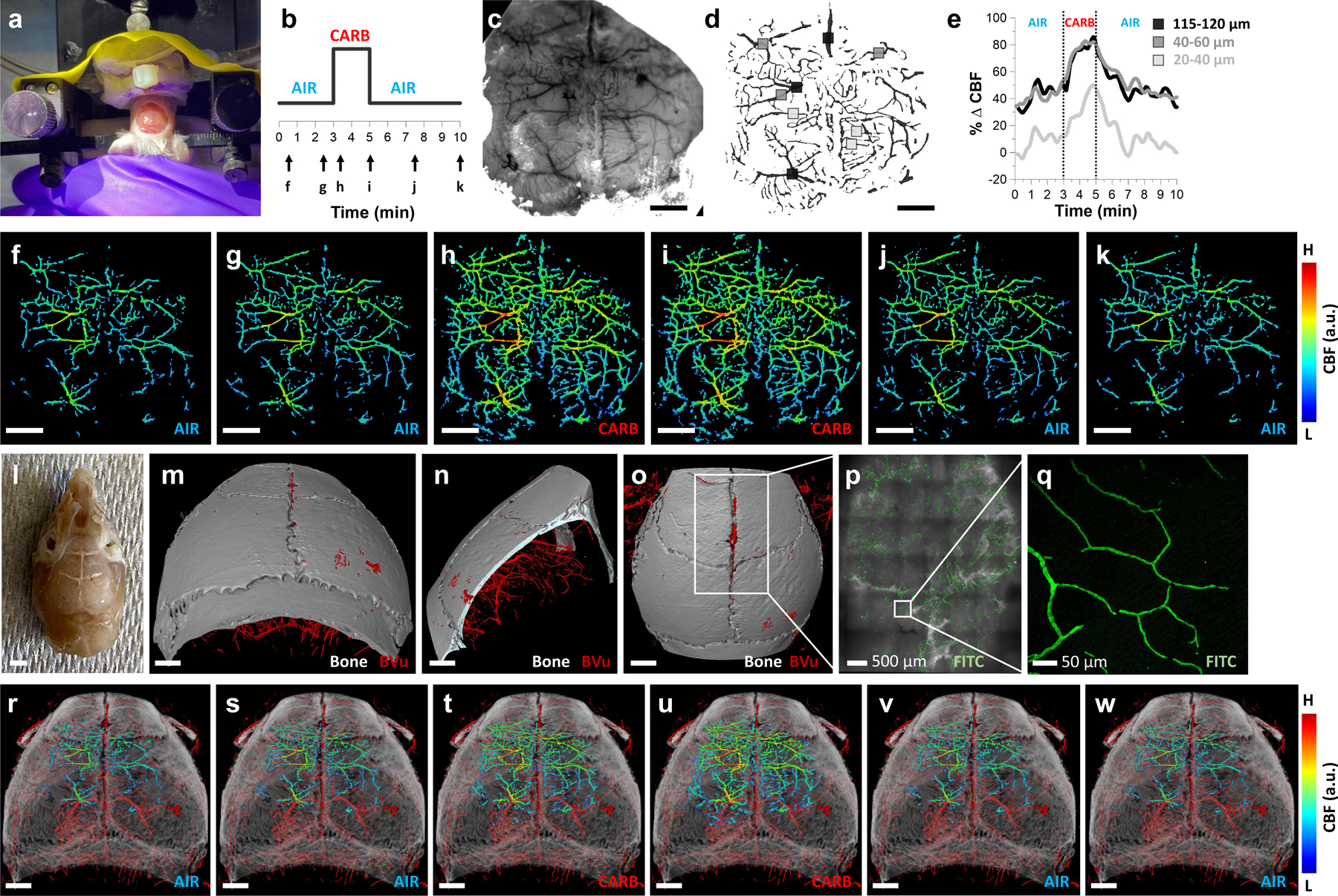
(a) A thinned-skull cranial window preparation for in vivo LSC and IOS imaging. (b) The animal was made to breathe room air (AIR), carbogen (95% oxygen/5% carbon dioxide) gas (CARB), and room air (AIR) for 3, 2 and 5 minutes, respectively during which dynamic CBF data was acquired. Black arrows in (b) correspond to the time points (i.e. 0.4 and 2.5 min, AIR; 3.3 and 5 min CARB; and 7.5 and 10 min, AIR) for which the corresponding CBF maps are shown in (f-k). Black, dark gray and light gray squares (d-e) indicate large (i.e. 115 μm < diameter < 120 μm), medium (i.e. 40 μm < diameter < 60 μm) and small (i.e. 20 μm < diameter < 40 μm) blood vessels that were identified using the IOS image (c) and from which the mean in vivo CBF traces shown in (e) computed. (e) Medium (dark gray trace) and large vessels (black trace) showed a larger peak increase in %ΔCBF w.r.t the global baseline than that exhibited by small vessels (light gray trace). (f-k) Spatio-temporal evolution of CBF corresponding to the experimental paradigm in (b) illustrating the significant response to carbogen inhalation (h, i). (l) The same sample prepared for ex vivo imaging in which the GalFITC-BVu perfused neurovasculature was visible (white contrast) in the intact skull. (m-o) Concurrent visualization of the 3D skull anatomy (gray) and underlying neurovasculature (red) using ex vivo CT imaging (7.5 μm) (m-o). (p) Fluorescent MIP images of perfused skull vessels from a tiled scan acquired at 10× with confocal microscopy (1.3 μm), and a 25× scan of the same sample acquired with MPM (0.6 μm) (q). The positive vascular contrast in CT data (m-o, r-w) was due to BVu while the GalFITC component provided fluorescence contrast in microscopy data (p-q). (r-w) Integrated 4D volume created by co-registering the 2D in vivo CBF maps (f-k) to 3D ex vivo neurovascular and skull anatomy data (m-o) illustrating the time evolution of the functional hyperemic response. The displayed intensity range for (f-k) was adjusted for visualization purposes without altering the original data. A 0.25 minute moving average filter was applied to (e) to reduce noise. Scale bars: 1 mm unless stated otherwise.
Extended Data Fig. 6. Validation of the image-based hemodynamic modeling approach.
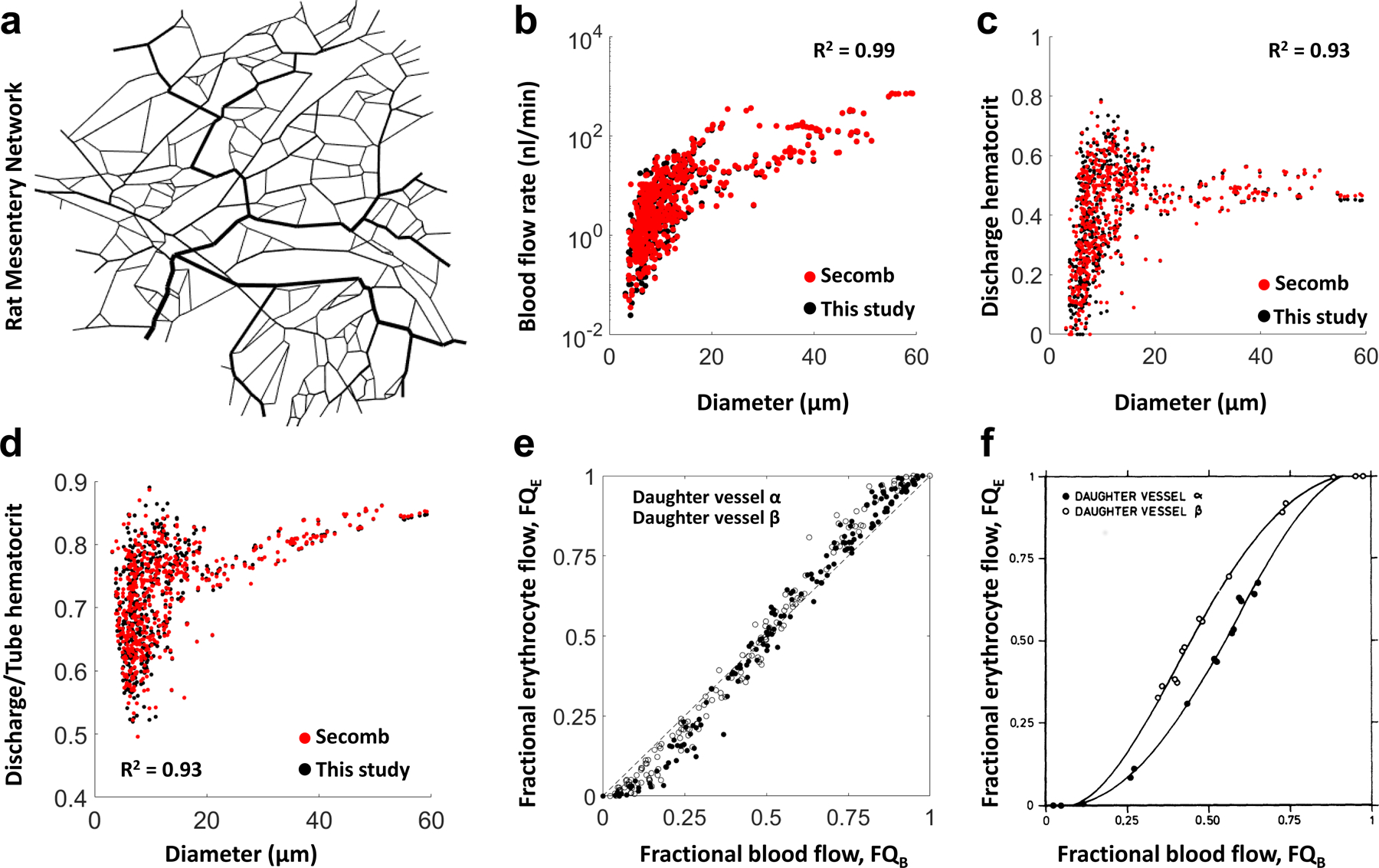
To validate our blood flow modeling approach, we employed a 546-segment vascular network of the rat mesentery (a) that was derived from high resolution intravital microscopy imaging data by Pries et al15. To simulate pressure and blood flow values in all segments of the network, experimentally obtained blood flow rates were prescribed at 35 boundary segments while one boundary node was subjected to a constant pressure boundary condition. Blood flow and discharge hematocrit distributions in all 546 segments simulated using our approach (black dots) are compared against the solution of the fully determined system as obtained by Pries et al15. We achieved an R2= 0.99 for blood flow rates (nl/min) and R2= 0.93 for discharge hematocrit distributions observing excellent agreement between these data. Moreover, the distribution of the ratio of discharge to tube hematocrit vs. vessel diameter satisfied the well-known Fahraeous effect and showed an R2= 0.94 against the simulated values reported by Pries et al15 (d). Finally, fractional erythrocyte flow (FQE) vs. fractional blood flow (FQB) distributions obtained using our approach satisfied the phase separation effect (e) in agreement with the distributions reported by Pries et al15 (f). Open and filled circles correspond to data points for daughter vessels α and β at diverging bifurcations. Collectively, these plots demonstrate the validity of our image-based hemodynamic modeling approach and its utility for predicting functional properties of micro-vascular networks. Panel (f) reproduced with permission from15.
Extended Data Fig. 7. The GalRh-BVu polymer remained stable for at least 11 months after sample preparation.
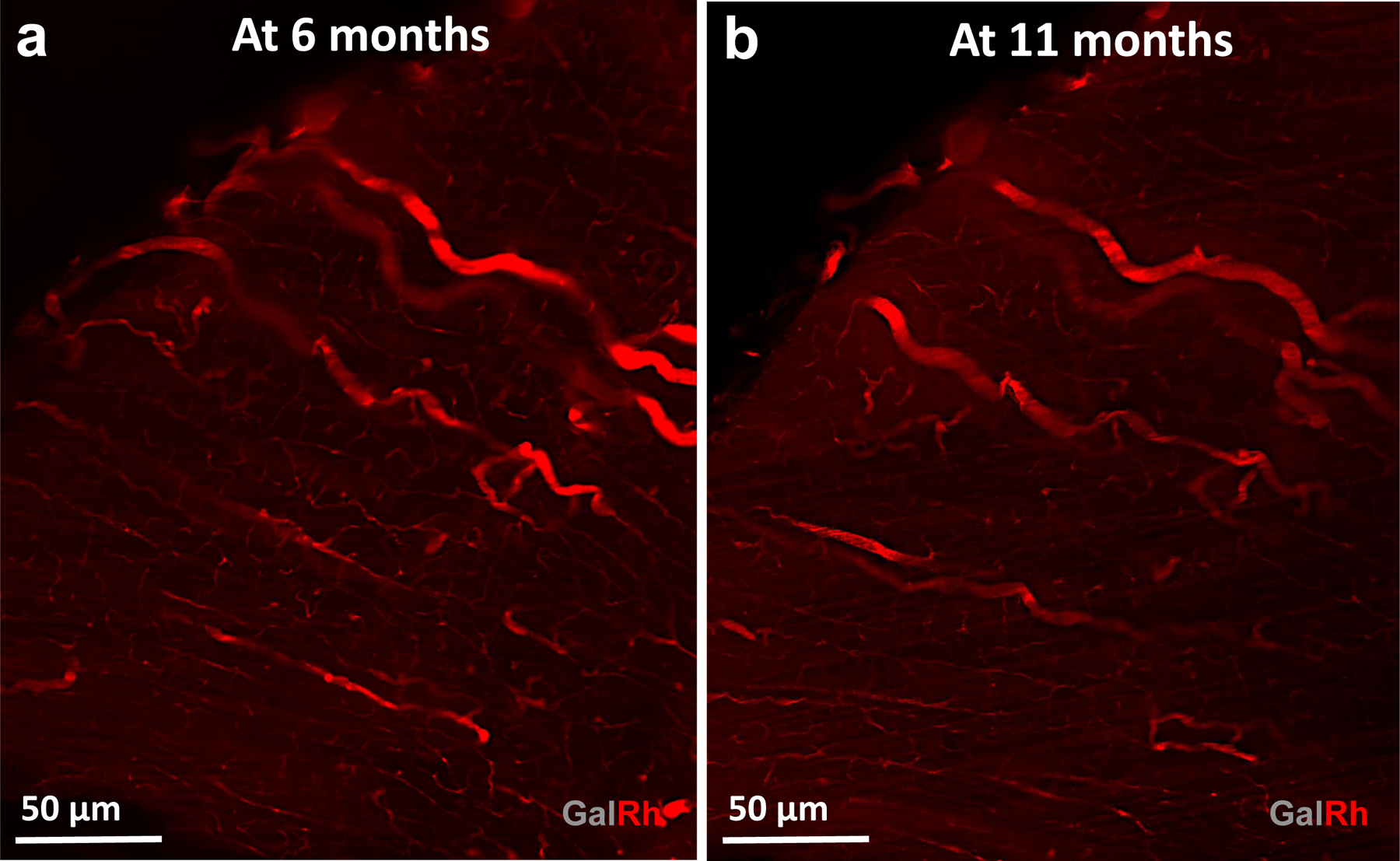
(a) LSM image of the thalamic vasculature in a murine brain acquired at 0.33 μm spatial resolution at 6 months after sample preparation. (b) LSM of the same field of view acquired at 0.56 μm spatial resolution at 11 months after sample preparation. Here, vascular contrast was enhanced by normalizing the image intensity to 0.1% of the dynamic range followed by 3D median filtering (radius = 2 voxels).
Supplementary Material
Supplementary Videos Video 1 Pathak_Supplementary_Video_1.mp4
Dynamic functional hyperemic response acquired using in vivo laser speckle imaging (blue-red color map) was integrated to the 3D skull anatomy (grey) and the underlying neurovasculature (red) data acquired ex vivo using CT imaging. The animal was made to breathe room air (AIR), carbogen (95% oxygen/5% carbon dioxide) gas (CARB), and room air (AIR) for 3, 2 and 5 minutes, respectively during which dynamic CBF data was acquired. Large (ROI 1) and medium blood vessels (ROI 2) showed a larger peak increase in % ΔCBF w.r.t the global baseline than that exhibited by small vessels (ROI 3).
ACKNOWLEDGEMENTS:
This work was supported by NIH/NCI grants 51R01CA196701-05, 1R01CA237597-01A1 and 5R01DE027957-02 (APP); NIH Instrumentation grant S10OD012287 (Cornell University); and Sidney Kimmel Comprehensive Cancer Center, Quantitative Sciences Pilot Project Grant (AB). We would like to thank Darren Yang for assistance with Supplementary Movie 1; Dr. Qihong Wang for assistance with perfusion experiments; Zhipeng Hou for assistance with image co-registration; Dr. Aleksander S. Popel for providing the 4T1 tumor model; and Rhea Pathak for assistance with data segmentation. This work is dedicated to the memories of Dr. Sanjeev Bhargava and Mr. Pratap I. Pathak.
Footnotes
COMPETING INTERESTS:
The authors have no competing interests to declare.
REFERENCES
- 1.Menshykau D & Tanaka S Mechanistic Image-based Modelling: Concepts and Applications. Handb Exp Pharmacol 260, 231–261 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Sbalzarini IF Modeling and Simulation of Biological Systems from Image Data. Bioessays 35, 482–490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gómez H, Georgieva L, Michos O & Iber D in Systems Biology (eds Nielsen Jens & Hohmann Stefan) 319–340 (2017). [Google Scholar]
- 4.Krucker T, Lang A & Meyer EP New Polyurethane-based Material for Vascular Corrosion Casting with Improved Physical and Imaging Characteristics. Microsc Res Tech 69, 138–147 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Schaad L et al. Correlative Imaging of the Murine Hind Limb Vasculature and Muscle Tissue by MicroCT and Light Microscopy. Sci Rep 7, 41842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim E, Zhang J, Hong K, Benoit NE & Pathak AP Vascular Phenotyping of Brain Tumors using Magnetic Resonance Microscopy (μMRI). J Cereb Blood Flow Metab 31, 1623–1636 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue S et al. Indian-Ink Perfusion Based Method for Reconstructing Continuous Vascular Networks in Whole Mouse Brain. PLoS One 9, e88067 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Giovanna AP et al. Whole-brain Vasculature Reconstruction at the Single Capillary Level. Sci Rep 8, 12573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayerich D, Abbott L & McCormick B Knife-edge Scanning Microscopy for Imaging and Reconstruction of Three-dimensional Anatomical Structures of the Mouse Brain. J Microsc 231, 134–143 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Erturk A et al. Three-dimensional Imaging of Solvent-cleared Organs using 3DISCO. Nat Protoc 7, 1983–1995 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Li Y et al. Direct Labeling and Visualization of Blood Vessels with Lipophilic Carbocyanine Dye DiI. Nat Protoc 3, 1703–1708 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Todorov MI et al. Machine Learning Analysis of Whole Mouse Brain Vasculature. Nat Methods 17, 442–449 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayerich D et al. Fast Macro-scale Transmission Imaging of Microvascular Networks using KESM. Biomed Opt Express 2, 2888–2896 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jing D et al. Tissue Clearing of Both Hard and Soft Tissue Organs with the PEGASOS Method. Cell Res 28, 803–818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pries AR, Secomb TW, Gaehtgens P & Gross JF Blood Flow in Microvascular Networks. Experiments and Simulation. Circ Res 67, 826–834 (1990). [DOI] [PubMed] [Google Scholar]
- 16.Aggarwal M, Mori S, Shimogori T, Blackshaw S & Zhang J Three-dimensional Diffusion Tensor Microimaging for Anatomical Characterization of the Mouse Brain. Magn Reson Med 64, 249–261 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H et al. Effect of Cranial Window Type on Monitoring Neurovasculature using Laser Speckle Contrast Imaging. Proc Spie 9690 (2016). [Google Scholar]
- 18.Mendez A et al. Phenotyping the Microvasculature in Critical-sized Calvarial Defects via Multimodal Optical Imaging. Tissue Eng Part C-Me 24, 430–440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg S et al. ilastik: Interactive Machine Learning for (Bio)image Analysis. Nat Methods 16, 1226–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Jiang H, van Zijl PC, Kim J, Pearlson GD & Mori S DtiStudio: Resource Program for Diffusion Tensor Computation and Fiber Bundle Tracking. Comput Methods Programs Biomed 81, 106–116 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Lorthois S, Cassot F & Lauwers F Simulation Study of Brain Blood Flow Regulation by Intra-cortical Arterioles in an Anatomically Accurate Large Human Vascular Network: Part I: Methodology and Baseline Flow. Neuroimage 54, 1031–1042 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Reichold J et al. Vascular Graph Model to Simulate the Cerebral Blood Flow in Realistic Vascular Networks. J Cereb Blood Flow Metab 29, 1429–1443 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Weber B, Keller AL, Reichold J & Logothetis NK The Microvascular System of the Striate and Extrastriate Visual Cortex of the Macaque. Cereb Cortex 18, 2318–2330 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Stamatelos SK, Bhargava A, Kim E, Popel AS & Pathak AP Tumor Ensemble-based Modeling and Visualization of Emergent Angiogenic Heterogeneity in Breast Cancer. Sci Rep 9, 5276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pries AR et al. Resistance To Blood Flow in Microvessels In Vivo. Circ Res 75, 904–915 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Bhargava A, Monteagudo B, Aggarwal M & Pathak A A Novel Vascular Fiducials-based Approach (VASFID) for Co-registering Multiscale Imaging Data for Microcirculation Systems Biology. The FASEB Journal 34, 1–1 (2020). [Google Scholar]
- 27.Schindelin J et al. Fiji: an Open-source Platform for Biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller MI, Trouve A & Younes L On the Metrics and Ruler-Lagrange Equations of Computational Anatomy. Annu Rev Biomed Eng 4, 375–405 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Aggarwal M, Zhang J, Miller MI, Sidman RL & Mori S Magnetic Resonance Imaging and Micro-computed Tomography Combined Atlas of Developing and Adult Mouse Brains for Stereotaxic Surgery. Neuroscience 162, 1339–1350 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charles JP, Cappellari O, Spence AJ, Hutchinson JR & Wells DJ Musculoskeletal Geometry, Muscle Architecture and Functional Specialisations of the Mouse Hindlimb. PLoS One 11, e0147669 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norton CR et al. Absence of a Major Role for the Snai1 and Snai3 Genes in Regulating Skeletal Muscle Regeneration in Mice. PLoS Curr 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senarathna J et al. A Miniature Multi-Contrast Microscope for Functional Imaging in Freely Behaving Animals. Nat Commun 10, 99 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plouraboue F et al. X-ray High-Resolution Vascular Network Imaging. J Microsc 215, 139–148 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Dyer EL et al. Quantifying Mesoscale Neuroanatomy Using X-ray Microtomography. eNeuro 4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirst C et al. Mapping the Fine-scale Organization and Plasticity of the Brain Vasculature. Cell 180, 780–795 e725 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Hossler FE & Douglas JE Vascular Corrosion Casting: Review of Advantages and Limitations in the Application of Some Simple Quantitative Methods. Microsc Microanal 7, 253–264 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Duvall CL, Taylor WR, Weiss D & Guldberg RE Quantitative Microcomputed Tomography Analysis of Collateral Vessel Development After Ischemic Injury. Am J Physiol Heart Circ Physiol 287, H302–310 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Partridge SC et al. Diffusion Tensor MRI: Preliminary Anisotropy Measures and Mapping of Breast Tumors. J Magn Reson Imaging 31, 339–347 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Jiang R et al. Diffusion Tensor Imaging of Breast Lesions: Evaluation of Apparent Diffusion Coefficient and Fractional Anisotropy and Tissue Cellularity. Br J Radiol 89, 20160076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim E et al. Vasculature-specific MRI Reveals Differential Anti-Angiogenic Effects of a Biomimetic Peptide in an Orthotopic Breast Cancer Model. Angiogenesis 18, 125–136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kakkad S et al. Collagen Fibers Mediate MRI-detected Water Diffusion and Anisotropy in Breast Cancers. Neoplasia 18, 585–593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cebulla J, Kim E, Rhie K, Zhang J & Pathak AP Multiscale and Multi-Modality Visualization of Angiogenesis in a Human Breast Cancer Model. Angiogenesis 17, 695–709 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim E et al. Multiscale Imaging and Computational Modeling of Blood Flow in the Tumor Vasculature. Ann Biomed Eng 40, 2425–2441 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raman V et al. Characterizing Vascular Parameters in Hypoxic Regions: a Combined Magnetic Resonance and Optical Imaging Study of a Human Prostate Cancer Model. Cancer Res 66, 9929–9936 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Riegler J et al. Tumor Elastography and Its Association with Collagen and the Tumor Microenvironment. Clin Cancer Res 24, 4455–4467 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Kakkad SM et al. Hypoxic Tumor Microenvironments Reduce Collagen I Fiber Density. Neoplasia 12, 608–617 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dewhirst MW & Secomb TW Transport of Drugs From Blood Vessels to Tumour Tissue. Nat Rev Cancer 17, 738–750 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chauhan VP et al. Angiotensin Inhibition Enhances Drug Delivery and Potentiates Chemotherapy by Decompressing Tumour Blood Vessels. Nat Commun 4, 2516 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain RK Normalization of Tumor Vasculature: an Emerging Concept in Antiangiogenic Therapy. Science 307, 58–62 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Wang H et al. Elastography Can Map the Local Inverse Relationship between Shear Modulus and Drug Delivery within the Pancreatic Ductal Adenocarcinoma Microenvironment. Clinical Cancer Research 25, 2136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito H et al. Arterial Fraction of Cerebral Blood Volume in Humans Measured by Positron Emission Tomography. Ann Nucl Med 15, 111–116 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Welsh AW et al. Three-dimensional US Fractional Moving Blood Volume: Validation of Renal Perfusion Quantification. Radiology 293, 460–468 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chugh BP et al. Measurement of Cerebral Blood Volume in Mouse Brain Regions using Micro-computed Tomography. Neuroimage 47, 1312–1318 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Xiong B et al. Precise Cerebral Vascular Atlas in Stereotaxic Coordinates of Whole Mouse Brain. Front Neuroanat 11, 128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stolp HB et al. Voxel-wise Comparisons of Cellular Microstructure and Diffusion-MRI in Mouse Hippocampus using 3D Bridging of Optically-clear histology with Neuroimaging Data (3D-BOND). Sci Rep 8, 4011 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pathak AP, Ward BD & Schmainda KM A Novel Technique for Modeling Susceptibility-based Contrast Mechanisms for Arbitrary Microvascular Geometries: the Finite Perturber Method. Neuroimage 40, 1130–1143 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meyer EP, Ulmann-Schuler A, Staufenbiel M & Krucker T Altered Morphology and 3D Architecture of Brain Vasculature in a Mouse model for Alzheimer’s Disease. Proc Natl Acad Sci U S A 105, 3587–3592 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Senarathna J et al. HemoSYS: A Toolkit for Image-based Systems Biology of Tumor Hemodynamics. Sci Rep 10, 2372 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pathak AP et al. In Vivo “MRI Phenotyping” Reveals Changes in Extracellular Matrix Transport and Vascularization That Mediate VEGF-driven Increase in Breast Cancer Metastasis. PLOS ONE 8, e63146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rege A, Thakor NV, Rhie K & Pathak AP In Vivo Laser Speckle Imaging Reveals Microvascular Remodeling and Hemodynamic Changes During Wound Healing Angiogenesis. Angiogenesis 15, 87–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maeda K, Mies G, Olah L & Hossmann KA Quantitative Measurement of Local Cerebral Blood Flow in the Anesthetized Mouse using Intraperitoneal [14C]Iodoantipyrine Injection and Final Arterial Heart Blood Sampling. J Cereb Blood Flow Metab 20, 10–14 (2000). [DOI] [PubMed] [Google Scholar]
- 62.Gertz K et al. Physical Activity Improves Long-Term Stroke Outcome via Endothelial Nitric Oxide Synthase-dependent Augmentation of Neovascularization and Cerebral Blood Flow. Circ Res 99, 1132–1140 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Lipowsky HH & Zweifach BW Network Analysis of Microcirculation of Cat Mesentery. Microvasc Res 7, 73–83 (1974). [DOI] [PubMed] [Google Scholar]
- 64.Gagnon L et al. Multimodal Reconstruction of Microvascular-flow Distributions using Combined Two-photon Microscopy and Doppler Optical Coherence Tomography. Neurophotonics 2, 015008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hillman EM Optical Brain Imaging In Vivo: Techniques and Applications from Animal to Man. J Biomed Opt 12, 051402 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fry BC, Lee J, Smith NP & Secomb TW Estimation of Blood Flow Rates in Large Microvascular Networks. Microcirculation 19, 530–538 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petzold GC & Murthy VN Role of Astrocytes in Neurovascular Coupling. Neuron 71, 782–797 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Puelles VG, Moeller MJ & Bertram JF We Can See Clearly Now: Optical Clearing and Kidney Morphometrics. Curr Opin Nephrol Hypertens 26, 179–186 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Williams MPI et al. A Novel Optical Tissue Clearing Protocol for Mouse Skeletal Muscle to Visualize Endplates in Their Tissue Context. Front Cell Neurosci 13, 49 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aggarwal M, Mori S, Shimogori T, Blackshaw S & Zhang J Three-dimensional Diffusion Tensor Microimaging for Anatomical Characterization of the Mouse Brain. Magn Reson Med 64, 249–261 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jing D et al. Tissue Clearing of Both Hard and Soft Tissue Organs with the PEGASOS Method. Cell Res 28, 803–818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu H et al. Effect of Cranial Window Type on Monitoring Neurovasculature using Laser Speckle Contrast Imaging. Proc SPIE 9690 (2016). [Google Scholar]
- 73.Mendez A et al. Phenotyping the Microvasculature in Critical-sized Calvarial Defects via Multimodal Optical Imaging. Tissue Eng Part C-Me 24, 430–440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berg S et al. ilastik: Interactive Machine Learning for (Bio)Image Analysis. Nat Methods 16, 1226–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Jiang H, van Zijl PC, Kim J, Pearlson GD & Mori S DTIStudio: Resource Program for Diffusion Tensor Computation and Fiber Bundle Tracking. Comput Methods Programs Biomed 81, 106–116 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Lorthois S, Cassot F & Lauwers F Simulation Study of Brain Blood Flow Regulation by Intra-cortical Arterioles in an Anatomically Accurate Large Human Vascular network: Part I: Methodology and Baseline Flow. Neuroimage 54, 1031–1042 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Reichold J et al. Vascular Graph Model to Simulate the Cerebral Blood flow in Realistic Vascular Networks. J Cereb Blood Flow Metab 29, 1429–1443 (2009). [DOI] [PubMed] [Google Scholar]
- 78.Weber B, Keller AL, Reichold J & Logothetis NK The Microvascular System of the Striate and Extrastriate Visual Cortex of the Macaque. Cereb Cortex 18, 2318–2330 (2008). [DOI] [PubMed] [Google Scholar]
- 79.Stamatelos SK, Bhargava A, Kim E, Popel AS & Pathak AP Tumor Ensemble-based Modeling and Visualization of Emergent Angiogenic Heterogeneity in Breast Cancer. Sci Rep 9, 5276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pries AR et al. Resistance to Blood Flow in Microvessels In Vivo. Circ Res 75, 904–915 (1994). [DOI] [PubMed] [Google Scholar]
- 81.Pries AR, Secomb TW, Gaehtgens P & Gross JF Blood Flow in Microvascular Networks. Experiments and Simulation. Circ Res 67, 826–834 (1990). [DOI] [PubMed] [Google Scholar]
- 82.Bhargava A, Monteagudo B, Aggarwal M & Pathak A A Novel Vascular Fiducials-based Approach (VASFID) for Co-registering Multiscale Imaging Data for Microcirculation Systems Biology. The FASEB Journal 34, 1–1 (2020). [Google Scholar]
- 83.Schindelin J et al. Fiji: An Open-source Platform for Biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aggarwal M, Zhang J, Miller MI, Sidman RL & Mori S Magnetic Resonance Imaging and Micro-computed Tomography Combined Atlas of Developing and Adult Mouse Brains for Stereotaxic Surgery. Neuroscience 162, 1339–1350 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Charles JP, Cappellari O, Spence AJ, Hutchinson JR & Wells DJ Musculoskeletal Geometry, Muscle Architecture and Functional Specializations of the Mouse Hindlimb. PLoS One 11, e0147669 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Senarathna J et al. A Miniature Multi-Contrast Microscope for Functional Imaging in Freely Behaving Animals. Nat Commun 10, 99 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Videos Video 1 Pathak_Supplementary_Video_1.mp4
Dynamic functional hyperemic response acquired using in vivo laser speckle imaging (blue-red color map) was integrated to the 3D skull anatomy (grey) and the underlying neurovasculature (red) data acquired ex vivo using CT imaging. The animal was made to breathe room air (AIR), carbogen (95% oxygen/5% carbon dioxide) gas (CARB), and room air (AIR) for 3, 2 and 5 minutes, respectively during which dynamic CBF data was acquired. Large (ROI 1) and medium blood vessels (ROI 2) showed a larger peak increase in % ΔCBF w.r.t the global baseline than that exhibited by small vessels (ROI 3).
Data Availability Statement
The authors declare that all the data supporting the findings of this study are included within the paper and its Extended Data and supplementary information files. This includes the availability of imaging data for: (i) the murine brain in Figs. 4 and 5, Supplementary Fig. 5–7 and Supplementary Tables 2 and 3; and Supplementary Movie 1, (ii) the breast tumor xenograft in Fig. 3 and Extended Data Figs. 2 and 3; (iii) the kidney and hind limb in Fig. 6. Finally, IHC and H&E data are available in Fig. 2 and Extended Data Fig. 1. The mouse hippocampal data that was used for the labeling of the CA and DG layers can be accessed freely from the Australian Mouse Brain Mapping Consortium (AMBMC) weblink: www.imaging.org.au/AMBMC.


